Abstract
The PhoP-PhoR two-component regulatory system controls the phosphate deficiency response in B. subtilis. A number of Pho regulon genes which require PhoP∼P for activation or repression have been identified. The studies reported here were initiated to understand the PhoP-DNA interaction necessary for Pho promoter regulation. The regulatory region of phoD was characterized in detail using oligo-directed mutagenesis, DNase I footprinting, and in vivo transcription assays. These data reveal basic principles of PhoP binding relevant to PhoP’s interaction with other Pho regulon promoters. Our results show that: (i) a dimer of PhoP∼P is able to bind two consensus repeats in a stable fashion; (ii) PhoP binding is highly cooperative within the core promoter region, which is located from −66 to −17 on the coding strand and contains four TT(A/T/C)ACA-like repeats; (iii) specific bases comprising the TT(A/T/C)ACA consensus are essential for transcriptional activation, but the specific base pairs of the intervening sequences separating the consensus repeats are not important for either PhoP binding or promoter activation; (iv) the spacing between two consensus repeats within a putative dimer binding site in the core region is important for both PhoP binding and promoter activation; (v) the exact spacing between two dimer binding sites within the core region is important for promoter activation but less so for PhoP binding affinity, as long as the repeats are on the same face of the helix; and (vi) the 5′ secondary binding region is important for coordinated PhoP binding to the core binding region, making it nearly essential for promoter activation.
Bacillus subtilis, a gram-positive bacterium, frequently encounters stress conditions in its natural environment, the soil. In order to survive, it has developed, through the course of evolution, several ways of detecting and responding to these pressures. One of the ways of accomplishing both is through the use of two-component signal transduction systems. Two-component systems are found in numerous bacteria and are composed of two main regulatory proteins, (i) the histidine-sensor kinase and (ii) the response regulator (22, 26, 27). When the histidine kinase senses a specific stimulus from the environment, it undergoes ATP-dependent autophosphorylation and then passes the phosphate to its cognate response regulator. The activated response regulator is able to elicit a response by interacting with specific proteins or by binding a specific set of promoters in order to activate or repress their transcription.
When B. subtilis is starved for phosphate, several genes of the phosphate or Pho regulon are activated or repressed through the coordination of the two-component regulatory proteins PhoP and PhoR (1, 3, 10, 11, 14, 24). These proteins have homology to DNA-binding response regulators and histidine kinases, respectively. Genetic evidence suggests that, of the three different response regulators previously shown to be involved in the Pho response, PhoP is the furthest downstream in the signaling pathway (7, 28).
Many of the Pho regulon genes and their promoters have been characterized through promoter fusion assays, in vitro transcription assays, and DNase I footprint analyses. These genes include phoA and phoB (9, 15, 17), which encode the two major vegetative alkaline phosphatases in the cell (98% of alkaline phosphatase activity); pstS, which encodes a high-affinity transport system to bring inorganic phosphate into the cell (24); and the tuaA operon, which encodes the proteins necessary for the biosynthesis of teichuronic acid, a phosphate-free anionic cell wall polymer used to replace the polyglycerol phosphate cell wall polymer, and teichoic acid, which is synthesized during phosphate-replete growth (13, 16).
Each of these genes’ promoters was bound by both PhoP and PhoP∼P but required PhoP∼P for activation. The predominant binding region, located at approximately the same position in every promoter (≈−21 to ≈−60 relative to the transcription start site) contained multiple TT(A/T)ACA-like repeats separated by approximately 5 bp (16). Sequence comparisons revealed that a minimum of four of these repeats existed in each promoter, and this conserved sequence arrangement was termed the core binding region. Deletion of only one repeat from the core binding region severely reduced transcriptional activation in vivo and in vitro. Further, gel retardation assays suggested that all four repeats were required for PhoP to bind efficiently (17, 23).
Two of the stronger Pho regulon promoters, phoA and pstS, contain a secondary PhoP binding region in addition to the core binding region (17). These secondary binding sites are in the coding sequences of their genes and consist of less than four TT(A/T)ACA-like repeats, and their deletion is deleterious to promoter activation.
Promoter sequence comparisons and limited promoter deletion analyses have led to the hypothesis that directly repeated sequences in the core binding region and secondary binding sites are important features for PhoP binding and Pho regulon promoter activation, a hypothesis which can now be tested.
The study reported here focuses on phoD, encoding a phosphodiesterase-alkaline phosphatase (29, 30), whose expression is dependent on PhoP and PhoR (3). PhoD has a putative role in cell wall teichoic acid turnover during phosphate deprivation, a novel Pho regulon function in B. subtilis and possibly other gram-positive bacteria.
