Abstract
Cardiomyocytes (CMs), endothelial cells (ECs), smooth-muscle cells (SMCs), and cardiac fibroblasts (CFs) differentiated from human induced-pluripotent stem cells (hiPSCs) are the fundamental components of cell-based regenerative myocardial therapy and can be used as in-vitro models for mechanistic studies and drug testing. However, newly differentiated hiPSC-CMs tend to more closely resemble fetal CMs than the mature CMs of adult hearts, and current techniques for improving CM maturation can be both complex and labor-intensive. Thus, the production of CMs for commercial and industrial applications will require more elementary methods for promoting CM maturity. CMs tend to develop a more mature phenotype when cultured as spheroids in a three-dimensional (3D) environment, rather than as two-dimensional monolayers, and the activity of ECs, SMCs, and CFs promote both CM maturation and electrical activity. Here, we introduce a simple and reproducible 3D-culture–based process for generating spheroids containing all four cardiac-cell types (i.e., cardiac spheroids) that is compatible with a wide range of applications and research equipment. Subsequent experiments demonstrated that the inclusion of vascular cells and CFs was associated with an increase in spheroid size, a decline in apoptosis, an improvement in sarcomere maturation and a change in CM bioenergetics.
Keywords: pluripotent stem cell, cardiomyocyte, suspension culture, maturation, organoids, biomanufacturing
Introduction
The development of efficient protocols for differentiating human induced-pluripotent stem cells (hiPSCs) into cardiomyocytes (CMs), endothelial cells (ECs), smooth-muscle cells (SMCs), and cardiac fibroblasts (CFs) (Adams et al., 2013; Liu et al., 2016; Palpant et al., 2017; Zhu et al., 2017; Kwong et al., 2019; Zhang et al., 2019; Kahn-Krell et al., 2021) has led to the establishment of an array of utilizations in the field of cardiac regeneration. Although early proposed applications focused on direct injections (Menasché et al., 2001; Laflamme et al., 2007; Sanganalmath and Bolli, 2013; Ye et al., 2014) and the transplantation of engineered cardiac tissues (Schaefer et al., 2017; Shadrin et al., 2017; Gao et al., 2018) and sheets (Ishigami et al., 2018; Bayrak and Gümüşderelioğlu, 2019; Tsuruyama et al., 2019) to resupply cardiomyocytes, lost to injuries such as myocardial infarction (MI), the uses of these cardiac surrogates have expanded to include in-vitro modeling of myocardial disease (Long et al., 2018; Giacomelli et al., 2021), drug screening (Mathur et al., 2015; Huebsch et al., 2016; Mills et al., 2017), and exosome production (Liu et al., 2017). A wide range of formats has been explored to fulfill the needs of each potential application including patches (Schaefer et al., 2017; Shadrin et al., 2017; Gao et al., 2018), sheets (Ishigami et al., 2018; Bayrak and Gümüşderelioğlu, 2019; Tsuruyama et al., 2019), spheres (Fischer et al., 2018; Chang et al., 2020), wires (Jackman et al., 2016; Sun and Nunes, 2016), and decellularized tissues (Das et al., 2019; Alexanian et al., 2020; Hochman-Mendez et al., 2020). The utilization of these products and the needs of each use play a key role in the ideal option for each situation. However, there is a crucial need to establish tools for multiple applications related to MI and heart failure that can be easily reproduced and scaled (Zhang et al., 2021a).
Although current differentiation techniques can produce beating hiPSC-derived CMs in as little as 9 days (Lian et al., 2012; Zhang et al., 2012), the biomolecular (Cao et al., 2008; Synnergren et al., 2008; Xu et al., 2009; Synnergren et al., 2010), electrical (Caspi et al., 2009; Kim et al., 2010; Lee et al., 2011), and mechanical (Binah et al., 2007; Zhang et al., 2021b) properties of these newly differentiated cells tend to resemble those of fetal CMs, rather than the mature CMs of adult hearts (Robertson et al., 2013). Electrical stimulation (Chan et al., 2013; Richards et al., 2016; Ruan et al., 2016; LaBarge et al., 2019a) and mechanical stretching (Lux et al., 2016; Ruan et al., 2016; Zhang et al., 2017; LaBarge et al., 2019a) have been used to increase the maturity of engineered cardiac tissue but are difficult to apply until after the tissue is manufactured. Thus, the production of CMs for commercial and industrial applications will require more elementary methods for promoting CM maturation (Zhang et al., 2021a), such as manipulating the conditions of hiPSC-CM culture (Haishuang Lin et al., 2017; Parikh et al., 2017; Jackman et al., 2018; Selvaraj et al., 2019).
Another method to increase complexity and functionality of cardiac tissue models is to use additional cell types that more accurately recreate the myocardial environment. When cultured in a three-dimensional (3D) environment, rather than as two-dimensional monolayers, CMs coalesce into spheroids (Fischer et al., 2018; Chang et al., 2020) and tend to develop a more mature phenotype (Jha et al., 2016; Sacchetto et al., 2020; Kai-Li Wang et al., 2021). ECs and SMCs also promote CM maturation (Pinto et al., 2016; Ayoubi et al., 2017; Giacomelli et al., 2017; Zhu et al., 2017; Kwong et al., 2019) while facilitating oxygen and nutrient delivery, which improves cell survival (Garzoni et al., 2009; Zhang et al., 2021a; Pretorius et al., 2021), and CFs contribute to myocardial development both by producing components of the extracellular matrix (ECM) and by forming gap junctions with CMs to support electronic signal transduction (Zhang et al., 2019; Beauchamp et al., 2020; Giacomelli et al., 2020; Li et al., 2020; Pretorius et al., 2021). A range of ratios between these different cell types have been explored in previous studies but a relationship of CM:EC:SMC:CF of 4:2:1:1 is predominantly used (Gao et al., 2018; Arai et al., 2020; Beauchamp et al., 2020; Daly et al., 2021; Pretorius et al., 2021) as it roughly recapitulates the relationships found in native myocardium of myocytes predominating with endothelial cells compromising the greatest non-myocyte population (Banerjee et al., 2007; Pinto et al., 2016).
Along with the functionality and maturity requirements for broad cardiac tissue surrogate application two important biomanufacturing considerations are system format and scalability. A uniform shape, size, and culture condition that can function in a range of uses would allow for centralized production (Abbasalizadeh et al., 2017; Adil and Schaffer, 2017; Li et al., 2017; Tomov et al., 2019). Spherical spheroids cultured in suspension provide a format that is broadly compatible with existing research equipment, is highly movable, and can function as a building block for larger construct needs (Mattapally et al., 2018; LaBarge et al., 2019a; LaBarge et al., 2019b; Kim et al., 2020; Daly et al., 2021; Polonchuk et al., 2021). Additionally, using previously established CM spheroid production processes combined with whole spheroid fusion, a scalable spheroid biomanufacturing platform can be established that does not require dissociation (Beauchamp et al., 2015; Bin Lin et al., 2017; Miwa et al., 2020; Kahn-Krell et al., 2021).
For the experiments described in this report, we differentiated hiPSCs into CMs, ECs, SMCs, and CFs and then combined the differentiated cells in a 3D culture environment, where they formed spheroids containing all four cardiac-cell types (i.e., cardiac microtissues). Subsequent analyses suggested that the inclusion of vascular cells and CFs increased spheroid size, reduced cellular apoptosis, and tended to promote sarcomere maturation and CM energy production. This process, with the potential to be scaled, advances on previous cardiac spheroid models (Giacomelli et al., 2017; Voges et al., 2017; Lee et al., 2019b; Helms et al., 2019; Keung et al., 2019; Beauchamp et al., 2020; Buono et al., 2020; Giacomelli et al., 2020; Israeli et al., 2020; Kupfer et al., 2020; Thomas et al., 2021) by producing large diameter tissues with cells from a single iPSC line, no exogenous matrix, and a reproducible product.
Materials and methods
hiPSC culture and expansion
All cells used in the study were differentiated from human induced pluripotent stem cell (hiPSC) line LZ-hiPSC5 which was reprogramed from human cardiac fibroblasts as described previously (Zhang et al., 2014). hiPSCs were cultured on 6-well plates in mTeSR Plus (STEMCELL Technologies) for 3 days, detached with gentle cell dissociation reagent (GCDR) (STEMCELL Technologies), resuspended in 40 ml TeSR E8 3D seed media (STEMCELL Technologies) supplemented with 10 µM Y27632 at a density of 1.5 × 105 cells/mL, and then cultured in a 125 ml Erlenmeyer flask on a Belly Dancer orbital shaker (IBI Scientific) at a speed of 4.75. On each of the following 2 days, 1.2 ml of feed medium was added to the flask, and after an additional day, half of the culture volume was replaced with fresh seed medium. After 4 days of culture on the orbital shaker, aggregates were dissociated with GCDR for 8 min, broken into smaller cell clumps via pipetting through a 37-µm reversible strainer (STEMCELL Technologies), and then diluted to a density of 1.5 × 105 cells/mL. Cells were cultured as previously described (Kahn-Krell et al., 2021) for four more days before differentiation was initiated.
CM differentiation
Cardiomyocyte (CM) differentiation was performed as described previously (Kahn-Krell et al., 2021). Briefly, differentiation was initiated on differentiation day (dD) 0 by replacing the culture media with RPMI 1640 supplemented with 1 × B27 without insulin (RPMI/B27–), 6 µM CHIR99021, and 10 µM Y-27632 at a density of 1.5 × 106 cells/mL. On dD1, the media was replaced with a 1.2-fold volume of RPMI/B27–, 1 µM CHIR99021, and 10 µM Y-27632, and the cells were cultured for two more days. On dD3, 70% of the culture media was replaced with RPMI/B27– supplemented with 10 µM IWR-1-endo; on dD5, the media was changed to RPMI/B27–; and on dD7, the media was changed to RPMI1640 supplemented with B27 with insulin (RPMI/B27+) in a volume equivalent to the volume used on dD0. Metabolic purification of the differentiated CMs was initiated on dD9 by changing the media to RPMI1640 without glucose supplemented with B27+ and 0.12% (w/w) sodium DL-lactate (Millipore Sigma). Three days later (on dD12), the purification media was replaced with RPMI/B27+, and the medium was refreshed every 5 days until spheroid assembly. Spheroid assembly was performed no more than 30 days after differentiation was initiated and as close to dD12 as possible.
EC differentiation
Endothelial-cell (EC) differentiation of hiPSCs was performed in monolayers of cultured hiPSCs with the STEMdiff Endothelial Differentiation Kit (STEMCELL Technologies) as directed by the manufacturer’s instructions. Briefly, hiPSCs were seeded into mTeSR Plus (STEMCELL Technologies) in a 6 well plate at a density of 5.0 × 104 per well and then on dD1 and dD2, the medium was changed to 3 ml STEMdiff Mesoderm Induction Medium (STEMCELL Technologies). The medium was replaced with 4 ml of STEMdiff Endothelial Induction Medium (STEMCELL Technologies) on dD3 and refreshed on dD5. On dD7, the cells were dissociated with ACCUTASE (Corning), transferred into a fibronectin-coated T75 flask, and cultured in EGM-2 MV (Lonza) supplemented with SB431542 (Fischer Scientific) until 100% confluent. Purification was performed by dissociating the cells, resuspending them in cold Dulbecco phosphate-buffered saline (DPBS) with 2% fetal bovine serum (FBS) at a density of 1 × 106 cells/100 μL, and then collecting cells that expressed both CD31 and CD144 via flow cytometry on a BD FACS Aria II instrument; 100-µL samples were labeled by incubating them with 5 µL of AlexaFluor-conjugated CD31 and 20 µL of phycoerythrin-conjugated CD144 antibodies for 45 min on ice and then washed in DPBS.
