Abstract
The hpt gene from the archaeon Methanobacterium thermoautotrophicum, encoding hypoxanthine (guanine) phosphoribosyltransferase, was cloned by functional complementation into Escherichia coli. The hpt-encoded amino acid sequence is most similar to adenine phosphoribosyltransferases, but the encoded enzyme has activity only with hypoxanthine and guanine. The synthesis of the recombinant enzyme is apparently limited by the presence of the rare arginine codons AGA and AGG and the rare isoleucine AUA codon on the hpt gene. The recombinant enzyme was purified to apparent homogeneity.
The purine salvage and interconversion pathways in Methanobacterium thermoautotrophicum have been characterized by determining purine enzyme levels and incorporation of purine bases in wild-type cells and in mutants resistant to purine analogs (36). Because of our interest in purine salvage in archaea and because there exists some controversy as to whether there are one or two purine phosphoribosyltransferases (PRTases) in methanogens (4, 6, 36), we decided to clone a PRTase gene from the thermophilic archaeon M. thermoautotrophicum. Based on activity with the normally occurring nucleobases, the purine PRTases can be divided into adenine and 6-oxopurine PRTases. The adenine PRTase (APRTase) typically reacts only with adenine (22, 23, 28, 35). There are three types of 6-oxopurine PRTases. The most abundant enzyme (HGPRTase) reacts with hypoxanthine and guanine only (1, 12, 23, 35). Another enzyme reacts with hypoxanthine, guanine, and xanthine (27, 32, 43, 46). Finally, an enzyme exists in Bacillus subtilis that reacts with xanthine only (7). Here we report on the cloning and expression in E. coli of a gene encoding a HGPRTase from M. thermoautotrophicum, a strictly anaerobic, thermophilic, obligatory autotrophic methanogenic archaeon.
Cloning and sequence analysis of the hpt gene.
Transformation and cloning procedures were done by standard techniques (26). DNA from M. thermoautotrophicum Marburg (DSM 2133) was isolated as previously described (16) and was partially digested with Sau3AI. Fragments in the range of 1 to 20 kb were subsequently ligated into the BamHI site of pUC19. For amplification of the library, E. coli DH5α cells (22) were transformed by electroporation. The cells were plated on Luria-Bertani ampicillin (100 mg/liter), and plasmid DNA was isolated from a culture of 1.5 × 105 pooled colonies. The library was transformed into strain SØ609 [Δ(gpt-pro-lac) thi hpt deoD purD rpsL] (17), and the cells were spread on selective minimal plates containing hypoxanthine (15 mg/liter). Five colonies appeared after 24 h of incubation, and all isolates required hypoxanthine or guanine for growth, indicating that the gene we have cloned encodes an HGPRTase. All isolates contained the same plasmid, called pSAUE1, which contained a putative hpt gene. The nucleotide sequence of an insert of 2,221 bp was determined by the dideoxy chain termination technique (27). Both strands were sequenced completely, and the sequence was analyzed by using the GCG sequence analysis software package (11). The cloned fragment contained the whole coding region of the hpt gene as well as parts of two other reading frames. That the cloned DNA fragment originated from M. thermoautotrophicum was verified by Southern blot analysis (26) using two restriction enzymes, StyI and PstI (data not shown). The 1,119-bp StyI-PstI fragment of pSAUE1 carries the complete hpt gene (Fig. 1) and encodes a protein of 193 amino acids. A search in the database for sequence homology using the BLAST algorithm package (2) revealed homology to both APRTases and HGPRTases. Amino acid sequence comparisons with PRTases indicated that the M. thermoautotrophicum HGPRTase showed 23 to 29% identity to APRTases from other organisms, 50% identity to a predicted APRTase of Methanococcus jannaschii (GenBank accession no. U67467), and 93% identity to a predicted APRTase from M. thermoautotrophicum ΔH (GenBank accession no. AE000666). In contrast, HGPRTases and HGXPRTases of other organisms show only 19 to 27% identity to the HGPRTase of M. thermoautotrophicum.
