Abstract
Caulobacter crescentus exhibits cell-type-specific control of chromosome replication and DNA methylation. Asymmetric cell division yields a replicating stalked cell and a nonreplicating swarmer cell. The motile swarmer cell must differentiate into a sessile stalked cell in order to replicate and execute asymmetric cell division. This program of cell division implies that chromosome replication initiates in the stalked cell only once per cell cycle. DNA methylation is restricted to the predivisional cell stage, and since DNA synthesis produces an unmethylated nascent strand, late DNA methylation also implies that DNA near the replication origin remains hemimethylated longer than DNA located further away. In this report, both assumptions are tested with an engineered Tn5-based transposon, Tn5Ω-MP. This allows a sensitive Southern blot assay that measures fully methylated, hemimethylated, and unmethylated DNA duplexes. Tn5Ω-MP was placed at 11 sites around the chromosome and it was clearly demonstrated that Tn5Ω-MP DNA near the replication origin remained hemimethylated longer than DNA located further away. One Tn5Ω-MP placed near the replication origin revealed small but detectable amounts of unmethylated duplex DNA in replicating stalked cells. Extra DNA synthesis produces a second unmethylated nascent strand. Therefore, measurement of unmethylated DNA is a critical test of the “once and only once per cell cycle” rule of chromosome replication in C. crescentus. Fewer than 1 in 1,000 stalked cells prematurely initiate a second round of chromosome replication. The implications for very precise negative control of chromosome replication are discussed with respect to the bacterial cell cycle.
Coordination between chromosome replication and the cell division cycle is an outstanding problem in cell biology. In Escherichia coli, chromosome replication is controlled by both positive and negative factors. RNA polymerase and the DnaA protein are positive factors that act at the earliest stages of chromosome replication (31). Both RNA polymerase transcription and DnaA protein binding prepare the chromosome replication origin for the replication system (2, 43). The Dam (GATC) DNA adenine methylase and the SeqA protein are negative factors that block extra chromosome replication (3, 22, 50). Dam methylation of parental DNA strands allows the SeqA protein to distinguish between parental DNA duplexes (unreplicated fully methylated DNA) and nascent-strand and parental-strand DNA duplexes (newly replicated hemimethylated DNA). The SeqA protein sequesters hemimethylated DNA, rendering it inaccessible to replication proteins until a later stage in the cell cycle (22, 50). Studies of these processes and many others have provided our best insights into bacterial chromosome replication and the bacterial cell cycle. However, it is still not well understood how chromosome replication is coordinated with the cell division cycle (31).
It is also not known whether model systems identified in E. coli are generally applicable to other bacteria, since it is now recognized that bacteria form a very diverse group of organisms (52). The genes for RNA polymerase (17) and DnaA (44) are highly conserved among eubacteria, but not archaebacteria and eukaryotes. This implies that eubacteria share similar positive factors, and presumably similar mechanisms, for initiating chromosome replication. However, this inference has not been critically tested, except for Bacillus subtilis, where it is clear that DnaA also acts to initiate chromosome replication (33). The Dam (GATC) DNA adenine methylase, though clearly essential to coordinate cell cycle chromosome replication in E. coli (3, 22, 50), is absent in most bacteria (4). These considerations argue that bacteria may have diverse negative factors to control chromosome replication.
Caulobacter crescentus presents an evolutionarily divergent, yet experimentally amenable, system for comparative cell cycle studies (6, 25, 27). C. crescentus exhibits cell-type-specific control of chromosome replication and DNA methylation (Fig. 1). Chromosome replication is restricted to the stalked-cell type (10, 23, 26), and DNA methylation is restricted to the predivisional cell (55). Asymmetric cell division yields a replicating stalked cell and a nonreplicating swarmer cell. These progeny cells have distinct morphologies and behaviors (6). The progeny stalked cell receives a replicating chromosome and directly proceeds to execute asymmetric cell division. On the other hand, the progeny swarmer cell receives a nonreplicating chromosome, and it must differentiate into a sessile stalked cell in order to replicate and execute asymmetric cell division (6, 25).
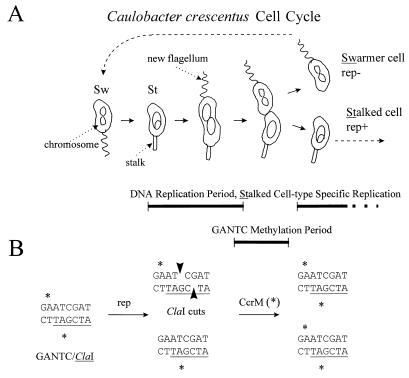
FIG. 1.
Chromosome replication and DNA methylation during the C. crescentus cell cycle. (A) Differentiation and the cell cycle are integrated. Cell division is asymmetric, forming distinct swarmer cell (Sw) and stalked-cell (St) progeny. The swarmer cell swims with its polar flagellum and delays chromosome replication until it differentiates into a stalked cell. This involves extensive cellular remodeling, such as flagellum ejection and stalk (cell wall) synthesis (6). Also, the nucleoid appears to be more compact in the swarmer cell (16), and this is suggested by the figure eight chromosome drawing versus the oval in the stalked cell. Bidirectional chromosome replication (rep) takes place only in the stalked-cell type along with a program of asymmetric cell division. A new polar flagellum is made, and two distinct chromosomes are formed and placed into the correct compartments. The expression of the CcrM enzyme that methylates the A of GANTC is restricted to a narrow period before cell division (55). (B) Cell cycle methylation assayed by conditional digestion with ClaI. GANTC methylation (∗) is present on both strands in swarmers. Replication creates asymmetric (top versus bottom) strand methylation in stalked cells. All strands become methylated by CcrM before cell division. Note that only methylation on the bottom strand blocks ClaI digestion, because only this methylated A overlaps the ClaI recognition site (shown underlined).
The molecular basis of the C. crescentus cell cycle is being analyzed. Studies suggest that the C. crescentus replication origin (Cori), like that of E. coli, employs both RNA polymerase and the DnaA protein as positive replication factors (24, 26). Cori also employs at least one novel negative factor, the CtrA (cell cycle transcription regulator) protein (38), which segregates to swarmer cells (12) and blocks chromosome replication (39). CtrA regulates many cell cycle processes, including flagellar synthesis (38), stalk synthesis (38), cell division (21, 38), and DNA methylation (38) (discussed below). CtrA is homologous to E. coli OmpR protein (38) and therefore belongs to a ubiquitous class of proteins known as two-component response regulators (35, 49). These proteins dominate bacterial adaptation and control systems (18). However, C. crescentus presents the first example where a response regulator protein controls DNA synthesis (39). This result certainly argues for the value of comparative cell cycle studies among diverse bacteria.
E. coli DNA adenine methylase, (GATC) Dam, and E. coli DNA replication studies, motivated a search for a comparable C. crescentus adenine methylase (GANTC) CcrM (55). However, CcrM is unrelated to Dam and shows the greatest homology to the cognate HinfI methylase (55). Experiments employing GANTC sites that overlap restriction endonuclease sites, illustrated in Fig. 1B, demonstrate conditional endonuclease digestion and imply cell cycle DNA methylation. The hemimethylated state is persistent, because GANTC (CcrM) methylation is restricted to the predivisional cell (55). CcrM protein accumulates only at this late stage of the cell cycle (47), by a combination of late cell cycle RNA synthesis (55) and selective protein degradation by a homologue of the E. coli Lon protease (53). Interestingly, the CtrA protein that represses chromosome replication also activates ccrM transcription (38).
