Abstract
Background
Germline genetic variation contributes to lung cancer (LC) susceptibility. Previous genome-wide association studies (GWAS) have implicated susceptibility loci involved in smoking behaviors and DNA repair genes, but further work is required to identify susceptibility variants.
Methods
To identify LC susceptibility loci, a family history-based genome-wide association by proxy (GWAx) of LC (48 843 European proxy LC patients, 195 387 controls) was combined with a previous LC GWAS (29 266 patients, 56 450 controls) by meta-analysis. Colocalization was used to explore candidate genes and overlap with existing traits at discovered susceptibility loci. Polygenic risk scores (PRS) were tested within an independent validation cohort (1 666 LC patients vs 6 664 controls) using variants selected from the LC susceptibility loci and a novel selection approach using published GWAS summary statistics. Finally, the effects of the LC PRS on somatic mutational burden were explored in patients whose tumor resections have been profiled by exome (n = 685) and genome sequencing (n = 61). Statistical tests were 2-sided.
Results
The GWAx–GWAS meta-analysis identified 8 novel LC loci. Colocalization implicated DNA repair genes (CHEK1), metabolic genes (CYP1A1), and smoking propensity genes (CHRNA4 and CHRNB2). PRS analysis demonstrated that these variants, as well as subgenome-wide significant variants related to expression quantitative trait loci and/or smoking propensity, assisted in LC genetic risk prediction (odds ratio = 1.37, 95% confidence interval = 1.29 to 1.45; P < .001). Patients with higher genetic PRS loads of smoking-related variants tended to have higher mutation burdens in their lung tumors.
Conclusions
This study has expanded the number of LC susceptibility loci and provided insights into the molecular mechanisms by which these susceptibility variants contribute to LC development.
Lung cancer (LC) is the most common cause of cancer-related deaths worldwide. Although most LC risk is attributable to exposure to tobacco smoke, a genetic basis for LC susceptibility was initially identified from familial aggregation studies after accounting for personal smoking habits (1-3), segregation-based analyses (4), and twin studies (5). Genome-wide association studies (GWAS) have subsequently identified multiple LC susceptibility loci in genes related to smoking behaviors (CHRNA5, CHRNA3, CHRNB4, CYP2A6) (6-8), DNA repair (CHEK2, BRCA2, ATM) (9-11), and genes related to telomere regulation (TERT, RTEL, OBFC1) (12,13) as well as many loci where the target genes are less obvious (13).
Although traditional GWAS approaches continue to expand in size, novel analytical approaches can leverage existing data from large, genotyped cohorts to identify additional susceptibility loci and explore candidate genes and potential mechanisms by which the susceptibility is mediated. In this current study, we undertook a genome-wide association by proxy (GWAx) of LC. This approach considers unaffected individuals with a first-degree relative diagnosed with the given trait as proxy patients and unaffected individuals without relatives diagnosed with the given trait as proxy controls in large genotyped biobanks (14,15). After meta-analyzing the results with existing LC GWAS, we explored potential candidate genes and functional mechanisms at the newly identified susceptibility loci by considering how the variants in candidate loci overlap (colocalize) with variants associated with other traits, such as tobacco-consumption, lung function, and gene expression quantitative trait loci (eQTL). We then performed polygenic risk scores (PRS) analyses based on the genome-wide significant variants from this expanded GWAS, as well as a novel variant method that selected variants (including subgenome-wide significant variants) that shared association with LC-related traits. Finally, we investigated how these LC PRSs affect the somatic mutation environment in patients whose tumors have been characterized by exome or whole genome sequencing (WGS).
Methods
Statistical Analysis
All statistical tests described below were 2-sided. A family history GWAx analysis of LC was undertaken using the method described by Liu et al. (14). After applying genotyping quality control metrics to exclude suboptimal genotypes and samples and limit genetic ancestry analysis to European decent (Supplementary Table 1 and Supplementary Materials, available online), 48 843 individuals with a family history of LC and 195 387 controls with no reported cancer diagnosed or reported family members with cancer were identified from the UK Biobank. A GWAx was performed using unconditional logistic regression model (with adjustment for age, sex, array type, number of siblings, and principal components from genetic-inferred ancestry). Association statistics were corrected to account for the genetic dilution related to the GWAx by doubling beta coefficients and standard error as described previously (14). Adjusted statistics from the UK Biobank GWAx and a previous LC GWAS (Transdisciplinary Research In Cancer of the Lung) cohort—29 266 patients and 56 450 controls—(Supplementary Materials, available online) were meta-analyzed using METASOFT under a fixed-effects assumption based on the inverse-variance-weighted effect size (16). Statistical significance (genome-wide significance) was defined as a P value less than 5 x 10-8. Independent genetic variants at a given locus were defined using linkage disequilibrium (LD) clumping (with a LD threshold of R2 < 0.1 and a window size of 10 000 kb). Genetic correlation (rg) between the GWAx and the GWAS was estimated using LD score regression, which was performed using the LDSC package (17).
