Abstract
Background
In the United States, cancer disproportionately impacts Black and African American individuals. Identifying genetic factors underlying cancer disparities has been an important research focus and requires data that are equitable in both quantity and quality across racial groups. It is widely recognized that DNA databases quantitatively underrepresent minorities. However, the differences in data quality between racial groups have not been well studied.
Methods
We compared the qualities of germline and tumor exomes between ancestrally African and European patients in The Cancer Genome Atlas of 7 cancers with at least 50 self-reported Black patients in the context of sequencing depth, tumor purity, and qualities of germline variants and somatic mutations.
Results
Germline and tumor exomes from ancestrally African patients were sequenced at statistically significantly lower depth in 6 out of the 7 cancers. For 3 cancers, most ancestrally European exomes were sequenced in early sample batches at higher depth, whereas ancestrally African exomes were concentrated in later batches and sequenced at much lower depth. For the other 3 cancers, the reasons of lower sequencing coverage of ancestrally African exomes remain unknown. Furthermore, even when the sequencing depths were comparable, African exomes had disproportionally higher percentages of positions with insufficient coverage, likely because of the known European bias in the human reference genome that impacted exome capture kit design.
Conclusions
Overall and positional lower sequencing depths of ancestrally African exomes in The Cancer Genome Atlas led to underdetection and lower quality of variants, highlighting the need to consider epidemiological factors for future genomics studies.
Ethnic minorities often experience cancer disparities in the United States. Black and African American individuals have higher death rates and shorter survival for many cancer types (https://www.cancer.gov/about-cancer/understanding/disparities). African American women have higher rates of aggressive breast cancer subtypes compared with White women, and African American men suffer higher incidence and mortality rates of prostate cancer compared with White men (1). In addition, American Indian and Alaskan Native individuals have higher death rates from kidney cancer and higher incidence of liver cancer than any other racial group. Furthermore, leukemia disproportionately impacts Black and Hispanic men. These disparities are proposed to reflect the complex interplay of nonbiological (socioeconomic, environmental, behavioral) and biological factors (eg, genetic and genomic features).
Many studies have been conducted to identify and define differences of genetic and genomic factors underlying cancer disparities, which requires data that are equitable in both quantity and quality across racial groups. It is now widely known that minority groups have historically been quantitatively underrepresented in DNA databases compared with those of European descent, resulting in a catalog of genetic variants that likely does not represent the full range of human genetic diversity (2,3). However, the differences of genetic and genomic data quality between racial and ancestral groups have not been well studied.
The Cancer Genome Atlas (TCGA), one of the largest and most widely used cancer sequencing projects, profiled 33 cancer types, 7 of which studied more than 50 self-reported Black patients. Recently, we discovered incidentally that for 6 out of these 7 cancers the sequencing read depths were statistically significantly lower for both tumor and patient-paired germline exomes from Blacks compared with those from self-reported White patients. This points to a potential disparity in genomic data quality, which has not been reported previously. Herein, we genetically inferred ancestries of the patients in these 7 cancers, explored possible reasons for these disparities, and studied the potential implications.
Methods
Genetic Inference of Ancestry
Patient germline exomes were used to infer ancestry using principal component analysis based on ancestry space calculated from the reference set of individuals in the 1000 Genomes Project (1000G) (4). The genetically inferred African and European ancestries are used in this manuscript instead of the self-reported race. Additional details of the genetic inference approach can be found in the Supplementary Methods (available online).
Read Depth Calculations
TCGA primary tumor and patient-paired germline exome binary alignment and map (BAM) files were downloaded from the Genomic Data Commons (https://portal.gdc.cancer.gov/). Reads were aligned by TCGA to human reference genome build GRCh38. The sequencing depths (total reads, total mapped reads, and total unmapped reads) of each exome were calculated using the SAMtools (5) version 1.10. The following parameters were used: 1) for total mapped reads: samtools view -c -f 2 -F 1024 -q 20; 2) for total reads: samtools view -c -f 1 -F 1024; and 3) for total unmapped reads: samtools view -c -f5 -F 1024.
For replicate BAM files, the BAM file with highest total reads was used. Read coverage per genomic position was calculated from germline BAM using the following command: samtools mpileup—incl-flags 2—excl-flags 1024—min-MQ 20—min-BQ 0 (mapped paired reads; no polymerase chain reaction (PCR) duplicates; mapping quality at least 20). Read coverage per somatic variant position was obtained directly from the somatic minor allele frequency (MAF) files from TCGA.
