Abstract
The serotype-specific, 5.9-kb region II of the Haemophilus influenzae type a capsulation locus was sequenced and found to contain four open reading frames termed acs1 to acs4. Acs1 was 96% identical to H. influenzae type b Orf1, previously shown to have CDP-ribitol pyrophosphorylase activity (J. Van Eldere, L. Brophy, B. Loynds, P. Celis, I. Hancock, S. Carman, J. S. Kroll, and E. R. Moxon, Mol. Microbiol. 15:107–118, 1995). Low but significant homology to other pyrophosphorylases was only detected in the N-terminal part of Acs1, whereas the C-terminal part was homologous to several short-chain dehydrogenases/reductases, suggesting that Acs1 might be a bifunctional enzyme. To test this hypothesis, acs1 was cloned in an expression vector and overexpressed in Escherichia coli. Cells expressing this protein displayed both ribitol 5-phosphate dehydrogenase and CDP-ribitol pyrophosphorylase activities, whereas these activities were not detectable in control cells. Acs1 was purified to near homogeneity and found to copurify with ribitol 5-phosphate dehydrogenase and CDP-ribitol pyrophosphorylase activities. These had superimposable elution profiles from DEAE-Sepharose and Blue-Sepharose columns. The dehydrogenase activity was specific for ribulose 5-phosphate and NADPH in one direction and for ribitol 5-phosphate and NADP+ in the other direction and was markedly stimulated by CTP. The pyrophosphorylase showed activity with CTP and ribitol 5-phosphate or arabitol 5-phosphate. We conclude that acs1 encodes a bifunctional enzyme that converts ribulose 5-phosphate into ribitol 5-phosphate and further into CDP-ribitol, which is the activated precursor form for incorporation of ribitol 5-phosphate into the H. influenzae type a capsular polysaccharide.
The production of a polysaccharide capsule is a common feature of many pathogenic bacteria that cause invasive disease (5). The capsule allows the invading organisms to escape the immune system by a number of mechanisms, such as impairment of phagocytosis, reduced opsonophagocytosis, and increased resistance to complement (31, 38, 47).
The gram-negative rod Haemophilus influenzae elaborates six structurally and serotypically different polysaccharide capsules designated types a to f (37). Until recently, serotype b capsulate strains were predominant among isolates from invasive infections. Introduction of the conjugate vaccine for H. influenzae type b (Hib) has led to a significant decline in the incidence of Hib invasive disease and a relative increase in the isolation of other capsular types (23, 34, 49).
Capsular polysaccharides are polymers of repeating units that consist of one to several different saccharides. Biosynthesis of a polysaccharide capsule is thought to start in the cytoplasm, where the individual sugars that constitute the repeating units are synthesized and converted into activated nucleotide derivatives. In a second phase, these activated sugars are polymerized. The final phase of capsule biosynthesis is the translocation of the polymerized polysaccharide from the inner membrane to the cell surface and its organization into a capsule (17).
The capsules of H. influenzae type a (Hia) and Hib both contain ribitol 5-phosphate, the polysaccharide of Hib being a polymer of -3-[β-d-ribose-(1-1)-d-ribitol-5-phosphate-] (7, 9) and that of Hia being a polymer of -4-[β-d-glucose-(1-4)-d-ribitol-5-phosphate-] (8).
The genes involved in H. influenzae capsule expression are clustered in the chromosomal capsulation locus (cap), which can be divided into three functionally distinct regions. A central serotype-specific region, called region II, is flanked by regions I and III, which are common to all capsular serotypes (21). This regional organization is also found in other organisms, like Escherichia coli (39), Neisseria meningitidis (13), Staphylococcus aureus (41), and Streptococcus pneumoniae (12). In encapsulated H. influenzae, region I contains four open reading frames, termed bexDCBA, which encode an ATP-driven polysaccharide export apparatus (20, 22). The function of region III has not yet been characterized but is likely to be found in postpolymerization events. In Hib, region II has been sequenced and found to contain four open reading frames. orf1 was shown to encode a CDP-ribitol pyrophosphorylase, and orf2 was hypothesized to code for a ribitol 5-phosphate dehydrogenase (46).
In this paper, we present the sequence of Hia cap locus region II and show data indicating that the gene product of the first open reading frame, which is 96% identical to that of Hib orf1, is a bifunctional ribulose 5-phosphate reductase–CDP-ribitol pyrophosphorylase.
MATERIALS AND METHODS
Materials.
Restriction enzymes and IPTG were from Life Technologies Inc. (Gaithersburg, Md.). NADPH, NADH, NADP+, NAD+, CTP, bovine serum albumin (BSA), phosphoglucomutase, and glucose 6-phosphate dehydrogenase from Leuconostoc mesenteroides were from Boehringer Mannheim GmbH (Mannheim, Germany). Antipain, leupeptin, ribulose 5-phosphate, ribose 5-phosphate, erythritol 4-phosphate, sorbitol 6-phosphate, arabitol 5-phosphate, xylulose 5-phosphate, glucose 1,6-bisphosphate, and UDP-glucose pyrophosphorylase were from Sigma (Sigma-Aldrich, Bornem, Belgium). Ribitol 5-phosphate was prepared as described previously (10). Oligonucleotides, DEAE-Sepharose and Blue-Sepharose were from Pharmacia (Uppsala, Sweden). Other chemicals were from Merck (Darmstadt, Germany), and were all of analytical grade.
DNA sequencing analysis.
Plasmids pAD2, pAD5, and pAD6 (kindly provided by A. Dhir) were used as the sources of cloned cap locus DNA from Hia RM107, a capsular type a isolate from a patient with respiratory infection, identical to ATCC 9006 (11). pAD2, pAD5, and pAD6 contain the 1.5-, 5.3-, and 11.0-kb EcoRI fragments of the Hia cap locus, respectively, cloned into pUC18 (Stratagene, La Jolla, Calif.).