Colocalization Analysis
Colocalization of genetic associations between LC, gene expression, and related traits was calculated using the COLOC package (https://github.com/anthony-aylward/coloc) (18) using default thresholds and a window size of 75 kb. eQTLs within lung tissues or brain regions related to addiction (substantia nigra, nucleus accumbens, frontal cortex, putamen, caudate) were obtained from the Genotype-Tissue Expression (GTEx) project (19). GWAS summary statistics were obtained from the GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN) for smoking behaviors, and summary statistics from additional related traits (forced vital capacity [FVC] and coffee consumption) were retrieved from OpenGWAS (20,21) (Supplementary Materials, available online). For this manuscript, we report the posterior probability of colocalization for a single shared variant responsible for the associations in both traits (posterior probability for hypothesis 4 [PP4]).
PRS Analysis
The PRSs were computed as the sum of the individuals weighted genotypes using PLINK and PRSice-2 software with the PRS subsequently scaled to a normalized distribution (Supplementary Materials, available online) (22,23). Genotyping weighting was derived from the estimates (log odd ratios) from the LC GWAx-GWAS meta-analysis. Variants selected for PRS inclusion were chosen by different criteria. First, we selected variants that achieved genome-wide significance (GWS) in the GWAx-GWAS meta-analysis (gwPRS). Second, a partial least squares (PLS) method was used to select variants involved with both LC and smoking (smPRS) or gene expression (eQTLPRS) based on previously published GWAS summary statistics and the GTEx summary statistics (Supplementary Materials, available online). Finally, a combined PRS of the unique variants from gwPRS, smPRS, and eQTLPRS was also considered (Supplementary Materials, available online).
The PRS construction considered LD between variants (any variant with R2 > 0.1 with a sentinel variant was excluded). The PRSs were assessed in a validation cohort of 1 666 LC patients and 6 664 controls from the UK Biobank that were not included in the GWAx described above. Unconditional logistic regression was used to test the association between PRSs and LC, with sex, array type, age of recruitment, and the first 5 principal components from genetic-inferred ancestry as covariates. Regression results are reported as an odds ratios unit change per a standard deviation in the PRS distribution.
Somatic Mutation Analysis
The association between the PRSs and somatic mutational burden was tested in The Cancer Genome Atlas (TCGA) cohorts of 685 LC samples of European ancestry and in an independent cohort of 61 LC patients samples, identified from central and eastern Europe (13). Germline genotypes were derived from Affymetrix arrays which was accessed via the database of Genotypes and Phenotypes (dbGAP) (application Project #2731) using single nucleotide polymorphism (SNP) imputation to the 1000 Genome phase 3 reference panel (Supplementary Materials, available online). TCGA somatic mutations (derived from WGS) were retrieved from the study of Ellrott et al. (24). In the replication cohort, germline genotypes and somatic mutations were derived from WGS undertaken in the patients paired normal tumor samples sequenced on a IlluminaX5 DNA sequencer (Supplementary Materials, available online). Total number of mutations was computed as a mutational feature and reported smoking tobacco-related signatures (cosmic signature annotation Single Base Substitution 4 (SBS4), Doublet Base Substitutions 2 (DBS2), and Indel signature 83 [A and B] (ID83A, ID83B) were generated using the SigProfiler pipeline (Supplementary Materials, available online). To test the association between the PRS and the tumor somatic mutational burden, the PRS were regressed against tumor somatic mutational burden as a continuous trait using Quasi-Poisson regression to account for overdispersed and left skewed distributions of the DNA mutational features. Covariates added in all PRS models included age, sex, the first 5 principal components from genetic inferred ancestry, tumor purity, and a categorical variable indicating the cohort type as appropriate. Effect estimates are given as incidence rate ratios (IRR). A P value less than .05 was considered as level of significance for PRS analyses. All statistical tests performed to calculate P values were 2-sided tests.
Results
The 8 Novel Susceptibility Loci
The family history GWAS (GWAx) on 48 843 self-reported family history LC patients and 195 387 controls from the UK Biobank (Supplementary Table 1, available online) identified 5 loci (5p15.33, 6p21.32, 12p13.33, 13q13.1, and 15q25.1) that had previously been discovered from the traditional GWAS previously performed on the Transdisciplinary Research In Cancer of the Lung cohort (Supplementary Table 2 and Supplementary Figure 1, available online). Genetic correlation using LD score regression confirmed a strong relationship between GWAx and the GWAS (rg = 1, SE = 0.066, P < .001) supporting the utility of the GWAx approach to detect susceptibility loci.
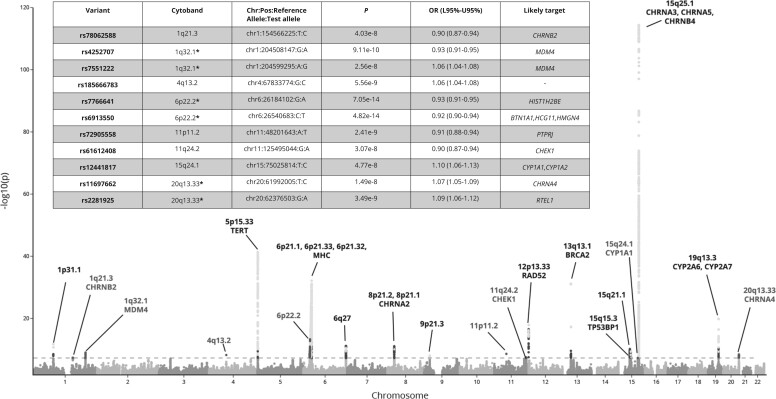
Meta-analysis between the GWAx and the traditional LC GWAS identified 65 variants that achieved a P value less than 5 x 10-8 across 23 distinct genomic loci defined by cytoband (Figure 1) after LD clumping genetic variants (Supplementary Table 2, available online). At previously described LC susceptibility loci, the meta-analysis also identified independent (R2 < 0.1) low-frequency (minor allele frequency [MAF] < 0.05) variants associated with LC at 5p15.33 (rs35812074), 19q13.2 (rs1801272), 15q25.1 (rs2229961, rs8192479, rs151118057), and 12p13.33 (rs7487683) in addition to the previously described common genetic variants (Supplementary Table 2, available online). At 13q13.1, where a rare LC susceptibility allele has been described (rs11571833, K3326X BRCA2; MAF = 0.01), an independent common susceptibility allele was also noted (rs11571734; MAF = 0.28).