Statistical Analysis
Statistical analyses were conducted using R version 3.6.2 (R Foundation, Vienna, Austria). All means comparisons used the Mann-Whitney U test. Proportions analysis by capture kit and ancestry used the 2-sample proportions z test. All statistical tests were 2-sided, and a P value less than .05 was considered statistically significant. In violin plots, the horizontal lines inside each box denote the median values. MAF at a variant position was calculated as count of minor alleles divided by total number of alleles at the position in a population. B allele concentration (BAC) is defined as the number of reads supporting the alternative allele divided by total read counts at the variant position. Area under the curve was computed from the BAC distribution using kernel density estimates from the density function in R.
Joint Genotyping of Germline Exomes
Variant calling of TCGA germline BAM files was performed per cancer type. Per-sample variant calling was performed using HaplotypeCaller from the Genome Analysis Toolkit (6), and multisample joint genotyping was performed using GenotypeGVCFs. For TCGA Breast Cancer (BRCA) germline exomes, variant calling was conducted only on the 31.51 Mb of exome regions common between all 3 NimbleGen kits (see Table 1). The exome capture bait positions in human reference genome build hg18 and hg19 were lifted over to the GRCh38 coordinates. The common capture region was identified using the command multiinter from the package bcftools (5) version 1.10. Variants passing Variant Quality Score Recalibration were annotated with population allele frequencies from 1000G using ANNOVAR tool (7).
Table 1.
Number of BRCA samples processed by each of the 3 NimbleGen kits with publicly accessible capture region files and the total capture size of each kit
| Exome capture kit | No. of primary tumor samples | No. of germline samples | Target size in Mb |
|---|---|---|---|
| NimbleGen hg18 exome v2 | 473 | 382 | 35.97 |
| NimbleGen SeqCap EZ Exome v2 | 114 | 114 | 80.59 |
| NimbleGen SeqCap EZ Exome v3 | 242 | 220 | 64.55 |
MAF Calculation
Biallelic germline variants were used for MAF calculations. Only variants that were detected in TCGA and reported by 1000G were included in the analyses. In addition, only positions detected in at least 25 TCGA patients were included. The variant MAFs from the 1000G European population were compared with those from ancestrally European TCGA patients, and MAFs from the 1000G African population were compared with those from ancestrally African TCGA patients.
Consent and Approval
The analyses and results reported in this paper are in whole or part based on data generated by TCGA Research Network (https://www.cancer.gov/tcga). All patients enrolled in TCGA germline and tumor exome sequencing were anonymized and deidentified. The data access was authorized under Mayo Clinic’s TCGA Project ID 4307.
Results
Exome Read Coverage
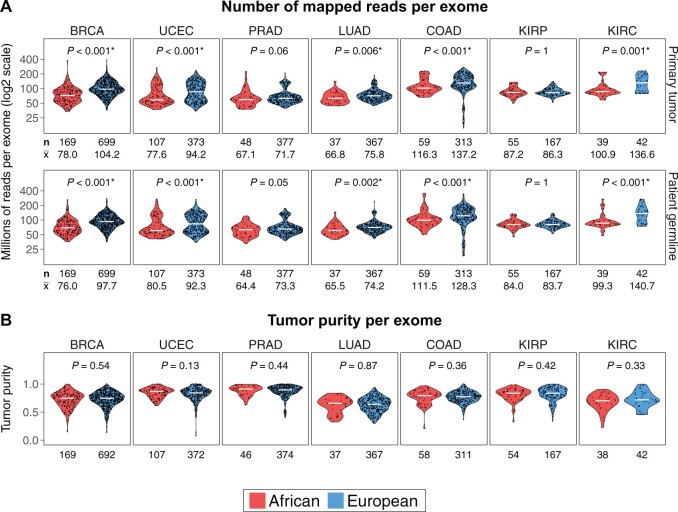
TCGA systematically studied more than 11 000 patients with cancer, among whom 985 (8.8%) were self-reported as Black or African American. Each patient was surveyed by multi-omic sequencing of tumor and patient-paired germline exomes, tumor RNA and microRNA transcriptomes, and tumor chromatinomes (chromatin modifications) (8). Seven cancer types included at least 50 tumor samples from self-reported Black patients: BRCA, uterine corpus endometrial (UCEC), kidney renal papillary cell, and kidney renal clear cell (KIRC) carcinomas and colon (COAD), prostate (PRAD), and lung (LUAD) adenocarcinomas. First, we observed statistically significantly lower read depths of both germline and tumor exomes from self-reported Black patients compared with those from self-reported White patients in 6 out of 7 cancer types (data not shown). Next, we performed genetic inference using the patients’ germline exomes to confirm African (Black) and European (White) ancestry and to assign ancestry to those with missing self-reported race and ethnicity information. Genetic inference of ancestries confirmed self-reported race with very few exceptions (Supplementary Figure 1, A and B, available online). The final numbers of ancestrally African and ancestrally European patients in each of the 7 cancer types are listed in the Supplementary Methods (Supplementary Figure 1, B, available online), and additional sample-level information is listed in the Supplementary Methods (Supplementary Table 1, available online). We discovered that 6 out of these 7 cancer types (all except kidney renal papillary cell carcinoma) had statistically significantly lower total numbers of sequencing reads of both tumor and germline exomes from ancestrally African patients compared with ancestrally Europeans (Supplementary Figure 2, A, available online). These 6 cancer types were included in further analyses. Five cancer types had lower numbers of mapped reads (Figure 1, A) from ancestrally African exomes.