Plasmid DNA was isolated by the procedure of Birnboim and Doly (4) or with the Nucleobond PC 100 plasmid extraction kit (Macherey Nagel, Düren, Germany). Subclones were sequenced on both strands by the dideoxy chain termination method of Sanger et al. (40) with the T7 sequencing kit (Pharmacia). Alternatively, the Cy5 AutoRead sequencing kit (Pharmacia) was used with Cy5-labeled primers and an ALFexpress DNA sequencer (Pharmacia).
Primer extension analysis.
Total cellular RNA was prepared from 80 ml of exponential-growth-phase culture of Hia RM107 (28). RNA quality was assessed by electrophoresis in 0.7% agarose gels with and without prior treatment with RNase. The primer extension study was done as described previously (46) with a 32P end-labeled oligonucleotide and 40 to 65 μg of total RNA.
Construction of pETacs1.
The Hia Acs1 protein was expressed by the T7 RNA polymerase-based system of Studier and coworkers (44). acs1 was PCR amplified from plasmid pAD5.10, which contains part of the pAD5 insert, using oligonucleotide primers with the following sequences: 5′ TAATCTGTTGGGATATCATATG and 5′ ACGGATCCGTATTAGCCATAACAGACTCACTC. The underlined sequences indicate the restriction sites for NdeI, which incorporates the start codon (boldface), and for BamHI, which flanks the stop codon. After digestion with NdeI and BamHI, the amplified DNA was cloned into pUC18 and named pUCacs1. The nucleotide sequence of the clone used in the expression experiments was confirmed by sequencing. The insert was excised from pUCacs1 with NdeI and BamHI and ligated into the expression vector pET3a (Promega, Madison, Wis.). This plasmid was amplified in E. coli DH5α, checked by restriction analysis with NdeI and BamHI, and used to transform E. coli Bl21(DE3)pLysS (Promega). This construct, designated pETacs1, contained the acs1 gene in the proper position and orientation for expression.
Overexpression of the recombinant protein Acs1 in E. coli.
A fresh E. coli Bl21(DE3)pLysS transformant colony harboring pETacs1 was grown at 37°C in 1 liter of M9 minimal medium supplemented with 0.4% glucose, 100 μg of ampicillin/ml, and 25 μg of chloramphenicol/ml until an optical density at 600 nm of 0.5 was reached. The culture was stored on ice for 15 min before addition of IPTG (isopropyl-β-d-thiogalactoside) to a final concentration of 0.4 mM and was subsequently incubated (unless otherwise indicated) at 15°C for 60 h. Protein extracts were prepared as described previously (48) by lysing the cells in 50 ml of lysing buffer (20 mM potassium phosphate, pH 7.4, 5 mM EDTA, 1 mM dithiothreitol [DTT], 1 mg of lysozyme/ml, 5 μg of leupeptin/ml, 5 μg of antipain/ml, and 0.5 mM phenylmethylsulfonyl fluoride) and submitting them to three cycles of freezing and thawing. DNA was digested by incubation for 1 h at 4°C with 0.1 mg of DNase I/ml and 10 mM MgSO4. The insoluble fraction, including cell debris and inclusion bodies, was removed by centrifugation at 40,000 × g at 4°C and was resuspended in 50 ml of lysing buffer. Both fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% [wt/vol]) (24) to detect insoluble and soluble recombinant protein. The gels were stained with Coomassie brilliant blue.
Purification of the Acs1 protein.
All purification steps were performed at 4°C. A soluble extract (48 ml) prepared from a 1-liter culture grown at 15°C was made 10% (wt/vol) in glycerol to prevent precipitation of proteins and was kept overnight at −80°C. After thawing, it was loaded at a flow rate of 2.5 ml/min onto a DEAE-Sepharose column (1.6 by 13 cm) equilibrated in 20 mM HEPES (pH 7.1), 3 μg of antipain/ml, 3 μg of leupeptin/ml, 1 mM DTT, and 10% glycerol (buffer A). The column was washed with 100 ml of buffer A and eluted with a linear salt gradient from 0 to 0.5 M KCl in 200 ml of the same buffer. All fractions were tested for their ability to reduce ribulose 5-phosphate or ribose 5-phosphate and were frozen overnight at −80°C. The fractions with the highest specific activities were thawed and loaded onto a Blue-Sepharose column (0.6 by 10 cm) equilibrated in buffer A. The column was washed with 6 ml of the same buffer, and elution was done by applying successively 6 ml of buffer A with 0.25 M NaCl, 1.5 M NaCl, and 1.5 M NaCl–5 mM NADP+. The protein concentrations in the active fractions were measured according to the method of Bradford (6) with bovine gamma globulin as a standard. After addition of BSA to a final concentration of 0.5% (wt/vol), the fractions were stored at −80°C. The purification was performed twice with similar results.
Enzyme assays.
Ribitol 5-phosphate dehydrogenase was assayed spectrophotometrically at 340 nm in a 1-ml reaction mixture containing, unless otherwise indicated, 25 mM HEPES (pH 7.1), 125 μM NADPH, 1 mM DTT, 50 μM CTP, and 200 μM ribulose 5-phosphate or ribose 5-phosphate. The reverse reaction was measured with purified protein in a mixture containing 25 mM Tris (pH 8.5), 1.12 mM NADP+, 1 mM DTT, 100 μM CTP, and 10 mM ribitol 5-phosphate in a final volume of 1 ml.
CDP-ribitol pyrophosphorylase activity was tested by a spectrophotometric assay at 340 nm, in which the inorganic pyrophosphate formed from ribitol 5-phosphate and CTP is used in a cascade of downstream reactions leading to the reduction of NAD+. The materials for this assay were 25 mM HEPES (pH 7.1), 200 μM ribitol 5-phosphate, 200 μM CTP, 5 mM MgCl2, 1 mM DTT, 1 μM glucose 1,6-bisphosphate, 500 μM UDP-glucose, 175 μM NAD+, 0.125 U of UDP-glucose pyrophosphorylase, 0.16 U of phosphoglucomutase, and 1 U of glucose 6-phosphate dehydrogenase (32). One unit of enzyme activity was defined as the amount of enzyme catalyzing the conversion of 1 μmol of substrate per min under standard assay conditions at 30°C.