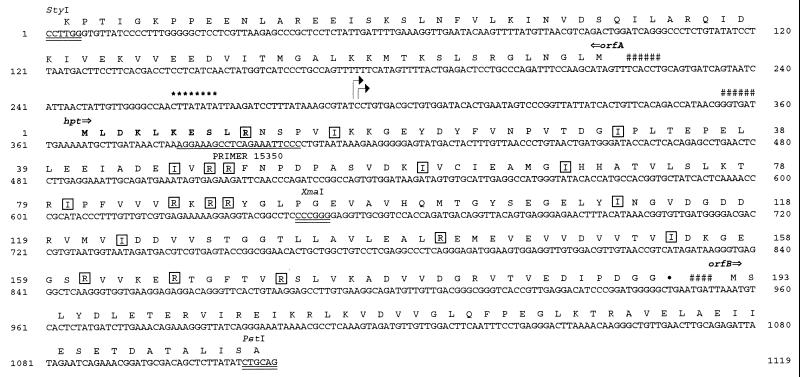
FIG. 1.
The nucleotide sequence of a 1,119-bp StyI-PstI DNA fragment containing part of orfA and orfB and the complete hpt gene of M. thermoautotrophicum Marburg. The deduced amino acid sequence of the HGPRTase, encoded by hpt, and the two open reading frames are shown above the sequence. The amino acids shown in bold have been verified by N-terminal sequencing of the purified HGPRTase. The potential ribosome binding sites in front of the open reading frames are marked with #. Relevant restriction sites are indicated and double underlined. The transcriptional start sites of the hpt gene, as identified by primer extension (Fig. 2) are marked with arrows, and the putative promoter region is marked with asterisks. Amino acids encoded by infrequently used arginine and isoleucine codons in E. coli are boxed.
Primer extension analysis.
To further characterize the hpt gene primer, extension analysis was performed. RNA was isolated from cells growing exponentially in minimal medium (14). Next, 400-ml cultures at an optical density at 436 nm of 0.3 to 0.4 were harvested, chilled quickly in liquid N2, and pelleted for 15 min at 8,000 × g. Cells were ruptured by using liquid N2 (16) and were suspended in 10 ml of RNA buffer (20 mM sodium acetate [pH 4.8], 0.5% sodium dodecyl sulfate, and 1 mM EDTA in diethylpyrocarbonate-treated distilled H2O). Next, 10 ml of phenol equilibrated with 20 mM sodium acetate (pH 4.8) was quickly added before incubating the suspension for 10 min at 65°C. The suspension was cooled for 10 min on ice and centrifuged at 15,000 × g for 5 min. The aqueous phase was extracted two times with phenol-chloroform-isoamylalcohol (24:24:1; pH 8.0) before the RNA was precipitated by adding 0.1 volumes of 3 M sodium acetate (pH 4.8) and 3 volumes of 96% ethanol followed by centrifugation for 20 min. The RNA pellet was dissolved in 0.5 ml of diethylpyrocarbonate-treated distilled H2O and extracted two times with phenol-chloroform-isoamylalcohol. The primer 15350 (5′-GGG AAT TTC TGA GGC TTT CC-3′) labelled with 32P at the 5′ end with T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) was used (Fig. 1). A 50-μg sample of total RNA prepared from M. thermoautotrophicum and 0.1 pmol of primer in 0.3 M KCl was incubated at 90°C for 2 min and allowed to hybridize by cooling the mixture to 42°C over a period of 1 h. Probes were then extended for 1 h at 42°C in extension buffer (50 mM Tris-HCl [pH 8.3], 150 mM KCl, 8 mM MgCl2, 5 mM dithiothreitol, dATP, dCTP, dGTP, and dTTP, each at 1 mM) with 100 U of Moloney murine leukemia virus reverse transcriptase (Gibco-BRL). The extension reaction was stopped with formamide sequencing dye, and the extension products were analyzed on a 7% sequencing gel and aligned versus a dideoxy sequencing ladder by using primer 15350 and pSAUE7 as DNA template.