SMC differentiation
hiPSCs were treated via the EC differentiation protocol through dD7 and then differentiated into smooth-muscle cells (SMCs) as described previously (Yang et al., 2016). Briefly, the cells were cultured until 80% confluent, and then the media was changed to high-glucose DMEM supplemented with 5% FBS, 5 ng/ml platelet-derived growth factor beta (PDGF-β), and 2.5 ng/ml transforming growth factor beta (TGFβ). The media was changed every 3 days for 9 days and then replaced with SmGM-2.
Cardiac fibroblast differentiation
When hiPSCs reached 100% confluency the medium was changed to RPMI/B27–insulin supplemented with 12 μM CHIR99021 (Tocris) for 24 h, RPMI/B27–insulin for 24 h, and then cardiac fibroblast differentiation basal (CFBM) medium (DMEM, high glucose with HAS, linoleic acid, lecithin, ascorbic acid, GlutaMAX, hydrocortisone hemisuccinate, rh insulin) supplemented with 75 ng/ml bFGF (WiCell Research Institute) for 18 days; the CFBM/bFGF medium was changed every other day.
CM, EC, SMC, and CF purity assessments
Cells were dissociated, washed in DPBS, fixed with 4% paraformaldehyde (PFA) for 15 min, washed three times, permeabilized with 0.1% Triton-X in DPBS, and blocked with 4% bovine serum albumin (BSA) and 4% FBS in DPBS for 30 min each; then, the cells were incubated with lineage-specific antibodies (Supplemental Table S1) for 1 h and analyzed on an Attune NxT flow cytometer (Thermo Fisher Scientific).
Spheroid fabrication and culture
A 2 ml sample of CM spheroids were treated with CM dissociation media (CMDM) (STEMCELL Technologies) for 15 min at 37°C with periodic mixing then counted to determine culture density. ECs, SMCs, and CFs were dissociated from monolayer cultures and 5.0 × 106, 2.5 × 106, and 2.5 × 106 of each respectively were combined and resuspended in 1 ml of spheroid media (OM) (Supplemental Table S3). Based on cell counts 1.0 × 107 total CMs were collected as whole spheroids in a 50 ml conical tube and the media was replaced with 19 ml OM. After addition of the other cell types the contents were mixed well and 200 µL was transferred to each well of a 96-Well, Nunclon Sphera-Treated, U-Shaped-Bottom Microplate (Thermo Fisher Scientific) to produce 96 spheroids. The media was refreshed every 2 days for 7 days, and then the spheroids were transferred to 6-well Ultra-Low Attachment Microplates (Corning), with no more than 16 spheroids per well. Plates were maintained on a belly dancer shaker (IBI Scientific) at speed of 4.75, and the media was exchanged every 4–7 days.
Spheroid size measurements
Spheroids were photographed with an Olympus CKX53 microscope at ×4 magnification, and spheroid diameters were determined with a modified ImageJ macro (Ivanov et al., 2014) as indicated in the Supplemental Information.
Quantitative polymerase chain reaction (qPCR)
RNA was extracted with TRIZOL (Thermo Fisher) and purified on Direct-zol RNA Miniprep Plus (Zymo Research) columns as directed by the manufacturer’s protocol. Reverse transcription was performed with Superscript IV VILO Master Mix (Invitrogen), and samples were diluted to 5 ng/μL. Each qPCR reaction was performed with 5 ng of cDNA, 0.5 µM primers (Supplemental Table S2), and PowerUp SYBR Green Master Mix (Applied Biosystems), and analysis was conducted on a QuantStudio Real-Time PCR Machine (Applied Biosystems). Results for each CT value were normalized to intrinsic glyceraldehyde phosphate dehydrogenase (GAPDH) abundance and to the CT value determined on the first day after spheroid assembly for each batch of spheroids. Data was collected from four independent batches of spheroids for each gene.
Western blotting
Spheroids were digested in 100 µL RIPA Buffer (Sigma-Aldrich); then, protein lysates were collected, mixed with ×1 Halt proteinase inhibitor cocktail mix (Thermo Scientific), and sonicated twice in 3-s intervals at 50% power. Sample concentrations were determined via Pierce BCA Protein Assay (Thermo Scientific), and then samples containing ×4 Laemmli Sample Buffer (Bio-Rad), 2 Mercaptoethanol (Bio-Rad), and 7.5 µg protein were run on an Expressplus PAGE 4–20% Gel (GenScript) at 200 V for 30 min. Proteins were transferred to nitrocellulose membranes with a Trans-Blot Turbo Transfer System (Bio-Rad); then, the membranes were blocked in 5% Blotting Grade Blocker Non Fat Dry Milk (Bio-Rad) for 30 min and stained with primary and secondary antibodies (Supplemental Table S1) for 1 h each. Blots were incubated for 5 min in Pierce SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific) and imaged with the ChemiDoc Touch Imaging System (Biorad). Protein bands were quantified with Image Lab Software (Bio-Rad); results for each sample were normalized to intrinsic beta actin abundance and to measurements made on the first day after spheroid assembly for each batch of spheroids. Data was collected from four independent batches of spheroids for each protein.
Spheroid preservation and sectioning
Spheroids were fixed in 4% formaldehyde (Pierce) for 15 min, placed in 30% sucrose at 4 °C overnight, and then then embedded in Tissue-Plus O.C.T Compound (Fisher Scientific) for histological analysis. Blocks were cut into 10-µm sections, mounted on charged glass microscope slides (Globe Scientific) and then stored at −20°C.
Masson-trichrome staining
Slides containing sections obtained at 100-µm intervals (i.e., every 10th section) were fixed in Bouin’s Solution at 55°C for 1 h and then treated with Weigerts iron hematoxylin working solution for 10 min, with Biebrich scarlet-acid fuchsin for 5 min, with phosphomolybdic acid-phosphotungstic acid for 5 min, with aniline blue solution for 5 min, and with 1% acetic acid for 1 min; then, the sections were dehydrated in 95% alcohol for 2 min, cleared with 2 changes of xylene, and mounted with permount and coverslips overnight. Sections were imaged with a ×10 objective on an Olympus B×51 Fluorescence Microscope; when multiple fields of view were required for a single section, the images were stitched together with Photoshop software.
Apoptosis
The spheroid core was identified by determining which Masson-trichrome–stained sections had the greatest surface area; then, sections from the core were stained using the In Situ Cell Death Detection Kit, TMR red (Sigma) according to the manufacturers protocol. Briefly, sections were fixed in 4% PFA for 15 min, permeabilized in 0.1% Triton X-100 and 0.1% sodium citrate for 2 min on ice, incubated with TUNEL reaction mixture and 4′, 6-diamidino-2-phenylindole (DAPI) for 1 h at 37°C, mounted in VECTASHIELD hardset Antifade Mounting Medium, and visualized via confocal laser scanning (Olympus FV3000 confocal microscope). Sections were evaluated for at least two spheroids in each of four batches, and positively stained cells or the size of stained regions were quantified with a modified ImageJ macro (Ivanov et al., 2014) as indicated in the Supplemental Information.
Immunostaining and apoptosis detection
The spheroid core was identified by determining which Masson-trichrome–stained sections had the greatest surface area; then, sections from the core were fixed in 4% PFA for 15 min, blocked, permeabilized in 10% donkey serum, 10% Tween20, 3% BSA, and 0.05% Triton-X for 30 min, incubated with primary antibodies (Supplemental Table S1) at room temperature for 45 min, washed with PBS (3 washes, 5 min per wash), incubated with DAPI and fluorescent secondary antibodies at room temperature for 45 min, mounted in VECTASHIELD hardset Antifade Mounting Medium, and visualized via confocal laser scanning (Olympus FV3000 confocal microscope). Sections were evaluated for at least two spheroids in each of four batches, and positively stained cells or the size of stained regions were quantified with a modified ImageJ macro (Ivanov et al., 2014) as indicated in the Supplemental Information.
ATP, NAD, and NADH quantification
ATP content was measured using a luminescent ATP detection Assay kit (ab113849; Abcam) following the manufacturer’s protocol. The amount of ATP was normalized to the total protein content, which was determined using a protein assay (23227, ThermoFisher). The data was presented as nanomoles per milligram protein. Total NAD+ and NADH levels were measured by colorimetric kit (ab65348; Abcam) according to manufacturer’s instruction. cAMP level was measured by Direct cAMP Enzyme-linked Immunosorbent Assay (ELISA) using a direct cAMP ELISA kit (ADI-900-066A, Enzo Life Sciences) following the manufacturer’s instructions. Tissue cAMP level was normalized to the total protein content and presented as picomole per milligram protein.
Microelectrode array (MEA)
CytoView MEA 24 well plates (Axion Biosystems) were precoated with 3 μg/ml fibronectin and incubated at 37°C for 1 h. Whole spheroids (1 per well) or CMs collected from dissociated spheroids were added to each well of the MEA plate. Isolated CMs were obtained by treating spheroids with CMDM and periodic pipetting for 30 min at 37°C, selected with the EasySep Human PSC-Derived Cardiomyocyte Enrichment Kit (StemCell), and resuspended at a concentration of 1.2 × 107 cells/mL in CM support media (CMSM) (StemCell Technologies); 5 µL of resuspended CMs were added to each well. Whole spheroids were analyzed 24–48 h after plating, and CMs were analyzed 7 days after plating. Field potential and contractility measurements were collected on a Maestro Edge apparatus (Axion Biosystems) and analyzed by using the Cardiac Module in Axion Navigator software. Action potential durations (APDs) were determined using the LEAP assay and characterized using the Cardiac Analysis Tool.
Transmission electron microscopy (TEM)
Spheroids were transferred onto a fibronectin-coated (1 μg/ml), 0.4-µm pore Transwell Polycarbonate Membrane and cultured for 3 days; then, the membranes were fixed in 2.5% glutaraldehyde solution for 1 h at 4°C and delivered to the UAB High-Resolution Imaging Facility. Sample blocks were sectioned along the width of the transwells with a diamond knife, and samples were mounted and viewed with a Tecnai Spirit T12 Transmission Electron Microscope. At least 4 images were collected for each spheroid, and sarcomere lengths and widths were determined for all sarcomeres in an image by using the line-measure tool in ImageJ software. Sarcomere lengths were determined as the distance from z-line to z-line and widths were determined as the distance from one end to the other of a continuous z-line.
Statistical analysis
Data are presented as mean ± SEM, as box-and-whisker plots, or as violin plots, and significance was evaluated via the Student’s t-test or analysis of variance (ANOVA). Analyses were performed with GraphPad Prism8 software (GraphPad Prism, RRID:SCR_002798), and p < 0.05 was considered significant.