Figure 1.
Comparison of exome sequencing depths between ancestrally European and African patients in 7 The Cancer Genome Atlas cancer types that each profiled at least 50 self-reported Black patients. A) Number of mapped reads per exome for primary tumor (upper panel) and patient-matched germline (lower panel). Mann-Whitney U test 2-sided P values are shown for all comparisons. Sequencing depths of both tumor and germline exomes from ancestrally European patients were statistically significantly higher than those of ancestrally African patients in breast cancer (BRCA; breast invasive carcinoma), LUAD (lung adenocarcinoma), UCEC (uterine corpus endometrial carcinoma), KIRC (kidney renal clear cell carcinoma), and COAD (colon adenocarcinoma). The number of patients in either ancestral group (n) and the average sequencing depth per sample in millions of reads () are displayed below the box plots. B) There were no differences in tumor purity between ancestrally African and European tumors in all 7 cancer types. The numbers of patients with available tumor purity data in either ancestral group are displayed below the box plots. KIRP = kidney renal papillary cell carcinoma; PRAD = prostate adenocarcinoma.
Source of the Coverage Disparity
We searched for the source of this sequencing depth disparity. There were no tumor purity differences between the 2 ancestral groups (Figure 1, B) based on metadata published by TCGA (9); therefore, the higher sequencing depths of ancestrally European exomes did not result from a necessity for deeper sequencing because of lower tumor purities. In addition, the lower sequencing depths in ancestrally African exomes persisted when we grouped samples according to sequencing center (Supplementary Figure 3, available online) and specimen collection site (Supplementary Figure 4, available online), indicating that neither of these factors could explain the depth disparity.
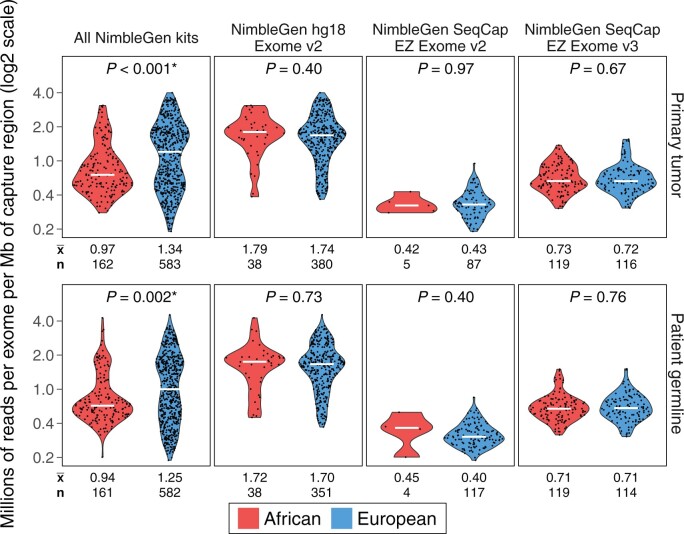
Next, we investigated sequencing depths between sample batches grouped by exome capture kits used to process patients’ DNA. We found that for 3 cancers (BRCA, UCEC, and KIRC) ancestrally European patients were statistically significantly enriched in batches processed by earlier or older capture kits and sequenced at higher depths (Figure 2; Supplementary Figure 5, available online). For example, the majority of BRCA exomes was processed using 3 versions of NimbleGen exome capture kits: 1) NimbleGen hg18 Exome v2; 2) NimbleGen (Roche NimbleGen, Inc., Pleasanton, CA) SeqCap EZ Exome v2; and 3) NimbleGen SeqCap EZ Exome v3. [Although the exome capture kits were designed based on older human genome reference builds, all reads were mapped to the GRCh38 reference as part of TCGA M3 project (8).] As shown in Figure 2, the oldest kit, NimbleGen hg18 v2 Exome, was used to process 65.2% (380 of 583) ancestrally European tumor specimens and 23.5% (38 of 162) ancestrally African tumors, as well as 60.3% (351 of 582) European germline and 23.6% (38 of 161) African germline specimens. The statistically significant enrichment of ancestrally European patients (2-sided P < .001; 2-sample proportions z test) and the statistically significantly higher sequencing depths (number of reads per exome per megabase of targeted capture