Nucleotide sequence accession number.
The EMBL accession number for the nucleotide sequence of Hia cap locus region II is Z 37516 (HIACAP).
RESULTS
Sequence analysis of the Hia cap locus region II.
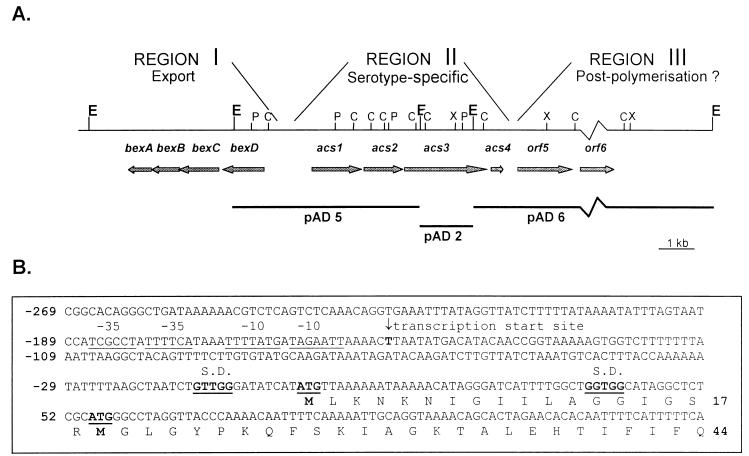
The central region (region II) of the Hia RM107 cap locus was sequenced on both strands, using cap locus-containing plasmids pAD2, -5, and -6 (11). Region II had a low G+C content (31%) compared to those of the common regions I (39%) (22) and III (40%) (our unpublished results). Four open reading frames, designated acs1 to acs4 (for type a capsule synthesis), were found on the opposite strand of the bex gene cluster of region I (Fig. 1A) (23). Within region II, the G+C contents of acs1 and acs2 (35.5 and 34.8%, respectively) were significantly higher than the G+C contents of acs3 and acs4 (28 and 26%).
FIG. 1.
(A) Regional organization of the cap locus in Hia RM107. Region I contains four genes called bexDCBA. On the opposite DNA strand, region II comprises four serotype-specific genes, designated acs1 to acs4. Region III has two open reading frames, orf5 and orf6 (our unpublished results). The arrows indicate genes and open reading frames. The horizontal bars indicate the 5.3-, 1.5-, and 11-kb plasmids, pAD5, -2, and -6, which were used as a source of cap locus DNA. The vertical lines show cleavage sites for restriction endonucleases: ClaI (C), EcoRI (E), PstI (P), and XbaI (X). (B) First 131 nucleotides and deduced amino acid sequence of acs1 and its 5′ untranslated region. Two possible ATG start codons and their respective Shine-Dalgarno sequences are indicated in boldface and underlined. The transcription start site at −149 bp from the first ATG codon is indicated with an arrow. Possible −10 and −35 consensus sequences are underlined.
For acs1, two possible ATG start codons were found, with the most 5′ located 1,502 bp upstream from the bexD start codon and the second located 54 nucleotides further downstream from the first (Fig. 1B). A stop codon terminating acs1 was found 1,425 bp downstream from the first ATG start codon. The deduced protein consists of 474 or 456 amino acids, with a predicted molecular mass of 52.4 or 50.6 kDa. The start codon of acs2 is 17 bp downstream from the acs1 stop codon. acs2 is 1,116 bp long and translates into a protein of 371 amino acids with a predicted molecular mass of 42.5 kDa. The start of acs3 is located 10 bp downstream from the stop codon of acs2. Three possible ATG start codons were identified, but the first was the only one preceded by a Shine-Dalgarno motif. acs3 is 2,370 bp long and codes for a protein of 789 amino acids with a predicted molecular mass of 92.7 kDa. The stop codon of acs3 is separated by 13 bp from the start codon of acs4, which is preceded by a possible Shine-Dalgarno motif 5 bp upstream. The 357-bp acs4 sequence encodes a protein of 118 amino acids with a calculated molecular mass of 14.6 kDa. Several stop codons were found in all three reading frames in the 230-bp sequence between the stop codon of acs4 and the ATG start codon of orf5, the first open reading frame of region III.
Primer extension analysis.
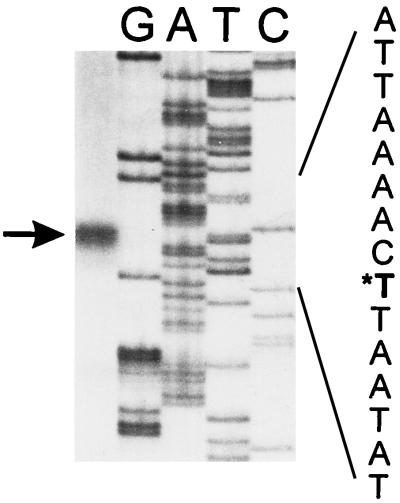
With oligo 5-1 (5′ CACCAGCCAAAATGATCC), complementary to acs1 bp 23 to 40, a major extension product was found starting 149 nucleotides upstream from the most 5′ acs1 start codon (Fig. 2). Two sequences, TAGAATT and TTTTATG, located 6 and 13 nucleotides upstream of the start of this transcript, matched the −10 consensus sequence. At an appropriate distance from the transcription start, the sequences TTTTCA and TCGCCT, separated by 1 nucleotide, could serve as −35 consensus sequences (Fig. 1B).
FIG. 2.
Primer extension analysis of acs1 with oligonucleotide 5-1 (5′ CACCAGCCAAAATGATCC). Lanes G, A, T, and C show the respective sequencing products resulting from a sequencing reaction with oligonucleotide 5-1 and with ddGTP, ddATP, ddTTP, and ddCTP, respectively. The first lane contains the primer extension product, which is indicated by an arrow. In the sequence represented on the right, the corresponding transcription start site is indicated with an asterisk.