Results
The inclusion of hiPSC-ECs, -SMCs, and CFs in CM spheroids increased spheroid size and improved cell viability
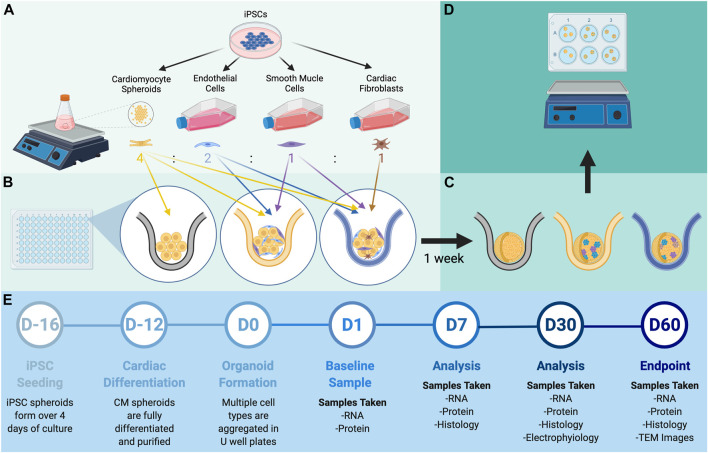
hiPSCs were differentiated into CMs, ECs, SMCs, and CFs via published protocols or the use of commercially available kits (Yang et al., 2016; Zhang et al., 2019; Kahn-Krell et al., 2021), and flow-cytometry analyses with lineage-specific antibodies [CMs: cardiac troponin T (cTnT), ECs: CD144 and CD31, SMCs: smooth-muscle actin (SMA), CFs: TE-7] confirmed that the purity of each differentiated cell population exceeded 95% (Supplementary Figures S1–S4). Spheroids containing CMs alone (C1) and spheroids containing a 4:2:1 ratio of CMs, ECs, and SMCs (C3) or a 4:2:1:1 ratio of CMs, ECs, SMCs, and CFs (C4) were produced by culturing the indicated proportions of cell types in low-attachment 96-well U plates for 1 week to promote aggregation and fusion (Figures 1A,B), and then in low-attachment 6-well plates on an orbital shaker for the remainder of the culture period (Figures 1C,D). D0 was defined as the day the spheroids/spheroids were initially assembled, and samples were obtained after one (D1), seven (D7), 14 (D14), 30 (D30), and 60 (D60) days of culture for characterization (Figure 1E); synchronized beating was observed in all constructs throughout the culture period. Size optimization studies determined that the mean diameter of C4 spheroids on D30 was larger (though not significantly) when the spheroids were generated from seeded CMs (∼801 µm) than from other initial CM population sizes (698–791 µm) (Figure 2A), with no significant increase in the proportion of apoptotic cells (Figure 2B). Thus, all spheroids and spheroids produced for subsequent experiments were constructed with 1.0 × 105 CMs.
FIGURE 1.
Experimental design. (A) hiPSCs were differentiated into CMs, ECs, SMCs, and CFs via established methods. (B) Spheroids containing all four cell types (C4 spheroids) were produced by combining a 4:2:1:1 ratio of CMs, ECs, SMCs, and CFs in 96-well U plates and allowing the cells to fuse for 7 days; then, (C) the spheroids were transferred to 6-well plates and (D) cultured with shaking for to 60 days. (E) Procedures for spheroid manufacture and experimental analyses are displayed on a timeline.
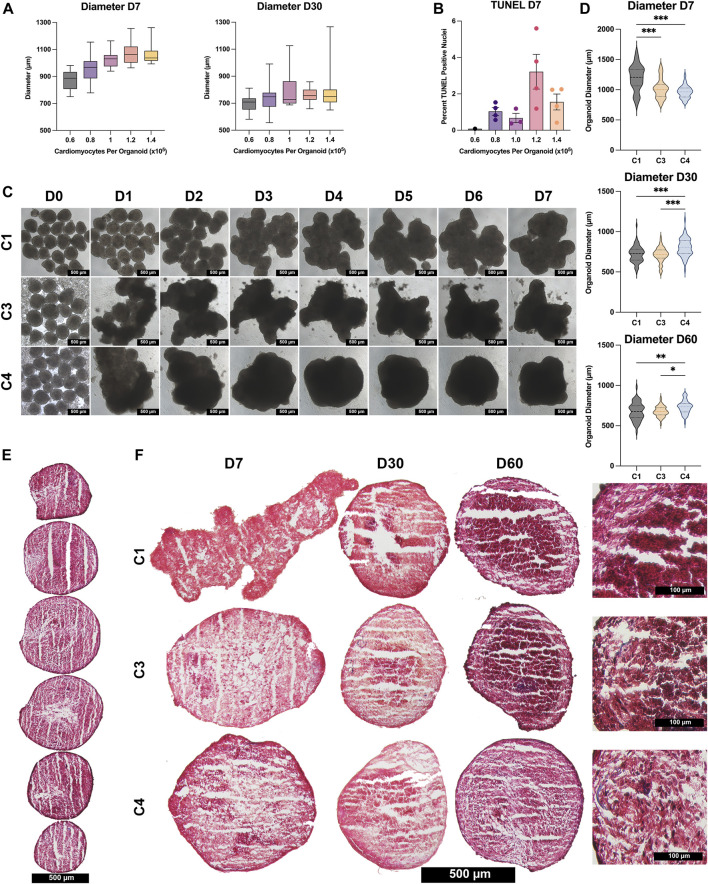
FIGURE 2.
Size and histological assessments. (A) C4 spheroids were generated from the indicated CM population sizes (0.6 × 105 black; 0.8 × 105 purple; 1.0 × 105 pink; , 1.2 × 105 orange; , 1.4 × 105 yellow), and spheroid diameters were measured on D7 and D30; results were displayed as box-whisker plots. (B) C4 spheroids were cut into 10-μm sections, and the sections were stained via terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL); then, apoptosis was quantified as the percentage of TUNEL-positive cells in 4 spheroids/group. (C) Images of C1, C3, and C4 spheroids generated from an initial population of 1 × 105 CMs were obtained each day from D0–D7 (bar = 500 μM). (D) Spheroid diameters of all 3 groups (C1 black; C3 yellow; C4 blue) were measured on D7, D30, and D60; results are displayed as violin plots. (E) Spheroids were cut into 10-µm sections and stained with Masson-trichrome; representative images are displayed for every 10th sequential section from a C4 spheroid on D60 (bar = 500 μM). (F) Representative images are displayed for the centermost sections from C1, C3, and C4 spheroids at the indicated time points (bar = 500 μM for the first three columns, and bar = 100 μM for the far-right column); muscle fibers appear red and collagen fibers appear blue. (*p < 0.05, **p < 0.01, ***p < 0.001; n > 30 per group; p-values determined as mean ± SEM).
C4 spheroids coalesced into a largely spherical shape within 3–4 days of seeding, while the architecture of the C3 spheroids and (especially) C1 spheroids remained irregular through at least D7 (Figure 2C). C4 spheroid diameters were also significantly smaller on D7 (C1: 1,198 ± 38.2 µm; C3: 1,016 ± 21.1 µm; C4: 964 ± 12.6 µm, p < 0.001 versus C1 and C3), but the trend was reversed on D30 (C1: 719 ± 11.9 µm; C3: 714 ± 10.4 µm; C4: 804 ± 13.9 µm, p < 0.001 versus C1 and C3) and D60 (C1: 676 ± 14.7 µm; C3: 678 ± 8.8 µm; C4: 730 ± 11.7 µm, p < 0.01 versus C1, p < 0.05 versus C3), when C4 spheroids were significantly larger than the other two constructs (Figure 2D). These differences may be accounted for by proliferation of non-myocytes in the C3 and C4 groups however, over time all groups showed trends of decreasing diameter and further immunostaining (Figure 3) did not show significant increases in ECs, SMCs, or CFs. Observations in Masson-trichrome stained sequential sections (Figure 2E) suggested that the density of muscle cells and fibers increased from D7 to D60 in all constructs, but only C3 and C4 spheroids displayed evidence of collagen formation (Figure 2F), which is consistent with the absence of ECM-producing ECs, SMCs, or CFs (Lee et al., 2019a; Strikoudis et al., 2019) in C1 spheroids. Furthermore, although assessments of apoptosis (TUNEL staining) did not differ significantly among groups on D7, apoptotic cells became increasingly common in C1 spheroids and (to a lesser extent) C3 spheroids during the culture period, while the proportion of apoptotic cells in C4 spheroids remained stable through at least D60, when apoptotic cells were significantly less common in C4 spheroids than in the other two constructs and in C3 spheroids than in C1 spheroids (Figure 3A). Apoptotic cells also tended to be located toward the center of C1 spheroids, which suggests that the vascular cells and CFs present in C4 spheroids facilitated access of the culture medium to the spheroid interior. These findings are consistent with previous work supporting the crucial role that non-myocytes play in promoting CM survival particularly where nutrient and oxygen diffusion is limited through release of paracrine factors (Ye et al., 2014; Hodgkinson et al., 2016; Giacomelli et al., 2017; Giacomelli et al., 2020; Munarin et al., 2020; Pretorius et al., 2021).
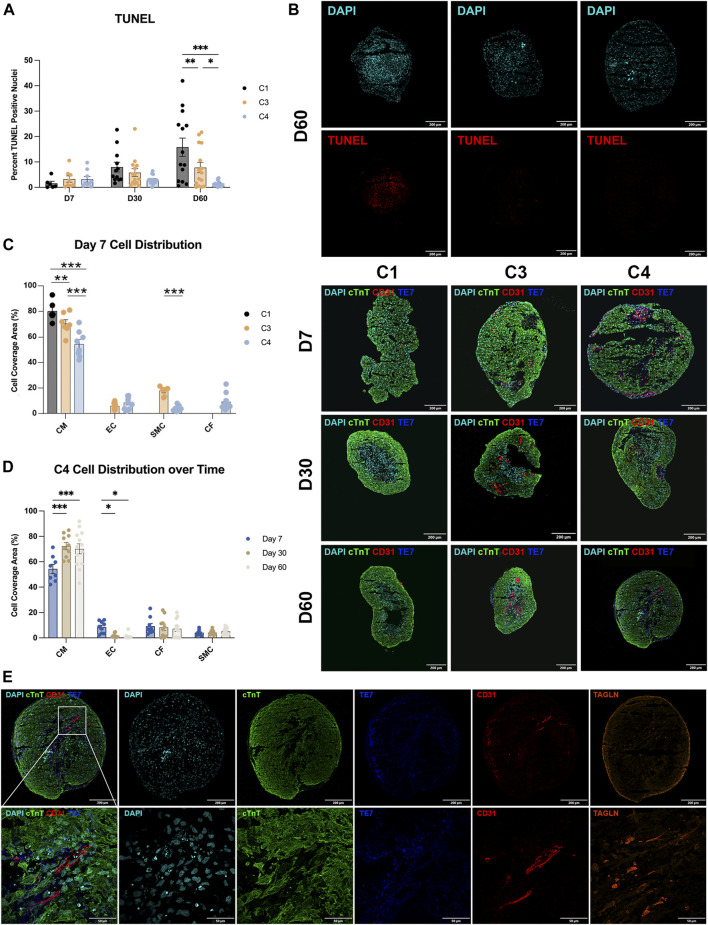
FIGURE 3.