This paper tests two key cell cycle parameters with experiments that systematically measure chromosome methylation by the GANTC (CcrM) methylase. The data shown in Fig. 1 imply that chromosome replication initiates in the stalked cell only once per cell cycle and that DNA near the replication origin remains hemimethylated longer than distant DNA. While these are reasonable inferences, neither has been critically tested and quantified. The results presented below demonstrate that C. crescentus produces two chromosomes bearing asymmetric hemimethylation along most of their circumferences. The period of DNA hemimethylation always decreases with the distance from the replication origin. These experiments also demonstrate that C. crescentus produces at most 0.1% unmethylated DNA near the replication origin. This parameter suggests a very precise mechanism(s) for ensuring that only two chromosomes are produced every cell cycle.
MATERIALS AND METHODS
Strains and genetic manipulations.
Original bacterial strains are listed in Table 1, and those C. crescentus strains used for methylation state experiments (see Fig. 4 to 7) are listed in Table 2. E. coli DH10B was used for plasmid construction, and E. coli S17-1 containing pGM1138 (Fig. 2) was use for conjugation with C. crescentus (13). The donor and the recipient were mixed and incubated at 30°C on PYE plates (19) and then spread on selective PYE plates. E. coli was counterselected by 20 μg of nalidixic acid/ml, and pGM1138 and/or its transposon Tn5Ω-MP (Fig. 2) was selected with 100 μg of spectinomycin/ml and 2.5 μg of streptomycin/ml. Integrated pGM1138 was counterselected on 3% sucrose PYE plates. All Tn5 insertions were transduced by φCr30 (15) into the NA1000 synchronizable genetic background (16) before the directed Tn5 replacements were performed (Fig. 2), as described under Results.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description or genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara, leu)7697 araD139 galU galK nupG rpsL | Gibco BRL |
| S17-1 | RP4-2 (Tc::Mu) (Km::Tn7); conjugation strain | 42 |
| C. crescentus | ||
| NA1000 | Wild-type, synchronizable form of CB15 | 16 |
| PC7070 | CB15 recA-526::(linked)Tn5 (Km Sr) | A. Newton |
| SC1090 | CB15 purA::Tn5 (Km Sr) | 5 |
| SC1107 | CB15 bla::Tn5 (Km Sr) | 5 |
| SC1140 | CB15 lac101::Tn5 (Km Sr) str-152 | B. Ely |
| SC1293 | CB15 trpE::Tn5 (Km Sr) | 5 |
| SC1321 | CB15 metF::Tn5 (Km Sr), Ts-140/pVS1 | 5 |
| SC1490 | CB15 leuA::Tn5 (Km Sr) | 5 |
| YBC2 | NA1000 rpoN::Tn5 (Km Sr) | 7 |
| GM53 | NA1000 × φCr30(SC1107) | This study |
| GM55 | NA1000 × φCr30(PC7070) recA+::Tn5 | This study |
| GM69 | NA1000 × φCr30(SC1090) | This study |
| GM84 | NA1000 × φCr30(SC1490) | This study |
| GM85 | NA1000 × φCr30(SC1293) | This study |
| GM214 | NA1000 × φCr30(SC1321) | This study |
| GM268 | NA1000 flaE184::Tn5 (Km Sr) | R. Bryan |
| GM269 | NA1000 cysD::Tn5 (Km Sr) | R. Bryan |
| GM340 | NA1000 × φCr30(SC1140) | This study |
| Plasmids | ||
| pIC20R | Ap; cloning vector | 29 |
| pGM104 | pIC20R BamHI 760 bp RK2 oriT | M. R. K. Alley |
| pGM1104 | pIC20R PstI sacB | M. R. K. Alley |
| pBluescript II | Ap; SK(+) or KS(+) cloning sites | 1 |
| pGM948 | pBluescript II SK; XbaI-to-SalI C. crescentus dnaA | 56 |
| pBR7-5 | pBR322 BamHI C. crescentus pbpA | P. J. Kang |
| pSUP5011 | pBR325 (Ap Cm)::Tn5 (Km oriT) | 2 |
| pHP45Ω | Ap; Ω fragment (Sp Sr) | 37 |
| pGM618 | pBluescript II KS; BamHI Cori | 26 |
| pGM1091 | pBluescript II KS; Cori MP-L and -R | This study |
| pGM1107 | pSUP5011; Tn5Ω-MP (Ap Cm Sp Sr) | This study |
| pGM1115 | pGM1107; Tn5Ω-MP (Ap Sp Sr) oriT | This study |
| pGM1138 | pGM1115; Tn5Ω-MP (Ap Sp Sr) sacB | This study |
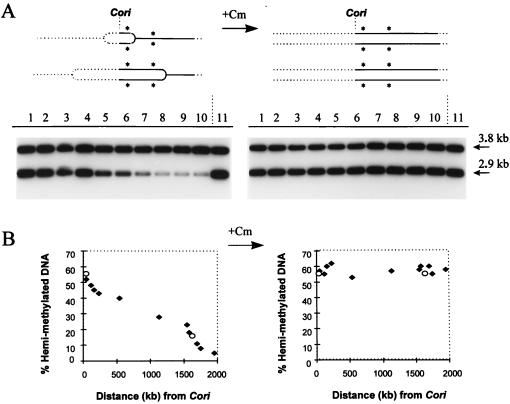
FIG. 4.
Chromosome hemimethylation gradient during exponential growth and its elimination by chloramphenicol (+Cm) treatment. (A) Experimental rationale and Southern blot analysis of methylation state. In a mixed population containing all cell types, DNA closest to Cori is most likely to have replicated and is therefore most likely to be hemimethylated, as indicated by the single asterisks following the forks. Chloramphenicol treatment allows replication forks to reach the Cori-distal DNA but blocks new chromosome replication and DNA methylation. The percentage of hemimethylated DNA across the whole chromosome becomes proportional to the percentage of replicating cells. Lanes 1 to 11 correspond to the separate Tn5Ω-MP strains (1 to 11) listed in Table 2. These were optimally grown in rich PYE medium and split into two cultures, with and without 20 μg of chloramphenicol/ml, and culturing continued for 2 h before their DNA was prepared. Southern blot analysis was performed as described in the legend to Fig. 3A and Materials and Methods. (B) Analysis of data in panel A by phosphorimaging (Molecular Dynamics). The percentage of the radiation in the 2.9-kb (hemimethylated) band was plotted versus the distance (compare the genome map in Fig. 2B) of the Tn5Ω-MP from Cori (solid diamonds). The hemimethylated states at dnaA and pbpA (open ovals), determined in separate experiments, are included for comparison.
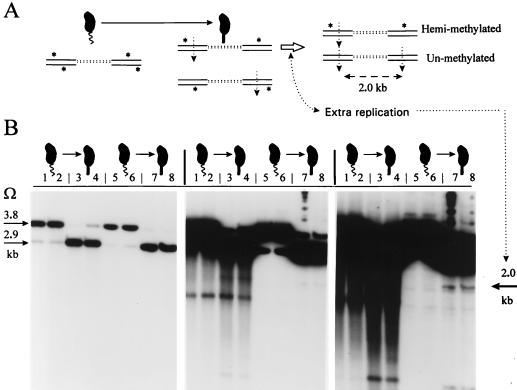
FIG. 7.