Several characteristics of phoD make it an interesting candidate for further study and representative of Pho regulon promoters for extensive mutagenesis. First, the phoD promoter is the strongest promoter in the Pho regulon. Second, the phoD promoter is the most tightly regulated promoter in the Pho regulon, having no promoter activity under phosphate-replete conditions or in the absence of PhoP or PhoR. Third, initial promoter studies revealed that the phoD promoter contains the elements characteristic of other Pho regulon promoters, (i) a core binding region with four TT(A/T)ACA-like repeats and (ii) a secondary binding site.
In an effort to experimentally define the PhoP consensus binding sequence and to investigate the interaction between the PhoP molecules bound to the core binding region with those bound at the secondary binding site, we mutated the phoD promoter extensively. By correlating the promoter-lacZ expression data of numerous mutant promoters with their PhoP binding data, we were able to experimentally define TT(A/T/C)ACA as the consensus binding sequence for PhoP and propose loop formation as part of the mechanism for the initiation of transcription from the stronger Pho regulon promoters.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. subtilis JH642 (pheA1 trpC2 [J.A. Hoch]) was the parent strain transformed with each of the promoter-lacZ fusions. Escherichia coli MV1190 [(Δlac-proAB) thi supE Δ(srl-recA)306::Tn10(Tetr) (F′:tra D36 proAB lacIqZΔM15)] and CJ236 [dut ung thi relA; pCJ105 (Cmr)], used during the oligo-directed mutagenesis of phoD, were supplied by Bio-Rad. E. coli lab strain DH5α [F− φ80dlacZΔM15 Δ(lacZY A-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 λ− thi-1 gyrA relA1] was used for cloning all other phoD constructions. B. subtilis MH5441 (pheA1 trpC2 amyE::phoD-lacZ Cmr) was used as the source of RNA for primer extension.
PCR was used to amplify the 365-bp promoter region of phoD from JH642 template DNA. The primers used were FMH208 and FMH209 containing EcoRI and BamHI sites, respectively. This product was then ligated to the pCRII vector provided with the TA cloning kit (Invitrogen) to form pSE6 and was sequenced using dye terminator cycle sequencing ready reaction mix (Perkin-Elmer) to ensure that the construction was the wild type. The insert was subcloned into the BamHI-EcoRI sites of either the phagemid, pTZ18U, creating pSE8 to be used as template for oligo-directed mutagenesis of the phoD promoter, or pDH32 (25), creating pSE7 to be transformed into JH642 for promoter-lacZ fusion studies.
Once the phoD promoters were mutated by oligo-directed mutagenesis, they were cut out from the pTZ18U vector by using the BamHI and EcoRI sites found initially on primers FMH208 and FMH209 and cloned into the BamHI and EcoRI sites of pDH32. These new plasmids were then sequenced to ensure the correct genotype and named according to the primer numbers used to mutate their inserts (Table 1).
TABLE 1.
phoD promoter mutations
| phoD-lacZa plasmid or strain | DNA sequenceb | Promoter β-galactosidase activityc |
|---|---|---|
| −35 −25 | ||
| pSE7/MH5801/ wild type | TTCACAGTCGTTTAACA | 1.00 |
| pSE271b/MH5802 | TTCACAGTCGTgTAACA | 0.60 |
| pSE272b/MH5803 | TTCACAGTCGTTgAACA | 0.16 |
| pSE273b/MH5804 | TTCACAGTCGTTTAACg | 0.58 |
| pSE274b/MH5805 | TTCACAGTCGTTTAgCA | 0.22 |
| pSE282b/MH5809 | TTCACAGTCGTTTgACA | 0.14 |