Apoptosis and cell-occupancy measurements. (A) C1, C3, and C4 spheroids were collected at the indicated time points and cut into 10-µm sections; then, the surface areas of every 10th section were measured, and the centermost section (i.e., the one with the largest surface area) was TUNEL-stained. Nuclei were counterstained with DAPI, and apoptosis was quantified as the percentage of TUNEL-positive cells. Representative images are displayed for the centermost sections from each group on D60. (B) Centermost sections were stained with cTnT, CD31, and TE-7 antibodies to visualize CMs, ECs, and CFs, respectively, and nuclei were counterstained with DAPI; representative images are displayed for each group on D7, D30, and D60. (C,D) CM, EC, and CF occupancy were quantified as the percentage of total surface area that was positive for expression of the corresponding marker protein and summarized (C) for C1, C3, and C4 on D7 and (D) for C4 on D7, D30, and D60. (E) Representative sections from a C4 spheroid on D60 are displayed at high magnification to demonstrate the presence of elongated ECs. (*p < 0.05, **p < 0.01, ***p < 0.001; n > 6 per group).
hiPSC-CMs occupied a progressively larger proportion of C4 spheroids over time
The distribution of cell types during the culture period was evaluated in sections stained for the expression of cTnT, CD31, and the fibroblast marker TE-7 (Figure 3B), as well as with SMC marker transgelin (TAGLIN). On D7, the proportion of cTnT-positive surface area (i.e., CM occupancy) was significantly lower in C4 spheroids than in the other two constructs, and in C3 spheroids than in C1 spheroids (Figure 3C), which is consistent with the initial composition of cell types used during spheroid construction. In C4 spheroids, CM occupancy increased from D7 to D30, while EC occupancy declined over the same period (Figure 3D), perhaps because the ECs grew from centrally located clumps on D7 into elongated structures that permeated the entire spheroid on D30 and D60 (Figure 3E). CM and EC occupancy remained largely stable in C4 spheroids from D30 to D60, while SMC and CF occupancy did not change significantly throughout the 60-day culture period. Although some restructuring occurred, immunofluorescent imaging studies illustrated sparse and non-uniform distribution of non-myocytes throughout the time course suggesting that even small amounts of these cells can provide the necessary benefit to CM survival. Low levels of CD31 expression were also observed in C1 spheroids, and both C3 spheroids and C1 spheroids contained a very small number of TE-7–positive cells, perhaps because growth factors present in the culture medium (e.g., VEGF, FGF) induced EC- and CF-like phenotypes in hiPSC-derived cells that were not fully differentiated before the constructs were assembled.
hiPSC-CMs in C4 spheroids matured during the culture period
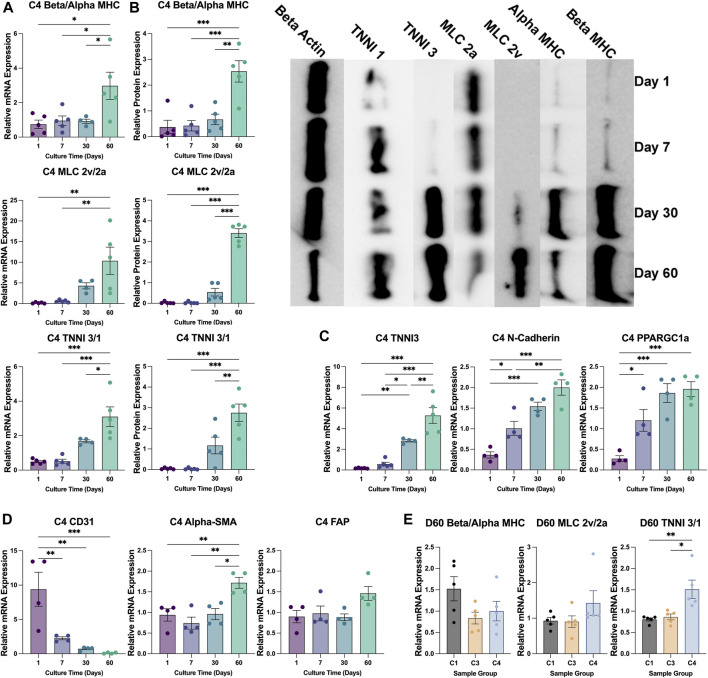
CM maturation and ventricular specification is associated with increases in the ratios of expression for beta versus alpha myosin heavy chain (beta:alpha MHC), ventricular versus atrial myosin light-chain 2 (MLC 2v:2a), and cardiac (type 3) versus slow-skeletal (type 1) troponin I (TNNI 3:1) (Guo and Pu, 2020; Karbassi et al., 2020). When calculated from measurements of either mRNA (Figure 4A) or protein (Figure 4B) abundance, each of the three ratios increased significantly in C4 spheroids throughout the culture period, including from D30 to D60 for all except the ratio of MLC 2v:2a mRNA, and the abundance of structural (TNNI3), electromechanical (N-cadherin), and metabolic [peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPARGC1a)] markers for CM maturation significantly increased through at least D30 (Figure 4C). Alpha-SMA expression also increased significantly throughout the culture period, while CD31 expression significantly declined, and the expression of fibroblast activating protein (FAP) was largely unchanged (Figure 4D). Collectively, these observations indicate that the maturation and modification of both CMs and vascular cells continues for at least 2 months in cultured C4 spheroids.
FIGURE 4.
Changes in patterns of gene expression during culture. (A,B) The abundance of beta MHC, alpha MHC, MLC 2v, MLC 2a, TNNI3, and TNNI1 (A) mRNA and (B) protein were evaluated via qPCR and Western blot, respectively, in C4 spheroids at the indicated time points (D1 purple; D7 blue; D30 teal; D60 green); then, the ratios of expression for the MHC (top), MLC (middle), and TNNI isoforms (bottom) were calculated. A representative Western blot of protein expression is displayed on the right of panel (B) (C,D) The abundance of (C) TNNI3, N-cadherin, and PPARGC1a mRNA and of (D) CD31, alpha-SMA, and FAP mRNA was measured via qPCR in C4 spheroids at the indicated time points. (E) The ratios of mRNA abundance for the MHC, MLC, and TNNI isoforms was calculated for C1, C3, and C4 spheroids on D60 (C1 black; C3 yellow; C4 blue). mRNA measurements were normalized to intrinsic GAPDH mRNA abundance, and protein measurements were normalized to intrinsic beta actin abundance. (*p < 0.05, **p < 0.01; n > 4 per group).
The inclusion of hiPSC-ECs, -SMCs, and CFs increased sarcomere maturation and CM energy production in cardiac spheroids
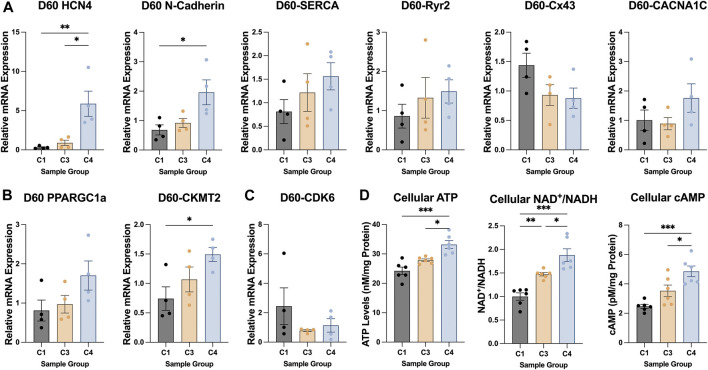
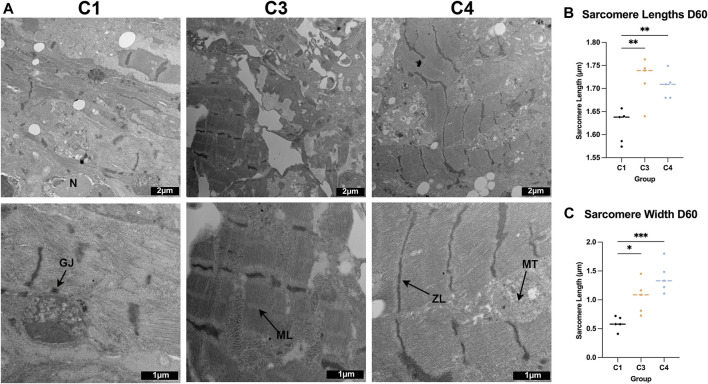
The TNNI 3:1 mRNA ratio was also significantly higher in C4 spheroids than in C3 spheroids or C1 spheroids on D60, but the ratios of beta:alpha MHC and MLC 2v:2a mRNA did not differ significantly among the three groups (Figure 4E). mRNA levels on D60 for a panel of genes that participate in CM electrical conduction [hyperpolarization activated cyclic nucleotide gated potassium channel 4 (HCN4), N-cadherin, sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA), ryanodine receptor 2 (Ryr2), connexin 43 (Cx43), calcium voltage-gated channel subunit alpha 1C (CACNA1C); Figure 5A] and metabolism [PPARGC1a, creatine kinase, mitochondrial 2 (CKMT2); Figure 5B] also tended to be more highly expressed in C4 spheroids; however, the only differences that reached statistical significance were for CKMT2 and N-cadherin, which were significantly greater in C4 spheroids than in C1 spheroids, and for HCN4, which was significantly greater in C4 spheroids than in either of the other two groups. These expression level differences observed on day 60 were found to be mostly absent when the same genes we examined on day 7 after formation (Supplementary Figure S5). Notably, mRNA levels for both Cx43 and the cell-cycle regulatory molecule cyclin-dependent kinase 6 (CDK6) (Figure 5C) appeared to be lower in C4 spheroids than in C1 spheroids, but not significantly, while ATP levels, the ratio of NAD:NADH, and cAMP levels were significantly greater in C4 spheroids than in C1 spheroids (Figure 5D). Furthermore, observations in transmission electron microscopy (TEM) images collected on D60 confirmed that all three spheroid constructs contained Z-lines and gap junctions, but more complex structures, such as M-lines, I-bands, and A-bands, were observed only in C3 and C4 spheroids. This was notably a qualitative improvement from images collected on spheroids at D30 (Supplementary Figure S6). Contractile fibers also appeared to be more organized, and mitochondria better aligned, in C4 spheroids (Figure 6A), and both sarcomere lengths (Figure 6B) and widths (Figure 6C) were significantly greater in C4 spheroids than C1 spheroids. Thus, the inclusion of ECs, SMCs, and CFs in cardiac spheroids tended to promote sarcomere maturation and CM energy production.
FIGURE 5.
Patterns of metabolic, electrical, and cell-cycle gene expression in C1, C3, and C4 spheroids. The magnitude of expression for genes that contribute to CM (A) electrical conduction (HCN4, N-cadherin, SERCA, Ryr2, Cx43, CACNA1C), (B) metabolism (PPARGC1a, CKMT2), and (C) cell-cycle activity (CDK6) was evaluated in C1, C3, and C4 spheroids on D60 (C1 black; C3 yellow; C4 blue) via qPCR; measurements were normalized to intrinsic GAPDH mRNA abundance. (D) ATP abundance, the NAD/NADH ratio, and cellular cAMP was evaluated in C1, C3, and C4 spheroids on D60 (C1 black; C3 yellow; C4 blue) via luminescent ATP detection assay, colorimetric assay, and direct cAMP ELISA respectively (*p < 0.05, **p < 0.01; n > 4 per group).