Sensitive detection of extra chromosome replication by assaying unmethylated DNA in stalked cells. (A) Rationale for assay. Swarmer cells differentiate into stalked cells, producing hemimethylated Tn5Ω-MP DNA, drawn as paired double lines where top or bottom asterisks denote GANTC methylation and vertical arrows denote susceptible ClaI or XhoI endonuclease sites (compare Fig. 2A). The open horizontal arrow indicates that one of the two replicating stalked-cell chromosomes has started a second round of replication, and in the absence of CcrM, this will produce sister hemimethylated and unmethylated DNA duplexes. The absence of methylation allows simultaneous left and right cleavage of the same Tn5Ω-MP, producing the 2.0-kb band (compare Fig. 2A). (B) Potentially unmethylated 2.0-kb chromosome DNA band, revealed during progressive 1- and 18-h and 4-day Southern blot exposures of the following experiment. DNA was prepared from the synchronous culture of Fig. 6 for T = 0 and T = 10 min (lanes 1 and 5, and 2 and 6) swarmer cells and from T = 50 and T = 63 min (lanes 3 and 7, and 4 and 8) stalked cells. Both standard phenol-chloroform extraction (lanes 1 to 4) and the agarose-embedding (lanes 5 to 8) methods for preparing DNA are compared. The Southern blotting protocol was performed as detailed in Materials and Methods and interpreted in Fig. 3A. DNA in each of lanes 1 to 8 was cut with ClaI and PstI, and the resulting blot was hybridized with >3 × 109 cpm of 32P-labeled Ω fragment/μg (Fig. 3A). Size standards, in the adjacent lanes (not shown), were the 1-kb ladder (Gibco BRL) and blotted chromosome DNA cut with HindIII, PstI, and XhoI. The arrows mark the 3.8-, 2.9-, and 2.0-kb bands corresponding to fully methylated, hemimethylated, and unmethylated DNAs (Fig. 3A).
TABLE 2.
C. crescentus strains derived from pGM1138
| No. | Strain | Method (precursor strain) | Genetic locus | Distance (kb) from Cori |
|---|---|---|---|---|
| 1 | GM1251 | Directed (GM69) | purA::Tn5Ω-MP | 120 |
| 2 | GM1252 | Directed (GM84) | leuA::Tn5Ω-MP | 160 |
| 3 | GM1259 | Directed (YBC2) | rpoN::Tn5Ω-MP | 230 |
| 4 | GM1253 | Directed (GM214) | metF::Tn5Ω-MP | 540 |
| 5 | GM1254 | Directed (GM55) | recA::Tn5Ω-MP | 1,130 |
| 6 | GM1255 | Directed (GM268) | flaE::Tn5Ω-MP | 1,550 |
| 7 | GM1256 | Directed (GM269) | cysD::Tn5Ω-MP | 1,580 |
| 8 | GM1249 | Directed (GM53) | bla::Tn5Ω-MP | 1,700 |
| 9 | GM1257 | Directed (GM340) | lac::Tn5Ω-MP | 1,750 |
| 10 | GM1258 | Directed (GM85) | trpE::Tn5Ω-MP | 1,950 |
| 11 | GM1261 | Random (NA1000) | zzz::Tn5Ω-MP | 50 |
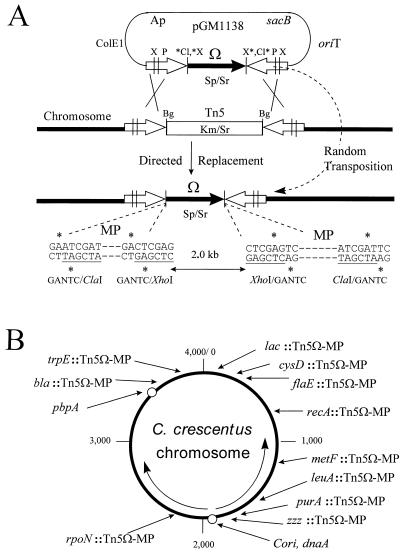
FIG. 2.
Random or directed chromosome placement of conditional methylation-sensitive cleavage sites on Tn5Ω-MP. (A) Strategy employing plasmid pGM1138 to deliver Tn5Ω-MP either by homologous recombination, e.g., with wild-type Tn5 (as shown), across the homologous IS50 elements (open arrows) or by standard random transposition. The solid arrow indicates the Ω antibiotic resistance fragment that provides selection for either random or directed placements. Genes for spectinomycin (Sp), streptomycin (Sr), kanamycin (Km), and ampicillin (Ap) resistance are shown. Additional genetic elements include sacB for counterselection (40), the pBR325 replication origin (ColE1) for propagation in E. coli, and the RK2 origin of transfer (oriT) for conjugation (42). Restriction sites for ClaI (Cl), XhoI (X), PstI (P), and BglII (Bg) are indicated. The asterisks denote sites conditionally blocked by GANTC methylation inside the oligonucleotide sequences, termed MP sequences, shown below the diagram. (B) Chromosome positions of conditional methylation-sensitive cleavage sites, both natural and those delivered by plasmid pGM1138. This map is based on the genetic map of C. crescentus compiled by B. Ely (12a). The positions of the transposons are based on physical (PFGE) criteria (14). The open circles denote the natural GANTC endonuclease site overlaps at dnaA and pbpA (described in the legend to Fig. 3A). The structure for Tn5Ω-MP is shown in Fig. 2A and 3A, and the strains bearing these genetic loci are listed in Table 2. zzz::Tn5Ω-MP is a random transposition in an apparently silent locus 50 kb from Cori (see the text for additional details). The arrows inside the circle indicate bidirectional replication (11, 26) from the C. crescentus origin (Cori).
Plasmid construction.
pGM1138 was derived from pSUP5011 (42) by the following steps. The methylation probe (MP) oligonucleotides listed in Table 3 were annealed and sequentially ligated into the HindIII and NruI sites (MP-L) and the XhoI and BamHI sites (MP-R) of pGM618. (The Cori DNA in pGM618 served as a convenient scaffold). The resulting plasmid, pGM1091, was cut with XhoI and blunt ended with Klenow polymerase, and the Cori DNA was replaced by the BamHI Ω fragment from pHP45Ω, likewise blunt ended by Klenow polymerase. The flanking sequences (shown in Fig. 2) were checked by double-stranded DNA sequencing with Ω primers 2 and 3 (Table 3). This new Ω-MP fragment with flanking MP sequences was removed by its flanking BamHI sites (designed into the MP oligonucleotides [Table 3]) and ligated between the BglII sites of Tn5 on pSUP5011 to form pGM1107. Subsequently, the RK2 oriT was added from the EcoRI fragment on pGM104, forming pGM1115, and the sacB gene was added as a BglII-to-BamHI fragment from pGM1104 into the unique BglII site of pGM1115, forming the final construct, pGM1138 (Fig. 2A).
TABLE 3.
Oligonucleotides
| Name | Sequence |
|---|---|
| MP-L top | 5′-AGCTTGGATCCTAGAATCGATACCGACTCGGAGCTGGACTCG |
| MP-L bottom | 5′-CGAGTCCAGCTCGAGTCGGTATCGATTCTAGGATCCA |
| MP-R top | 5′-TCGAGTCAGGCTCATCGATTCGATCGCAGCG |
| MP-R bottom | 5′-GATCCGCTGCGATCGAATCGATGAGCCTGAC |
| Ω primer 2 | 5′-GGGCTTTACTAAGCTGATC |
| Ω primer 3 | 5′-GCGAGCAGGGGAATTGATCC |
Growth and synchrony.
All C. crescentus strains were grown in liquid PYE medium (19) at 30°C without selection. Synchronized cultures were obtained by growing asynchronous 500-ml PYE cultures to an optical density at 660 nm (OD660) of 0.6 to 1.0 and applying the Ludox (Dupont) density gradient technique to isolate pure swarmer cells (16). These were released into fresh PYE medium at an OD660 of 0.3 to 0.6 and incubated with vigorous shaking at 30°C. At subsequent times, covering the entire 90-min cell division cycle from swarmer to stalked cell to asymmetric cell (Fig. 1), 2- to 5-ml samples were withdrawn, adjusted to 0.1% sodium azide, and chilled on ice. For DNA preparations, these cells were concentrated, resuspended in 0.1 ml of 10 mM Tris, pH 7.5, adjusted to 50 mM NaEDTA, frozen by dry ice and ethanol, and stored at −20°C.
