Abstract
We have identified LB-AUT7, a gene differentially expressed 6 h after ectomycorrhizal interaction between Laccaria bicolor and Pinus resinosa. LB-Aut7p can functionally complement its Saccharomyces cerevisiae homolog, which is involved in the attachment of autophagosomes to microtubules. Our findings suggest the induction of an autophagocytosis-like vesicular transport process during ectomycorrhizal interaction.
Ectomycorrhizal fungi such as Laccaria bicolor are ubiquitous symbiotic fungi found all over the world (16, 29, 30). The host range of these fungi includes most gymnosperm and angiosperm trees as well as economically important timber-producing species (27). Mycorrhizal fungi play an important role in the health and survival of many tree species. Mycorrhizal symbiosis is especially important for transplanted trees and seedlings and when the soil conditions are unfavorable (6, 34). The fungi form a network of hyphae called the Hartig network, penetrating between the cortical cells of the root system. The ectomycorrhizal fungi provide the plant several benefits, which include the enhanced ability to absorb water and important ions such as phosphorus and nitrogen (8, 13, 33), and protection from soilborne root pathogens such as Fusarium oxysporum (10, 11) and heavy metals (9, 14). Mycorrhizal fungi form diverse interactions, and their beneficial effects also differ from species to species and under various soil and environmental conditions (20, 21, 39). In order to better utilize ectomycorrhizal fungi for forest tree health and increased yield of timber, it is important to understand the mechanisms by which the fungi and plants recognize and respond to each other for the formation of symbiotic ectomycorrhizas.
The molecular mechanisms that control the formation of symbiotic ectomycorrhizas are not well understood. The interaction between the fungus and roots must involve a series of events resulting in an integrated and functioning structure, namely, the mycorrhiza. These interaction events are likely mediated by the switching on and off of several genes in both the fungus as well as the host plant (7, 15). The root cells, presumably, produce certain elicitors that regulate the expression of fungal genes involved in the establishment of symbiosis (2, 18). This includes turning on genes responsible for attachment to the root surface and developmental processes, such as the formation of the Hartig network and hyphal mantle (7, 31, 32). It may also include turning off fungal genes encoding factors that elicit host plant defense reactions during early stages of interaction. Salzer et al. (31) have shown that specific elicitors present in ectomycorrhizal fungi were inactivated by chitinases secreted by roots during interaction, without affecting the fungus, thus enabling formation of ectomycorrhizas without eliciting a defense response from the plant.
Our objective in this study was to identify and characterize symbiosis-regulated genes from the ectomycorrhizal fungus L. bicolor that are differentially expressed in interaction with red pine seedling roots. Primarily we are interested in genes whose expression is triggered during very early stages of the interaction. Towards this goal, we have developed an in vitro L. bicolor × Pinus resinosa interaction model system (17) that allows us to identify symbiosis-regulated genes by the mRNA differential display technique (24). We describe here cDNA cloning and characterization of LB-AUT7, an L. bicolor gene that is turned on as early as 6 h after interaction with pine. LB-AUT7 has significant homologies with AUT7 from Saccharomyces cerevisiae. AUT7 in yeast has been identified as a gene essential for autophagocytosis (23). Autophagocytosis is a degradative pathway, transporting proteins from the cytoplasm to the lysosome (vacuole) (12, 37). During autophagy cytosol-containing, double-membrane layered vesicles (autophagosomes) are formed and delivered to the vacuole, where they are degraded together with their cytosolic content. Most recently it has been shown that Aut7p forms a protein complex with the microtubule-associated Aut2p. Aut7p and Aut2p are necessary for the attachment of autophagosomes (vesicles) to microtubules, to allow their directed movement to the vacuole (23).
mRNA differential display and identification of symbiosis-regulated genes from L. bicolor.