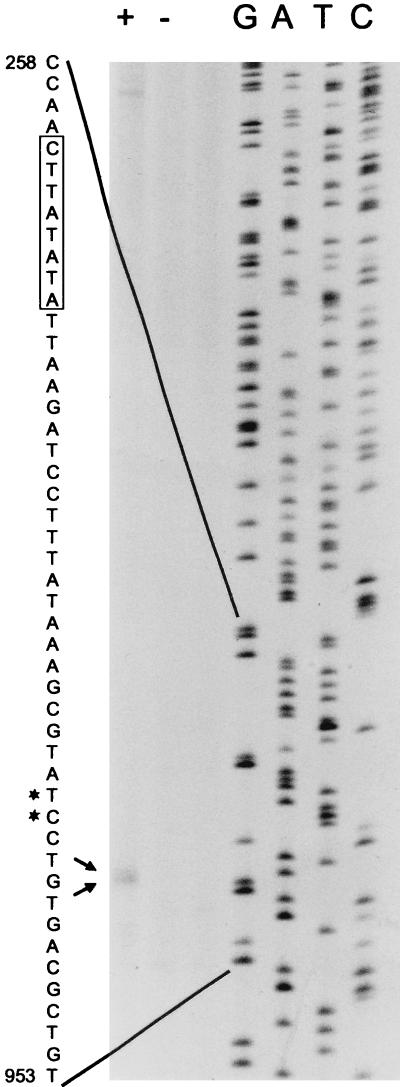
The primer extension analyses (Fig. 1 and 2) mapped two transcriptional start sites 73 bp upstream of the translational start at a position 23 bp downstream of a putative box A promoter element (5, 33). A sequence complementary to 6 bp of the 3′ end of 16S rRNA from M. thermoautotrophicum that could function as ribosome binding site was found 6 bp upstream of the ATG start codon. Located 152 bp upstream of the start codon of the hpt gene starts was an open reading frame, orfA, with a possible ATG start codon preceded by a 6-bp match to 16S rRNA. This reading frame codes for 275 amino acids of the N-terminal part of a protein which shows 95% identity to an open reading frame (no. 1321) from M. thermoautotrophicum ΔH and 61% identity to an open reading frame from M. jannaschii which code for a putative signal recognition particle. Downstream of the TGA stop codon of the hpt gene there is no apparent poly T stretch or secondary structures indicating a terminator. Rather, there is an open reading frame, orfB, encoding 218 amino acids of the N-terminal part of a putative protein. The reading frame starts with an ATG codon 8 bp downstream of the stop codon of the hpt gene and is preceded by a 4-bp match to 16S rRNA. In a search for sequence homology to this putative protein, we found an open reading frame with 82% identity to an open reading frame (no. 1319) from M. thermoautotrophicum ΔH and 36% identity from M. jannaschii, which shows homology to a diphtheria toxin resistance receptor. The organization orfA-hpt-orfB seen in M. thermoautotrophicum Marburg was the same as in M. thermoautotrophicum ΔH but differs from that of M. jannaschii, where the homologues to orfA, hpt, and orfB are scattered on the genome.
FIG. 2.
Primer extension analysis of the hpt-specified mRNA. The numbering of the nucleotide sequence is as in Fig. 1, with the coding strand displayed. A 50-μg quantity of RNA prepared from M. thermoautotrophicum was used in each experiment, and results obtained with complete extension mixture (+) and with an extension mixture without reverse transcriptase (as a control) (−) are shown. Lanes G, A, T, and C show the sequences generated using primer 15350 together with plasmid pSAUE 7 as template. The putative transcriptional start sites are marked on the gel with arrows and on the sequence to the left of the gel with asterisks. A putative box A promoter element is boxed on the sequence.
Subcloning of the hpt gene.
Plasmid pSAUE7 was constructed by inserting an 890-bp PstI fragment into the PstI site of pUC18 and was transformed into SØ609. pSAUE11 was constructed by ligating a 475-bp XmaI-PstI fragment and a 379-bp XmaI-cleaved PCR product into the vector pTrc99A (3), which was cleaved with NcoI and treated with Klenow polymerase before cleaving with PstI. The PCR product was generated by using the oligodeoxynucleotides 5′-CTT GAT AAA CTA AAG GAA AGC CTC-3′ (positions 370 to 393; Fig. 1) and 5′-TCA CGA CGT CAT CTA TTA CC-3′ (positions 750 to 731). pSAUE12 was constructed by ligating a 586-bp PCR product generated by using the oligodeoxynucleotides 5′-TAG TTA TTT AGC CCC CAT CCG GGA TGT CCT CAA C-3′ into pTrc99A treated with Klenow polymerase before cleaving with SmaI. The primer contains a 9-base 5′ nonmatching sequence which replaces the UGA stop codon of the methanogenic hpt with the more efficient stop codon UAA (24) and inserts stop codons in all three reading frames.
Purification and characterization of the recombinant hypoxanthine guanine phosphoribosyltransferase.