region) in this earlier sample batch were likely the reasons for the overall higher sequencing coverages of ancestrally European exomes in the BRCA dataset. Later sample batches using the NimbleGen SeqCap EZ Exome v2 and v3 kits were enriched with ancestrally African patients (76.5% of African tumors and 76.4% of the African germline) and sequenced at statistically significantly lower coverages (Figure 2). Sequencing depths did not differ by ancestry for specimens processed by the same exome capture kit. Note that the read lengths across all sample batches were comparable (between 48 and 50 base pairs). Therefore, the imbalanced allocations of ancestrally African and European patients among sample batches sequenced at different depths are likely the reason for overall lower sequencing depths of ancestrally African patients in BRCA, UCEC, and KIRC (Figure 2; Supplementary Figure 5, available online). For KIRC, we could not map all exome capture kit names provided by TCGA to known kits and could not establish a clear timeline of sample processing. However, the 2 sample batches processed by Custom V2 Exome Bait and NimbleGen SeqCap EZ Exome v2 kits were statistically significantly enriched with ancestrally European samples and sequenced at statistically significantly higher depths, whereas the samples captured by the Roche SeqCap EZ HGSC VCRome kit, which was presumably designed later by scientists at the Human Genome Sequencing Center at Baylor College of Medicine to use as a clinical research kit, were all ancestrally African and were sequenced at lower depths (Supplementary Figure 5, available online).
Figure 2.
Number of reads per exome per megabase of capture region in The Cancer Genome Atlas breast cancer patients. Sequencing depths are illustrated as the number of reads per exome per megabase of the capture region. From left to right are sequencing coverage data from all individuals captured by NimbleGen kits, followed by those captured by NimbleGen hg18 Exome v2, NimbleGen SeqCap EZ Exome v2, and NimbleGen SeqCap EZ Exome v3 kits. Mann-Whitney U test 2-sided P values are shown for all comparisons. The earlier sample batch using the older NimbleGen hg18 Exome v2 kit was sequenced at 1.70-1.78 million reads per megabase of targeted region on average, which was statistically significantly higher compared with exome batches captured by the 2 more recent kits: NimbleGen SeqCap EZ Exome v2 and v3. Sequencing depths did not differ between ancestrally African and European exomes processed by the same capture kit. Patient allocations by race were unbalanced among 3 exome capture kits: ancestrally European patients were statistically significantly enriched, whereas ancestrally African patients were statistically significantly underrepresented (2-sided P < .001; 2-sample proportions z test) in the NimbleGen hg18 Exome v2 batch in which the sequencing depths were statistically significantly higher.
The reasons underlying lower sequencing coverages among African exomes for COAD, PRAD, and LUAD remain unknown. Regardless of the cause, lower sequencing depths of tumor and germline exomes likely resulted in less-complete data, underdetection of both germline variants and somatic mutations, and inferior variant quality among patients with African ancestry.