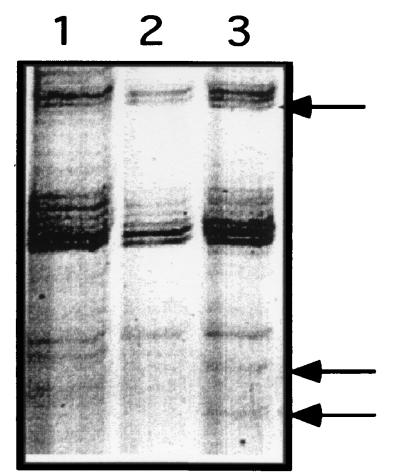
The in vitro ectomycorrhizal model system (17) and mRNA differential display (24) were used to study the plant-fungus interaction versus that with free-living fungus. In this system, fungal genes that are differentially expressed during the early stages of fungus-pine interaction were identified. Figure 1 shows part of a differential-display reverse transcriptase PCR (DDRT-PCR) gel showing L. bicolor cDNA clones which are differentially expressed after 6 h of interaction with red pine seedlings. Based on preliminary results from Northern hybridizations using DDRT-PCR clones and DNA sequence analysis, three clones were selected, PF6.1, PF6.2, and PF6.3 (Fig. 1). Of these three DDRT-PCR clones, PF6.3 (Fig. 1) was selected for further characterization because of its sequence characteristics and significant homology to yeast AUT7. Thus, this system provided us with a powerful tool to study the molecular basis of ectomycorrhizal symbiosis during the very early stages of interaction. Since the establishment of mycorrhizal roots is a long-term process, soil-based or other solid-medium-based systems can be used to establish mycorrhizal roots as we have done and can be used to study gene expression in synthesized ectomycorrhizal roots. A solid-agar-based system was used by Tagu and colleagues (35, 36) to study gene expression from mycorrhizal roots formed by interaction of the ectomycorrhizal fungus Pisolithus tinctorius and its plant host, eucalyptus.
FIG. 1.
Autoradiogram of part of an mRNA differential display gel showing the differences between fungus induced in response to pine signals and free-living mycelium as a control at 6 h. Shown are RNA from L. bicolor at the beginning of experiment, (lane 1), RNA of free-living fungus at 6 h (lane 2), and RNA of fungus after 6 h of interaction with red pine seedlings (lane 3). Arrows indicate selected differentially expressed cDNAs after 6 h of interaction. The arrow at the bottom corresponds to the PF6.3 clone.
Isolation and characterization of LB-AUT7 cDNA.
RNA extracted from L. bicolor cocultivated with pine seedlings for 6 to 48 h was pooled and used to isolate poly(A)+ RNA by employing the Oligo-dT mRNA purification system (New England Biolabs, Beverly, Mass.). This poly(A)+ RNA was used to construct a cDNA library in the phagemid vector lambda-ZAP (Stratagene, La Jolla, Calif.). The cDNA library was screened with a cDNA fragment, corresponding to LB-AUT7, isolated by DDRT-PCR. A total of 105 recombinant phages were screened, and seven positive plaques were isolated; the largest cDNA clone was selected and named LB-AUT7. DNA sequence analysis of LB-AUT7 cDNA and a subsequent open reading frame search using MacDNAsis software (Hitachi Corp.) have indicated that LB-AUT7 cDNA contained an open reading frame encoding a 197-amino-acid protein with a predicted molecular mass of 22.8 kDa and a pI of 8.2. The LB-AUT7 cDNA clone also contained 57 nucleotides of the 5′ untranscribed region and 84 nucleotides of the 3′ untranscribed region, for a total length of 732 bases (GenBank accession no. U93506).
Homologies of LB-AUT7 to known sequences.
Sequences similar to those of LB-AUT7 have not been previously isolated from ectomycorrhizal fungi. However, LB-Aut7p has significant homologies to a number of proteins (Fig. 2), including Aut7p from Saccharomyces cerevisiae (23), which is 77% identical (Fig. 2) based on analysis performed with CLUSTAL software. The hydrophobicity plot of LB-Aut7 protein also shows a potential membrane insertion domain starting at amino acid 175.
