Abstract
The highly conserved DnaK chaperones consist of an N-terminal ATPase domain, a central substrate-binding domain, and a C-terminal domain whose function is not known. Since Bacillus subtilis dnaK was not able to complement an Escherichia coli dnaK null mutant, we performed domain element swap experiments to identify the regions responsible for this finding. It turned out that the B. subtilis DnaK protein needed approximately normal amounts of the cochaperone DnaJ to be functional in E. coli. The ATPase domain and the substrate-binding domain form a species-specific functional unit, while the C-terminal domains, although less conserved, are exchangeable. Deletion of the C-terminal domain in E. coli DnaK affected neither complementation of growth at high temperatures nor propagation of phage λ but abolished degradation of ς32.
Hsp70 chaperones are a highly conserved group of proteins which participate in various cellular processes, including the folding of nascent polypeptides, assembly and disassembly of multimeric protein structures, membrane translocation of secreted proteins, and protein degradation (3, 11, 12). The DnaK protein, the prokaryotic Hsp70 homologue, has been found in all of the eubacterial species examined so far. Its chaperone activity relies on the transient association of DnaK with substrates in a process controlled by ATP and the cochaperones DnaJ and GrpE (12, 18). In the ATP conformation, DnaK binds and releases substrates very rapidly (18, 21, 25, 27). Upon ATP hydrolysis, DnaK is switched into the ADP conformation, which exchanges substrates slowly. The ATPase and substrate-binding activities of DnaK are divided into two separable functional units: the N-terminal ATPase domain (amino acid [aa] 1 to 385; ∼44 kDa) and the central substrate-binding domain (aa 386 to 540; ∼17 kDa). These two domains are followed by a C-terminal domain (aa 543 to 637; ∼10 kDa) whose function is unknown. During our work on Bacillus subtilis dnaK, we found that the Escherichia coli ΔdnaK52 mutant could not be complemented by the dnaK gene of B. subtilis for growth defects at high temperatures and for propagation of phage λ. Therefore, this study was performed to identify the region(s) within the B. subtilis DnaK protein which is responsible for this failure to complement the ΔdnaK52 mutant of E. coli for growth at high temperatures and for propagation of phage λ. For this purpose, a series of hybrid genes have been constructed based upon the domain model of the Hsp70 proteins.
Construction of hybrid dnaK genes.
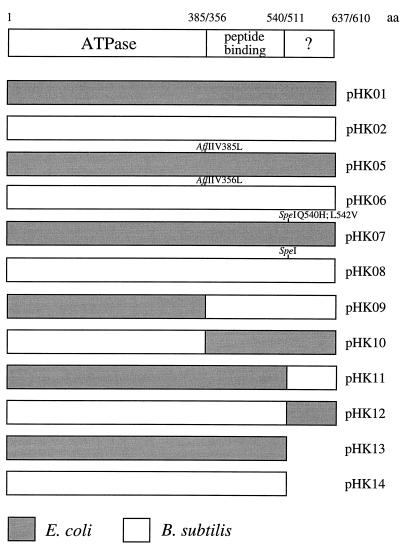
To allow reciprocal exchange of the different domains, a restriction site was first introduced into the interdomain region separating the ATPase and the peptide-binding domains (AflII site) and that separating the peptide-binding domain and the C-terminal domain (SpeI site). The amino acid exchanges caused by the introduction of a restriction site in pHK05, pHK06, and pHK07 did not change the complementation profiles in comparison to the corresponding wild-type counterparts (data not shown). The different hybrid genes shown in Fig. 1 were then constructed. All dnaK genes, the two wild types and the mutant alleles, were ligated into plasmid pUHE21-2fdΔ12 (1) downstream of an isopropyl-β-d-thiogalactopyranoside (IPTG)-controllable promoter and transformed into MC4100 (24) and isogenic dnaK derivatives containing plasmid pDMI,1 (15) coding for the LacI repressor. Upon addition of 1 mM IPTG, all dnaK genes from plasmids pHK01 through pHK14 were stably expressed to equal levels, as visualized by examining cell lysates separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue, and reached levels about fivefold higher than that of chromosomally expressed wild-type DnaK (data not shown).
FIG. 1.