Sequence homology searches.
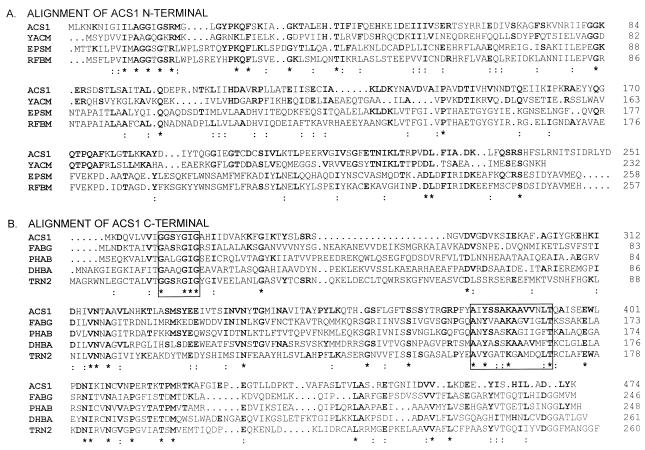
Comparison of acs1 to Hib orf1, the first open reading frame in the Hib cap locus region II (46), revealed 96% identity at the nucleotide and deduced amino acid sequence levels. The deduced amino acid sequence of Acs1 was compared to other known sequences, using the search programs FASTA (36) and BLAST (2). Homology with several dehydrogenases was detected (Fig. 3B). All of these were about 25% identical to Acs1 and were members of the short-chain dehydrogenase/reductase (SDR) family (18). Remarkably, this homology was restricted to the 250 C-terminal residues of Acs1.
FIG. 3.
Alignment of the predicted amino acid sequence of Acs1 with homologous proteins detected by BLAST and FASTA searches. Multiple sequence alignments were performed with the CLUSTAL W program (45). Conserved residues are in boldface; ∗, identical residues; :, amino acids belonging to the same physicochemical group. (A) Alignment of Acs1 amino acids 1 to 251. YACM, B. subtilis hypothetical 25.8-kDa enzyme, unidentified protein family UPF0007 (33% identity) (35); EpsM, Acinetobacter calcoaceticus bifunctional phosphomannose-isomerase–GDP-mannose-pyrophosphorylase (unpublished; TrEMBL accession no. Q43941); RFBM, Salmonella typhimurium mannose 1-P-guanylyltransferase (25). (B) Alignment of Acs1 amino acids 252 to 474. FABG, B. subtilis 3-oxoacyl acylcarrier protein reductase (30); PHAB, acetoacetyl-CoA reductase from Acinetobacter sp. strain RA3849 (42); DHBA, B. subtilis 2,3-dihydro-2,3-dihydroxylbenzoate dehydrogenase (1); TRN2, Hyoscyamus niger tropinone reductase II (33). All homologous proteins are members of the SDR family. The NAD(P) binding site is indicated by a box at amino acids 261 to 267. The sequence motif typical of the SDR family (18) is contained in a box at amino acids 380 to 393.
A separate homology search with the 250 N-terminal amino acids of Acs1 showed 21 to 33% identity to six of the seven members of the unidentified protein family UPF0007 (Prosite accession no. PDOC00997). In addition, a weak but significant homology to several pyrophosphorylases was found (Fig. 3A), particularly in the conserved region (G111G112G114T115R116L117P122K123) of UDP-N-acetylglucosamine pyrophosphorylases (29). Interestingly, several of the conserved amino acid residues are located between the two possible start methionines of Acs1, suggesting that translation starts most likely at the first start codon. At the nucleotide level, no difference was seen in codon usage or in G+C contents between the 5′ half and the 3′ half of acs1.
Comparison of Acs2 to Hib Orf2 revealed 67.1% identity. This identity was particularly pronounced in the N-terminal half of the proteins (88.4% in the first 190 amino acids). An ATP-GTP binding motif was found at amino acids 152 to 159. No significant homologies to other proteins were found.
There was no overall similarity between Acs3 and Hib Orf3. However, the C-terminal 400 amino acids were homologous to those of several teichoic acid biosynthesis-related proteins (all with about 48% similarity), like TasA (OrfX) from S. pneumoniae (19) and TagB and -F from Bacillus subtilis (16, 27). Interestingly, these proteins share a conserved motif at amino acids 692 to 705 of Acs3, which is also found in Hib Orf3, in a H. influenzae type c capsulation protein (our unpublished results), and in a teichoic acid biosynthesis protein from Methanobacterium thermoautotrophicum (accession no. O26465). In addition, the 100 N-terminal amino acids of Acs3 show a significant identity (all about 38%) to several sugar transferases, like EpsI from S. thermophilus (43) and Cps14I and -J from S. pneumoniae (19).
Acs4 was not homologous to Hib Orf4 or to any protein in the databases.
Expression of Acs1.
The results of the sequence comparisons indicated that Acs1 could be a bifunctional protein capable not only of forming CDP-ribitol but also of catalyzing a dehydrogenase reaction specifically required for the synthesis of the capsular polysaccharide, most likely a reduction of ribulose 5-phosphate or ribose 5-phosphate into ribitol 5-phosphate. To test this hypothesis, we expressed the protein in E. coli.
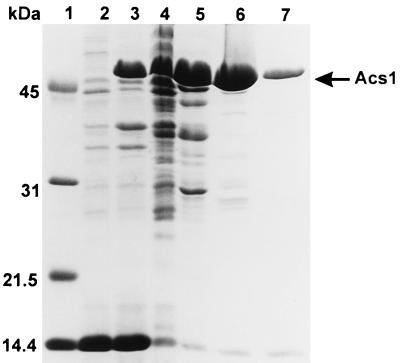
acs1 was amplified by PCR with a primer corresponding to the acs1 5′ end containing the start codon as part of a NdeI restriction site and a 3′ primer flanking the stop codon and including a BamHI restriction site. The coding sequence was inserted in a pET3a expression vector. The resulting plasmid, pETacs1, was then used to transform E. coli Bl21(DE3)pLysS. Addition of IPTG to a growing culture in M9 minimal medium resulted in the expression of an approximately 53-kDa protein in the cells harboring the recombinant plasmid. In cells containing the expression vector without insert, no 53-kDa band was found (Fig. 4). SDS-PAGE showed that approximately 50% of the overexpressed protein was in soluble form when the expression was carried out at 15°C, whereas at 22 and 27°C the proportions of soluble recombinant protein were only about 20 and 5%, respectively.