FIG. 2.
Multiple sequence alignment of LB-Aut7p by the CLUSTAL method with PAM250 residue weight table. Residues identical with LB-Aut7p are shown on a black background. From U80885 only amino acids 586 to 715 are included in the alignment. A. thaliana, Arabidopsis thaliana; C. elegans, Caenorhabditis elegans.
Differential expression and genomic organization of LB-AUT7.
In order to confirm the patterns of gene expression obtained from differential display for LB-AUT7, a full-length cDNA clone of LB-AUT7 was used as a probe to perform Northern analysis. RNA from free-living mycelium and/or mycelium after interaction with red pine seedlings was isolated as described previously (17). To obtain RNA from mycorrhizal roots, red pine seeds were surface sterilized and allowed to germinate under conditions described by Richter and Bruhn (30). After seedlings had formed fine roots (4 to 6 weeks), synthesis experiments between L. bicolor and red pine seedlings were conducted as described previously (5). After 3 to 4 months, ectomycorrhiza formation was observed under a microscope prior to extraction of RNA for analysis. For northern analysis, 10 μg each of total RNA was electrophoresed on 1% agarose gels with formaldehyde and transferred to Hybond N+ membranes (Amersham Corp., Arlington Heights, Ill.). Prehybridization, hybridization, and washings were done as described before (38). RNA gels were also stained with SYBR Green II (Molecular Probes, Eugene, Oreg.) to confirm the quality of RNA and also equality of loadings in each lane. As seen in Fig. 3A, the expression of LB-AUT7 was dependent upon interaction with red pine seedlings, and its expression was not detectable in free-living mycelium. We also confirmed the in vivo relevance of the L. bicolor LB-AUT7 cDNA clone as symbiosis regulated. RNA from red pine-L. bicolor ectomycorrhizal roots showed differential expression of LB-AUT7 compared to RNA isolated from free-living L. bicolor. As shown in Fig. 3B, LB-AUT7 transcript was detected only from mycorrhizal roots and not from free-living mycelium or from nonmycorrhizal pine roots.
FIG. 3.
(A) Northern analysis of LB-AUT7 expression. Shown is an RNA blot of L. bicolor RNA isolated from free-living mycelium (lane F) and after 6 h of interaction with red pine seedlings (lane PF). Ten micrograms of total RNA was used per lane. Full-length 32P-labeled LB-AUT7 cDNA was used as a probe for hybridization. (B) Northern analysis using total RNAs from nonmycorrhizal red pine roots (lane R), free-living L. bicolor (lane F), and mycorrhizal roots (lane M) was performed as described for panel A. (C) Southern analysis of LB-AUT7 from L. bicolor. Ten micrograms each of L. bicolor genomic DNA digested with BamHI (lane 1), EcoRI (lane 2), and PstI (lane 3) was used per lane. Hybridization was done with 32P-labeled LB-AUT7 cDNA. Molecular sizes are shown in kilobases.
The fact that LB-AUT7 is differentially expressed even in mycorrhizal roots indicates that this gene is regulated by interaction with a plant host and that a continual expression of this gene is needed for the maintenance of ectomycorrhizae in red pine by L. bicolor. As per the eucalyptus × P. tinctorius model system proposed by Tagu and colleagues (35, 36), the preinfection stage (0 to 12 h) may have an important role in signal exchange and continuation to the next stage of symbiotic interaction. We have identified differentially expressed cDNA clones from L. bicolor after 24 h of interaction also and are currently characterizing them. These clones may represent determinants of establishment of symbiotic association between L. bicolor and red pine. Clones of 6-h interaction may serve as essential genes in the establishment of recognition events and developmental processes for ectomycorrhizae formation as well as maintenance of the symbiotic interaction.