Impact of Disparate Sequencing Depths on Variant Calling and Variant Quality
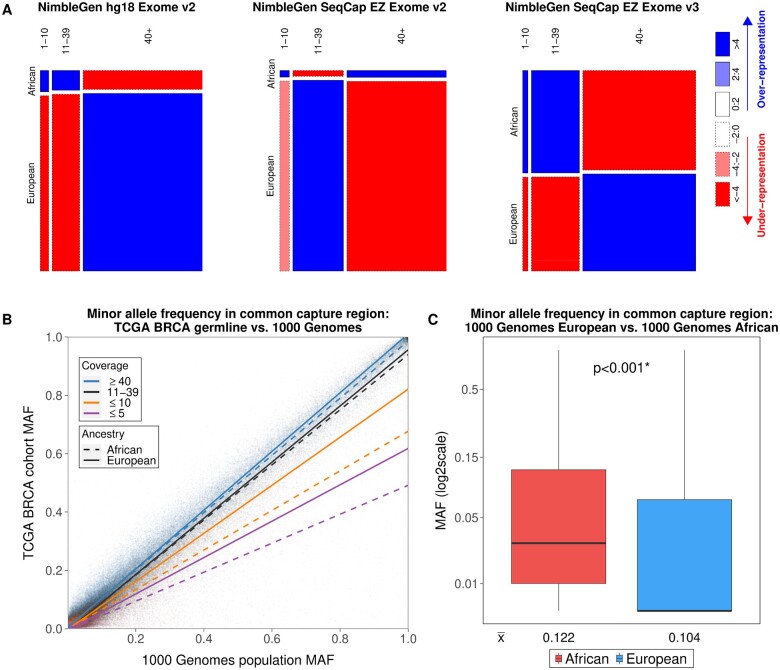
First, we investigated the consequences of disparate sequencing depths on germline variant calling, focusing on TCGA BRCA patients because the BRCA cohort enrolled the largest number of self-reported Black patients (confirmed by genetic ancestral inference) and had the biggest differences in sequencing depths between ancestrally European and African exomes. TCGA did not report germline variants at the individual level, so we performed multisample joint genotype calling as described in the methods. Previous studies have reported that the sensitivity for singleton variant detection declines statistically significantly once read depth falls below 10 reads (10). Thus, we categorized sequencing depths into 3 groups: low (positions covered by 1-10 reads), medium (11-39 reads), and high (≥40 reads). Interestingly, even though overall sequencing depths did not differ between ancestrally African and European exomes within each of the capture kit batches as shown in Figure 2 (lower panel), germline exomes from ancestrally African patients still had a higher than expected percentage of genomic positions with low coverages by all 3 kit batches (Figure 3, A;Supplementary Table 2, available online). The observed overrepresentation of low-coverage positions within batch as well as the overall lower sequencing depth across all batches in ancestrally African patients suggested that variant detection could be impaired compared with ancestrally European patients. Indeed, the BRCA cohort MAFs of the germline variants located at low-coverage positions were substantially lower than the population MAFs documented by 1000G (11) for both ancestral groups but more so for that of African (Figure 3, B;Supplementary Figure 6, available online). These data suggest that germline variants would have been underreported among ancestrally African patients in TCGA.
Figure 3.
Proportions of germline exome positions from ancestrally African and European patients at different levels of coverage in The Cancer Genome Atlas (TCGA) breast cancer (BRCA) dataset. A) Mosaic plots of read coverage levels. The common target regions of the 3 NimbleGen exome capture kits were included in the analyses. For all 3 sample batches, the low-coverage positions (1-10 reads) were enriched in ancestrally African exomes, whereas the high-coverage positions (≥40 reads) were enriched in ancestrally European exomes in the NimbleGen hg18 Exome v2 and NimbleGen SeqCap EZ Exome v3 kits. B) Low coverage resulted in underdetection of germline variants in both ancestrally African and European patients but was worse for ancestrally African patients. The variant minor allele frequencies (MAF) from TCGA BRCA patients were compared with those from the 1000 Genomes Project. For the variants located at positions with medium (11-39 reads) or high (≥40 reads) coverage, the agreement of the MAFs between 1000 Genomes Project and TCGA BRCA cohort was high, with the slopes of the corresponding regression lines close to 1. The MAFs of the variants with low coverage (1-10 reads) in TCGA BRCA cohort were statistically significantly lower compared with population MAFs from the 1000 Genomes Project in both ancestrally European and African patients, and the slopes of their regression lines substantially deviated from 1 and were substantially lower for ancestrally African compared with European exomes. C) The exome capture regions are more polymorphic in ancestrally African populations. Germline variant MAFs from the 1000 Genomes Project were statistically significantly higher (Mann-Whitney U test 2-sided P < .001) in the African population than those in the European population at the 280 714 variant positions overlapping with the NimbleGen common capture region. The horizonal lines inside the boxplot are median MAF values for either population, and the numbers below each boxplot denote the mean. BAC = B allele concentrations.
We further studied the observation that even with comparable sequencing depths between ancestrally African and European exomes (samples processed in the same batch by the same exome capture kit), disproportionally higher numbers of exome positions from patients of African ancestry had insufficient or low coverages (Figure 3, A). We found that the capture regions were more polymorphic for ancestrally African compared with European exomes. As shown in Figure 3 (Figure 3, C), the MAFs of variants located within the exome capture regions were statistically significantly higher among the African population of 1000G compared with those of the European population. We speculate that the exome capture probes might be less effective for ancestrally African genomes.