Construction of hybrid dnaK genes. The domain structures of the DnaK proteins and the positions of the domain boundaries (E. coli/B. subtilis) are given. Amino acid exchanges caused by the introduction of restriction sites are indicated. By using sequence-specific mutagenesis (14), two different restriction sites were introduced into the two dnaK genes. An AflII site was introduced in the region separating the ATPase and peptide-binding domains, leading to V385L and V356L exchanges in the DnaK proteins of E. coli and B. subtilis, respectively. An SpeI site was created within the interdomain region between the peptide-binding and C-terminal domains of both genes, leading to Q540H and L542V exchanges in the E. coli protein only. The hybrid genes shown here were constructed by using these modified prototype genes.
Complementation experiments with ΔdnaK52 and dnaK756 E. coli mutants.
First, we tested the hybrid dnaK genes for complementation of the ΔdnaK52 null mutant for growth defects at two different temperatures. In this mutant, an about 1-kb internal fragment of dnaK has been replaced with a cat cassette (23), which causes cold and heat sensitivity, and thus the mutant possesses a very narrow temperature range for growth (5, 6). The different pHK plasmids were transformed into strain BB2414, a ΔdnaK52 derivative of MC4100 (6), and plated on Luria-Bertani agar plates containing IPTG at a final concentration of 250 μM to induce the dnaK alleles; these plates were incubated at either 30 or 40°C, and growth was recorded. In the presence of either the empty vector pUHE21-2fdΔ12 (data not shown) or the different recombinant plasmids, strain BB2414 formed colonies at 30°C (Fig. 2A). When plated at 40°C, it was unable to form colonies in the absence of IPTG with either plasmid (data not shown). Growth occurred only in the presence of the inducer with a plasmid expressing either wild-type E. coli DnaK (pHK01), hybrid DnaK with the ATPase and the peptide-binding domain from E. coli and the C-terminal domain from B. subtilis (pHK11), or a truncated version of E. coli DnaK lacking the C-terminal domain (pHK13; Fig. 2A). We conclude from these results that B. subtilis DnaK cannot substitute for the E. coli homologue in the ΔdnaK52 strain and that the C-terminal domain is completely dispensable for growth at least up to 42°C.
FIG. 2.
Complementation of temperature-sensitive phenotypes and phage λ propagation by plasmid-encoded hybrid dnaK genes. ΔdnaK52, dnaK756 and ΔdnaK52 mutant strains carrying the dnaJ gene under arabinose and IPTG control and containing the different dnaK hybrids were tested for growth at 30 and 40 or 46°C (A) and for propagation of bacteriophage λ at 30°C (B). Growth was assayed by determining the ability of the cells to form colonies or plaques on Luria Bertani agar plates. +, wild-type number and size of colonies or plaques; - -, no colonies or plaques. All strains were grown in the presence of 250 μM IPTG to induce the expression of the dnaK genes, and those carrying pdnaJ also received 0.5% arabinose. Plates were scored for colonies and plaques after 20 h of incubation.
As already observed for the ΔdnaK52 allele, E. coli BB2362, an MC4100 derivative carrying the dnaK756 allele (6), was able to form colonies at 30°C in the presence of the vector plasmid pUHE21-2fdΔ12 (data not shown) or any of the hybrid plasmids (Fig. 2A). When these different strains were incubated at 46°C, they were able to form colonies only in the presence of 50 μM IPTG. Here, both the E. coli and B. subtilis wild-type alleles were able to complement the dnaK756 mutant for growth (Fig. 2A, pHK01 and pKH02). Proteins consisting of the ATPase domain from one species and the two other domains from the second species turned out also to be active (pHK09 and pKH10). As already observed for the ΔdnaK52 allele, the small C-terminal domain was exchangeable without significantly influencing the complementing activity of the DnaK proteins (pHK11 and pHK12) and was even dispensable (pHK13 and pHK14).