FIG. 4.
SDS-PAGE analysis of crude extracts and of purified fractions. Expression was carried out at 15°C for 60 h, and the extracts were prepared as described in Materials and Methods. Lane 1, molecular mass markers; lane 2, insoluble proteins prepared from control cells (58 μg); lanes 3 and 4, insoluble proteins (70 μg) and soluble proteins (156 μg), respectively, present in extracts from cells expressing Acs1; lane 5, DEAE-Sepharose fraction 27 (135 μg); lane 6, Blue-Sepharose fraction 24 (72 μg); lane 7, Blue-Sepharose fraction 26 (20 μg).
Purification of the recombinant Acs1.
An extract was prepared from a culture of pETacs1-containing E. coli Bl21(DE3)pLysS that was grown in 1 liter of M9 medium at 15°C and induced with IPTG for 60 h. This crude extract was shown to oxidize NADPH in the presence of ribulose 5-phosphate or ribose 5-phosphate at a rate of 0.034 and 0.012 μmol min−1 mg−1, respectively, indicating ribitol 5-phosphate dehydrogenase activity. Due to the presence of ribose 5-phosphate isomerase in crude extracts, it was not possible to determine at this stage whether ribulose 5-phosphate or ribose 5-phosphate was the true substrate for Acs1. Interestingly, the dehydrogenase activity was found to be markedly stimulated by CTP, which, at 50 μM, increased the activity with ribulose 5-phosphate and with ribose 5-phosphate to 1.88 and 0.44 μmol min−1 mg−1, respectively. When the expression was carried out at higher temperatures (18, 22, and 27°C), lower specific activities were observed in comparison with an expression at 15°C (1.28, 0.08, and 0.03 μmol min−1 mg−1, respectively, of ribitol 5-phosphate dehydrogenase activity measured with ribulose 5-phosphate in the presence of 50 μM CTP).
CDP-ribitol pyrophosphorylase was also measured in the extracts of cells induced at 15°C and was found to amount to 0.22 μmol min−1 mg−1. Neither ribitol 5-phosphate dehydrogenase activity nor CDP-ribitol pyrophosphorylase activity could be detected in a control extract prepared from an E. coli culture containing the pET3a vector without acs1 (less than 0.5% of the activities measured in an extract of pETacs1-containing cells incubated at 15°C).
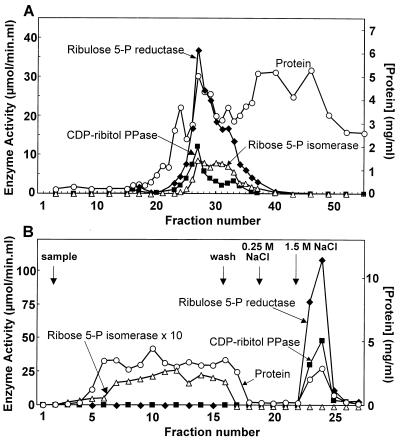
The overexpressed protein was purified by chromatography on DEAE-Sepharose and on Blue-Sepharose. As shown in Fig. 5, ribitol 5-phosphate dehydrogenase and CDP-ribitol pyrophosphorylase had nearly superimposable elution profiles from both columns. Furthermore, they coeluted with the overexpressed 53-kDa protein, which was nearly homogeneous after the Blue-Sepharose step (Fig. 4). In the DEAE-Sepharose fractions, ribitol 5-phosphate dehydrogenase displayed an activity that was about twofold higher with ribulose 5-phosphate as the substrate than with ribose 5-phosphate. After the Blue-Sepharose step, the activity was entirely specific for ribulose 5-phosphate. This suggested that ribulose 5-phosphate was the true substrate and that DEAE-Sepharose fractions were still contaminated with ribose 5-phosphate isomerase while Blue-Sepharose fractions no longer were. Measurement of ribose 5-phosphate isomerase in the eluate of both columns entirely confirmed this view (Fig. 5). The overall purification yield and recovery were about fourfold higher in the case of CDP-ribitol pyrophosphorylase than in the case of ribitol 5-phosphate dehydrogenase (Table 1), most likely because the former activity was underestimated in the crude extract due to the presence of active pyrophosphatases. To test this hypothesis, CDP-ribitol pyrophosphorylase activity was measured in the presence of KF, known to inhibit inorganic pyrophosphatases (3). When this was done, pyrophosphorylase activity in the crude extract increased from 0.22 to 0.46 μmol min−1 mg−1, whereas in the Blue-Sepharose fractions, no effect was observed.
FIG. 5.
Purification of ribulose 5-phosphate reductase (ribotol 5-phosphate dehydrogenase) and CDP-ribitol pyrophosphorylase by chromatography on DEAE-Sepharose (A) and Blue-Sepharose (B). (A) A bacterial extract containing about 300 mg of protein was loaded onto the DEAE-Sepharose column, which was developed with a linear KCl gradient. (B) Fractions 26 to 32 of the DEAE-Sepharose column were loaded onto a Blue-Sepharose column; proteins were eluted with a stepwise NaCl gradient. Ribulose 5-phosphate (5-P) reductase (⧫) was measured in the presence of 50 μM CTP. CDP-ribitol pyrophosphorylase (PPase) (■), ribose 5-phosphate isomerase (▵), and the protein concentration (○) were also measured.
TABLE 1.