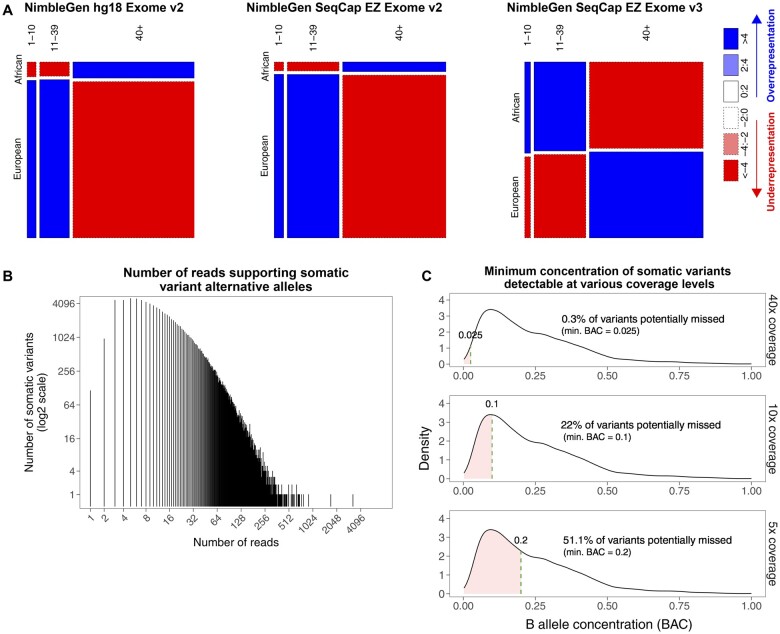
Lower sequencing depths among ancestrally African exomes also impacted somatic mutation detection. Even though the overall sequencing depths between the tumor exomes of ancestrally African and European patients were not different within batches (Figure 2, upper panel), the tumor exomes from ancestrally African patients were enriched with low-coverage positions (≤10 reads) (Figure 4, A) in the sample batch containing the majority of ancestrally African patients (processed by the NimbleGen SeqCap EZ Exome v3 kit). This enrichment of low-coverage positions might have contributed to the overall lower confidence of somatic mutation calls in ancestrally African patients (Supplementary Figure 7, available online). Notably, TCGA made every effort to report all somatic mutations, including those covered by as few as 1, 2, or 3 alternative allele supporting reads (Figure 4, B). However, even with this strategy of high-sensitivity mutation calling, substantial numbers of somatic mutations would still have been missed at the low-coverage positions. For example, as shown in Figure 4 (Figure 4, C, lower panel), assuming 5x coverage (with 5 reads at the mutation position), if only 1 alternative allele supporting read is required to call a somatic mutation, only the mutations with BAC (fraction of reads supporting alternative alleles) of at least 20.0% would have been detected. According to BAC values recorded in TCGA mutation reports, 51.1% of detected somatic mutations had BACs less than 20.0% and therefore would have been missed if the read coverage were no more than 5 at those positions. Similarly, 22.0% of TCGA-reported somatic mutations had BAC of no more than 10.0%, which would not have been detected if the sequencing depths were below 10x (Figure 4, C, middle panel). Supplementary Figures 8 and 9 (available online) illustrate the same issue for the low-coverage positions in all 6 cancers, combined or separately. These observations imply that because of the lower overall coverage as well as the disproportionally higher number of positions with insufficient coverages in ancestrally African patients, many somatic mutations were undetected, including some functionally important ones.
Figure 4.
Proportions of tumor exome positions from ancestrally African and European patients at different levels of coverage in The Cancer Genome Atlas (TCGA) breast cancer dataset. A) Mosaic plots of read coverage levels. The common target regions of the 3 NimbleGen exome capture kits were included in the analyses. Genomic positions were categorized into 3 groups based on read coverage: low-coverage positions (positions covered by 1-10 reads per patient), medium-coverage positions (11-39 reads), and high-coverage positions (≥40 reads). For samples processed by the NimbleGen SeqCap EZ Exome v2 and v3 kits, which processed the majority of ancestrally African samples, the low-coverage positions were enriched in ancestrally African tumor exomes, whereas the high-coverage positions were enriched in ancestrally European tumor exomes. B) Histogram of the number of alternative allele supporting reads per mutation position from all TCGA-reported protein-altering somatic mutations, including nonsynonymous single nucleotide mutations, splicing site mutations, and frame-shifting small insertions and deletions. C) Hypothetical undercalling of somatic mutations if only 1 alternative allele supporting read is required at depths of coverage of 40x (upper), 10x (middle), and 5x (lower). The distribution of B allele concentrations (BAC, the fraction of reads supporting alternative alleles) of protein-altering somatic mutations were calculated using TCGA mutation reports for the breast cancer samples captured by NimbleGen kits.