We have also extracted RNA from mycorrhizal roots of aspen (Populus tremuloides) colonized by L. bicolor and larch (Larix decidua) colonized by Sullius spp. and subjected the RNA to Northern analysis using LB-AUT7 cDNA as a probe. A weak signal was detected with aspen and larch (data not shown). Thus, it seems that the LB-AUT7 expression may be specific to or regulated by symbiotic interaction with various plant hosts. Further screening of ectomycorrhizal roots formed by various ectomycorrhizal fungi on their respective plant hosts needs to be performed to determine the expression of LB-AUT7 homologues.
To determine the gene copy number, we performed Southern analysis. Genomic DNA from L. bicolor was isolated as described by Reymond (28), and 10 μg each of genomic DNA was digested with restriction enzymes BamHI, EcoRI, and PstI. These digested DNA fragments were electrophoresed, transferred to a Hybond N+ membrane (Amersham Corp.), and cross-linked by using a UV cross-linker (Fisher Biotech, Itasca, Ill.). Hybridization to the probe and washing of the filter were carried out under relatively high stringency conditions as described previously (17). Genomic Southern blotting of LB-AUT7 has indicated that there is possibly only one copy of this gene (Fig. 3C). We also detected a second, faint band in the EcoRI digest at the 6-kb range, which may be due to hybridization to another related gene with limited homology in L. bicolor. In S. cerevisiae no additional Aut7p homologues encoded by the genome are found, but since in rat two Aut7p homologues (LC3 and GEF-2) have been detected (also in Arabidopsis spp. there are several homologous proteins), this suggests the existence of several related proteins in the same organism. All these proteins are most likely involved in vesicle transport or the attachment of vesicles to microtubules. We could detect only one type of transcript for LB-AUT7 under the experimental conditions used or from the mycorrhizal roots. Further characterization of genomic clones of LB-AUT7 is currently in progress.
LB-AUT7 can functionally substitute for its yeast homologue during autophagy.
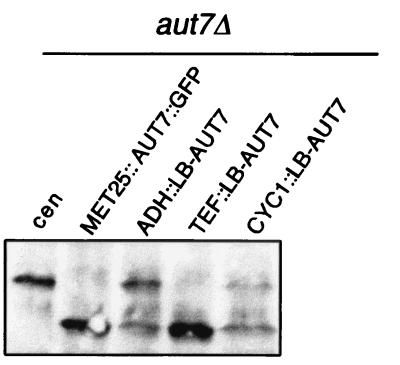
In yeast, Aut7p is implicated in autophagy. It functions by binding to microtubules via Aut2p and thus mediates attachment of autophagosomes for their transport to the vacuole (23). We looked for the ability of LB-AUT7 to complement the autophagic defects seen in aut7Δ yeast cells (23). LB-AUT7 was expressed from centromeric plasmids under the control of the CYC1, ADH, and TEF promoters (26). LB-AUT7 cDNA was inserted as an EcoRI-SpeI fragment into the respective sites of pRS416-TEF (26). EcoRI-XbaI fragments from LB-AUT7 were similarly inserted in the respective sites of pRS416-CYC1 and pRS416-ADH (26). Cells were grown to the stationary phase and after Western blotting were analyzed with antibodies directed against aminopeptidase I as described previously (23). Under control of the TEF promoter an almost complete complementation of the maturation defect of proaminopeptidase I was detected (Fig. 4, lane 4). Expression using the weaker CYC1 and ADH promoters resulted in only partially processed aminopeptidase I (Fig. 4, lanes 3 and 5). Expression of LB-AUT7 in aut7Δ yeast cells with the TEF promoter further resulted in complementation of the reduced survival rate during starvation for nitrogen and the defect in accumulation of autophagic vesicles in the vacuole during starvation in the presence of phenylmethylsulfonyl fluoride (data not shown).
FIG. 4.
Complementation of the maturation defect of proaminopeptidase I in aut7Δ yeast cells by LB-AUT7 expressed under control of the TEF promoter. LB-AUT7 cDNA was inserted in pRS416-TEF, pRS416-CYC1, or pRS416-ADH. Cells were grown to the stationary phase and after Western blotting were analyzed with antibodies directed against aminopeptidase I.