Purification tablea
| Purification step | Vol (ml) | Protein (mg/ml) | Ribulose 5-P reductase sp act (μmol/min/mg) | CDP-ribitol PPase sp act (μmol/min/mg) | Recovery (%)
|
|
|---|---|---|---|---|---|---|
| Reductase | PPase | |||||
| Crude extract | 48 | 6.25 | 1.88 | 0.22 | 100.0 | 100.0 |
| DEAE-Sepharose | 14 | 4.57 | 5.60 | 1.42 | 63.5 | 137.7 |
| Blue-Sepharose | 4 | 2.48 | 37.50 | 15.70 | 65.8 | 235.5 |
Elution of DEAE- and Blue-Sepharose columns was performed as described in Materials and Methods and the legend to Fig. 5. PPase, pyrophosphorylase; 5-P, 5-phosphate.
Stability of Acs1.
Acs1 was found to be a rather unstable protein. Thus, when the homogeneous enzyme was incubated at a concentration of 0.3 mg/ml in the presence of 20 mM HEPES (pH 7.1), 1 mM DTT, and 0.5 mg of BSA/ml at 23°C, its ribitol 5-phosphate dehydrogenase activity decreased to about 50% of the initial activity after 2 h. This decrease in activity was completely prevented by the addition of 50 μM CTP to the dilution buffer.
Characterization and kinetic properties of Acs1.
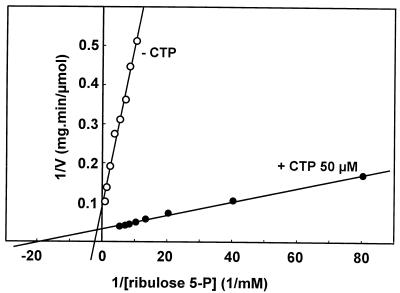
Acs1 showed a broad pH optimum of pH 7 to 8.4 for ribulose 5-phosphate reductase activity. The reaction was strictly NADPH dependent; no activity was observed with NADH. Double-reciprocal plots showed that the addition of 50 μM CTP decreased the Km value for ribulose 5-phosphate from 400 to 50 μM and increased the Vmax from 9.55 to 27.9 μmol min−1 mg of protein−1 (Fig. 6). The Ka for CTP was 2 μM, and the enzyme was not stimulated by UTP, ATP, GTP, ADP, or dCTP. The Km value for NADPH was about 10 μM. No activity was observed with xylulose 5-phosphate as the substrate. At pHs 7.1 and 8.4, ribulose 5-phosphate reductase activity was not inhibited either by 0.5 mM ribitol 5-phosphate or by 0.5 mM NADP+.
FIG. 6.
Double-reciprocal plot showing the effect of CTP on ribulose 5-phosphate reductase activity. The enzyme was assayed with 125 μM NADPH and 0 or 50 μM CTP.
The opposite reaction (oxidation of ribitol 5-phosphate to ribulose 5-phosphate) was measured at pH 8.5, with elevated concentrations of NADP+ (1.12 mM) and ribitol 5-phosphate (10 mM). Under these conditions, an activity of 0.73 μmol min−1 mg−1 was detected in the absence of CTP and an activity of 2.87 μmol min−1 mg−1 was detected in the presence of 100 μM CTP. In this reverse reaction, arabitol 5-phosphate could not substitute for ribitol 5-phosphate.
CDP-ribitol pyrophosphorylase activity was determined at two pH values (7.1 and 8.0), but no difference in activity was detected. The Km for ribitol 5-phosphate was 37 μM, and the Vmax was 15.7 μmol min−1 mg−1. Activity with arabitol 5-phosphate was also detected, with similar kinetic constants. In contrast, no activity was detected with erythritol 4-phosphate or sorbitol 6-phosphate. The Km value for CTP was 150 μM, and no activity was detected when CTP was replaced by UTP.
DISCUSSION
The almost-complete identity between acs1 of Hia and the first gene in Hib region II, termed orf1, confirms prior hybridization data showing homology between parts of the Hia and Hib regions II (15). Moreover, it is highly suggestive of a common function in Hia and Hib capsule synthesis. Since biochemical experiments with Hib mutants had shown that orf1 encodes a CDP-ribitol pyrophosphorylase (46), Acs1 was expected to have the same function. Sequence comparisons indicated that Acs1 and Hib Orf1 apparently each have two distinct domains: an N-terminal domain, homologous to those of several pyrophosphorylases, and a C-terminal domain, with homology to short-chain alcohol dehydrogenases. Proof that Acs1 was indeed a bifunctional enzyme came from expression experiments with E. coli showing that both ribitol 5-phosphate dehydrogenase and CDP-ribitol pyrophosphorylase activities were induced by expression of the acs1 gene. Furthermore, these two activities were shown to copurify with the overexpressed 53-kDa protein. The purification recovery of CDP-ribitol pyrophosphorylase was higher than 100%, indicating that its activity was underestimated in the crude extract. This was most likely due to the presence of contaminating inorganic pyrophosphatases, leading to hydrolysis of inorganic pyrophosphate, the formation of which was measured in the pyrophosphorylase assay. This was confirmed by the finding that fluoride, an inhibitor of inorganic pyrophosphatases (3), did indeed increase the CDP-ribitol pyrophosphorylase activity measured in the crude extract but not that of the pure enzyme.
Calculations indicate that Acs1 could easily support the rate of capsule synthesis in vivo. Assuming that (i) protein and capsule make up about 50 and 10%, respectively of the dry weight, (ii) that 59% of the dry weight of the capsule is contributed by ribitol 5-phosphate, and (iii) that the division time of H. influenzae is 30 min, we calculate that the rate of ribitol 5-phosphate incorporation is roughly equal to 0.1 mg of ribitol 5-phosphate/(30 min · mg), i.e., 15 nmol min−1 mg of protein−1. Such a specific activity would be accounted for if Acs1 represented 0.1% of the total protein content, which seems to be a reasonable assumption.