Discussion
We studied exome sequencing data qualities in 7 TCGA cancer types with more than 50 self-reported Black patients. The genetic inference of ancestries confirmed self-reported race with very few exceptions. Ancestrally African exomes were covered at statistically significantly lower sequencing depths compared with ancestrally European exomes for BRCA, PRAD, LUAD, UCEC, KIRC, and COAD. Using BRCA as an example, we demonstrated that this ancestral difference likely resulted from sequencing exomes in different temporal batches using different exome capture kits. Exomes processed using the oldest kit were sequenced at higher depths regardless of ancestry; however, the overrepresentation of ancestrally European patients in the early batches led to higher depths overall among the European exomes. Note that ideally a multivariate regression model simultaneously evaluating the contribution to sequencing depth from tumor purity, capture kit, sample collection and sequencing centers is preferred. Unfortunately there are many missing values in each variable, especially for tumor purity and capture kit. Therefore we instead chose to evaluate each factor separately.
In addition, higher polymorphisms among ancestrally African genomes within the targeted exome regions might further hinder the ability to sufficiently capture and sequence African-specific variants. It has been reported that the human reference genome is racially biased (12). The human reference genome was derived from 13 anonymous volunteers from Buffalo, New York, where the ethnic populations are almost all European (German, Irish, Polish, and others). The human reference genome, therefore, is mostly European as well. Because the exome capture baits were designed based on the reference genome, the capture efficiency would be subjected to this known racial or ancestral bias. The statistically significantly higher MAFs from ancestrally African exomes may lead to a higher degree of mismatches and lower capture efficiency between the baits and their targeted regions, which may result in higher percent of variant positions in ancestrally African patients covered by a lower number of reads. This speculation is based on MAF data alone. We compared the coverage differences of the regions surrounding high vs low polymorphisms and did not observe a statistically significant difference (data not shown). Because the capture efficiency and coverage is also heavily influenced by other factors such as the GC content, the role of high MAF in determining the final sequencing coverage of a region may not be directly visible.
We also examined the impact of the biased reference genome on variant calling and read mapping. For somatic mutation calls, the overall variant qualities were lower in tumors from patients of African ancestry compared with those of European ancestry. For germline variant calls, we did not observe differences between ancestrally African and European patients in the overall variant quality using variant GQ values (data not shown). The current best practice for germline genotyping is multisample joint genotyping, which calls variants using reads from all samples. This approach helps increase calling sensitivity and alleviate some of the impact from lower sequencing depth. We did not observe a higher proportion of misaligned reads in ancestrally African exomes (data not shown), probably because the read aligner scored alignment confidence based on specificity. Even if there are a small number of mismatches and gaps between the read and the reference genome, the read is still considered aligned with high confidence if it cannot be placed elsewhere in the genome.
For the other 26 cancer types in TCGA, fewer self-reported Black patients were studied and therefore not included in this analysis. Similarly, we did not study the other racial minority groups (Asian, Pacific Islanders, etc) because of small cohort sizes. Our findings serve as a reminder to those who utilize TCGA datasets in their research that there is potential underdetection of both germline variants and somatic mutations among ancestrally African individuals. These data highlight the need to consider epidemiological factors when designing and conducting future genomic and genetic studies. The scientific community and TCGA might consider resequencing residual DNAs, if still available, from these Black cancer patients with confirmed African ancestry. Alternatively, additional ancestrally African patients could be sequenced at high coverage to compensate for the current disparity.
Funding
This work was supported by the Mayo Clinic Center for Individualized Medicine at Jacksonville Florida and by Dr Yan Asmann’s base budget from Mayo Clinic Research Committee.
Notes
Role of the funders: The Center for Individualized Medicine, one of the funders, had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. Dr Asmann also partially funded this project and was involved in all activities listed above.
Disclosures: The authors declare that they have no competing interests. MES, who is a JNCI associate rditor and co-author on this article, was not involved in the editorial review or decision to publish the manuscript.
Author contributions: DPW - Writing—Original Draft Preparation, Formal Analysis, Investigation, Visualization, Methodology, Software, Data Curation, Writing—Review & Editing. MES - Conceptualization, Supervision, Writing—reviewing and editing. DCR - Conceptualization, Writing—reviewing and editing. ASM - Conceptualization, Writing—reviewing and editing. YWA - Conceptualization, Funding Acquisition, Supervision, Resources, Writing—reviewing and editing.