DNA analysis.
C. crescentus total chromosome DNA was usually prepared by standard phenol and chloroform extraction. The frozen cells described above were thawed at 37°C and mixed with 0.4 ml of a solution of 10 mM Tris (pH 7.5), 40 mM NaEDTA, 0.1% Triton X-100, and 2 mg of lysozyme/ml. After 1 to 2 h at 37°C, 0.2 mg of proteinase K (Gibco BRL)/ml was added and the mixture was incubated at 55°C for 12 to 16 h. This was adjusted to 0.2 M ammonium acetate and extracted three times with phenol-chloroform-isoamyl alcohol (25:24:1) and once by pure chloroform before precipitation with isopropanol. The pellets were resuspended in 50 μl of a solution of 10 mM Tris (pH 7.5), 1 mM NaEDTA, and 10 μg of RNase A/ml. C. crescentus total chromosome DNA was also prepared by embedding the cells in agarose. These cells were not frozen but were mixed directly with an equal volume of 1% agarose at 42°C, pipetted into 0.2-ml molds, and otherwise processed as for analysis by pulsed-field gel electrophoresis (PFGE) (14). DNA samples were digested with restriction endonucleases under the conditions specified by the supplier (Gibco BRL or BioLabs). The agarose-embedded DNA, approximately 40-μl samples, was first rinsed with sterile water and then equilibrated with endonuclease buffer and digested with at least a 10-fold excess of enzyme. PFGE of C. crescentus DNA has been described previously (14). Tn5Ω-MP has both DraI and AseI sites that allow its placement on the PFGE map by selective genomic band cleavage (14). Southern blot analysis employed Hybond+ membranes (Amersham), UV light cross-linkage (Stratalinker; Stratagene), and the hybridization and wash conditions employed for genomic footprinting (8). 32P DNA probes were labeled with [α-32P]dCTP (Amersham) (6,000 Ci/mmol) with the T7 Quickprime kit (Pharmacia).
RESULTS
Strategy for measuring chromosome methylation.
A special Tn5-based transposon, Tn5Ω-MP, was created to probe DNA methylation at random or preselected sites on the chromosome. The methylation state is assayed by restriction endonuclease digestion that is conditionally blocked by overlapping GANTC methylation (as described above [Fig. 1B]). Such sites are rare on C. crescentus DNA. Tn5Ω-MP solves this problem by providing two sets of GANTC overlapping ClaI and XhoI sites (Fig. 2A), termed MPs. These sequences are further described and applied in the chromosome methylation experiments presented below.
Plasmid pGM1138 delivers Tn5Ω-MP to the C. crescentus chromosome by the system outlined in Fig. 2. Tn5Ω-MP is selected in C. crescentus by the spectinomycin and streptomycin antibiotic resistance (Sp-Sr) gene of the Ω fragment (37). The sacB gene allows for the screening and counterselection of vector DNA (40). Plasmid pGM1138 also contains a ColE1 replication origin and the ampicillin resistance (Ap) genes, both derived from pBR325 (42). ColE1 and Ap allow replication and antibiotic selection in E. coli but not in C. crescentus (13). The RK2-derived origin of transfer (oriT) allows pGM1138 to be mobilized by conjugation between E. coli S17-1 and C. crescentus (13, 42). Since pGM1138 cannot replicate in C. crescentus, spectinomycin and streptomycin antibiotic selection demands either that Tn5Ω-MP transpose or that pGM1138 integrate into the C. crescentus chromosome. As described below, transposition and integration can be easily distinguished by the genetic markers on pGM1138 without the need for molecular analysis of the DNA.
Random or directed Tn5Ω-MP insertions into the chromosome are determined by the choice of C. crescentus recipient strains in the conjugation experiment. The C. crescentus wild-type strain NA1000 lacks homology with Tn5Ω-MP and receives it by random transposition mediated by Tn5 transposase (5, 42). Tn5Ω-MP retains a functional transposase in IS50R and can transpose in C. crescentus and, presumably, in most gram-negative bacteria. In a standard conjugation experiment between E. coli S17-1 containing pGM1138 and C. crescentus NA1000, Sp-Sr colonies were recovered at frequencies of between 10−7 and 10−8. Tn5 transposes randomly into chromosome sites, and the delivery vector, often termed the “suicide” plasmid, is not recovered in the chromosome DNA (5, 42). In this system (Fig. 2), random transposition was confirmed by Southern blot hybridization experiments. Total cellular DNA was prepared from 20 separate Sp-Sr colonies, digested with BamHI, and processed for Southern blot hybridization. Since BamHI does not cut inside Tn5Ω-MP, hybridization with internal Ω fragment sequences will reveal the pattern of flanking DNA. This experiment produced unique Ω-hybridizing DNA bands ranging from 5 to 12 kb, indicating a different transposition event for each of these 20 isolates (data not shown). Also, the absence of vector DNA was confirmed by rehybridizing these blots with pBR325 DNA. However, this molecular analysis is not obligatory, since the sacB gene can be scored by sensitivity on 3% sucrose plates, and all 20 isolates showed sucrose resistance growth compared with an integrated pGM1138 plasmid control (data not shown).
Directed Tn5Ω-MP insertions into the C. crescentus chromosome occurs by homologous recombination between IS50 elements on pGM1138 and the IS50 elements of Tn5 transposons at previously characterized sites (5, 14). Lists of such Tn5 strains are presented in Tables 1 and 2. In a standard conjugation experiment between E. coli S17-1 containing pGM1138 and Tn5-containing C. crescentus strains (e.g., GM69 [Table 1]), Sp-Sr colonies were recovered at frequencies between 10−4 and 10−5. Therefore, compared with C. crescentus NA1000 lacking Tn5 (described above), these conjugation experiments produced colonies at frequencies that are typically 100- to 1,000-fold higher. This implies an alternative pathway for establishing Tn5Ω-MP by homologous recombination, a conclusion supported by the following observations. For example, in a standard conjugation experiment, comparable recA+ and recA Tn5 C. crescentus strains (GM55 and PC7070) produced Sp-Sr C. crescentus colonies at 10−4 and 10−7 frequencies, respectively (data not shown).
Directed Tn5 replacement is a two-step recombination process requiring crossover events at both flanking IS50 elements, as shown in Fig. 2. Since double-crossover events are rare in C. crescentus (32), Sp-Sr selection causes only one recombination between pGM1138 and Tn5. A second recombination is needed to exchange the DNA sequences between both IS50 elements. This is done by counterselection on 3% sucrose plates against the vector (sacB) DNA and scoring for the exchange of antibiotic markers. Typically between 10−3 and 10−4 cells survive sucrose selection. With the Tn5 strains listed in Table 2, among the survivors following sucrose counterselection, approximately 50% exchanged Km for Sp-Sr phenotypes and approximately 50% retained Km but lost Sp-Sr phenotypes. This result is theoretically expected, assuming equal recombination frequencies at both IS50L and IS50R. Also, among the survivors, from 1 to 10% retained both Km and Sp-Sr phenotypes, indicating that sacB function was lost without homologous recombination. This is probably due to endogenous C. crescentus insertion element inactivation of sacB. Southern blot experiments clearly indicated that these Km and Sp-Sr cells retained altered vector DNA sequences while their sibling Sp-Sr cells did not (data not shown). Therefore, molecular analysis is not essential, and an antibiotic screen is sufficient to confirm the double crossover shown in Fig. 2.
Temporal gradient of chromosome DNA methylation.