The kinetic properties of Acs1 indicate that the dehydrogenase is specific for ribulose 5-phosphate. The activity observed with ribose 5-phosphate in crude extracts and in partially purified preparations can easily be explained by the conversion of ribose 5-phosphate to ribulose 5-phosphate via ribose 5-phosphate isomerase, an enzyme of the pentose phosphate pathway. In theory, ribulose 5-phosphate could be reduced to either arabitol 5-phosphate or ribitol 5-phosphate. Since only ribitol 5-phosphate is used in the opposite reaction, it seems unlikely that arabitol 5-phosphate is a reaction product.
In contrast to the NAD(H)-specific ribitol 5-phosphate dehydrogenase from Lactobacillus casei (14), Hia Acs1 is specific for NADP(H). Due to the different ratios of the oxidized over the reduced forms of these nucleotides (26), the use of NAD(H) permits oxidation of ribitol 5-phosphate whereas NADP(H) favors reduction. This is in keeping with the physiological role of these enzymes, on the one hand in a catabolic pathway consuming ribitol in L. casei (14) and on the other hand in a biosynthetic pathway leading to a ribitol-containing polymer in H. influenzae. Thus, in H. influenzae, the enzyme truly functions as a ribulose 5-phosphate reductase rather than as a ribitol 5-phosphate dehydrogenase.
An intriguing property of this reductase is that it is markedly stimulated by CTP, causing a higher affinity for ribulose 5-phosphate and a higher Vmax. The very low Ka value for CTP (2 μM) suggests that the enzyme is constantly saturated and therefore that CTP does not play a regulatory role in vivo.
The pyrophosphorylase was shown to act on both arabitol 5-phosphate and ribitol 5-phosphate. Since no arabitol is found in the capsule of Hia or Hib (8, 9), arabitol 5-phosphate is presumably not a physiologically relevant substrate. It is not known if binding of CTP to the pyrophosphorylase catalytic site also mediates its effect on the reductase or if a distinct allosteric site is involved. The very different values of Ka (2 μM) and Km (150 μM) for CTP could suggest two different sites, but one should remain aware of the different experimental conditions for determining both values.
ACKNOWLEDGMENTS
We thank A. Dhir for the gift of plasmids pAD2, pAD5, and pAD6. We also thank Kate Peel for technical assistance.
This work was supported by a Glaxo Wellcome Grant in Infectiology and Clinical Microbiology and by the Belgian Federal Service for Scientific, Technical and Cultural Affairs.
REFERENCES
- 1.Adams R, Schumann W. Cloning and mapping of the Bacillus subtilis locus homologous to Escherichia coli ent genes. Gene. 1993;133:119–121. doi: 10.1016/0378-1119(93)90235-u. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Baykov A A, Artjukov A A, Avaeva S M. Fluoride inhibition of inorganic pyrophosphatase. I. Kinetic studies in a Mg2+-PPi system using a new continuous enzyme assay. Biochim Biophys Acta. 1976;13:982–992. doi: 10.1016/0005-2744(76)90343-0. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulnois G J, Jann K. Bacterial polysaccharide capsule synthesis, export and evolution of structural diversity. Mol Microbiol. 1989;3:1819–1823. doi: 10.1111/j.1365-2958.1989.tb00168.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Branefors-Helander P, Erbing C, Kenne L, Linberg B. The structure of the capsular antigen from Haemophilus influenzae type b. Acta Chem Scand B. 1976;30:276–277. [Google Scholar]
- 8.Branefors-Helander P, Erbing C, Kenne L, Linberg B. The structure of the capsular antigen from Haemophilus influenzae type a. Carbohydr Res. 1977;56:117–122. doi: 10.1016/s0008-6215(00)84242-1. [DOI] [PubMed] [Google Scholar]
- 9.Crisel R M, Baker R S, Dorman D E. Capsular polymer of Haemophilus influenzae, type b. J Biol Chem. 1975;250:4926–4930. [PubMed] [Google Scholar]
- 10.Detheux M, Vandercammen A, Van Schaftingen E. Effectors of the regulatory protein acting on liver glucokinase: a kinetic investigation. Eur J Biochem. 1991;200:553–561. doi: 10.1111/j.1432-1033.1991.tb16218.x. [DOI] [PubMed] [Google Scholar]
- 11.Dhir A. Molecular studies on the contribution of capsular polysaccharide to the virulence of Haemophilus influenzae. Ph.D. thesis. Oxford, United Kingdom: University of Oxford; 1989. [Google Scholar]
- 12.Dillard J P, Yother J. Genetic and molecular characterization of capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 3. Mol Microbiol. 1994;12:959–972. doi: 10.1111/j.1365-2958.1994.tb01084.x. [DOI] [PubMed] [Google Scholar]
- 13.Frosch M, Weisgerber C, Meyer T F. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc Natl Acad Sci USA. 1989;86:1669–1673. doi: 10.1073/pnas.86.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hausman S Z, London J. Purification and characterization of ribitol-5-phosphate and xylitol-5-phosphate dehydrogenases from strains of Lactobacillus casei. J Bacteriol. 1987;169:1651–1655. doi: 10.1128/jb.169.4.1651-1655.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoiseth S K, Connelly C J, Moxon E R. Genetics of spontaneous, high-frequency loss of b capsule expression in Haemophilus influenzae. Infect Immun. 1985;49:389–395. doi: 10.1128/iai.49.2.389-395.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honeyman A L, Stewart G S. The nucleotide sequence of the rodC operon in Bacillus subtilis. Mol Microbiol. 1989;3:1257–1268. doi: 10.1111/j.1365-2958.1989.tb00276.x. [DOI] [PubMed] [Google Scholar]
- 17.Jann B, Jann K. Structure and biosynthesis of the capsular antigens of Escherichia coli. Curr Top Microbiol Immunol. 1990;150:19–42. doi: 10.1007/978-3-642-74694-9_2. [DOI] [PubMed] [Google Scholar]