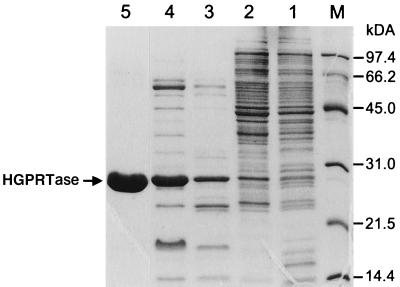
The enzyme was purified from strain SØ609 harboring plasmid pSAUE12 grown in 1 liter of Luria-Bertani medium (100 mg/liter) with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation, resuspended in 10 ml of extraction buffer (10 mM Tris-HCl [pH 7.6], 2 mM EDTA) at 0 to 4°C, disrupted in a French press at 3,810 lb/in2, and centrifuged (10 min, 10,000 × g). Nucleic acids were precipitated by adding streptomycin sulfate to 1% followed by incubation for 1 h and centrifugation. The supernatant was subjected to heat denaturation for 30 min at 70°C followed by the removal of denatured protein by centrifugation and was dialyzed for 16 h against 2 liters of TM buffer (10 mM Tris-HCl [pH 7.6], 10 mM MgCl2). The dialysate was applied to a diethylaminoethyl cellulose-52 column (2.5 by 24 cm; Sigma, St. Louis, Mo.) equilibrated with TM buffer and run at a flow rate of 1.0 ml/min. HGPRTase was eluted with a gradient of 0 to 1 M NaCl over a period of 30 min. The activity-containing fractions (36 ml) were dialyzed for 16 h against 2 liters of TM buffer. An aliquot of 5 ml was applied on a 1-ml GTP-agarose column (Boehringer, Mannheim, Germany). The column was washed with 3 ml of TM buffer containing 1 M NaCl. The enzyme was eluted with 1 ml of TM buffer containing 1 mM PRPP. The purification of the HGPRTase is summarized in Table 1 and visualized in Fig. 3. The activity of the purified enzyme with guanine (0.1 mM) was 66% of that with hypoxanthine, while activity with adenine, xanthine, uracil, orotate, or cytosine was <0.01%. PRTase activity in crude extract of M. thermoautotrophicum revealed 16 nmol/min/mg of protein with hypoxanthine, 14 nmol/min/mg of protein with guanine, and <0.01 nmol/min/mg of protein with adenine and xanthine. The purified HGPRTase was subjected to N-terminal analysis by Edman degradation done with an Applied Biosystems 477A Protein Sequencer with on-line phenylthiohydanthoin-amino acid detection. N-terminal sequencing yielded the amino acid sequence Met-Leu-Asp-Lys-Leu-Lys-Glu-Ser-Leu-Arg, which is in perfect agreement with the N-terminal end predicted from the nucleotide sequence (Fig. 2). The purified enzyme was stable for at least 6 months at 4°C in the TM buffer. A broad pH optimum of between pH 7.6 and 8.2 with an optimal activity between 70 and 80°C was found. The enzyme activity at 37°C and at 85°C was 1 to 2% of that measured at 65°C.
TABLE 1.
Purification of the recombinant HGPRTase from M. thermoautotrophicum expressed in E. coli SØ609 from plasmid pSAUE12
| Fraction | Protein (mg) | Specific activitya (nmol/min/mg of protein) | Total activity (nmol/min) | Fold purifi-cation | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 196 | 253 | 49,588 | 1 | 100 |
| 1% Streptomycin precipitation | 94 | 420 | 39,480 | 1.7 | 80 |
| Heat denaturation | 17 | 2,315 | 39,355 | 9.2 | 79 |
| DEAE(52)-chroma-tography | 3.6 | 7,308 | 26,309 | 29 | 53 |
| Affinity-chromatog-raphy (GTP-agarose) | 0.36 | 39,400 | 14,184 | 156 | 29 |
FIG. 3.
Sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis of the purification steps of the recombinant HGPRTase from M. thermoautotrophicum. A total of 10 μg of protein was applied at each purification step (Table 1). Lanes: 1, crude extract; 2, streptomycin step; 3, heat denaturation step; 4, DEAE step; 5, GTP-agarose affinity step. The molecular mass markers used were as follows: Phosphorylase b (97.4 kDa); serum albumin (66.2 kDa); ovalbumin (45.0 kDa); carbonic anhydrase (31.0 kDa); soybean trypsin inhibitor (21.5 kDa); and lysozyme (14.5 kDa). The position of the purified HGPRTase is marked with an arrow.
Controlled expression of the hpt gene of M. thermoautotrophicum in E. coli.