The Tn5Ω-MP system (Fig. 2) was employed to confirm the general validity of the temporal methylation gradient and to quantify the average degree of hemimethylation across the length of the chromosome. Presumably, DNA near the replication origin will remain hemimethylated longer than distant DNA. The genetic loci chosen for these experiments are presented in Table 2 and positioned on the circular C. crescentus chromosome in Fig. 2B. These loci span the approximately 2,000 kb traveled by the replication forks (11). As listed in Table 2, Tn5Ω-MPs 1 through 10 were created by directed replacement of Tn5 transposons at well-characterized locations. Tn5Ω-MP 11 was created by random transposition and selected for its proximity to Cori (described below).
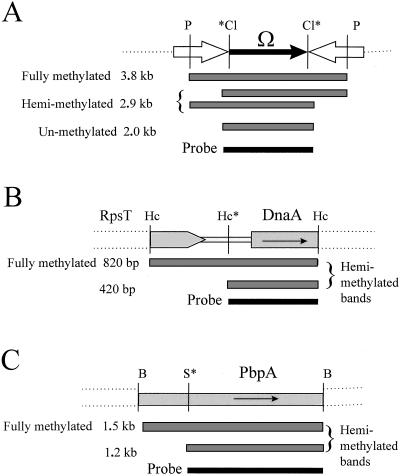
The configuration of Tn5Ω-MP allows a Southern blot assay that distinguishes fully methylated, hemimethylated, and unmethylated DNAs. Note that GANTC overlaps the left sides of ClaI and XhoI at IS50L and the right sides of ClaI and XhoI at IS50R (Fig. 2). Therefore, hemimethylated DNA can be cleaved at the left or the right end but not at both ends of the same molecule. Only when both DNA strands are unmethylated can ClaI or XhoI cleave both ends of the same molecule. The expected banding patterns from Southern blot analysis are diagrammed in Fig. 3. For example, Southern blot analysis employing PstI and ClaI will produce 3.8-, 2.9-, and 2.0-kb bands from fully methylated, hemimethylated, and unmethylated Tn5Ω-MP DNAs, respectively.
FIG. 3.
Methylation state assay and anticipated Southern blot bands. (A) Tn5Ω-MP cut with PstI (P) and ClaI (Cl) and hybridized with intervening Ω-homologous DNA (probe), the 2.0-kb SmaI band isolated from pHLP45Ω. Compare this with Fig. 2A and note that a comparable banding pattern is obtained with XhoI. The alternate left (∗Cl) then right (Cl∗) GANTC overlap with these two ClaI sites ensures that they cannot both be cut on the same molecule unless it is unmethylated on both strands. (B) The natural HincII (Hc) and GANTC overlap (Hc∗) in the 5′ region of the dnaA gene has been described before (55). The probe was the 420-bp HincII band isolated from plasmid pGM948. (C) The natural SalI and GANTC overlap (S∗; GTCGACTC) is present inside the pbpA gene. The probe was the 1.2-kb SalI-to-BamHI fragment from plasmid pBR7-5. Note that for the natural GANTC restriction site overlaps, as illustrated in Fig. 1B, only one of the two sister hemimethylated duplexes can be cut, so unlike the situation shown in panel A, in those shown in both panels B and C two different hemimethylated bands are produced, and one comigrates with the fully methylated band (compare the results shown in Fig. 6A with those shown in Fig. 6B and C). Note also that the DNA probes have equal homology with all of the bands, and this allows quantitative measurements of their respective methylation states.
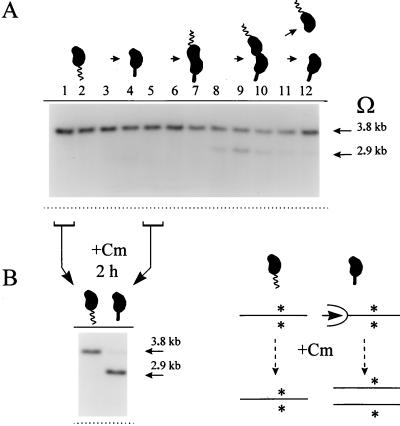
A chromosome methylation gradient was demonstrated by the experiments shown in Fig. 4, utilizing the Southern blot assays diagrammed in Fig. 3 for each of the strains listed in Table 2 and Fig. 2B. Each strain was grown exponentially in rich PYE medium at 30°C with approximately 90-min doubling times. These are maximum C. crescentus growth rates. When the cells reached a low OD of between 0.2 and 0.3, the cultures were split; one received 20 μg of chloramphenicol/ml, and both were grown for an extra 2 h before their DNA was extracted. Chloramphenicol blocks both cell growth and protein synthesis. This antibiotic treatment, diagrammed in Fig. 4A, allows ongoing replication forks to finish DNA synthesis but blocks new DNA replication and the induction of CcrM (GANTC) DNA methylation, since both processes require new protein synthesis. These consequences of chloramphenicol treatment are also demonstrated in the control experiments described below (Fig. 5). Southern blot analysis of these Tn5Ω-MP strains shows both 3.8-kb fully methylated (unreplicated) and 2.9-kb hemimethylated (replicated) bands. Note that lanes 1 through 11 correspond to the entries in Table 2. A plot of the percentage of hemimethylated DNA versus distance from the C. crescentus replication origin (Cori) reveals a decreasing gradient of DNA hemimethylation in exponentially growing cells (Fig. 4B). Near Cori the DNA is 55% hemimethylated, and this drops to about 5% near the terminus of replication, approximately 2,000 kb away (Fig. 4B and Table 2). Presumably, this 55% hemimethylated DNA measurement near Cori reflects the percentage of stalked cells in S phase. The remaining fully methylated DNA comes from swarmer cells or predivisional cells expressing CcrM. The gradient clearly requires chromosome replication, since it is abolished by chloramphenicol treatment (Fig. 4). The DNA from the chloramphenicol-treated cells show approximately uniform (50 to 60%) DNA hemimethylation, reflecting only the percentage of stalked cells in the population and independent of the distance between Tn5Ω-MP and Cori. Note also that the naturally occurring methylation probes, dnaA and pbpA, that are proximal and distal, respectively, to Cori (Fig. 2B) also lie among the points plotted by the Tn5Ω-MP strains (Fig. 4B).
FIG. 5.
Late replication and hemimethylation of the Cori-distal bla gene marked by Tn5Ω-MP. (A) Methylation state during the cell cycle. Synchronous swarmer cells, strain GM1249 (Table 2), were released into rich PYE medium and sampled for analysis at T = 0, 5, 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100 min during the cell cycle (corresponding to lanes 1 to 12). Southern blot analysis was performed as described in the legend to Fig. 3A and Materials and Methods. The 3.8- and 2.9-kb arrows mark fully methylated and hemimethylated bands. (B) Chloramphenicol blocks new replication in swarmer cells and allows continued replication (fork progression) in stalked cells. The culture described above was sampled and treated with chloramphenicol (as for Fig. 4) at T = 0 (swarmer cells) and at T = 30 min (stalked cells). The Southern blot analysis and interpretation was otherwise the same as above and for Fig. 4. Only the stalked-cell DNA was chased into the hemimethylated state. The asterisks indicate methylated top and bottom strands.
The hemimethylation gradient hypothesis is further supported by experiments with synchronized cells. These experiments confirm that a Cori-distal Tn5Ω-MP becomes hemimethylated later than a Cori-proximal Tn5Ω-MP. Swarmer cells were isolated and released into fresh rich (PYE) medium, where they synchronously differentiated into stalked cells and initiated the program of asymmetric cell division. A strain with bla::Tn5Ω-MP (GM1249 [Table 2]) located 1,700 kb away from Cori started showing Tn5Ω-MP hemimethylation 60 min into the cell cycle (Fig. 5A, lane 8), when the cells were clearly predivisional and pinched in the middle. By contrast, a strain with zzz::Tn5Ω-MP (GM1261 [Table 2]) located only 50 kb away from Cori started showing Tn5Ω-MP hemimethylation just 10 to 15 min into the cell cycle (Fig. 6A, lane 4, and D), when only a few stalked cells (among the swarmer cells) were visible. Also note the difference between the maximal degrees of hemimethylation of these two chromosome sites. A maximum of only 20% hemimethylation was observed at bla::Tn5Ω-MP (Fig. 5A, lane 9) compared with 100% hemimethylation at zzz::Tn5Ω-MP (Fig. 6A, lanes 8 to 10, and D). Perhaps only 20% of the replication forks reach bla::Tn5Ω-MP, and this may happen when only 20% of the cells initiate chromosome replication. Alternatively, the induction period of the CcrM (GANTC) methylation enzyme may overlap the late replicating period, supporting the gradient hypothesis (Fig. 4) that late-replicating DNA is only transiently hemimethylated.