Figure 1.
Manhattan plot of the meta-analysis of the genome-wide by proxy (GWAx) with genome-wide association study (GWAS) into lung cancer. The Manhattan plot displays the results of the meta-analysis of the GWAx (48 843 proxy patients and 195 387 controls without a family history of any cancer) and the Transdisciplinary Research In Cancer of the Lung GWAS (29 266 patients and 56 450 controls) with already identified and novel loci noted with the likely candidate gene name presented. The table represents the newly 11 independent loci across 8 distinct cytoband regions (sites with 2 independent hits are denoted by * within the cytoband column). The x-axis is the chromosome position across the autosomal chromosomes, and the y-axis contains the association level displayed as the -log10(P value), derived by a multivariate logistic regression model. The dotted line displays the genome-wide significance threshold (5 x 10-8). L95% = lower bound confidence interval; OR = odds ratio; U95% = upper bound confidence interval.
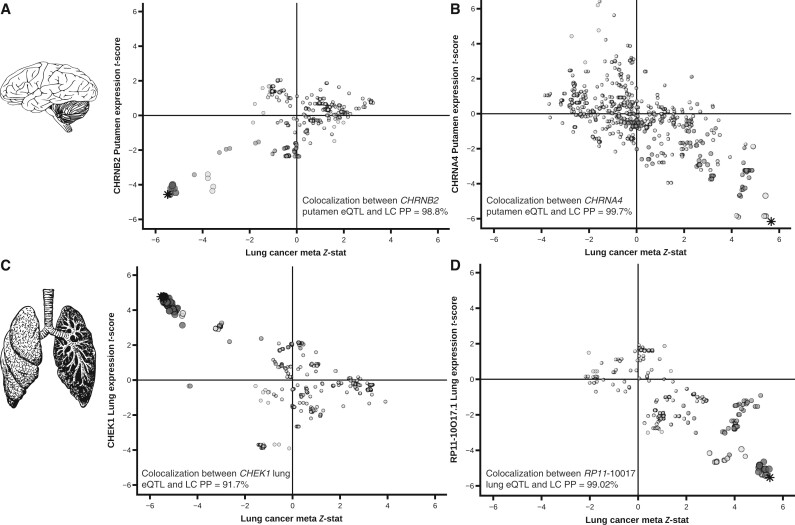
Eleven LC susceptibility variants at 8 loci have previously not been associated with LC at genome-wide (GW) significance (Figure 1). We explored these loci using colocalization with traits related to LC; smoking behavior; gene expression, particularly in the lung epithelium and brain (addiction); and lung function. Using the GSCAN summary statistics, we observed that the LC susceptibility variants at 1q21.3-rs78062588, 6p22.2-rs7766641, and 20q13.33-rs11697662 were also associated at GW significance with traits related to propensity to smoke tobacco (Supplementary Table 2, available online). The sentinel variants at 1q21.3-rs78062588 and 20q13.33-rs11697662 are also eQTLs for the nicotinic acetylcholine receptors (nAChRs) subunits CHRNB2 and CHRNA4 (Figure 2, A and B; Supplementary Tables 2 and 3, available online). At 6p22.2, LC susceptibility loci were noted (Supplementary Table 2, available online), typified by 2 sentinel variants: rs6913550 and rs7766641. rs7766641 was also associated with propensity to smoke (colocalization between GSCAN cigarettes per day [CPD] and LC: PP4 = 99%), whereas, curiously, rs6913550 was not (colocalization between CPD and LC: PP4 = 0%) (Supplementary Table 2, available online).
Figure 2.
Brain and lung eQTLs discovered within the 8 novel loci. Colocalization plots between lung cancer (x-axis) and CHRNB2 putamen expression (1q21.3) A) CHRNA4 putamen expression (20q13.33); (B) CHEK1 lung expression (11q24.2); (C) RP11-10O17.1 lung gene expression (15q24); (D) (y-axis). Each variant and eQTL status were compared using COLOC for colocalization to confirm that the lung cancer SNP was the same SNP driving the eQTL effect in both brain and lung tissues, the Bayesian posterior probability (PP4) of each gene was tested. Stars indicate the variant of interest and shading scaled representing the level of LD shared between other markers with sentinel variant (r2 > 0.8; r2 > 0.4; r2 > 0.1). eQTL = expression quantitative trait loci; LC = lung cancer; LD = linkage disequilibrium; SNP = single nucleotide polymorphism.