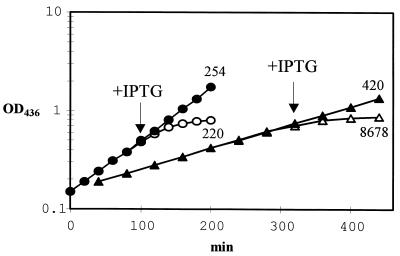
The orientation of the insert in pSAUE1 and pSAUE7 was arranged so that the transcription of the hpt gene could not be driven by the lac promoter in the pUC vector. In both situations, the expression resulted in an enzyme with an activity corresponding to 210 to 230 nmol/min/mg of protein driven from the hpt promoter. To obtain a higher level of expression, the hpt gene was inserted into the expression vector pTRC99A, which contains the strong trc E. coli promoter under lacIq control and a ribosome binding site at an optimal distance from the ATG start codon of the hpt gene. Unfortunately, this construct (pSAUE12) resulted in enzyme levels of the same magnitude as those found in SØ609 containing pSAUE1 or pSAUE7. All constructs grew at a rate 10 to 12% below that of SØ609 containing only the vector. Growth of SØ609/pSAU12 was further reduced by the addition of IPTG, and no increase in enzyme level was obtained (Fig. 4). In the early reports on codon usage in archaea (15, 29), it was noticed that the codon usage was different from that of E. coli. Archaea appears to have a strong preference for the AGA and AGG arginine codons (8), which are infrequently used in E. coli. The minor tRNAArg, encoded by argU and recognizing AGG and AGA, and tRNAIle, encoded by ileX and recognizing AUA have been shown to be limiting factors in bacterial expression of foreign genes (10). Analysis of the codon usage of the hpt gene showed that there was a high number of the rare codons—nine AUA isoleucine codons, six AGG codons, and four AGA arginine codons. When the rare codons are arranged in clusters or in tandems, the expression is further perturbed. This was in fact the arrangement found here (Fig. 1). Introduction of the plasmid pRI952 (10), containing the genes coding for the tRNAs which decode these codons, into SØ609 already containing pSAUE12 resulted in a 35-fold increase in the enzymatic level when the cells were induced with 0.2 mM IPTG (Fig. 4). However, the pRI952 plasmid did not relieve the observed growth inhibition. The doubling time for SØ609/pRI952 was 1 h. This indicates that the observed growth inhibition (Fig. 4) is most likely a result of the synthesis of the recombinant enzyme. When an extract of cells containing the high level of HGPRTase (8,678 nmol/min/mg of protein) was analyzed on sodium dodecyl sulfate-polyacrylamide gels, a major band which ran in the same position as the purified enzyme was observed (data not shown).
FIG. 4.
Growth of E. coli SØ609/pSAUE12 and HGPRTase levels. Effects of IPTG and plasmid pRI952, containing genes that encode tRNAs for the rare codons AGA, AGG, and AUA. Closed symbols indicate growth (optical density at 436 nm [OD436]) in control cultures, and open symbols indicate growth after the addition of IPTG. Time of IPTG addition is indicated. Symbols: ○ and ●, SØ609/pSAUE12; ▵ and ▴, SØ609/pSAUE12 + pRI952. Numbers shown above and below the growth curves are HGPRTase levels given as nanomoles per minute per milligram of protein determined in cells isolated at the last point of the growth curve.
Conclusions.
The successful isolation of genes of archaeal origin by functional complementation in E. coli has been reported for genes from thermophiles and methanogens encoding enzymes of biosynthetic pathways (15, 20, 21, 25, 29, 31, 34). We cloned a HGPRTase gene from M. thermoautotrophicum, although the amino acid sequence is most similar to an APRTase. Crystallographic studies of the human HGPRTase (13) indicated that the part of the protein that differentiates between adenine and 6-oxopurines is situated in the carboxy-terminal part of the protein. Lys-165 is thought to determine whether adenine is discriminated against as substrate. Alignment of sequences of various HGPRTases shows that a lysine occupies a similar position in the HGPRTase of M. thermoautotrophicum Marburg. Val-187 and Asp-193 in the C-terminal end of the human HGPRTase also occupy a similar position in the methanogenic enzymes, and both have been proposed to play a role in the discrimination against xanthine as substrate. However, the putative PRPP binding fold of the HGPRTase from M. thermoautotrophicum differs from those of other HGPRTases, which are characterized by two adjacent carboxylic acids, Glu-133 and Asp-134 (13), where APRTases has two aspartate residues (9). The methanogenic enzyme, however, has two aspartate residues at this position. Two other amino acid residues, Thr-129 and Gly-131, found in the methanogenic HGPRTase are highly conserved in the PRPP binding fold of APRTases. Thr-138 and Thr-141 of the human HGPRTase which form hydrogen bonds to the phosphate oxygen of GMP are conserved in the methanogenic HGPRTase. Thus, the important residues of HGPRTase are fairly conserved in the methanogenic enzyme, while other residues are not conserved, a property that may reflect the fact that the enzyme has its highest activity at 75°C. Particularly in the C-terminal end of the methanogenic enzyme, a large number of amino acids that are completely different from those of other HGPRTases are found. From our data, it appears that methanogens possess only a single purine PRTase, namely HGPRTase. In contrast, halophilic archaea (32) and Sulfolobus species (23a) contain both an APRTase and an HGPRTase.