Since symbiotic initiation is a process of developmental regulation and differential gene expression in response to plants (35, 36), LB-AUT7 may play an important role in the differentiation and development of L. bicolor in response to plant signals. In addition, since there will be changes in the nutritional status of the fungus during the establishment of symbiosis, expression of proteins such as LB-Aut7p may serve as a means to recycle the cellular components to synthesize new macromolecules for growth of the fungus. It has been shown with several ectomycorrhizal fungi that there is an increase in the production and turnover of vesicles during symbiotic interaction with their plant hosts (19), and proteins such as LB-Aut7p are probably involved in this vesicular transport. Also in yeast an essential function of Aut7p was found for the cell differentiation process of sporulation (23). Motif searches using PRODOM and PSORTII indicated that the LB-Aut7p also has several tyrosine kinase phosphorylation sites. Thus, it is possible that LB-Aut7p activity is regulated through phosphorylation state and that in turn regulates other proteins involved in vesicle transport and differentiation. This may also involve cyclic AMP-mediated pathways which are regulated by the nutritional status of the fungus during interaction. Further analysis through in situ localization will allow the unraveling of the definitive role LB-Aut7p plays during and after the establishment of symbiotic interaction of L. bicolor with red pine.
Isolation of LB-AUT7 out of a cDNA library prepared from the RNA of symbiotic fungal cells confirms expression of this gene during symbiosis. Since the expression of the LB-AUT7 gene in free-living fungus cannot be detected (Fig. 3B), its expression seems to be dependent on a symbiotic interaction with its plant host. Due to the 77% identity between LB-Aut7p and yeast Aut7p (Fig. 2), together with the functionality of LB-AUT7 in aut7 deletion mutant yeast cells during autophagy (Fig. 4), we also expect a function of LB-AUT7 in the transport of vesicular intermediates or the attachment of vesicles to microtubules. Taken together this might lead to the hypothesis that autophagy itself or a related vesicle transport process is induced during symbiosis.
Initiation of ectomycorrhizal development might involve activation of primary or universal regulator genes to transcribe specific target genes for mycorrhiza formation. This may include expression of genes for signal transduction as well as genes involved in metabolism and biosynthesis. The ectomycorrhizal signal transduction pathway at the very early stages of symbiosis in response to plant signals may include signaling and transcription cascades which can be modulated by feedback modulation as seen in yeast and other fungal systems (3, 25). Once the initial stages of interaction have occurred through the primary cascade of gene expression between both partners, further development of ectomycorrhizae may proceed through expression of secondary gene expression cascades, resulting in the formation of ectomycorrhizae (4). This may also result in limited host defense response to limit further infection by other mycorrhizal fungi (1, 22). Thus, involvement of genes such as LB-AUT7 may play a key role in preparing the fungus for entry into the plant host root by providing necessary components for hyphal biogenesis and differentiation. Currently we are in the process of characterization of genomic clones of LB-AUT7 and its regulatory elements to establish its functional significance for ectomycorrhiza formation as well as to determine the regulation of this gene by signals from red pine seedlings.
Acknowledgments
This work was supported by USDA NRICGP grant 95-37107-1665, U.S. Forest Service Cooperative grant #23-081, and a grant from the State of Michigan Research Excellence Fund to G.K.P. This work was further supported by the DFG, Bonn, Germany (grant Wo210/12-1), to M.T.
We are grateful to D. Mumberg, Marburg, Germany, for providing plasmids pRS416-CYC1, pRS-ADH, and pRS416-TEF; to D. J. Klionsky Davis for antibodies directed against aminopeptidase I; and to D. H. Wolf for helpful discussions.