At 1q32.1, 11p11.2, 11q24.2, and 15q24, the sentinel variants (rs4252707, rs72905558, rs61612408, rs12441817, respectively) were not associated with smoking behaviors (colocalization between CPD and LC for all 4 variants: PP4 = 0%). 11q24.2-rs61612408 was associated with the expression of the CHEK1 gene in multiple tissues including lung epithelia. Further, there was evidence for colocalization between these associations (colocalization between CHEK1 lung eQTL and LC: PP4 = 91.7%), with the allele associated with increased expression correlating with decreased risk of LC (Figure 2, C). The 15q24-CYP1A1 locus has been associated with multiple traits, including coffee consumption (20) and FVC (21). In this study, there was evidence for colocalization with LC but only for FVC (colocalization for rs12441817 between coffee consumption and LC: PP4 = 0.00%; colocalization between FVC and LC: PP4 = 97.05%) (Supplementary Figure 2, available online). There was also colocalization for rs12441817 and CYP1A1 expression in the nucleus accumbens (colocalization: PP4 = 93.52%) (Supplementary Figure 3, available online) and an eQTL effect with the processed pseudogene RP11-10O17.1 in lung tissue (colocalization between eQTL RP11-10O17.1 and LC: PP4 = 99.02%) (Figure 2, D). At 4q13.2-rs185666783, the candidate genes remain ambiguous.
PRS Evaluation
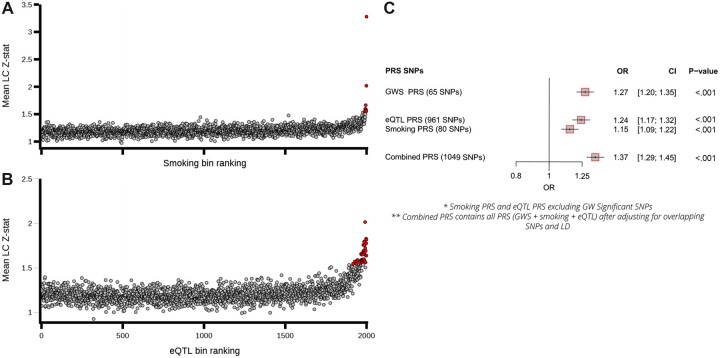
Next, we constructed PRSs from variants selected from our meta-analysis and tested their ability to predict LC risk in a validation cohort of 1 666 LC patients and 6 664 matched controls from the UK Biobank. First, we selected 65 independent variants that reached a GWS threshold from our meta-analysis. This PRS was associated with LC (gwPRS: odds ratio [OR] per standard deviation increase in PRS = 1.27, 95% confidence interval [CI] = 1.20 to 1.35; P < .001) (Figure 3). We also sought to select relevant variants that did not pass the GWS threshold. As many of LC GWS variants identified by our meta-analysis also tended to be associated with smoking behaviors and/or eQTLs (Supplementary Table 2, available online), we used PLS of published GWAS summary statistics (GSCAN and GTEx) to identify variants associated with LC as well as smoking behaviors and/or eQTL features (see Supplementary Materials). There appeared to be an excess of evidence for association with LC in an important number of the variants selected by this approach (Figure 3, A). Two polygenic risk scores, smPRS (80 variants) and eQTLPRS (961 variants), were constructed using the non-GWS variants selected by this PLS approach, with number of variants selected guided by the degree of enrichment observed (see Supplementary Materials, available online; Figure 3, A and B). Both PRSs were associated with LC in the validation LC patient-control cohort implying these non-GWS genetic variants are enriched for susceptibility alleles and add value to risk prediction (Figure 3, C). Lastly, we constructed a combined PRS (1049 variants) from the GWS variants, eQTL, and the smoking propensity variants, which improved LC risk prediction in this independent series (OR = 1.37, 95% CI = 1.29 to 1.45; P < .001).
Figure 3.
Germline polygenic risk score construction using smoking and eQTL related SNPs and performance testing within the UK Biobank lung cancer cohort. A) The mean lung cancer association statistics calculated by variant bins (100 variants per bin) ranked by partial least squares (PLS) components. Variants (clumped on LD based on lung cancer P values) were ranked based on PLS components for smoking propensity (Component1_smoking, top) and eQTLs (Component1_eQTL, [B]) (x-axis) and plotted against the mean lung cancer Z statistics calculated across variants in each bin (y-axis). Bin values that exceed 3 SDs from the mean are noted, with the excess observed (number of bins smoking propensity = 9, number of bins eQTL = 37) implying that the variants within these bins are enriched for LC-susceptibility alleles. C) A forest plot of the performance of the constructed PRSs in comparison to the PRS based on the 65 GWS independent loci as a baseline which included array type, sex, age of recruitment and the first 5 principal components from genetic-inferred ancestry). CI = confidence interval; eQTL = expression quantitative trait loci; LC = lung cancer; LD = linkage disequilibrium; GWS = genome-wide significant; OR = odds ratio; PRS = polygenic risk scores; SNP = single nucleotide polymorphism.