Nucleotide sequence accession number.
The sequence reported in this paper was deposited in the GenBank database under accession no. AF007759.
Acknowledgments
This work was supported by a grant from the Danish Centre for Microbiology.
We thank Kenneth Harlow for performing the N-terminal amino acid sequence analysis.
REFERENCES
- 1.Allen T E, Ullman B. Molecular characterization and overexpression of the hypoxanthine-guanine phosphoribosyltransferase gene from Trypanosoma cruzi. Mol Biochem Parasitol. 1994;65:233–245. doi: 10.1016/0166-6851(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amann E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 4.Bowen T L, Lin W C, Whitman W B. Characterization of guanine and hypoxanthine phosphoribosyltransferases in Methanococcus voltae. J Bacteriol. 1996;178:2521–2526. doi: 10.1128/jb.178.9.2521-2526.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown J W, Daniels C J, Reeve J N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16:287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1017–1140. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Christiansen L C, Schou S, Nygaard P, Saxild H H. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalgaard J Z, Garrett R A. Archaeal hyperthermophile genes. In: Kates M, et al., editors. The biochemistry of Archaea (Archaebacteria). Amsterdam, The Netherlands: Elsevier Science Publishers B. V.; 1993. pp. 535–563. [Google Scholar]
- 9.De Boer J G, Glickman B W. Mutational analysis of the structure function of the adenine phosphoribosyltransferase enzyme of Chinese hamster. J Mol Biol. 1991;221:163–174. doi: 10.1016/0022-2836(91)80212-d. [DOI] [PubMed] [Google Scholar]
- 10.Del Tito B J, Jr, Ward J M, Hodgson J, Gershater C J, Edwards H, Wysocki L A, Watson F A, Sathe G, Kane J F. Effects of a minor isoleusyl tRNA on heterologous protein translation in Escherichia coli. J Bacteriol. 1995;177:7086–7091. doi: 10.1128/jb.177.24.7086-7091.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux J P, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donald R G K, Carter D, Ullman B, Roos D S. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- 13.Eads, J. S., G. Scapin, Y. Xu, C. Grubmeyer, and J. C. Sacchettini. The crystal structure of human hypoxanthine-guanine phosphoribosyltransferase with bound GMP. Cell 78:325–334. [DOI] [PubMed]
- 14.Gast D A, Jenal U, Wasserfallen A, Leisinger T. Regulation of tryptophan biosynthesis in Methanobacterium thermoautotrophicum Marburg. J Bacteriol. 1994;176:4590–4596. doi: 10.1128/jb.176.15.4590-4596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton P T, Reeve J N. Sequence divergence of an archaebacterial gene cloned from a mesophilic and a thermophilic methanogen. J Mol Evol. 1985;22:351–360. doi: 10.1007/BF02115691. [DOI] [PubMed] [Google Scholar]
- 16.Jarrell K F, Faguy D, Hebert A M, Kalmokoff M L. A general method of isolation high molecular weight DNA from methanogenic archaea (archaebacteria) Can J Microbiol. 1992;38:65–68. doi: 10.1139/m92-010. [DOI] [PubMed] [Google Scholar]
- 17.Jochimsen B, Nygaard P, Vestergaard T. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Adenosine deaminase (add), guanosine kinase (gsk) and hypoxanthine phosphoribosyltransferase (hpt) Mol Gen Genet. 1975;143:85–91. doi: 10.1007/BF00269424. [DOI] [PubMed] [Google Scholar]
- 18.Lee C C, Craig III S P, Eakin A E. A single amino acid substitution in the human and bacterial hypoxanthine phosphoribosyltransferase modulates specificity for the binding of guanine. Biochemistry. 1998;37:3491–3498. doi: 10.1021/bi9720179. [DOI] [PubMed] [Google Scholar]