| pSE283b/MH5810 | TTCACAGTCGTTTAAgA | 0.11 |
| pSE287b/MH5811 | TTCACAGTCGTTTAAtA | 0.62 |
| pSE288b/MH5812 | TTCACAGTCGTTcAACA | 0.41 |
| pSE289b/MH5813 | TTCACAGTCGTcTAACA | 0.20 |
| pSE290b/MH5814 | TTCACgGTCGTTTAACA | 0.42 |
| pSE291b/MH5815 | TTCAtAGTCGTTTAACA | 0.88 |
| pSE292b/MH5816 | TTCgCAGTCGTTTAACA | 0.28 |
| pSE293b/MH5817 | TTtACAGTCGTTTAACA | 1.00 |
| pSE294b/MH5818 | TcCACAGTCGTTTAACA | 0.35 |
| pSE295b/MH5819 | cTCACAGTCGTTTAACA | 0.45 |
| pSE330b/MH5827 | TTCAgAGTCGTTTAACA | 0.20 |
| pSE331b/MH5828 | TTgACAGTCGTTTAACA | 0.64 |
| pSE342b/MH5838 | TgCACAGTCGTTTAACA | 0.08 |
| pSE343b/MH5839 | gTCACAGTCGTTTAACA | 0.38 |
| pSE345b/MH5840 | TTCgCAGTCGTTTAgCA | 0.00 |
| pSE296b/MH5820 | TTCAGAGTCGcTTAACA | 0.92 |
| pSE297b/MH5821 | TTCACAGTCaTTTAACA | 0.90 |
| pSE298b/MH5822 | TTCACAGTtGTTTAACA | 1.00 |
| pSE299b/MH5823 | TTCACAGcCGTTTAACA | 0.89 |
| pSE300b/MH5824 | TTCACAaTCGTTTAACA | 0.77 |
| pSE332b/MH5829 | TTCACAGTgGTTTAACA | 1.06 |
| pSE333b/MH5830 | TTCACAGgCGTTTAACA | 0.99 |
| pSE336b/MH5831 | TTCACAGTCGgTTAACA | 1.08 |
| −55 −45 | ||
| Wild type | TTACAATCAGTTCACA | 1.00 |
| pSE348B/MH5843 | TTACAATCAGTTCgCA | 0.29 |
| pSE346b/MH5841 | TTAgAATCAGTTCACA | 0.24 |
| pSE347b/MH5842 | TTgCAATCAGTTCACA | 0.30 |
| −35 −25 | ||
| pSE374b/MH5846 | TTCACAGTCgatcgatcgaGTTTTAACA | 0.03 |
| −45 −35 | ||
| pSE344b/MH5837 | TTCACACTgatcgatcgaTCTTCACA | 0.14 |
| −35 −25 | ||
| pSE373b/MH5845 | TTCACAGTCgatcgGTTTAACA | 0.00 |
| −45 −35 | ||
| pSE341b/MH5836 | TTCACACTgatcgTCTTCACA | 0.00 |
| pSE275b/MH5806 | 5-bp insertion gacag at −90 | 0.17 |
| pSE301b/MH5825 | 10-bp insertion gacaggacag at −90 | 0.52 |
| pSE375b/MH5847 | 3′-199AAAATTCTAGTACACTT-183 5′ to | 0.02 |
| 3′-199AAGTGTACTAGAATTTT-183 5′ | ||
| pSE276b/MH8507 | 5′ Binding region deletion | 0.03 |
| pSE367 | Core binding region deletion |
Transformation of the parent strain, JH642, with the plasmids shown in the table resulted in the strains with wild-type or mutant phoD-lacZ fusions in single copy at the amyE locus.
The phoD nucleotide sequences from −22 to −38, −43 to −58, and −33 to −48 are shown. The wild-type consensus repeats are underlined. The nucleotide substitutions and insertions are in bold and lower case.
Cells containing the various phoD-lacZ fusions were grown in LPDM where β-galactosidase activity was determined every hour for 12 h. The highest level of induction attained before repression was used to calculate the specific activity of each promoter. The table gives an average of three independent assays and expresses them as fractions of wild-type activity.
Deletion of the 5′ binding site of the phoD promoter was accomplished by using PCR to amplify a 289-bp fragment using primers FMH276 (5′ EcoRI site) and FMH209. This fragment was then cloned into pDH32 using the same methods necessary to clone the wild-type phoD promoter construction creating the plasmid pSE276b. Deletion of the core binding region was done by using primers FMH367 (3′ BamHI site) and FMH208 to amplify a 245-bp fragment and ligating it to pCRII creating the plasmid pSE367.
Oligo-directed mutagenesis.
Oligo-directed mutagenesis (Mutagene; Bio-Rad) was done according to the instructions of the manufacturer. Each promoter was then sequenced using dye terminator cycle sequencing ready reaction mix (Perkin-Elmer) for confirmation of the mutant construction.
Growth conditions and β-galactosidase activity.