FIGURE 6.
TEM assessments of the ultrastructure of C1, C3, and C4 spheroids. (A) Whole C1, C3, and C4 spheroids on day 60 were sectioned and imaged via TEM (bar = 2 μM for the top row, and bar = 1 μM for the bottom row); ZL: Z-line, ML: M-line, MT: mitochondria, GJ: gap junction, N: nucleus. (B) Sarcomere lengths were calculated by measuring the distance between adjacent Z-lines. (C) Sarcomere widths were measured as the length of each uninterrupted Z-line. (*p < 0.05, **p < 0.01, ***p < 0.001; n > 5 spheroids per group).
Field-potential duration and conduction velocity were greater in CMs from C4 spheroids than from C1 spheroids
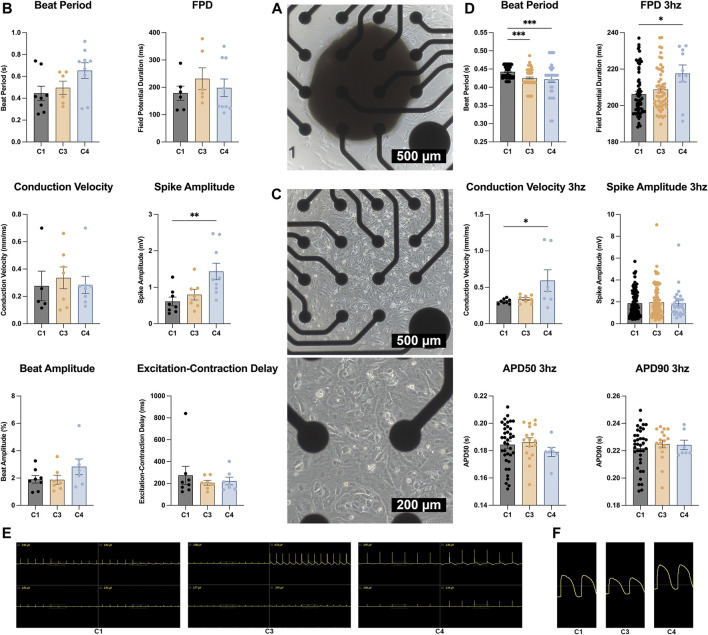
MEA assessments conducted on D30 of whole spheroids (Figure 7A) indicated that the electromechanical properties of all three constructs were generally similar: spike amplitude measurements were significantly greater in C4 spheroids than in C1 spheroids, but variations among groups in beat period, beat amplitude, field-potential duration, conduction velocity, and excitation-contraction delay were small and not significant (Figure 7B). However, when CMs were isolated from the constructs (Figure 7C), beat period was significantly shorter for CMs from C3 or C4 spheroids (i.e., C3 or C4 CMs) than from C1 spheroids (C1 CMs), and field-potential durations were significantly longer in C4 than in C1 CMs when the cells were paced at 3 hz (Figure 7D). Conduction velocities were also significantly greater in C4 than C1 CMs during pacing, likely because the variability among measurements in C4 CMs was exceptionally high, while measurements of spike amplitudes and action-potential durations were similar in CMs from all three constructs.
FIGURE 7.
MEA assessments. (A) Whole C1, C3, and C4 spheroids (C1 black; C3 yellow; C4 blue) were attached to a 4 × 4 MEA; then, field potentials and impedance were measured and used to calculate (B) beat period, spike amplitude, field potential duration (FPD), conduction velocity, beat amplitude, and excitation. (C) C1, C3, and C4 spheroids (C1 black; C3 yellow; C4 blue) were dissociated on D30; then, CMs were collected and plated on the MEA electrodes. Field potentials and impedance were measured with and without pacing at 3 hz (330 ms or 180 bpm) and used to calculate (D) beat period, spike amplitude, FPD, conduction velocity, and the action potential duration until 50 and 90% recovery (APD50 and APD90), respectively. (E) MEA field potential traces for C1, C3, and C4 whole organoids only showing the 4 electrodes that were used for analysis. (F) MEA field potential traces of dissociated C1, C3, and C4 organoids after undergoing LEAP induction (*p < 0.05, **p < 0.01, ***p < 0.001; n > 6 per group).
Discussion
The biological properties and activity of cells in the human heart are reproduced with greater fidelity by cardiac spheroids than by other in-vitro cell-culture systems. Thus, spheroids tend to provide a more accurate platform for modeling cardiac disease or drug development, and the individual cell populations present in cardiac spheroids may be more suitable for tissue engineering and other therapeutic applications. Here, we show that spheroids composed of hiPSC-derived CMs, ECs, SMCs, and CFs can be readily assembled and cultured for up to 60 days in solution; that C4 spheroids were significantly larger than C1 spheroids or C3 spheroids; and that the inclusion of vascular cells and CFs tended to promote sarcomere maturation and CM energy production. Furthermore, whereas apoptotic cells became increasingly common from D7 to D60 in C1 spheroids and C3 spheroids, the proportion in C4 spheroids remained stable throughout the culture period, which suggests that C4 spheroids are sufficiently durable for long-term studies. Notably, all four cardiac-cell types were differentiated from the same line of hiPSCs and, consequently, had the same genetic background, and the ratio of CMs, ECs, SMCs, and CFs (4:2:1:1) in C4 spheroids roughly followed the distribution trends found in human myocardium (Banerjee et al., 2007; Pinto et al., 2016; Gao et al., 2018; Arai et al., 2020; Beauchamp et al., 2020; Daly et al., 2021; Pretorius et al., 2021), but whether this ratio also produces the most native-like cardiac-cell phenotypes in cultured spheroids has yet to be conclusively demonstrated. The present study illustrates that additional optimization of this seeding ratio may be necessary to account for cell loss and restructuring that occurs and techniques for determining the composition in the fully matured tissue may be necessary. Additionally, the ideal ratio for different applications may vary depending on the region or state of myocardium that is being modeled.
The suspension culture system used in this report is broadly compatible with existing research equipment, and quality control checkpoints can be incorporated to maximize reproducibility; thus, our protocol can be readily adapted for a wide range of applications (Mattapally et al., 2018; LaBarge et al., 2019a; LaBarge et al., 2019b; Kim et al., 2020; Daly et al., 2021; Polonchuk et al., 2021) and production facilities (Abbasalizadeh et al., 2017; Adil and Schaffer, 2017; Li et al., 2017; Tomov et al., 2019). Furthermore, techniques for promoting spheroid fusion (Kim et al., 2018; Mattapally et al., 2018) and for the use of spheroids in 3D bioprinting (Duan, 2016; Maiullari et al., 2018; Aguilar et al., 2019; Alonzo et al., 2019; LaBarge et al., 2019b; Qasim et al., 2019; Lu Wang et al., 2021) have already been established, so the spheroids generated via this protocol could serve as building blocks for even larger and more sophisticated cardiac-tissue constructs, and because ECM production occurred spontaneously in C4 spheroids, exogenously administered ECM components may be unnecessary. However, only ∼50 spheroids per batch were produced in this study, and several of the steps were performed manually, so additional automation will be necessary to maximize productivity on an industrial scale.
The wide application of cardiac spheroid systems has led many groups to develop a range of models for both in vitro and in vivo uses. The microtissues in the present study achieved a greater overall diameter (500–1000 µm) than were previously produced (50–400 µm) with maintained cellular complexity and viability (Beauchamp et al., 2020; Giacomelli et al., 2020; Giacomelli et al., 2021). Although decreased central necrosis was observed in the C4 spheroids, the large diameter and fusion process raises concerns for nutrient and oxygen diffusion (Langan et al., 2016) which will require further assessment in future studies. The use of bioreactor culture environment rather than static culture may provide partial mitigation. Spheroids were also shown to maintain beating and viability over a 60 days period which provides greater longitudinal analysis than the single time points at 28, 20, 15, 30, and 12 days used by others (Ravenscroft et al., 2016; Giacomelli et al., 2017; Israeli et al., 2020; Polonchuk et al., 2021; Thomas et al., 2021). Finally, the protocol of fusing the multiple cell types into larger cardiac spheroids described here uses 96 well plates, a widely used format, and doesn’t require CM dissociation, a challenging and labor intensive step, while still maintaining consistent ratios of cell types. This is in comparison to complicated fabrication processes involving complete CM dissociation, hanging drop methods, agarose mold production, and additional centrifugation steps (Nugraha et al., 2019; Buono et al., 2020; Kupfer et al., 2020; Giacomelli et al., 2021; Thomas et al., 2021) and high variability due to spontaneous differentiation compared with the combination of purified cell types (Hoang et al., 2018; Schulze et al., 2019; Israeli et al., 2020) found in other studies.
Although the current study showed improvements in key biomolecular and functional markers a limitation in the data was the lack of functional cardiac metrics at each time point. Particularly, the lack of electromechanical testing at D60 when other biomarkers appeared to show the greatest improvement leaves a gap in maturation characterization. Future iterations of the cardiac spheroid model should consider incorporating data from both early and late time points for all analyses to provide greater longitudinal understanding. Additionally, the media composition chosen was based on previous work with cardiac cell coculture (Giacomelli et al., 2017; Gao et al., 2018; Noor et al., 2019; Pretorius et al., 2021) but was not optimized or fully defined. The use of FBS, although routine, has previously been shown to lead to dedifferentiation of ECs (Kaupisch et al., 2012; Sriram et al., 2015) and an alternative media formulation may be considered for future model iterations that is cGMP compliant (Zimmermann, 2020).
In conclusion, the experiments in this report demonstrate that C4 spheroids composed of four different hiPSC-derived cell populations (CMs, ECs, SMCs, and CFs) can be efficiently manufactured via a 3D suspension culture that is compatible with a wide range of applications and research equipment. Measures of spheroid size and cell viability were significantly greater in C4 spheroids than in spheroids lacking CFs after 60 days of culture, and the inclusion of vascular cells and CFs tended to promote sarcomere maturation and CM energy production. Future work will continue to investigate methods for improving CM maturity and spheroid yield while incorporating automated processes that are suitable for large-scale production facilities (Table 1).
TABLE 1.