- 19.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 20.Meile L, Stettler R, Banholzer R, Kotik M, Leisinger T. Tryptophan gene cluster of Methanobacterium thermoautotrophicum Marburg: Molecular cloning and nucleotide sequence of a putative trpEGCFBAD operon. J Bacteriol. 1991;173:5017–5023. doi: 10.1128/jb.173.16.5017-5023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metcalf W W, Zhang J, Shi X, Wolfe R S. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J Bacteriol. 1996;178:5797–5802. doi: 10.1128/jb.178.19.5797-5802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naguib F N M, Iltzsch M H, el Kouni M M, Panzica R P, el Kouni M H. Structure-activity relationships for the binding of ligands to xanthine or guanine phosphoribosyltransferase from Toxoplasma gondii. Biochem Pharmacol. 1995;50:1685–1693. doi: 10.1016/0006-2952(95)02070-5. [DOI] [PubMed] [Google Scholar]
- 23.Nygaard P. Utilization of preformed purine bases and nucleosides. In: Munch-Petersen A, editor. Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. London, United Kingdom: Academic Press Ltd.; 1983. pp. 27–93. [Google Scholar]
- 23a.Nygaard, P. Unpublished observations.
- 24.Poole E S, Brown C M, Tate W P. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcarea C, Hervé G, Ladjimi M M, Cunin R. Aspartate transcarbamylase form the deep-sea hyperthermophilic archaeon Pyrococcus abyssi: genetic organization, structure, and expression in Escherichia coli. J Bacteriol. 1997;179:4143–4157. doi: 10.1128/jb.179.13.4143-4157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnorr K M, Gaillard C, Biget E, Nygaard P, Laloue M. A second form of adenine phosphoribosyltransferase in Arabidopsis thaliana with relative specificity towards cytokinins. Plant J. 1996;9:891–898. doi: 10.1046/j.1365-313x.1996.9060891.x. [DOI] [PubMed] [Google Scholar]
- 29.Sibold L, Henriquet M. Cloning of the trp genes from the archaebacterium Methanococcus voltae: nucleotide sequence of the trp BA genes. Mol Gen Genet. 1988;214:439–450. doi: 10.1007/BF00330478. [DOI] [PubMed] [Google Scholar]
- 30.Somoza J R, Chin M S, Focia P J, Wang C C, Fletterick R J. Crystal structure of the hypoxanthine-guanine-xanthine phosphoribosyltransferase from the protozoan parasite Tritrichomonas foetus. Biochemistry. 1996;35:7032–7040. doi: 10.1021/bi953072p. [DOI] [PubMed] [Google Scholar]
- 31.Sørensen I S, Dandanell G. Identification and sequence analysis of Sulfolobus solfataricus purE and purK genes. FEMS Microbiol Lett. 1997;154:173–180. doi: 10.1111/j.1574-6968.1997.tb12640.x. [DOI] [PubMed] [Google Scholar]
- 32.Stuer-Lauridsen B, Nygaard P. Purine salvage in two halophilic archaea: characterization of salvage pathways and isolation of mutants resistant to purine analogs. J Bacteriol. 1998;180:457–463. doi: 10.1128/jb.180.3.457-463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wich G, Hummel H, Jarsch M, Bar U, Bock A. Transcription signals for stable RNA genes in Methanococcus. Nucleic Acids Res. 1986;14:2459–2479. doi: 10.1093/nar/14.6.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood A G, Redborg A H, Cue D R, Whitman W B, Konisky J. Complementation of argG and hisA mutations of Escherichia coli by DNA cloned from the archaebacterium Methanococcus voltae. J Bacteriol. 1983;156:19–29. doi: 10.1128/jb.156.1.19-29.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woods R A, Roberts D G, Friedman T, Jolly D, Filpula D. Hypoxanthine: guanine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1983;191:407–412. doi: 10.1007/BF00425755. [DOI] [PubMed] [Google Scholar]
- 36.Worrell V E, Nagle D P., Jr Genetic and physiological characterization of the purine salvage pathway in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J Bacteriol. 1990;172:3328–3334. doi: 10.1128/jb.172.6.3328-3334.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]