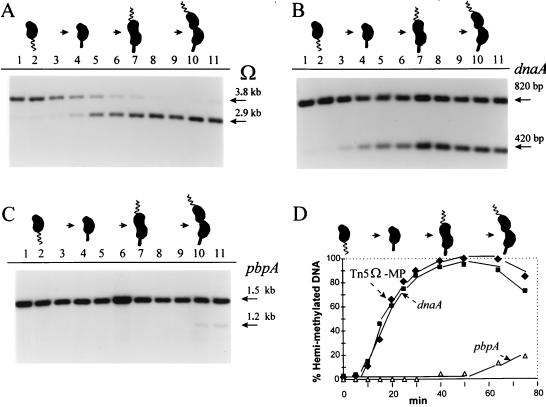
FIG. 6.
Early replication and hemimethylation of Cori-proximal chromosome DNA marked by a randomly inserted Tn5Ω-MP. (A) Methylation state during the cell cycle at zzz::Tn5Ω-MP (50 kb from Cori). Synchronous swarmer cells, strain GM1261 (Table 2), were released into rich PYE medium and sampled for analysis at T = 0, 5, 10, 15, 20, 25, 30, 40, 50, 63, and 75 min during the cell cycle (corresponding to lanes 1 to 11). Southern blot analysis was performed as described in the legend to Fig. 3A and Materials and Methods. (B and C) These same DNA samples were analyzed as described in the legends to Fig. 3B and C for Cori-proximal dnaA and Cori-distal pbpA genes, respectively. (D) Quantitation of all three replication and methylation markers in panels A, B, and C. The same culture was used for the experiment shown in Fig. 7.
The control experiment shown in Fig. 5B confirms that at least 95% of the chromosomes initiated replication during the experiment shown in Fig. 5A, as well confirming as the efficacy of the chloramphenicol treatment used to support the methylation gradient hypothesis. In the experiments shown in Fig. 4, it was argued that chloramphenicol blocks the initiation of replication (in swarmer cells) but not the elongation of replication forks (in stalked cells) and that it also blocks the induction of CcrM DNA methylation. These consequences of chloramphenicol treatment caused the collapse of the methylation gradient in unsynchronized cultures (Fig. 4). These inferences were confirmed by chloramphenicol treatment of synchronized swarmer and stalked cells (Fig. 5B), sampled as indicated from the synchronous culture shown in Fig. 5A. Before chloramphenicol treatment, both swarmer and stalked cells had 100% fully methylated (3.8-kb) DNA at bla::Tn5Ω-MP (Fig. 5A, lanes 1 and 5). After 2 h of chloramphenicol treatment, the swarmer cells retained fully methylated (3.8-kb) DNA while the stalked cells acquired ∼95% hemimethylated (2.9-kb) DNA (Fig. 5B). The drawing in Fig. 5B interprets these results. Chloramphenicol treatment blocked chromosome replication in the swarmer cells. In the stalked cells, the initiated replication forks reached bla::Tn5Ω-MP, and this DNA remained hemimethylated, because chloramphenicol blocked CcrM expression. Compare Fig. 5B with Fig. 5A, lanes 9 through 12, where in the absence of chloramphenicol CcrM methylation was induced, as it is normally induced, in the predivisional cells (55).
Measurement of extra chromosome replication by measuring unmethylated chromosome DNA.
The fidelity of chromosome replication can be measured by the Tn5Ω-MP Southern blot assay. Following the first initiation of chromosome replication in stalked cells, an extra round of chromosome replication will produce one unmethylated and one hemimethylated duplex from a hemimethylated duplex DNA molecule. This situation (see Fig. 7A) would produce a 2.0-kb band in the Tn5Ω-MP Southern blot assay (Fig. 3A). This band was not readily apparent in any of the experiments shown in Fig. 4A, 5, and 6A. Therefore, no or very few C. crescentus chromosomes break the once and only once rule for the initiation of replication during the cell cycle that is implied in Fig. 1A.
To maximize the detection of extra replication forks reaching Tn5Ω-MP, randomly placed transposons were screened for their proximity to Cori. This screen employed the methylation gradient data presented in Fig. 4B. A total of 50 random Tn5Ω-MP strains were grown exponentially in rich PYE medium, and their DNA was analyzed as described for Fig. 4A. The zzz::Tn5Ω-MP (GM1261 [Table 2]) chosen for the experiments described below (Fig. 6 and 7) lies 50 kb to the right of Cori (Fig. 2B) and has a wild-type 90-min doubling time in PYE medium.
To determine the optimal time during the cell cycle to detect extra rounds of chromosome replication, the following control experiment was performed (Fig. 6). Strain GM1261 was synchronized, and DNA was prepared at the indicated cell cycle times spanning the swarmer, stalked, and predivisional cell stages. The Southern blot assays diagrammed in Fig. 3 were applied to zzz::Tn5Ω-MP, dnaA, and pbpA (Fig. 6A, B, and C, respectively), and their quantitation is plotted in Fig. 6D. The natural GANTC methylation sites at dnaA and pbpA mark Cori-proximal and -distal sites, respectively (Fig. 2). Therefore, the passage of replication forks through dnaA and pbpA defines the early and late S-phase times. Note that the zzz::Tn5Ω-MP and dnaA plots closely coincide (Fig. 6D), indicating their proximity to Cori. The GANTC site at pbpA becomes only partially hemimethylated at later times, as was bla::Tn5Ω-MP in Fig. 5A. The declining hemimethylation at zzz::Tn5Ω-MP and dnaA (Fig. 6D) demonstrates the leading edge of the GANTC remethylation period caused by induced CcrM activity in the predivisional cells.
The Tn5Ω-MP Southern blot experiment shown in Fig. 7B presents strong evidence for rare extra chromosome replication that occurs at most only once per 1,000 cell divisions. Two samples of swarmer cells and two samples of late-S-phase stalked cells from the cell cycle experiment shown in Fig. 6 were analyzed for unmethylated chromosome DNA. Figure 7B also compares the quality of chromosome DNA prepared by the standard phenol and chloroform extraction method (lanes 1 to 4) and the superior agarose-embedding method (lanes 5 to 8). Progressively longer membrane exposures to X-ray film (1 and 18 h and 4 days [Fig. 7B]) demonstrate the appearance of the 2.0-kb Tn5Ω-MP band diagnostic of unmethylated DNA. This band is present in stalked cells (Fig. 7B, lanes 7 and 8) but not in swarmer cells (Fig. 7B, lanes 5 and 6), implying that it was produced by chromosome replication. Other minor bands are also present, but the 2.0-kb band is the only band uniquely produced from stalked-cell DNA digested with ClaI (Fig. 7B) and with XhoI (data not shown). This indicates that the 2.0-kb band is derived from Tn5Ω-MP and specific endonuclease digestion flanking the Ω fragment (Fig. 2A). This 2.0-kb band was quantified by X-ray film densitometry and by phosphorimaging of progressively longer membrane exposures. These two methods revealed that the 2.0-kb band was 1 part in 2,000 and 1 part in 3,000, respectively, of total Tn5Ω-MP DNA. This corresponds to at most 1 of 1,000 stalked cells initiating an extra round of replication from one of its two chromosomes.