PRS Germline Influences on Mutational Burden and Mutational Signatures
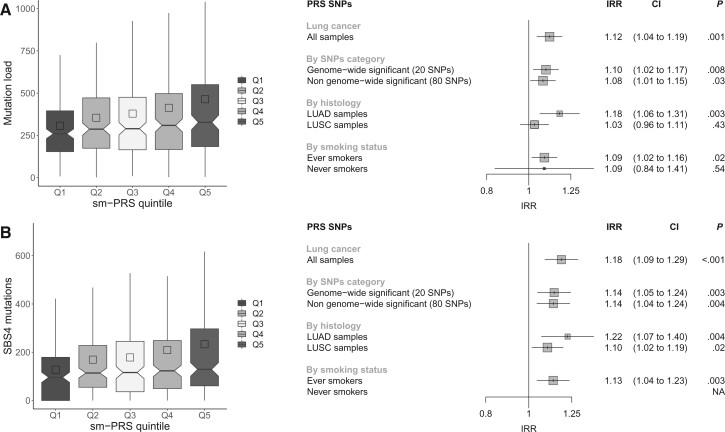
Finally, we evaluated the association of the GWS PRS (gwPRS), smoking propensity PRS (smPRS), eQTLPRS, and the combined PRS with somatic mutational burden. In the analysis of 685 lung tumors from TCGA, there was no evidence for association involving the GWS PRS (IRR = 1.03, 95% CI = 0.96. to 1.10; P = .44), eQTLPRS (IRR = 1.05, 95% CI = 0.9 to 1.13; P = .14) and the combined PRS (IRR = 1.03, 95% CI = 0.96 to 1.10; P = .37) on mutation burden (Supplementary Figure 5, available online), however smPRS was associated with somatic mutation load (IRR = 1.12, 95% CI =1.04 to 1.19; P = < .001) (Figure 4, A). The smPRS was similarly associated with burden of mutational signatures attributed to tobacco smoke (SBS4 : IRR = 1.18, 95% CI = 1.09 to 1.29; P < .001) (Figure 4, B) and was also observed in somatic insertions and deletions mutation signatures related to tobacco smoke: DBS2 (IRR = 1.17, 95% CI = 1.05 to 1.29; P = .003), ID83A (IRR = 1.21, 95% CI = 1.07 to 1.37; P = .002), and ID83B (IRR = 1.13, 95% CI = 1.01 to 1.27; P = .04), respectively (Supplementary Figure 6, available online). These associations were observed more prominently in patients with Lung Adenocarcinoma (IRR = 1.18, 95% CI = 1.06 to 1.31; P = .002) (Figure 4, A). The 15q25 CHRNA5 LC sentinel variant rs55781567 had the most striking effect (average total mutation count of 327 for homozygous risk carriers of the G allele compared to 283 in homozygous nonrisk carriers of the C allele) (Supplementary Figure 7, available online), but the associations remained significant after excluding GW variants for LC (Figure 4, A). The association between the smPRS and somatic mutation burden was replicated in 61 patients who have undergone WGS (IRR = 1.39, 95% CI = 1.03 to 1.89; P = .04) (Supplementary Figure 8, available online).
Figure 4.
Polygenic risk scores for smoking (smPRS) associations with total number of mutations and mutations attributable to SBS4 in TCGA cohort. A) Associations with total number of mutations. B) Associations with SBS4 mutations. The left panels represent the distribution of the number of mutations in the smPRS quintiles. The right panels correspond, respectively, to the forest plots of smPRS associations with total mutational burden (panel A) and SBS4 mutations (panel B). For each PRS, the association was tested 1) in all lung cancer patients when considering all SNPs in the smPRS SNPs selection, 2) in all lung cancer patients when considering different subsets of SNPs in the PRS computation, 3) stratifying by histology, and 4) stratifying by smoking status. CI = confidence interval; IRR = incidence rate ratios; LUAD = Lung adenocarcinoma; LUSC = Lung Squamous Cell Carcinoma; NA = Not available; Q = quintile; TCGA = The Cancer Genome Atlas; SNP = single nucleotide polymorphism.
Discussion
This study identified 23 LC susceptibility loci, including 8 novel loci, by combining large, genotyped biobank data and traditional GWAS. Of the 8 novel loci, 3 were also associated with propensity to smoke. This included brain eQTLs variants for both subunits of the neuronal nAChRs α4β2 receptor. Variants in LD with the α4 subunit (rs2373500) have been described in nicotine dependency and LC risk, albeit not at GW significance for LC (25), whereas the β2 receptor association with LC risk has not been previously described. The neuronal nAChRs α4β2 receptor is the most abundant nAChR subtype within the human brain and important within the dopaminergic signaling pathway. The α4β2 receptor has a key role in nicotine dependence behaviors (26) and is a major target in nicotine addiction intervention (27,28). The third novel locus related to LC and propensity to smoke is telomeric to the major histocompatibility complex (MHC) region, where the target candidate gene(s) is less obvious. The MHC region was among the first susceptibility loci to be associated with LC (6,29-31). However, rs7766641 is not in LD with these previously described variants (R2 < 0.001) and associated with the number of CPD, implying that these are distinct associations.