Data collected from MEA analysis of whole and dissociated spheroids.
| Format | Group | Beat period (s) | Spike amplitude (mV) | FPD (ms) | Conduction velocity (cm/s) | Beat amplitude (%) | ECD (ms) | APD50 (ms) | APD90 (ms) |
|---|---|---|---|---|---|---|---|---|---|
| Whole Spheroid | C1 | 0.45 ± 0.07 | 0.61 ± 0.12 | 179 ± 26 | 27.6 ± 10.9 | 1.90 ± 0.27 | 275 ± 83 | ||
| C3 | 0.50 ± 0.06 | 0.80 ± 0.15 | 231 ± 40 | 33.6 ± 8.0 | 1.88 ± 0.33 | 207 ± 22 | |||
| C4 | 0.65 ± 0.07 | 1.43 ± 0.23 | 198 ± 32 | 28.5 ± 6.3 | 2.83 ± 0.57 | 222 ± 35 | |||
| Isolated CMs (Paced at 3 Hz) | C1 | 0.44 ± 0.01 | 1.87 ± 0.12 | 206 ± 2 | 30.7 ± 1.1 | 2.02 ± 0.13 | 142 ± 3 | 184 ± 3 | 222 ± 2 |
| C3 | 0.43 ± 0.01 | 1.95 ± 0.15 | 209 ± 2 | 34.0 ± 1.9 | 2.10 ± 0.19 | 144 ± 3 | 186 ± 3 | 225 ± 3 | |
| C4 | 0.42 ± 0.01 | 1.85 ± 0.24 | 218 ± 5 | 59.3 ± 14.7 | 1.74 ± 0.17 | 146 ± 3 | 179 ± 3 | 224 ± 4 |
Acknowledgments
The authors would like to thank the UAB Comprehensive Flow Cytometry Core and the UAB High-Resolution Imaging Facility for their assistance with sample preparation. The authors would also like to thank W. Kevin Meisner, PhD, ELS, for editorial assistance.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
AK-K developed the fabrication protocol, designed the experiments, performed histology and imaging, sample preparation, and wrote the manuscript. DP assisted in experimental design, collected TEM images, and completed manuscript edits. BG performed cell culture and sample analysis. XL assisted in experimental design and performed cell culture. YW assisted in experimental design and performed cell culture. JhZ differentiated and characterized the CFs. AQ collected data on bioenergetics. YN and TK assisted in experimental design and manuscript edits. LY helped with manuscript preparation and revisions. JyZ provided project leadership, funding acquisition, method development and manuscript revisions.
Funding
This research was funded by the National Heart, Lung, and Blood Institute (Grant Numbers 5R01HL114120, 5R01HL131017, 5U01HL134764); the American Heart Association Pre-doctoral Award (Grant Number 19PRE34380402) (AK-K); the National Institute of General Medical Sciences (Grant Number T32GM008361); and the National Institute of Biomedical Imaging and Bioengineering (Grant Number T32EB023872).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.908848/full#supplementary-material
References
- Abbasalizadeh S., Pakzad M., Cabral J., Baharvand H. (2017). Allogeneic cell therapy manufacturing: Process development technologies and facility design options. Expert Opin. Biol. Ther. 17 (10), 1201–1219. 10.1080/14712598.2017.1354982 [DOI] [PubMed] [Google Scholar]
- Adams W. J., Zhang Y., Cloutier J., Kuchimanchi P., Newton G., Sehrawat S., et al. (2013). Functional vascular endothelium derived from human induced pluripotent stem cells. Stem Cell Rep. 1 (2), 105–113. 10.1016/j.stemcr.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adil M. M., Schaffer D. V. (2017). Expansion of human pluripotent stem cells. Curr. Opin. Chem. Eng. 15, 24–35. 10.1016/j.coche.2016.11.002 [DOI] [Google Scholar]
- Aguilar I. N., Smith L. J., Olivos D. J., Chu T.-M. G., Kacena M. A., Wagner D. R., et al. (2019). Scaffold-free bioprinting of mesenchymal stem cells with the regenova printer: Optimization of printing parameters. Bioprinting 15, e00048. 10.1016/j.bprint.2019.e00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexanian R. A., Mahapatra K., Lang D., Vaidyanathan R., Markandeya Y. S., Gill R. K., et al. (2020). Induced cardiac progenitor cells repopulate decellularized mouse heart scaffolds and differentiate to generate cardiac tissue. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1867 (3), 118559. 10.1016/j.bbamcr.2019.118559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo M., AnilKumar S., Roman B., Tasnim N., Joddar B. (2019). 3D bioprinting of cardiac tissue and cardiac stem cell therapy. Transl. Res. 211, 64–83. 10.1016/j.trsl.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K., Murata D., Takao S., Nakamura A., Itoh M., Kitsuka T., et al. (2020). Drug response analysis for scaffold-free cardiac constructs fabricated using bio-3D printer. Sci. Rep. 10 (1), 8972. 10.1038/s41598-020-65681-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoubi S., Sheikh S. P., Eskildsen T. V. (2017). Human induced pluripotent stem cell-derived vascular smooth muscle cells: Differentiation and therapeutic potential. Cardiovasc. Res. 113 (11), 1282–1293. 10.1093/cvr/cvx125 [DOI] [PubMed] [Google Scholar]
- Banerjee I., Fuseler J. W., Price R. L., Borg T. K., Baudino T. A. (2007). Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. J. Physiology-Heart Circulatory Physiology 293 (3), H1883–H1891. 10.1152/ajpheart.00514.2007 [DOI] [PubMed] [Google Scholar]
- Bayrak G. K., Gümüşderelioğlu M. (2019). Construction of cardiomyoblast sheets for cardiac tissue repair: Comparison of three different approaches. Cytotechnology 71 (4), 819–833. 10.1007/s10616-019-00325-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp P., Jackson C. B., Ozhathil L. C., Agarkova I., Galindo C. L., Sawyer D. B., et al. (2020). 3D Co-culture of hiPSC-derived cardiomyocytes with cardiac fibroblasts improves tissue-like features of cardiac spheroids. Front. Mol. Biosci. 7, 14. 10.3389/fmolb.2020.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp P., Moritz W., Kelm J. M., Ullrich N. D., Agarkova I., Anson B. D., et al. (2015). Development and characterization of a scaffold-free 3D spheroid model of induced pluripotent stem cell-derived human cardiomyocytes. Tissue Eng. Part C. Methods 21 (8), 852–861. 10.1089/ten.tec.2014.0376 [DOI] [PubMed] [Google Scholar]
- Bin Lin B., Lin X., Stachel M., Wang E., Luo Y., Lader J., et al. (2017). Culture in glucose-depleted medium supplemented with fatty acid and 3, 3', 5-Triiodo-l-Thyronine facilitates purification and maturation of human pluripotent stem cell-derived cardiomyocytes. Front. Endocrinol. 8, 253. 10.3389/fendo.2017.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binah O., Dolnikov K., Sadan O., Shilkrut M., Zeevi-Levin N., Amit M., et al. (2007). Functional and developmental properties of human embryonic stem cells–derived cardiomyocytes. J. Electrocardiol. 40 (6), S192–S196. 10.1016/j.jelectrocard.2007.05.035 [DOI] [PubMed] [Google Scholar]
- Buono M. F., Boehmer L. v., Strang J., Hoerstrup S. P., Emmert M. Y., Nugraha B., et al. (2020). Human cardiac organoids for modeling genetic cardiomyopathy. Cells 9 (7), 1733. 10.3390/cells9071733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F., Wagner R. A., Wilson K. D., Xie X., Fu J.-D., Drukker M., et al. (2008). Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE 3 (10), e3474. 10.1371/journal.pone.0003474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi O., Itzhaki I., Kehat I., Gepstein A., Arbel G., Huber I., et al. (2009). In vitro electrophysiological drug testing using human embryonic stem cell derived cardiomyocytes. Stem Cells Dev. 18 (1), 161–172. 10.1089/scd.2007.0280 [DOI] [PubMed] [Google Scholar]
- Chan Y.-C., Ting S., Lee Y.-K., Ng K.-M., Zhang J., Chen Z., et al. (2013). Electrical stimulation promotes maturation of cardiomyocytes derived from human embryonic stem cells. J. Cardiovasc. Transl. Res. 6 (6), 989–999. 10.1007/s12265-013-9510-z [DOI] [PubMed] [Google Scholar]
- Chang S., Finklea F., Williams B., Hammons H., Hodge A., Scott S., et al. (2020). Emulsion‐based encapsulation of pluripotent stem cells in hydrogel microspheres for cardiac differentiation. Biotechnol. Prog. 36 (4), e2986. 10.1002/btpr.2986 [DOI] [PubMed] [Google Scholar]
- Daly A. C., Davidson M. D., Burdick J. A. (2021). 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun. 12 (1), 753. 10.1038/s41467-021-21029-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Kim S.-W., Choi Y.-J., Lee S., Lee S.-H., Kong J.-S., et al. (2019). Decellularized extracellular matrix bioinks and the external stimuli to enhance cardiac tissue development in vitro . Acta Biomater. 95, 188–200. Acta Biomater 70 2018. 10.1016/j.actbio.2019.04.026 [DOI] [PubMed] [Google Scholar]
- Duan B. (2016). State-of-the-Art review of 3D bioprinting for cardiovascular tissue engineering. Ann. Biomed. Eng. 45 (1), 195–209. 10.1007/s10439-016-1607-5 [DOI] [PubMed] [Google Scholar]
- Fischer B., Meier A., Dehne A., Salhotra A., Tran T. A., Neumann S., et al. (2018). A complete workflow for the differentiation and the dissociation of hiPSC-derived cardiospheres. Stem Cell Res. 32, 65–72. 10.1016/j.scr.2018.08.015 [DOI] [PubMed] [Google Scholar]
- Gao L., Gregorich Z. R., Zhu W., Mattapally S., Oduk Y., Lou X., et al. (2018). Large cardiac muscle patches engineered from human induced-pluripotent stem cell–derived cardiac cells improve recovery from myocardial infarction in swine. Circulation 137 (16), 1712–1730. 10.1161/circulationaha.117.030785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzoni L. R., Rossi M. I. D., Barros A. P. D. N. d., Guarani V., Keramidas M., Balottin L. B. L., et al. (2009). Dissecting coronary angiogenesis: 3D co-culture of cardiomyocytes with endothelial or mesenchymal cells. Exp. Cell Res. 315 (19), 3406–3418. 10.1016/j.yexcr.2009.09.016 [DOI] [PubMed] [Google Scholar]
- Giacomelli E., Bellin M., Sala L., Meer B. J. v., Tertoolen L. G. J., Orlova V. V., et al. (2017). Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Dev. Camb. Engl. 144 (6), 1008–1017. 10.1242/dev.143438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E., Meraviglia V., Campostrini G., Cochrane A., Cao X., Helden R. W. J. v., et al. (2020). Human-iPSC-Derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell 26 (6), 862–879.e11. e811. 10.1016/j.stem.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E., Sala L., Oostwaard D. W.-v., Bellin M. (2021). Cardiac microtissues from human pluripotent stem cells recapitulate the phenotype of long-QT syndrome. Biochem. Biophysical Res. Commun. 572, 118–124. 10.1016/j.bbrc.2021.07.068 [DOI] [PubMed] [Google Scholar]
- Guo Y., Pu W. T. (2020). Cardiomyocyte maturation. Circ. Res. 126 (8), 1086–1106. 10.1161/circresaha.119.315862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haishuang Lin H., Li Q., Lei Y. (2017). Three-dimensional tissues using human pluripotent stem cell spheroids as biofabrication building blocks. Biofabrication 9 (2), 025007. 10.1088/1758-5090/aa663b [DOI] [PubMed] [Google Scholar]
- Helms H. R., Jarrell D. K., Jacot J. G. (2019). Generation of cardiac organoids using cardiomyocytes, endothelial cells, epicardial cells, and cardiac fibroblasts derived from human induced pluripotent stem cells. FASEB J. 33 (S1), lb170. 10.1096/fasebj.2019.33.1_supplement.lb170 [DOI] [Google Scholar]