These results seem to be in contrast to those obtained with the ΔdnaK52 mutant strain and raise the question of why there are allele-specific differences in complementation. Two different possibilities can be envisaged: (i) the amount of DnaJ protein in the dnaK null mutant is reduced due to a polar effect of the Cmr marker on the downstream dnaJ gene compared to the wild type or the dnaK756 allele (27), or (ii) the mutant DnaK756 protein retains some residual activity which can be increased in the presence of the wild-type protein. To distinguish between these two possibilities, we constructed plasmid pdnaJ, which carries the dnaJ gene under the control of an arabinose- and IPTG-inducible promoter. This plasmid was transformed into all BB2414 derivate strains containing wild-type and hybrid dnaK genes. The amounts of DnaJ in the different E. coli strains used here was determined by immunoblotting. They were below the level of detection in the ΔdnaK52 mutant and in the same strain carrying the pdnaJ plasmid in the absence of the inducer and restored to wild-type or dnaK756 levels when the latter strain was grown in the presence of 0.5% arabinose and 1 mM IPTG (data not shown). Under dnaJ-inducing conditions, wild-type B. subtilis DnaK (pHK02) and the hybrid derivates containing the ATPase and substrate-binding domains of B. subtilis were now able to complement E. coli ΔdnaK52 for growth at high temperature (Fig. 2A). Therefore, the B. subtilis DnaK protein is able to complement ΔdnaK52 for growth at high temperature, provided that sufficient amounts of DnaJ are present. Quite recently, the exact ratios of DnaK and DnaJ have been determined to be 30:1 in E. coli (28) and 3:1 in B. subtilis (21). The fact that these ratios differ by a factor of 10 might indicate that the B. subtilis DnaK protein needs this large amount of DnaJ to stimulate its ATPase activity.
Plating efficiency of bacteriophage λ in the presence of the different hybrid DnaK proteins.
The dnaK gene has been discovered to be a conditional-lethal mutation which does not allow replication of phage λ DNA (10), and DnaK is involved in the dissociation of the complex consisting of the λ P and E. coli DnaB proteins (8). Therefore, we asked which hybrid DnaK proteins would allow the propagation of phage λ in dnaK mutant strains. All three of the strains described above (dnaK756, ΔdnaK52, and ΔdnaK52 plus pdnaJ) were infected with λ vir, and growth was recorded in the presence of 250 μM IPTG at 30°C. In principle, we observed a growth pattern of phage λ comparable to that observed for complementation of growth defects at high temperature (Fig. 2B). Deletion of the C-terminal domain of E. coli DnaK did not abolish λ propagation but caused inactivation of B. subtilis DnaK in the dnaK null mutant, even in the presence of wild-type levels of DnaJ. These results confirm those for complementation for growth at high temperature and highlight that B. subtilis DnaK needs increased amounts of DnaJ to be active and, in addition, that DnaK756 must possess some residual activity.
Degradation of ς32.
In E. coli, the genes encoding cytosolic heat shock proteins form a regulon that is positively controlled by the rpoH gene product, the heat shock promoter-specific ς32 subunit of RNA polymerase (2, 20, 29). A key aspect of this regulation, the sensing of stress and transmission of this information to ς32, involves the DnaK chaperone machine (9). The DnaK chaperone system was shown to physically interact with ς32 (7, 16, 17), and two sites with high affinity for DnaK have been located within ς32 (19). Therefore, we asked whether hybrid DnaK proteins would be able to interact with ς32, causing destabilization and thereby a reduction in its total amount within the cells. The amount of ς32 was determined in the three E. coli indicator strains containing the different DnaK-expressing plasmids in the absence or presence of 250 μM IPTG to induce the dnaK genes from the plasmids. While induction of the E. coli wild-type dnaK gene clearly reduced the amount of ς32 in all three indicator strains, the B. subtilis wild-type homologue needed larger amounts of the DnaJ protein to lower the intracellular level of ς32 (Fig. 3). Hybrid DnaK proteins containing the ATPase domain of one species and the peptide-binding and C-terminal domains of the other species exhibited only slight activity in the ΔdnaK52 mutant strain expressing DnaJ (pHK09 and pHK10; Fig. 3). The C-terminal domains were again exchangeable (pHK11 and pHK12) without changing the complementation profile in comparison to that of the wild-type counterpart. Deletion of this domain in E. coli DnaK (pHK13) rendered it inactive in the dnaK null mutant and caused a reduction of its activity in dnaK756. The truncated B. subtilis DnaK protein (pHK14) was no longer active, even in the presence of sufficient DnaJ levels. In summary, exchange of the ATPase domain resulted in nearly inactive proteins, as already described for complementation for growth defects at high temperature and λ propagation. The deletion of the C-terminal domain of E. coli or B. subtilis DnaK resulted in strongly reduced complementation in both the dnaK756 and the ΔdnaK52 mutants independent of the amount of DnaJ. The major conclusion from these results is that the C-terminal domain is essential for degradation of ς32 in vivo. It might be involved in the binding of ς32, its unfolding, or both. Since this domain is not involved in binding of the λ P protein and of thermosensitive cytoplasmic proteins, ς32 might represent a specific substrate.