This meta-analysis also identified additional LC susceptibility loci that appear to be independent of smoking propensity. These included variants at 15q24 near CYP1A1, CYP1A2, and CYP11A1 that participate in the metabolism of many different xenobiotics and some endogenous substrates. Variants at the 15q24 CYP1A1 and CYP1A2 locus have been linked with multiple traits, notably other forms of propensity coffee consumption (20) and FVC (21), although these variants associated to each trait appear to be distinct. Colocalization appears to implicate FVC as more likely to be involved in the LC association, and the etiological link also seems more plausible considering aspects of lung function and LC risk. For tissue expression, rs12441817 colocalized with lung tissue expression of the processed pseudogene RP11-10O17.1 (Figure 2, D) although how this pseudogene relates to LC susceptibility is unclear.
An additional novel LC susceptibility variant, rs61612408, was a lung tissue eQTL for the DNA repair gene CHEK1 (Figure 2, C). We additionally noted the variant rs4252707 impacting the MDM4 gene, which is an important p53 regulator. This variant was previously associated with nonglioblastoma tumors (32) and more recently squamous cell carcinomas of the lung and head and neck (33). At 11p11.2, despite evidence for an eQTL effect, colocalization analysis showed little evidence for involvement with genes C1QTNF4 (lung) and MTCH2 (brain-cortex), suggesting that these signals are unlikely to explain the LC association. At 4q13.2, the finding remains ambiguous, but from histological subtypes analysis performed from the previous reported GWAS study, it appears that this signal is mostly found in lung adenocarcinoma.
We additionally sought to use the shared genetic etiology between LC susceptibility, smoking-related traits, and gene expression annotations (eQTL) to explore variants that did not achieve GW significance. We used the PLS method to select variants related to these traits for the PRS analyses and demonstrated that such variants are indeed enriched for susceptibility alleles. Although the role of these individual variants remains to be confirmed, these sub-GWS variants were located near relevant candidate genes (propensity to smoke candidates like CHNRA6 and DBH, and eQTLs for ERCC2, RAD51C, XRCC3, and CASP8). Combining both sub-GWS PRS lists (smPRS and eQTL PRS) with GW-significant results reached an odds ratio of 1.37 per standard deviation unit increase in score improving on previous PRS predictions (OR = 1.17 and 1.26, respectively) (34,35), despite the conservative clumping approach (R2 < 0.1) employed. This suggests that integrating functional annotations may be of interest for PRS analysis.
Lastly, the analysis of the smPRS demonstrated an association between a person’s genetic risk load and mutation burden and/or burden of tobacco-related somatic mutational signatures, within 2 independent patient cohorts and using different sequencing methods (exome sequencing and WGS). These associations appear consistent with the notion that genetic variants influence individuals’ smoking behavior, which in turn influences their carcinogenic exposure, and consequently, their somatic mutation burden (36).
In conclusion, this work has increased the number of variants associated with LC susceptibility, with the identification of novel susceptibility loci. PRS analysis highlighted that many additional variants remain to be discovered and provided insights into the carcinogenic mechanisms.
Funding
This work was supported by the Institut National du Cancer (INCa) (GeniLuc 2017-1-TABAC-03-CIRC-1 - [TABAC 17‐022],) National Institutes of Health (NIH) / National Cancer Institute (NCI), Integral NIH 5U19CA203654-03, Cancer Research UK (grant number C18281/A29019), the France Génomique National infrastructure, funded as part of the (Investissements d’Avenir) program managed by the Agence Nationale pour la Recherche (contract ANR-10-INBS-09). Christopher Amos is a Research Scholar of the Cancer Prevention Institute of Texas and supported by RR170048. Ricardo Penha is supported by a IARC Postdoctoral Fellowship at the International Agency for Research on Cancer.
Notes
Role of the funders: The funders provided financial assistance for personnel and the genomic analysis for this work. The funders did not play a role in the design; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclosures: The authors have no conflicts of interest regarding the present study.
Author contributions: AG: Conceptualization, Formal analysis, Data curation, Methodology, Investigation, Software, Validation, Writing—Draft, review and editing, Visualization. JA: Conceptualization, Formal analysis, Data curation, Methodology, Investigation, Software, Validation, Writing—Draft, review and editing, Visualization. RP: Formal analysis, Data curation, Validation, Investigation, Writing-Review & Editing. KSB: Formal analysis, Writing-Review & Editing. VG: Formal analysis, Data curation. CV: Formal analysis, Data curation. BA: Formal analysis, Data curation. MM: Formal analysis, Data curation RO: Data curation VM: Data curation. AB: Data curation. JF: Data curation. DZ: Resources. AM: Resources. BS: Resources. VJ: Resources. MS: Resources. DM: Resources. JS: Resources. MO: Resources. ILCCO consortium: Resources, Funding acquisition JW: Data curation. SR: Data curation LK: Writing—Review and Editing. RH: Writing—Review and Editing. SK: Writing—Review and Editing. PB: Resources. ASS: Data curation. AF: Data curation. AV: Data curation. MJ: Writing—Review and Editing. CA: Conceptualization, Writing—Review and Editing, Funding acquisition, Resources. MF: Conceptualization, Supervision, Methodology, Writing—Draft, review and editing. JM: Conceptualization, Supervision, Data curation, Methodology, Writing—Draft, review and editing, Funding acquisition.