Data Availability
Germline and primary tumor exome BAM files aligned to the GRCh38 reference assembly, as well as protected somatic MAF files containing TCGA-called somatic mutations, were downloaded from TCGA Repository of the Genomic Data Commons (GDC) (https://portal.gdc.cancer.gov/). Patient and sample metadata (including race, sample collection sites and sequencing centers, etc.) were obtained using the R package TCGAbiolinks. Exome capture kit data and sequencing dates were downloaded using the GDC API (https://gdc.cancer.gov/developers/gdc-application-programming-interface-api). Tumor purity values were taken from a published TCGA paper (9). BED files for three NimbleGen kits were downloaded from https://sequencing.roche.com/en/support-resources/discontinued-products/seqcap-ez-exome-v3-kit.html (SeqCap EZ Exome v3), https://web.archive.org/web/20161106062747/ (hg18 Exome v2), and https://web.archive.org/web/20141010064923/ (SeqCap EZ Exome v2). The Jupyter notebook, bash scripts, and R scripts used to conduct the analyses are available at the GitHub repository https://github.com/dpwickland/Racial_Disparities_in_Exome_Read_Depth.
Supplementary Material
Contributor Information
Daniel P Wickland, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA.
Mark E Sherman, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA.
Derek C Radisky, Department of Cancer Biology, Mayo Clinic, Jacksonville, FL, USA.
Aaron S Mansfield, Division of Medical Oncology, Department of Oncology, Mayo Clinic, Rochester, MN, USA; Precision Cancer Therapeutics, Mayo Clinic Center for Individualized Medicine, Rochester, MN, USA.
Yan W Asmann, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA; Precision Cancer Therapeutics, Mayo Clinic Center for Individualized Medicine, Rochester, MN, USA.
References
- 1. Zavala VA, Bracci PM, Carethers JM, et al. Cancer health disparities in racial/ethnic minorities in the United States. Br J Cancer. 2021;124(2):315–332. doi: 10.1038/s41416-020-01038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bentley AR, Callier SL, Rotimi CN.. Evaluating the promise of inclusion of African ancestry populations in genomics. NPJ Genom Med. 2020;5(1):1.-. doi: 10.1038/s41525-019.0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asmann YW, Parikh K, Bergsagel PL, et al. Inflation of tumor mutation burden by tumor-only sequencing in under-represented groups. NPJ Precis Oncol. 2021;5(1):3–6. doi: 10.1038/s41698-021-00164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patterson N, Price AL, Reich D.. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Danecek P, Bonfield JK, Liddle J, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2):1–4. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43(1110):11.10.1–11.10.33. [DOI] [PMC free article] [PubMed]
- 7. Wang K, Li M, Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellrott K, Bailey MH, Saksena G, et al. ; for the Cancer Genome Atlas Research Network. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6(3):271–281.e7. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aran D, Sirota M, Butte AJ.. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6(1):12. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rashkin S, Jun G, Chen S, Abecasis GR; for the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO). Optimal sequencing strategies for identifying disease-associated singletons. PLoS Genet. 2017;13(6):e1006811. doi: 10.1371/journal.pgen.1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Auton A, Brooks LD, Durbin RM, et al. ; for the 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levy-Sakin M, Pastor S, Mostovoy Y, et al. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun. 2019;10(1):1–14. doi: 10.1038/s41467-019-08992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Germline and primary tumor exome BAM files aligned to the GRCh38 reference assembly, as well as protected somatic MAF files containing TCGA-called somatic mutations, were downloaded from TCGA Repository of the Genomic Data Commons (GDC) (https://portal.gdc.cancer.gov/). Patient and sample metadata (including race, sample collection sites and sequencing centers, etc.) were obtained using the R package TCGAbiolinks. Exome capture kit data and sequencing dates were downloaded using the GDC API (https://gdc.cancer.gov/developers/gdc-application-programming-interface-api). Tumor purity values were taken from a published TCGA paper (9). BED files for three NimbleGen kits were downloaded from https://sequencing.roche.com/en/support-resources/discontinued-products/seqcap-ez-exome-v3-kit.html (SeqCap EZ Exome v3), https://web.archive.org/web/20161106062747/ (hg18 Exome v2), and https://web.archive.org/web/20141010064923/ (SeqCap EZ Exome v2). The Jupyter notebook, bash scripts, and R scripts used to conduct the analyses are available at the GitHub repository https://github.com/dpwickland/Racial_Disparities_in_Exome_Read_Depth.