- 18.Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;32:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 19.Kolkman M A, Wakarchuk W, Nuijten P J, van der Zeijst B A. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 20.Kroll J S, Hopkins I, Moxon E R. Capsule loss in H. influenzae type b occurs by recombination-mediated disruption of a gene essential for polysaccharide export. Cell. 1988;53:347–356. doi: 10.1016/0092-8674(88)90155-9. [DOI] [PubMed] [Google Scholar]
- 21.Kroll J S, Zamze S, Loynds B, Moxon E R. Common organization of chromosomal loci for production of different capsular polysaccharides in Haemophilus influenzae. J Bacteriol. 1989;171:3343–3347. doi: 10.1128/jb.171.6.3343-3347.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroll J S, Loynds B, Brophy L N, Moxon E R. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol. 1990;4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 23.Kroll J S, Moxon E R, Loynds B M. Natural genetic transfer of a putative virulence-enhancing mutation to Haemophilus influenzae type a. J Infect Dis. 1994;169:676–679. doi: 10.1093/infdis/169.3.676. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee S J, Romana L K, Reeves P R. Sequence and structural analysis of the rfb (O antigen) gene cluster from a group C1 Salmonella enterica strain. J Gen Microbiol. 1992;138:1843–1855. doi: 10.1099/00221287-138-9-1843. [DOI] [PubMed] [Google Scholar]
- 26.Matin A, Gottschal J C. Influence of dilution rate on NAD(P) and NAD(P)H concentrations and ratios in a Pseudomonas sp. grown in continuous culture. J Gen Microbiol. 1976;94:333–341. doi: 10.1099/00221287-94-2-333. [DOI] [PubMed] [Google Scholar]
- 27.Mauël C, Young M, Karamata D. Genes concerned with synthesis of poly(glycerol phosphate), the essential teichoic acid in Bacillus subtilis strain 168, are organized in two divergent transcription units. J Gen Microbiol. 1991;137:929–941. doi: 10.1099/00221287-137-4-929. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Mio T, Yabe T, Arisawa M, Yamada-Okabe H. The eukaryotic UDP-N-acetylglucosamine pyrophosphorylases. Gene cloning, protein expression, and catalytic mechanism. J Biol Chem. 1998;273:14392–14397. doi: 10.1074/jbc.273.23.14392. [DOI] [PubMed] [Google Scholar]
- 30.Morbidoni H R, de Mendoza D, Cronan J E., Jr Bacillus subtilis acyl carrier protein is encoded in a cluster of lipid biosynthesis genes. J Bacteriol. 1996;178:4794–4800. doi: 10.1128/jb.178.16.4794-4800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moxon E R, Kroll J S. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbiol Immunol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- 32.Nakae T, Nikaido H. Multiple molecular forms of uridine diphosphate glucose pyrophosphorylase from Salmonella typhimurium. I. Catalytic properties of various forms. J Biol Chem. 1971;246:4386–4396. [PubMed] [Google Scholar]
- 33.Nakajima K, Hashimoto T, Yamada Y. cDNA encoding tropinone reductase-II from Hyoscyamus niger. Plant Physiol. 1993;103:1465–1466. doi: 10.1104/pp.103.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nitta D M, Jackson M A, Burry V F, Olson L C. Invasive Haemophilus influenzae type f disease. Pediatr Infect Dis J. 1995;14:157–160. [PubMed] [Google Scholar]
- 35.Ogasawara N, Nakai S, Yoshikawa H. Systematic sequencing of the 180 kilobase region of the Bacillus subtilis chromosome containing the replication origin. DNA Res. 1994;1:1–14. doi: 10.1093/dnares/1.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pittman M. Variation and type specificity in the bacterial species Haemophilus influenzae. J Exp Med. 1931;53:471–493. doi: 10.1084/jem.53.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbins J B. Vaccines for the prevention of encapsulated bacterial diseases: current status, problems and prospects for the future. Immunochemistry. 1978;15:839–854. doi: 10.1016/0161-5890(78)90117-7. [DOI] [PubMed] [Google Scholar]
- 39.Roberts I S, Mountford R, Hodge R, Jann K B, Boulnois G J. Common organization of gene clusters for production of different capsular polysaccharides (K antigens) in Escherichia coli. J Bacteriol. 1988;170:1305–1310. doi: 10.1128/jb.170.3.1305-1310.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sau S, Lee C Y. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J Bacteriol. 1996;178:2118–2126. doi: 10.1128/jb.178.7.2118-2126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schembri M A, Bayly R C, Davies J K. Phosphate concentration regulates transcription of the Acinetobacter polyhydroxyalkanoic acid biosynthetic genes. J Bacteriol. 1995;177:4501–4507. doi: 10.1128/jb.177.15.4501-4507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 45.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Eldere J, Brophy L, Loynds B, Celis P, Hancock I, Carman S, Kroll J S, Moxon E R. Region II of the Haemophilus influenzae type b capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol Microbiol. 1995;15:107–118. doi: 10.1111/j.1365-2958.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 47.Van Oss C J, Gillman C F. Phagocytosis as a surface phenomenon. 3. Influence of C1423 on the contact angle and on the phagocytosis of sensitized encapsulated bacteria. Immunol Commun. 1973;2:415–419. doi: 10.3109/08820137309022812. [DOI] [PubMed] [Google Scholar]
- 48.Veiga-da-Cunha M, Detheux M, Watelet N, Van Schaftingen E. Cloning and expression of a Xenopus liver cDNA encoding a fructose-phosphate-insensitive regulatory protein of glucokinase. Eur J Biochem. 1994;225:43–51. doi: 10.1111/j.1432-1033.1994.00043.x. [DOI] [PubMed] [Google Scholar]
- 49.Waggoner-Fountain L A, Hendley J O, Cody E J, Perriello V A, Donowitz L G. The emergence of Haemophilus influenzae types e and f as significant pathogens. Clin Infect Dis. 1995;21:1322–1324. doi: 10.1093/clinids/21.5.1322. [DOI] [PubMed] [Google Scholar]