Acknowledgements: We would like to acknowledge TCGA Research Network (https://www.cancer.gov/tcga) and the contribution of specimen donors and research groups involved in this resource. We also would like to acknowledge the GTEx project and the supporting bodies (https://commonfund.nih.gov/GTEx), specimen donors, and research groups. Additionally, we would like to acknowledge the work carried about by the Benjamin Neale lab for their work on the UK Biobank (http://www.nealelab.is/uk-biobank/).
We thank the International LC Case Control consortium (ILCCO) for contributing lung cancer summary statistics. The ILCCO researchers are listed in the Supplementary Materials (available online) with affiliations.
Disclaimer: Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article, and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Data Availability
Additional information, including summary statistics, PRS lists, code used in this study can be found on the project webpage here: https://iarc-genetics.github.io/GWAx_lung_cancer/.
Oncoarray data and summary statistics can be accessed by the database of Genotypes and Phenotypes (dbGAP) under accession phs000876.v1.p1. TCGA data was accessed by dbGAP through Project #2731: Investigation of tobacco-related cancer susceptibility genes following a 2-hit carcinogenic model.
Supplementary Material
Contributor Information
Aurélie A G Gabriel, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Joshua R Atkins, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Ricardo C C Penha, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Karl Smith-Byrne, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France; Cancer Epidemiology Unit, Nuffield Department of Population Health, University of Oxford, Oxford, England.
Valerie Gaborieau, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Catherine Voegele, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Behnoush Abedi-Ardekani, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Maja Milojevic, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Robert Olaso, Université Paris-Saclay, The French Alternative Energies and Atomic Energy Commission (CEA), Centre National de Recherche en Génomique Humaine (CNRGH), Evry, France.
Vincent Meyer, Université Paris-Saclay, The French Alternative Energies and Atomic Energy Commission (CEA), Centre National de Recherche en Génomique Humaine (CNRGH), Evry, France.
Anne Boland, Université Paris-Saclay, The French Alternative Energies and Atomic Energy Commission (CEA), Centre National de Recherche en Génomique Humaine (CNRGH), Evry, France.
Jean François Deleuze, Université Paris-Saclay, The French Alternative Energies and Atomic Energy Commission (CEA), Centre National de Recherche en Génomique Humaine (CNRGH), Evry, France.
David Zaridze, Russian N.N. Blokhin Cancer Research Centre, Moscow, Russian Federation.
Anush Mukeriya, Russian N.N. Blokhin Cancer Research Centre, Moscow, Russian Federation.
Beata Swiatkowska, Department of Environmental Epidemiology, Nofer Institute of Occupational Medicine, Lodz, Poland.
Vladimir Janout, Faculty of Medicine, Palacky University, Olomouc, Czech Republic.
Miriam Schejbalová, First Faculty of Medicine, Charles University, Prague, Czech Republic.
Dana Mates, National Institute of Public Health, Bucharest, Romania.
Jelena Stojšić, Department of Thoracic Pathology, Service of Pathology, University Clinical Centre of Serbia, Belgrade, Serbia.
Miodrag Ognjanovic, International Organisation for Cancer Prevention and Research, Belgrade, Serbia.
John S Witte, Department of Epidemiology & Biostatistics, University of California San Francisco, San Francisco, CA, USA.
Sara R Rashkin, Department of Epidemiology & Biostatistics, University of California San Francisco, San Francisco, CA, USA; Department of Hematology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Linda Kachuri, Department of Epidemiology & Biostatistics, University of California San Francisco, San Francisco, CA, USA.
Rayjean J Hung, Lunenfeld-Tanenbaum Research Institute, Sinai Health, Toronto, Canada.
Siddhartha Kar, MRC Integrative Epidemiology Unit, University of Bristol, Bristol, UK; Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
Paul Brennan, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Anne-Sophie Sertier, Fondation Synergie Lyon Cancer, Plateforme de bioinformatique Gilles Thomas, Lyon, France.
Anthony Ferrari, Fondation Synergie Lyon Cancer, Plateforme de bioinformatique Gilles Thomas, Lyon, France.
Alain Viari, Fondation Synergie Lyon Cancer, Plateforme de bioinformatique Gilles Thomas, Lyon, France; Inria Centre de Recherche Grenoble Rhone-Alpes, Grenoble, France.
Mattias Johansson, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
Christopher I Amos, Institute for Clinical and Translational Research, Baylor College of Medicine, Houston, USA.
Matthieu Foll, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
James D McKay, Genomic Epidemiology Branch, International Agency for Research on Cancer/World Health Organization (IARC/WHO), Lyon, France.