- Hoang P., Wang J., Conklin B. R., Healy K. E., Ma Z. (2018). Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat. Protoc. 13 (4), 723–737. 10.1038/nprot.2018.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman-Mendez C., Campos D. B. P. d., Pinto R. S., Mendes B. J. d. S., Rocha G. M., Monnerat G., et al. (2020). Tissue-engineered human embryonic stem cell-containing cardiac patches: Evaluating recellularization of decellularized matrix. J. Tissue Eng. 11, 204173142092148. 10.1177/2041731420921482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson C. P., Bareja A., Gomez J. A., Dzau V. J. (2016). Emerging concepts in paracrine mechanisms in regenerative cardiovascular medicine and biology. Circ. Res. 118 (1), 95–107. 10.1161/circresaha.115.305373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N., Loskill P., Deveshwar N., Spencer C. I., Judge L. M., Mandegar M. A., et al. (2016). Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci. Rep. 6 (1), 24726. 10.1038/srep24726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigami M., Masumoto H., Ikuno T., Aoki T., Kawatou M., Minakata K., et al. (2018). Human iPS cell-derived cardiac tissue sheets for functional restoration of infarcted porcine hearts. PLoS ONE 13 (8), e0201650. 10.1371/journal.pone.0201650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli Y., Gabalski M., Ball K., Wasserman A., Zou J., Ni G., et al. (2020). Generation of heart organoids modeling early human cardiac development under defined conditions. Laurel Hollow: bioRxiv. 10.1101/2020.06.25.171611 [DOI] [Google Scholar]
- Ivanov D. P., Parker T. L., Walker D. A., Alexander C., Ashford M. B., Gellert P. R., et al. (2014). Multiplexing spheroid volume, resazurin and acid phosphatase viability assays for high-throughput screening of tumour spheroids and stem cell neurospheres. PLoS ONE 9 (8), e103817. 10.1371/journal.pone.0103817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman C. P., Carlson A. L., Bursac N. (2016). Dynamic culture yields engineered myocardium with near-adult functional output. Biomaterials 111, 66–79. Circ. Res. 109 2011. 10.1016/j.biomaterials.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman C., Li H., Bursac N. (2018). Long-term contractile activity and thyroid hormone supplementation produce engineered rat myocardium with adult-like structure and function. Acta Biomater. 78, 98–110. Current opinion in chemical engineering 7 2015). 10.1016/j.actbio.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha R., Wu Q., Singh M., Preininger M. K., Han P., Ding G., et al. (2016). Simulated microgravity and 3D culture enhance induction, viability, proliferation and differentiation of cardiac progenitors from human pluripotent stem cells. Sci. Rep. 6 (1), 30956. 10.1038/srep30956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn-Krell A., Pretorius D., Ou J., Fast V. G., Litovsky S., Berry J., et al. (2021). Bioreactor suspension culture: Differentiation and production of cardiomyocyte spheroids from human induced pluripotent stem cells. Front. Bioeng. Biotechnol. 9, 674260. 10.3389/fbioe.2021.674260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai-Li Wang K.-L., Xue Q., Xu X.-H., Hu F., Shao H. (2021). Recent progress in induced pluripotent stem cell-derived 3D cultures for cardiac regeneration. Cell Tissue Res. 384 (2), 231–240. 10.1007/s00441-021-03414-x [DOI] [PubMed] [Google Scholar]
- Karbassi E., Fenix A., Marchiano S., Muraoka N., Nakamura K., Yang X., et al. (2020). Cardiomyocyte maturation: Advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 17 (6), 341–359. 10.1038/s41569-019-0331-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupisch A., Kennedy L., Stelmanis V., Tye B., Kane N. M., Mountford J. C., et al. (2012). Derivation of vascular endothelial cells from human embryonic stem cells under GMP-compliant conditions: Towards clinical studies in ischaemic disease. J. Cardiovasc. Transl. Res. 5 (5), 605–617. 10.1007/s12265-012-9379-2 [DOI] [PubMed] [Google Scholar]
- Keung W., Chan P. K. W., Backeris P. C., Lee E. K., Wong N., Wong A. O. T., et al. (2019). Human cardiac ventricular‐like organoid chambers and tissue strips from pluripotent stem cells as a two‐tiered assay for inotropic responses. Clin. Pharmacol. Ther. 106 (2), 402–414. 10.1002/cpt.1385 [DOI] [PubMed] [Google Scholar]
- Kim C., Majdi M., Xia P., Wei K. A., Talantova M., Spiering S., et al. (2010). Non-cardiomyocytes influence the electrophysiological maturation of human embryonic stem cell-derived cardiomyocytes during differentiation. Stem Cells Dev. 19 (6), 783–795. 10.1089/scd.2009.0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. Y., Kofron C. M., King M. E., Markes A. R., Okundaye A. O., Qu Z., et al. (2018). Directed fusion of cardiac spheroids into larger heterocellular microtissues enables investigation of cardiac action potential propagation via cardiac fibroblasts. PLoS ONE 13 (5), e0196714. 10.1371/journal.pone.0196714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. j., Kim E. M., Yamamoto M., Park H., Shin H. (2020). Engineering multi‐cellular spheroids for tissue engineering and regenerative medicine. Adv. Healthc. Mat. 9 (23), 2000608. 10.1002/adhm.202000608 [DOI] [PubMed] [Google Scholar]
- Kupfer M. E., Lin W.-H., Ravikumar V., Qiu K., Wang L., Gao L., et al. (2020). In situ expansion, differentiation, and electromechanical coupling of human cardiac muscle in a 3D bioprinted, chambered organoid. Circ. Res. 127 (2), 207–224. 10.1161/circresaha.119.316155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong G., Marquez H. A., Yang C., Wong J. Y., Kotton D. N. (2019). Generation of a purified iPSC-derived smooth muscle-like population for cell sheet engineering. Stem Cell Rep. 13 (3), 499–514. 10.1016/j.stemcr.2019.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBarge W., Mattappally S., Kannappan R., Fast V. G., Pretorius D., Berry J. L., et al. (2019a). Maturation of three-dimensional, hiPSC-derived cardiomyocyte spheroids utilizing cyclic, uniaxial stretch and electrical stimulation. PLoS ONE 14 (7), e0219442. 10.1371/journal.pone.0219442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBarge W., Morales A., Pretorius D., Kahn-Krell A. M., Kannappan R., Zhang J., et al. (2019b). Scaffold-Free bioprinter utilizing layer-by-layer printing of cellular spheroids. Micromachines 10 (9), 570. 10.3390/mi10090570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M. A., Chen K. Y., Naumova A. V., Muskheli V., Fugate J. A., Dupras S. K., et al. (2007). Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 25 (9), 1015–1024. 10.1038/nbt1327 [DOI] [PubMed] [Google Scholar]
- Langan L. M., Dodd N. J., Owen S. F., Purcell W. M., Jackson S. K., Jha A. N., et al. (2016). Direct measurements of oxygen gradients in spheroid culture system using electron parametric resonance oximetry. PLoS One 11 (2), e0149492. 10.1371/journal.pone.0149492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-K., Ng K.-M., Lai W.-H., Chan Y.-C., Lau Y.-M., Lian Q., et al. (2011). Calcium homeostasis in human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Rev. Rep. 7 (4), 976–986. 10.1007/s12015-011-9273-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A., Hudson A. R., Shiwarski D. J., Tashman J. W., Hinton T. J., Yerneni S., et al. (2019a). 3D bioprinting of collagen to rebuild components of the human heart. Science 365 (6452), 482–487. 10.1126/science.aav9051 [DOI] [PubMed] [Google Scholar]
- Lee M.-O., Jung K. B., Jo S.-J., Hyun S.-A., Moon K.-S., Seo J.-W., et al. (2019b). Modelling cardiac fibrosis using three-dimensional cardiac microtissues derived from human embryonic stem cells. J. Biol. Eng. 13 (1), 15. 10.1186/s13036-019-0139-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li L., Chen Z.-N., Gao G., Yao R., Sun W., et al. (2017). Engineering-derived approaches for iPSC preparation, expansion, differentiation and applications. Biofabrication 9 (3), 032001. 10.1088/1758-5090/aa7e9a [DOI] [PubMed] [Google Scholar]
- Li Y., Song D., Mao L., Abraham D. M., Bursac N. (2020). Lack of Thy1 defines a pathogenic fraction of cardiac fibroblasts in heart failure. Biomaterials 236, 119824. 10.1016/j.biomaterials.2020.119824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L. B., Azarin S. M., et al. (2012). Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 109 (27), E1848–E1857. 10.1073/pnas.1200250109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Qi J., Xu X., Zeisberg M., Guan K., Zeisberg E. M., et al. (2016). Differentiation of functional endothelial cells from human induced pluripotent stem cells: A novel, highly efficient and cost effective method. Differentiation 92 (4), 225–236. 10.1016/j.diff.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Liu L., Jin X., Hu C.-F., Li R., Zhou Z. e., Shen C.-X., et al. (2017). Exosomes derived from mesenchymal stem cells rescue myocardial ischaemia/reperfusion injury by inducing cardiomyocyte autophagy via AMPK and akt pathways. Cell. Physiol. biochem. 43 (1), 52–68. 10.1159/000480317 [DOI] [PubMed] [Google Scholar]
- Long C., Li H., Tiburcy M., Rodriguez-Caycedo C., Kyrychenko V., Zhou H., et al. (2018). Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 4 (1), eaap9004. 10.1126/sciadv.aap9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Wang L., Serpooshan V., Zhang J. (2021). Engineering human cardiac muscle patch constructs for prevention of post-infarction LV remodeling. Front. Cardiovasc. Med. 8, 621781. 10.3389/fcvm.2021.621781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux M., Andrée B., Horvath T., Nosko A., Manikowski D., Hilfiker-Kleiner D., et al. (2016). In vitro maturation of large-scale cardiac patches based on a perfusable starter matrix by cyclic mechanical stimulation. Acta Biomater. 30, 177–187. 10.1016/j.actbio.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Maiullari F., Costantini M., Milan M., Pace V., Chirivì M., Maiullari S., et al. (2018). A multi-cellular 3D bioprinting approach for vascularized heart tissue engineering based on HUVECs and iPSC-derived cardiomyocytes. Sci. Rep. 8 (1), 13532. 10.1038/s41598-018-31848-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur A., Loskill P., Shao K., Huebsch N., Hong S., Marcus S. G., et al. (2015). Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 5 (1), 8883. 10.1038/srep08883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapally S., Zhu W., Fast V. G., Gao L., Worley C., Kannappan R., et al. (2018). Spheroids of cardiomyocytes derived from human-induced pluripotent stem cells improve recovery from myocardial injury in mice. Am. J. Physiology-Heart Circulatory Physiology 315 (2), H327–H339. 10.1152/ajpheart.00688.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menasché P., Hagège A. A., Scorsin M., Pouzet B., Desnos M., Duboc D., et al. (2001). Myoblast transplantation for heart failure. Lancet 357 (9252), 279–280. 10.1016/s0140-6736(00)03617-5 [DOI] [PubMed] [Google Scholar]