FIG. 3.
Amounts of ς32 in the presence of different hybrid proteins. ΔdnaK52 or dnaK756 mutant strains carrying hybrid dnaK genes were grown to mid-exponential phase at 30°C and induced with 250 μM IPTG. Whole-cell fractions corresponding to identical amounts of cell culture were collected before (−) or 2 h after (+) the addition of IPTG, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to nitrocellulose. The membranes were probed with anti-ς32 antibodies (1:10,000 dilution) and developed by a colorimetric assay as previously described (13).
Acknowledgments
We thank Hermann Bujard for providing plasmids and Elke Deuerling and Thomas Laufen for comments on the manuscript.
Financial support was provided by the Deutsche Forschungsgemeinschaft (Schwerpunkt-Programm Cellular Stress response, Schu 414/9-4) and the Fonds der Chemischen Industrie to W.S.
REFERENCES
- 1.Buchberger A, Valencia A, McMacken R, Sander C, Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukau B. Regulation of the heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B, Walker G C. ΔdnaK52 mutants of Escherichia coli have defects in chromosome segregation and plasmid maintenance at normal growth temperatures. J Bacteriol. 1989;171:6030–6038. doi: 10.1128/jb.171.11.6030-6038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukau B, Walker G C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukau B, Walker G C. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamer J, Bujard H, Bukau B. Physical interactions between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor ς32. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-m. [DOI] [PubMed] [Google Scholar]
- 8.Georgopoulos C, Ang D, Liberek K, Zylicz M. Properties of the Escherichia coli heat shock proteins and their role in bacteriophage λ growth. In: Morimoto R I, Tissières A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 191–221. [Google Scholar]
- 9.Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Morimoto R I, Tissières A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- 10.Georgopoulos C P, Herskowitz I. Escherichia coli mutants blocked in lambda DNA synthesis. In: Hershey A D, editor. The bacteriophage lambda. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1971. pp. 553–564. [Google Scholar]
- 11.Gething M-J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 12.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 13.Homuth G, Heinemann M, Zuber U, Schumann W. The genes lepA and hemN form a bicistronic operon in Bacillus subtilis. Microbiology. 1996;142:1641–1649. doi: 10.1099/13500872-142-7-1641. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–383. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 15.Lanzer M, Bujard H. Promoters determine largely the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85:6632–6636. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberek K, Galitski T P, Zylicz M, Georgopoulos C. The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the ς32 transcription factor. Proc Natl Acad Sci USA. 1992;89:3516–3520. doi: 10.1073/pnas.89.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberek K, Georgopoulos C. Autoregulation of the Escherichia coli heat shock response by the DnaK and DnaJ heat shock proteins. Proc Natl Acad Sci USA. 1993;90:11019–11023. doi: 10.1073/pnas.90.23.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarty J S, Buchberger A, Reinstein J, Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 19.McCarty J S, Rüdiger S, Schönfeld H J, Schneider-Mergener J, Nakahigashi K, Yura T, Bukau B. Regulatory region C of the E. coli heat shock transcription factor, ς32, constitutes a DnaK binding site and is conserved among eubacteria. J Mol Biol. 1996;256:829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 20.Missiakas D, Raina S, Georgopoulos C. Heat shock regulation. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. London, England: Chapman & Hall, Ltd.; 1996. pp. 481–502. [Google Scholar]
- 21.Mogk, A. Unpublished data.
- 22.Palleros D R, Reid K L, Shi L, Fink A L. DnaK ATPase activity revisited. FEBS Lett. 1993;336:124–128. doi: 10.1016/0014-5793(93)81624-9. [DOI] [PubMed] [Google Scholar]
- 23.Peak K-H, Walker G C. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol. 1987;169:283–290. doi: 10.1128/jb.169.1.283-290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 26.Sell S M, Eisen C, Ang D, Zylicz M, Georgopoulos C. Isolation and characterization of dnaJ null mutants of Escherichia coli. J Bacteriol. 1990;172:4827–4835. doi: 10.1128/jb.172.9.4827-4835.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl F U. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system—DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomoyasu T, Ogura T, Tatsuta T, Bukau B. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol Microbiol. 1998;30:567–581. doi: 10.1046/j.1365-2958.1998.01090.x. [DOI] [PubMed] [Google Scholar]
- 29.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]