References
- 1. Tokuhata GK, Lilienfeld AM.. Familial aggregation of lung cancer in humans. J Natl Cancer Inst. 1963;30(2):289-312. [PubMed] [Google Scholar]
- 2. Schwartz AG, Yang P, Swanson GM.. Familial risk of lung cancer among nonsmokers and their relatives. Am J Epidemiol. 1996;144(6):554-562. doi: 10.1093/oxfordjournals.aje.a008965 [DOI] [PubMed] [Google Scholar]
- 3. Ooi WL, Elston RC, Chen VW, Bailey-Wilson JE, Rothschild H.. Increased familial risk for lung cancer. J Natl Cancer Inst. 1986;76(2):217-222. doi: 10.1093/jnci/76.2.217. [DOI] [PubMed] [Google Scholar]
- 4. Sellers TA, Bailey-Wilson JE, Elston RC, et al. Evidence for mendelian inheritance in the pathogenesis of lung cancer. J Natl Cancer Inst. 1990;82(15):1272-1279. doi: 10.1093/jnci/82.15.1272. [DOI] [PubMed] [Google Scholar]
- 5. Mucci LA, Hjelmborg JB, Harris JR, et al. Familial risk and heritability of cancer among twins in Nordic countries. JAMA. 2016;315(1):68-76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633-637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 7. Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616-622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patel YM, Park SL, Han Y, et al. Novel association of genetic markers affecting CYP2A6 activity and lung cancer risk. Cancer Res. 2016;76(19):5768-5776. doi: 10.1158/0008-5472.CAN-16-0446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brennan P, McKay J, Moore L, et al. Uncommon CHEK2 mis-sense variant and reduced risk of tobacco-related cancers: case control study. Hum Mol Genet. 2007;16(15):1794-1801. doi: 10.1093/hmg/ddm127 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46(7):736-741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ji X, Mukherjee S, Landi MT, et al. Protein-altering germline mutations implicate novel genes related to lung cancer development. Nat Commun. 2020;11(1):2220.doi: 10.1038/s41467-020-15905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40(12):1404-1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McKay JD, Hung RJ, Han Y, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017;49(7):1126-1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu JZ, Erlich Y, Pickrell JK.. Case-control association mapping by proxy using family history of disease. Nat Genet. 2017;49(3):325-331. doi: 10.1038/ng.3766. [DOI] [PubMed] [Google Scholar]
- 15. Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51(3):404-413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han B, Eskin E.. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88(5):586-598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291-295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383.doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318-1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sulem P, Gudbjartsson DF, Geller F, et al. Sequence variants at CYP1A1-CYP1A2 and AHR associate with coffee consumption. Hum Mol Genet. 2011;20(10):2071-2077. doi: 10.1093/hmg/ddr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J, Elsworth B, Wade KH, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408. doi:10.7554/eLife.34408. [DOI] [PMC free article] [PubMed]
- 22. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi SW, O’Reilly PF.. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. 2019;8(7):giz082. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ellrott K, Bailey MH, Saksena G, et al. ; for the Cancer Genome Atlas Research Network. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6(3):271-81.e7. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu M, Jiang Y, Wedow R, et al. ; for the HUNT All-In Psychiatry. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51(2):237-244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McGranahan TM, Patzlaff NE, Grady SR, Heinemann SF, Booker TK.. Α4β2 nicotinic acetylcholine receptors on dopaminergic neurons mediate nicotine reward and anxiety relief. J Neurosci. 2011;31(30):10891-10902. doi: 10.1523/JNEUROSCI.0937-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walsh RM Jr, Roh SH, Gharpure A, Morales-Perez CL, Teng J, Hibbs RE.. Structural principles of distinct assemblies of the human α4β2 nicotinic receptor. Nature. 2018;557(7704):261-265. doi: 10.1038/s41586-018-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gonzales D, Rennard SI, Nides M, et al. Varenicline, an alpha4beta2 nicotinic acetylcholine receptor partial agonist, vs sustained-release bupropion and placebo for smoking cessation: a randomized controlled trial. JAMA. 2006;296(1):47-55. doi: 10.1001/jama.296.1.47. [DOI] [PubMed] [Google Scholar]
- 29. Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40(12):1407-1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferreiro-Iglesias A, Lesseur C, McKay J, et al. Fine mapping of MHC region in lung cancer highlights independent susceptibility loci by ethnicity. Nat Commun. 2018;9(1):3927.doi: 10.1038/s41467-018-05890-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69(16):6633-6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. ; for the GliomaScan Consortium. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789-794. doi: 10.1038/ng.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lesseur C, Ferreiro-Iglesias A, McKay JD, et al. Genome-wide association meta-analysis identifies pleiotropic risk loci for aerodigestive squamous cell cancers. PLoS Genet. 2021;17(3):e1009254.doi: 10.1371/journal.pgen.1009254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kachuri L, Graff RE, Smith-Byrne K, et al. Pan-cancer analysis demonstrates that integrating polygenic risk scores with modifiable risk factors improves risk prediction. Nat Commun. 2020;11(1):6084.doi: 10.1038/s41467-020-19600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hung RJ, Warkentin MT, Brhane Y, et al. Assessing lung cancer absolute risk trajectory based on a polygenic risk model. Cancer Res. 2021;81(6):1607-1615. doi: 10.1158/0008-5472.CAN-20-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X, Ricciuti B, Nguyen T, et al. Association between smoking history and tumor mutation burden in advanced non-small cell lung cancer. Cancer Res. 2021;81(9):2566-2573. doi: 10.1158/0008-5472.CAN-20-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional information, including summary statistics, PRS lists, code used in this study can be found on the project webpage here: https://iarc-genetics.github.io/GWAx_lung_cancer/.
Oncoarray data and summary statistics can be accessed by the database of Genotypes and Phenotypes (dbGAP) under accession phs000876.v1.p1. TCGA data was accessed by dbGAP through Project #2731: Investigation of tobacco-related cancer susceptibility genes following a 2-hit carcinogenic model.