- Mills R. J., Titmarsh D. M., Koenig X., Parker B. L., Ryall J. G., Quaife-Ryan G. A., et al. (2017). Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 114 (40), E8372–E8381. 10.1073/pnas.1707316114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa T., Idiris A., Kumagai H. (2020). A novel cardiac differentiation method of a large number and uniformly-sized spheroids using microfabricated culture vessels. Regen. Ther. 15, 18–26. 10.1016/j.reth.2020.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munarin F., Kant R. J., Rupert C. E., Khoo A., Coulombe K. L. K. (2020). Engineered human myocardium with local release of angiogenic proteins improves vascularization and cardiac function in injured rat hearts. Biomaterials 251, 120033. 10.1016/j.biomaterials.2020.120033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor N., Shapira A., Edri R., Gal I., Wertheim L., Dvir T., et al. (2019). 3D printing of personalized thick and perfusable cardiac patches and hearts. Adv. Sci. (Weinh). 6 (11), 1900344. 10.1002/advs.201900344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugraha B., Buono M. F., Boehmer L., Hoerstrup S. P., Emmert M. Y. (2019). Human cardiac organoids for disease modeling. Clin. Pharmacol. Ther. 105 (1), 79–85. 10.1002/cpt.1286 [DOI] [PubMed] [Google Scholar]
- Palpant N. J., Pabon L., Friedman C. E., Roberts M., Hadland B., Zaunbrecher R. J., et al. (2017). Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells. Nat. Protoc. 12 (1), 15–31. 10.1038/nprot.2016.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S. S., Blackwell D. J., Gomez-Hurtado N., Frisk M., Wang L., Kim K., et al. (2017). Thyroid and glucocorticoid hormones promote functional T-tubule development in human-induced pluripotent stem cell–derived cardiomyocytes. Circ. Res. 121 (12), 1323–1330. 10.1161/circresaha.117.311920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A. R., Ilinykh A., Ivey M. J., Kuwabara J. T., D’Antoni M. L., Debuque R., et al. (2016). Revisiting cardiac cellular composition. Circ. Res. 118 (3), 400–409. 10.1161/circresaha.115.307778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonchuk L., Surija L., Lee M. H., Sharma P., Ming C. L. C., Richter F., et al. (2021). Towards engineering heart tissues from bioprinted cardiac spheroids. Biofabrication 13 (4), 045009. 10.1088/1758-5090/ac14ca [DOI] [PubMed] [Google Scholar]
- Pretorius D., Kahn-Krell A. M., Lou X., Fast V. G., Berry J. L., Kamp T. J., et al. (2021). Layer-by-layer fabrication of large and thick human cardiac muscle patch constructs with superior electrophysiological properties. Front. Cell Dev. Biol. 9, 670504. 10.3389/fcell.2021.670504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasim M., Haq F., Kang M.-H., Kim J.-H. (2019). 3D printing approaches for cardiac tissue engineering and role of immune modulation in tissue regeneration. Int. J. Nanomedicine 14, 1311–1333. 10.2147/ijn.s189587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft S. M., Pointon A., Williams A. W., Cross M. J., Sidaway J. E. (2016). Cardiac non-myocyte cells show enhanced pharmacological function suggestive of contractile maturity in stem cell derived cardiomyocyte microtissues. Toxicol. Sci. 152 (1), 99–112. 10.1093/toxsci/kfw069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards D. J., Tan Y., Coyle R., Li Y., Xu R., Yeung N., et al. (2016). Nanowires and electrical stimulation synergistically improve functions of hiPSC cardiac spheroids. Nano Lett. 16 (7), 4670–4678. 10.1021/acs.nanolett.6b02093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C., Tran D. D., George S. C. (2013). Concise review: Maturation phases of human pluripotent stem cell‐derived cardiomyocytes. STEM CELLS 31 (5), 829–837. 10.1002/stem.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J.-L., Tulloch N. L., Razumova M. V., Saiget M., Muskheli V., Pabon L., et al. (2016). Mechanical stress conditioning and electrical stimulation promote contractility and force maturation of induced pluripotent stem cell-derived human cardiac tissue. Circulation 134 (20), 1557–1567. 10.1161/circulationaha.114.014998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetto C., Vitiello L., Windt L. J. d., Rampazzo A., Calore M. (2020). Modeling cardiovascular diseases with hiPSC-derived cardiomyocytes in 2D and 3D cultures. Int. J. Mol. Sci. 21 (9), 3404. 10.3390/ijms21093404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanganalmath S. K., Bolli R. (2013). Cell therapy for heart failure. Circ. Res. 113 (6), 810–834. 10.1161/circresaha.113.300219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J. A., Guzman P. A., Riemenschneider S. B., Kamp T. J., Tranquillo R. T. (2017). A cardiac patch from aligned microvessel and cardiomyocyte patches. J. Tissue Eng. Regen. Med. 12 (2), 546–556. 10.1002/term.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze M. L., Lemoine M. D., Fischer A. W., Scherschel K., David R., Riecken K., et al. (2019). Dissecting hiPSC-CM pacemaker function in a cardiac organoid model. Biomaterials 206, 133–145. 10.1016/j.biomaterials.2019.03.023 [DOI] [PubMed] [Google Scholar]
- Selvaraj S., Mondragon-Gonzalez R., Xu B., Magli A., Kim H., Lainé J., et al. (2019). Screening identifies small molecules that enhance the maturation of human pluripotent stem cell-derived myotubes. eLife 8, e47970. 10.7554/elife.47970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrin I. Y., Allen B. W., Qian Y., Jackman C. P., Carlson A. L., Juhas M. E., et al. (2017). Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat. Commun. 8 (1), 1825. 10.1038/s41467-017-01946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram G., Tan J. Y., Islam I., Rufaihah A. J., Cao T. (2015). Efficient differentiation of human embryonic stem cells to arterial and venous endothelial cells under feeder- and serum-free conditions. Stem Cell Res. Ther. 6 (1), 261. 10.1186/s13287-015-0260-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strikoudis A., Cieślak A., Loffredo L., Chen Y.-W., Patel N., Saqi A., et al. (2019). Modeling of fibrotic Lung disease using 3D organoids derived from human pluripotent stem cells. Cell Rep. 27 (12), 3709–3723.e5. 10.1016/j.celrep.2019.05.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Nunes S. S. (2016). Biowire platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Methods 101, 21–26. 10.1016/j.ymeth.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Synnergren J., Åkesson K., Dahlenborg K., Vidarsson H., Améen C., Steel D., et al. (2008). Molecular signature of cardiomyocyte clusters derived from human embryonic stem cells. STEM CELLS 26 (7), 1831–1840. 10.1634/stemcells.2007-1033 [DOI] [PubMed] [Google Scholar]
- Synnergren J., Heins N., Brolén G., Eriksson G., Lindahl A., Hyllner J., et al. (2010). Transcriptional profiling of human embryonic stem cells differentiating to definitive and primitive endoderm and further toward the hepatic lineage. Stem Cells Dev. 19 (7), 961–978. 10.1089/scd.2009.0220 [DOI] [PubMed] [Google Scholar]
- Thomas D., Kim H., Lopez N., Wu J. C. (2021). Fabrication of 3D cardiac microtissue arrays using human iPSC-derived cardiomyocytes, cardiac fibroblasts, and endothelial cells. J. Vis. Exp. 169 (169). 10.3791/61879 [DOI] [PubMed] [Google Scholar]
- Tomov M. L., Gil C. J., Cetnar A., Theus A. S., Lima B. J., Nish J. E., et al. (2019). Engineering functional cardiac tissues for regenerative medicine applications. Curr. Cardiol. Rep. 21 (9), 105. 10.1007/s11886-019-1178-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruyama S., Matsuura K., Sakaguchi K., Shimizu T. (2019). Pulsatile tubular cardiac tissues fabricated by wrapping human iPS cells-derived cardiomyocyte sheets. Regen. Ther. 11, 297–305. 10.1016/j.reth.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges H. K., Mills R. J., Elliott D. A., Parton R. G., Porrello E. R., Hudson J. E., et al. (2017). Development of a human cardiac organoid injury model reveals innate regenerative potential. Development 144 (6), 1118–1127. 10.1242/dev.143966 [DOI] [PubMed] [Google Scholar]
- Xu X. Q., Soo S. Y., Sun W., Zweigerdt R. (2009). Global expression profile of highly enriched cardiomyocytes derived from human embryonic stem cells. STEM CELLS 27 (9), 2163–2174. 10.1002/stem.166 [DOI] [PubMed] [Google Scholar]
- Yang L., Geng Z., Nickel T., Johnson C., Gao L., Dutton J., et al. (2016). Differentiation of human induced-pluripotent stem cells into smooth-muscle cells: Two novel protocols. PLoS ONE 11 (1), e0147155. 10.1371/journal.pone.0147155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Chang Y.-H., Xiong Q., Zhang P., Zhang L., Somasundaram P., et al. (2014). Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell Stem Cell 15 (6), 750–761. 10.1016/j.stem.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Klos M., Wilson G. F., Herman A. M., Lian X., Raval K. K., et al. (2012). Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: The matrix sandwich method. Circ. Res. 111 (9), 1125–1136. 10.1161/CIRCRESAHA.112.273144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Guo J., Zhang P., Xiong Q., Wu S. C., Xia L., et al. (2014). Derivation and high engraftment of patient-specific cardiomyocyte sheet using induced pluripotent stem cells generated from adult cardiac fibroblast. Circ. Heart Fail. 8 (1), 156–166. 10.1161/circheartfailure.114.001317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kong C. W., Tong M. H., Chooi W. H., Huang N., Li R. A., et al. (2017). Maturation of human embryonic stem cell-derived cardiomyocytes (hESC-CMs) in 3D collagen matrix: Effects of niche cell supplementation and mechanical stimulation. Acta Biomater. 49, 204–217. 10.1016/j.actbio.2016.11.058 [DOI] [PubMed] [Google Scholar]
- Zhang J., Tao R., Campbell K. F., Carvalho J. L., Ruiz E. C., Kim G. C., et al. (2019). Functional cardiac fibroblasts derived from human pluripotent stem cells via second heart field progenitors. Nat. Commun. 10 (1), 2238. 10.1038/s41467-019-09831-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Bolli R., Garry D. J., Marbán E., Menasché P., Zimmermann W.-H., et al. (2021a). Basic and translational research in cardiac repair and regeneration. J. Am. Coll. Cardiol. 78 (21), 2092–2105. 10.1016/j.jacc.2021.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Z., Zhao S. R., Tu C., Pang P., Zhang M., Wu J. C., et al. (2021b). Protocol to measure contraction, calcium, and action potential in human-induced pluripotent stem cell-derived cardiomyocytes. Star. Protoc. 2 (4), 100859. 10.1016/j.xpro.2021.100859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Gao L., Zhang J. (2017). Pluripotent stem cell derived cardiac cells for myocardial repair. J. Vis. Exp. 120, 55142. 10.3791/55142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W.-H. (2020). Tissue engineered heart repair from preclinical models to first-in-patient studies. Curr. Opin. Physiology 14, 70–77. 10.1016/j.cophys.2020.02.001 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.