DISCUSSION
How accurate is the two-chromosome rule during the C. crescentus cell cycle?
C. crescentus chromosome replication is probably more faithful than the 0.1% error frequency calculated from the experiment shown in Fig. 7B. This calculation depends on the interpretation, diagrammed in Fig. 7A, that the 2.0-kb band is unmethylated DNA resulting from an extra round of chromosome replication. Although this conclusion is reasonable, based on the arguments presented above, it is certainly not definitive, and the 2.0-kb band may result from other mechanisms. For example, some DNA may escape being fully methylated by CcrM in the predivisional cell or ClaI and XhoI digestion may not be 100% blocked by methylation. A number of unexplained faint bands appear in Fig. 7B, implying some spurious site-specific endonuclease activities. It is possible that C. crescentus produces no unmethylated chromosome DNA, or more likely much less than 0.1%, which is near the detection limit of our method.
The Tn5Ω-MP Southern blotting technique for measuring extra C. crescentus chromosome replication (Fig. 7) is probably more sensitive than other techniques that employ DNA density shift or DNA fluorometry protocols. For example, DNA density shift experiments, based on the original Meselson and Stahl experiments (30, 54), distinguish unreplicated (heavy-heavy) DNA from once-replicated (heavy-light) DNA and twice-replicated (light-light) DNA. In such experiments, sensitivity is limited by the inherently poor resolution of CsCl gradients. DNA fluorometry has also proven to be exceptionally valuable for cell cycle studies, since current techniques allow single-cell analysis (9, 45, 51). When combined with antibiotic treatment, DNA fluorometry allows one to count the chromosomes inside a bacterial cell (9, 45, 51). Treatment with chloramphenicol (for example, as described above [Fig. 4 and 5]) allows chromosome replication to completion without initiation, and cells arrest with integral numbers of chromosomes. DNA fluorometry of wild-type C. crescentus demonstrated that cells have either one or two chromosomes, and no obvious three-chromosome peak was observed (51). However, the signal-to-noise ratios for such experiments suggest that a subpopulation of three-chromosome C. crescentus cells would not be detected unless it represented at least 1% of the total population (personal observations). Also, spurious three-chromosome peaks could be produced by two cells sticking together. By contrast, the Southern blotting technique is not influenced by a cell aggregation artifact, and it is at least an order of magnitude (less than 0.1% of cells detected) more sensitive.
An additional, and theoretically more interesting, advantage of the Tn5Ω-MP Southern blotting technique is that it does not require the whole chromosome to replicate before a third chromosome is detected. It could be argued that there is no space to accommodate a third chromosome, and therefore no need to repress extra chromosome replication, inside a wild-type C. crescentus cell. The Tn5Ω-MP employed in the experiment shown in Fig. 7 was placed only 50 kb away from Cori. Detecting a third chromosome requires only 2.5% chromosome replication and presents no obvious spatial barrier. Also, previous experiments demonstrate no inherent barrier to producing unmethylated C. crescentus plasmid DNA, because stable incP plasmids accumulate as much as 20% unmethylated plasmid DNA in stalked cells (55). Therefore, a very precise system for restricting the initiation of chromosome replication to only one round per cell cycle, and not restricted cytoplasmic volume, is the most likely explanation for the low frequency of unmethylated chromosome DNA in stalked cells.
What mechanism(s) might repress extra chromosome replication?
E. coli has at least two systems to repress the initiation of chromosome replication: the Dam-SeqA system, described in the introduction, and the DnaA-DnaN system. Recent in vitro data demonstrate that DnaN contacts and quenches DnaA activity by stimulating its intrinsic ATPase (20). Therefore, DnaN, a replication fork assembly component, could provide a negative feedback to block DnaA immediately after the initiation of replication (20). Such a system could be common to many bacteria, including C. crescentus, which has both DnaA (26, 56) and DnaN (41) homologues.
Whether C. crescentus has a repression system analogous to the E. coli Dam-SeqA system remains an open question. Dam is unrelated to CcrM (55), and unlike Dam, CcrM is essential for cell viability (47). Interestingly, CcrM methylation represses its own transcription in swarmer cells (48), suggesting a wider role for CcrM in the repression of other cell cycle processes. The E. coli replication origin contains a statistically significant clustering of Dam (GATC) sites (57). The C. crescentus replication origin likewise has five CcrM sites (24, 26), but this number is not statistically significant for an intergenic region. The E. coli replication origin (250 bp) supports autonomous plasmid replication (31), and it is repressed by the Dam-SeqA system (22, 46, 50). The C. crescentus replication origin (Cori; approximately 500 bp) also supports autonomous plasmid replication (27), but Cori plasmids are significantly more promiscuous than the chromosome, and Cori plasmids accumulate unmethylated DNA in stalked cells (unpublished results). This implies that in C. crescentus, and apparently unlike in E. coli, extra negative control elements are positioned outside those DNA sequences that are essential for autonomous replication. Therefore, if C. crescentus has a system analogous to Dam-SeqA, this system probably has significant mechanistic differences as well. For example, Nathan et al. suggested that the initiation of C. crescentus chromosome replication requires an execution point prior to cell division (34). Recent results argue that the response regulator CtrA is a swarmer cell-specific repressor of chromosome replication (39). However, the bulk degradation of CtrA protein in the new stalked cells suggests that CtrA may not be available to repress chromosome replication at the start of S phase (12, 39).
What might be the function(s) of a chromosome methylation gradient?
The results shown in Fig. 4 expand the results in previous publications (47, 55) and clearly demonstrate that the period of DNA hemimethylation always decreases with the distance from the replication origin. In growing cells, DNA close to Cori is hemimethylated for nearly 60% of the cell cycle but DNA near the terminus of replication is only briefly hemimethylated. Template-directed mismatch repair is one established system in E. coli, in which Dam methylation marks the template strand and directs repair enzymes to resynthesize the new unmethylated strand (28). Although this possibility has not been explored, a comparable mismatch repair role may be played by CcrM. The E. coli mismatch repair system has a restricted opportunity to recognize mistakes, because E. coli rapidly remethylates most of its Dam (GATC) sites (28). Perhaps C. crescentus, with its well-developed cell cycle timing mechanisms, has expanded the window of opportunity for its DNA surveillance system(s) by restricting CcrM expression to the end of the cell cycle. Chromosome methylation is apparently not used for chromosome partitioning. Replication produces distinctly methylated top and bottom DNA duplexes (Fig. 1), but top and bottom strands are segregated randomly to swarmer and stalked cells at cell division (23, 36).
In summary, the chromosome methylation gradient and the very low incidence of secondary chromosome replication, documented in this report, are clearly fundamental properties of the C. crescentus asymmetric cell division cycle. Current molecular studies offer clues, but the details of how C. crescentus produces exactly two (but functionally different) chromosomes and places one into the swarmer cell compartment and the other into the stalked-cell compartment remain to be understood.
ACKNOWLEDGMENTS
I thank R. Siam, A. Reisenauer, and the reviewers for critically reading the manuscript.
This study was supported by a Medical Research Council of Canada (MRC) Scholarship (SH-50791-AP007403) and Operating Grant MT-13453 to G.T.M. The initial stages of this study were conducted in L. Shapiro’s laboratory at Stanford with support from National Institutes of Health grant GM 51426 to L.S.