REFERENCES
- 1.Albrecht C, Laurent P, Lapeyrie F. Eucalyptus root and shoot chitinases, induced following root colonization by pathogenic and ectomycorrhizal fungi, compared on one and two dimensional activity gels. Plant Sci. 1994;100:157–164. [Google Scholar]
- 2.Anderson A J. Mycorrhizae-host specificity and recognition. Phytopathology. 1988;78:375–378. [Google Scholar]
- 3.Bardwell L, Cook J G, Inouye C J, Thorner J. Signal propagation and regulation in the regulation of mating pheromone response pathway of the yeast Saccharomyces cerevisiae. Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- 4.Barker S J, Tagu D, Delp G. Regulation of root and fungal morphogenesis in mycorrhizal symbioses. Plant Physiol. 1998;116:1201–1207. doi: 10.1104/pp.116.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bills S N, Richter D L, Podila G K. Genetic transformation of the ectomycorrhizal fungus Paxillus involutus by particle bombardment. Mycol Res. 1995;99:557–561. [Google Scholar]
- 6.Boyle C D, Hellenbrand K E. Assessment of the effect of mycorrhizal fungi on drought tolerance of conifer seedlings. Can J Bot. 1990;69:1764–1771. [Google Scholar]
- 7.Burgess T, Laurent P, Dell B, Malajczuk N, Martin F. Effect of the fungal isolate aggressivity on the biosynthesis of symbiosis-related polypeptides in differentiating eucalypt ectomycorrhiza. Planta. 1995;195:408–417. [Google Scholar]
- 8.Cairney J W G, Smith S E. Influence of intracellular phosphorus concentration on phosphate absorption by the ectomycorrhizal basidiomycete Pisolithus tinctorius. Mycol Res. 1992;96:673–676. [Google Scholar]
- 9.Colpaert J V, Assche J A V. Zinc toxicity in ectomycorrhizal Pinus sylvestris. Plant Soil. 1992;143:201–211. [Google Scholar]
- 10.Duchesne L C, Peterson R L, Ellis B E. The accumulation of plant-produced antimicrobial compounds in response to ectomycorrhizal fungi; a review. Phytoprotection. 1987;68:17–27. [Google Scholar]
- 11.Duchesne L C, Peterson R L, Ellis B E. The time course of disease suppression and antibiosis by the ectomycorrhizal fungus Paxillus involutus. New Phytol. 1989;111:693–698. doi: 10.1111/j.1469-8137.1989.tb02364.x. [DOI] [PubMed] [Google Scholar]
- 12.Dunn W J. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 13.Farquhar M L, Peterson R C. Later events in suppression of Fusarium root rot of red pine seedlings by the ectomycorrhizal fungus Paxillus involutus. Can J Bot. 1991;69:1372–1383. [Google Scholar]
- 14.Galli V, Schuepp H, Brunold C. Heavy metal binding by mycorrhizal fungi. Physiol Plant. 1994;92:364–368. [Google Scholar]
- 15.Gianinazzi-Pearson V, Gianinazzi S. Cellular and genetical aspects of interactions between hosts and fungal symbionts in mycorrhizae. Genome. 1989;31:336–341. [Google Scholar]
- 16.Harley J L, Smith S E. Mycorrhizal symbiosis. London, United Kingdom: Academic Press; 1983. [Google Scholar]
- 17.Kim S J, Hiremath S T, Podila G K. Cloning and identification of symbiosis-regulated genes from ectomycorrhizal fungus Laccaria bicolor. Mycol Res. 1999;103:168–172. [Google Scholar]
- 18.Koske R E, Gemma J N. Fungal reactions to plants prior to mycorrhizal formation. In: Allen M F, editor. Mycorrhizal functioning: an integrative plant-fungal process. New York, N.Y: Chapman & Hall, Inc.; 1992. pp. 3–36. [Google Scholar]
- 19.Kottke I, Oberwinkler F. Root-fungus interactions observed on initial stages of mantle formation and Hartig net establishment in mycorrhizas of Amanita muscaria on Picea abies in pure culture. Can J Bot. 1986;64:2348–2354. [Google Scholar]
- 20.Kropp B R. Variable interactions between non-Mycorrhizal and ectomycorrhizal strains of the basidiomycete Laccaria bicolor. Mycol Res. 1990;94:412–415. [Google Scholar]
- 21.Kropp B R, McAfee B J, Fortin J A. Variable loss of ectomycorrhizal ability in monokaryotic and dikaryotic cultures of Laccaria bicolor. Can J Bot. 1987;64:1224–1226. [Google Scholar]
- 22.Lamb C J. Plant disease resistance genes in signal perception and transduction. Cell. 1994;76:419–422. doi: 10.1016/0092-8674(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 23.Lang T, Schaeffler E, Bernreuther D, Bredschneider M, Wolf D H, Thumm M. Aut2 and Aut7, two novel microtubule associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 25.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 26.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 27.Peterson R L, Piche Y, Plenchette C. Mycorrhizae and their potential use in the agricultural and forestry industries. Biotechnol Adv. 1984;2:101–120. doi: 10.1016/0734-9750(84)90243-x. [DOI] [PubMed] [Google Scholar]
- 28.Reymond C D. A rapid method for the preparation of multiple samples of eukaryotic DNA. Nucleic Acids Res. 1987;15:817–818. doi: 10.1093/nar/15.19.8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter D L, Bruhn J N. Field survival of containerized red and jack pine seedlings inoculated with mycelial slurries of ectomycorrhizal fungi. New For. 1989;3:247–258. [Google Scholar]
- 30.Richter D L, Bruhn J N. Pinus resinosa ectomycorrhizae: seven host-fungus combinations synthesized in pure culture. Symbiosis. 1990;7:211–228. [Google Scholar]
- 31.Salzer P, Hubner B, Sirrenberg A, Hager A. Differential effect of purified spruce chitinases and β-1,3 glucanases on the activity of elicitors from ectomycorrhizal fungi. Plant Physiol. 1997;114:957–968. doi: 10.1104/pp.114.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirrenberg A, Salzer P, Hager A. Induction of mycorrhiza like structures and defense reactions in dual cultures of spruce callus and ectomycorrhizal fungi. New Phytol. 1995;130:149–156. [Google Scholar]
- 33.Smith S E, Gianinazzi-Pearson V, Koide R, Cairney J W G. Nutrient transport in mycorrhizas: structure, physiology and consequences for efficiency of the symbiosis. Plant Soil. 1994;159:103–113. [Google Scholar]
- 34.Smith S E, Read D J. Mycorrhizal symbiosis. 2nd ed. London, United Kingdom: Academic Press; 1997. [Google Scholar]
- 35.Tagu D, Martin F. Expressed sequence tags of randomly selected cDNA clones from Eucalyptus globus-Pisolithus tinctorius ectomycorrhizae. Mol Plant-Microbe Interact. 1995;8:781–783. [PubMed] [Google Scholar]
- 36.Tagu D, Phthon P, Cretin C, Martin F. Cloning symbiosis-related cDNAs from eucalypt ectomycorrhiza by PCR-assisted differential screening. New Phytol. 1993;125:339–343. doi: 10.1111/j.1469-8137.1993.tb03884.x. [DOI] [PubMed] [Google Scholar]
- 37.Thumm M, Wolf D H. From proteasome to lysosome: studies on yeast demonstrate the principles of protein degradation in the eukaryote cell. In: Rivett A J, editor. Advances in Molecular Cell Biology. Vol. 27. Greenwich, Conn: Jai Press; 1998. pp. 41–67. [Google Scholar]
- 38.Varley D, Hiremath S T, Podila G K. Cutinase gene expression in Cryphonectria parasitica, the chest nut blight fungus: effect of hypovirulence agents on expression. Mol Cell Biol. 1992;12:4539–4544. doi: 10.1128/mcb.12.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong K K, Piche Y, Fortin J A. Differential development of root colonization among four closely related genotypes of ectomycorrhizal Laccaria bicolor. Mycol Res. 1990;94:876–884. [Google Scholar]