REFERENCES
- 1.Alting-Mees M A, Short J M. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker T A, Wickner S H. Genetics and enzymology of DNA replication in Escherichia coli. Annu Rev Genet. 1992;26:447–477. doi: 10.1146/annurev.ge.26.120192.002311. [DOI] [PubMed] [Google Scholar]
- 3.Bakker A, Smith D W. Methylation of GATC sites is required for precise timing between rounds of DNA replication in Escherichia coli. J Bacteriol. 1989;171:5738–5742. doi: 10.1128/jb.171.10.5738-5742.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbeyron T, Kean K, Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J Bacteriol. 1984;160:586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett J T, Croft R H, Ferber D M, Gerardot C J, Schoenlein P V, Ely B. Genetic mapping with Tn5-derived auxotrophs of Caulobacter crescentus. J Bacteriol. 1982;151:888–898. doi: 10.1128/jb.151.2.888-898.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun Y V, Marczynski G T, Shapiro L. The expression of asymmetry during Caulobacter cell differentiation. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 7.Brun Y V, Shapiro L. A temporally controlled ς-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 8.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey H, Kell D B. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degnen S T, Newton A. Chromosome replication during development in Caulobacter crescentus. J Mol Biol. 1972;64:671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- 11.Dingwall A, Shapiro L. Rate, origin, and bidirectionality of Caulobacter chromosome replication as determined by pulsed-field gel electrophoresis. Proc Natl Acad Sci USA. 1989;86:119–123. doi: 10.1073/pnas.86.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domian I J, Quon K C, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1 to S transition in a bacterial cell-cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 12a.Ely, B. 12 Novermber 1997, posting date. Caulobacter crescentus. [Online.] http://www.cosm.sc.edu/caulobacter/map.html. [15 February 1999, last date accessed.]
- 13.Ely B. Vectors for transposon mutagenesis of non-enteric bacteria. Mol Gen Genet. 1985;200:302–304. doi: 10.1007/BF00425440. [DOI] [PubMed] [Google Scholar]
- 14.Ely B, Ely T W. Use of pulsed field gel electrophoresis to estimate the total number of genes required for motility in Caulobacter crescentus. Genetics. 1989;123:649–654. doi: 10.1093/genetics/123.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ely B, Johnson R C. Generalized transduction in Caulobacter crescentus. Genetics. 1977;87:391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber T M, Bryant D A. Molecular systematic studies of eubacteria, using ς70-type sigma factors of group 1 and group 2. J Bacteriol. 1997;179:1734–1747. doi: 10.1128/jb.179.5.1734-1747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoch J A, Silhavy T J. Two-component signal transduction. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 19.Johnson R C, Ely B. Isolation of spontaneously derived mutants from Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosome replicase. Cell. 1998;94:61–71. doi: 10.1016/s0092-8674(00)81222-2. [DOI] [PubMed] [Google Scholar]
- 21.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;15:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu M, Campbell J L, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 23.Marczynski G T, Dingwall A, Shapiro L. Plasmid and chromosomal DNA replication and partitioning during the Caulobacter crescentus cell cycle. J Mol Biol. 1990;212:709–722. doi: 10.1016/0022-2836(90)90232-B. [DOI] [PubMed] [Google Scholar]
- 24.Marczynski G T, Lentine K, Shapiro L. A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 1995;9:1543–1557. doi: 10.1101/gad.9.12.1543. [DOI] [PubMed] [Google Scholar]
- 25.Marczynski G T, Shapiro L. Bacterial chromosome origins of replication. Curr Opin Genet Dev. 1993;3:775–782. doi: 10.1016/s0959-437x(05)80098-x. [DOI] [PubMed] [Google Scholar]
- 26.Marczynski G T, Shapiro L. Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J Mol Biol. 1992;226:959–977. doi: 10.1016/0022-2836(92)91045-q. [DOI] [PubMed] [Google Scholar]
- 27.Marczynski G T, Shapiro L. The control of asymmetric gene expression during Caulobacter cell differentiation. Arch Microbiol. 1995;163:313–321. doi: 10.1007/BF00404203. [DOI] [PubMed] [Google Scholar]
- 28.Marinus M G. Methylation of DNA. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 782–791. [Google Scholar]
- 29.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 30.Meselson M, Stahl F W. The replication of DNA in Escherichia coli. Proc Natl Acad Sci USA. 1958;44:671–682. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messer W, Weigel C. Initiation of chromosome replication. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1579–1601. [Google Scholar]
- 32.Minnich S A, Ohta N, Taylor N, Newton A. Role of the 25-, 27-, and 29-kilodalton flagellins in Caulobacter crescentus cell motility: method for construction of deletion and Tn5 insertion mutants by gene replacement. J Bacteriol. 1988;170:3953–3960. doi: 10.1128/jb.170.9.3953-3960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriya S, Kato K, Yoshikawa H, Ogasawara N. Isolation of a dnaA mutant of Bacillus subtilis defective in initiation of replication: amount of DnaA protein determines cells’ initiation potential. EMBO J. 1990;9:2905–2910. doi: 10.1002/j.1460-2075.1990.tb07481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathan P, Osley M A, Newton A. Circular organization of the DNA synthetic pathway in Caulobacter crescentus. J Bacteriol. 1982;151:503–506. doi: 10.1128/jb.151.1.503-506.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nixon B T, Ronson C W, Ausubel F M. Two-component regulatory systems responsive to environmental stimuli share conserved domains with nitrogen assimilation regulatory genes. Proc Natl Acad Sci USA. 1986;83:7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osley M A, Newton A. Chromosome segregation and development in Caulobacter crescentus. J Mol Biol. 1974;90:359–370. doi: 10.1016/0022-2836(74)90379-9. [DOI] [PubMed] [Google Scholar]
- 37.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 38.Quon K C, Marczynski G T, Shapiro L. Cell-cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 39.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ried J L, Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 41.Roberts R C, Shapiro L. Transcription of genes encoding DNA replication proteins is coincident with cell cycle control of DNA replication in Caulobacter crescentus. J Bacteriol. 1997;179:2319–2330. doi: 10.1128/jb.179.7.2319-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Puler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 43.Skarstad K, Baker T A, Kornberg A. Strand separation required for initiation of replication at the chromosome origin of E. coli is facilitated by a distant RNA-DNA hybrid. EMBO J. 1990;9:2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skarstad K, Boye E. The initiator protein DnaA: evolution, properties and function. Biochim Biophys Acta. 1994;1217:111–130. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 45.Skarstad K, Von Meyenburg K, Hansen F G, Boye E. Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J Bacteriol. 1988;170:852–858. doi: 10.1128/jb.170.2.852-858.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 47.Stephens C M, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephens C M, Zweiger G, Shapiro L. Coordinate cell-cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol. 1995;177:1662–1669. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Freiesleben U, Rasmussen K V, Schaechter M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol. 1994;14:763–772. doi: 10.1111/j.1365-2958.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- 51.Winzeler E, Shapiro L. Use of flow cytometry to identify a Caulobacter 4.5 S RNA temperature-sensitive mutant defective in the cell-cycle. J Mol Biol. 1995;251:346–365. doi: 10.1006/jmbi.1995.0439. [DOI] [PubMed] [Google Scholar]
- 52.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright R, Stephens C, Zweiger G, Shapiro L, Alley M R K. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 54.Zakian V A, Brewer B J, Fangman W L. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell. 1979;17:923–934. doi: 10.1016/0092-8674(79)90332-5. [DOI] [PubMed] [Google Scholar]
- 55.Zweiger G, Marczynski G T, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the pre-divisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- 56.Zweiger G, Shapiro L. Expression of Caulobacter dnaA as a function of the cell cycle. J Bacteriol. 1994;176:401–408. doi: 10.1128/jb.176.2.401-408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zyskind J W, Cleary J M, Brusilow W S A, Harding N E, Smith D W. Chromosome replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc Natl Acad Sci USA. 1983;80:1164–1168. doi: 10.1073/pnas.80.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]