The inocula for all B. subtilis cultures were grown overnight in high-phosphate defined medium containing 5 mM phosphate (24). The strains were used to inoculate low-phosphate defined medium (LPDM) (8), where optical density at 540 nm growth readings were recorded and β-galactosidase specific activity levels were assayed every hour for 12 h. β-Galactosidase activity was detected using the method of Ferrari et al. (4). One unit was defined as 0.33 μM of o-nitrophenol produced min−1 at 37°C. The specific activity was calculated as activity per milligram of protein. These results are expressed in terms of the fraction of activity observed compared to that of the wild-type strain (Table 1; see Fig. 3a). The final readings were taken at the point in the growth curve where activity was at its highest level before repression of the Pho response by Spo0A∼P (12).
FIG. 3.
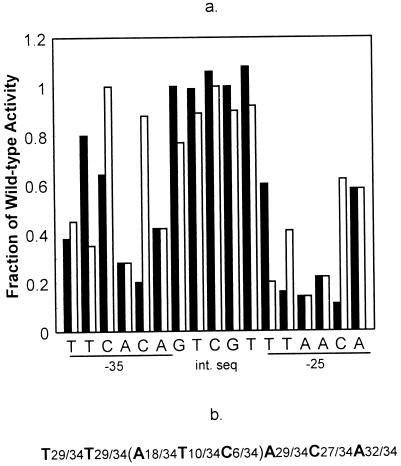
(a) Point mutational analysis of the phoD promoter. The black bars represent the bases in the 3′ half of the core binding region (from the −25 to the −35 consensus repeat) that were individually changed to guanines. The white bars represent all the bases which were changed from purines to purines or pyrimidines to pyrimidines. Plasmids containing these various phoD-lacZ promoter fusions in pDH32 were linearized and integrated into B. subtilis JH642 chromosome at the amyE locus as a result of a double crossover. The strains carrying these various promoter constructions were then grown in LPDM, and the promoter activity was detected every hour for a 12-h growth period. The highest level of induction attained before repression was used to calculate the specific activity of each promoter (12). The figure gives an average of three independent assays. The results are expressed in terms of the fraction of activity observed compared to the wild-type strain. (b) Consensus PhoP binding sequence based on the comparison of 34 TT(A/T)ACA-like repeats in seven different Pho regulon promoters.
Purification of PhoP and *PhoR.
Purification of PhoP and *PhoR (the cytoplasmic region of PhoR) was performed as described previously (15).
DNase I footprinting experiments.
DNase I footprinting experiments were done as previously described by Liu and Hulett (15), except for the concentration of PhoP and PhoP∼P used in the various experiments (see figure legends). Labeling of all probes (the phoD promoter and all mutant derivatives) on the coding strand was accomplished by digesting each plasmid construction with BamHI and filling in the sticky ends with Klenow fragment in the presence of [α-32P]dATP and [α-32P]dCTP. Subsequently, the plasmid was digested with EcoRI, and the insert was separated from the vector using a 6% polyacrylamide gel. The noncoding strands were labeled in the reverse order using [α-32P]dATP.
General methods.
Transformation of E. coli was done according to the method of Hanahan (5). Transformants were selected for drug resistance and color on Luria-Bertani plates containing 150 μg of penicillin ml−1 and 120 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) ml−1. Transformation of B. subtilis was done by the two-step transformation method of Cutting and Vander Horn (2). Transformants were selected for drug resistance on Tryptose Blood Agar Base (Difco) plates containing 5 μg of chloramphenicol ml−1.
Primer extension.
Strain MH5441 was grown in LPDM to stimulate the activation of phosphate starvation-inducible promoters (8). RNA was extracted, and primer extension of the phoD transcript was performed using the method described previously by Chesnut et al. (1). Two primers were used. One was complementary to the bases +63 to +78 (FMH 231), and the other was complementary to the bases +94 to +112 (FMH 232). A sequencing ladder was produced by end labeling each primer with [α-32P]dATP, annealing to pSE9, and using Sequenase (United States Biochemical) according to the instructions of the manufacturer.
RESULTS
Determination of the phoD transcription start site.
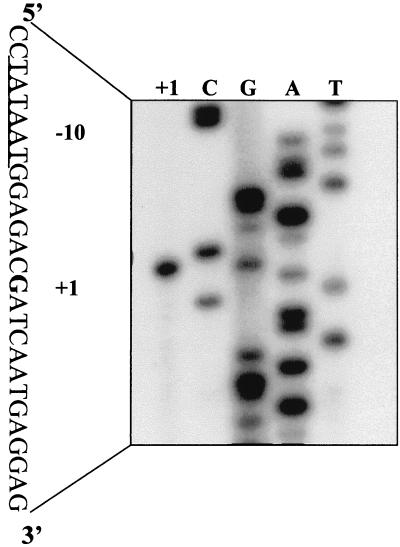
Prior investigation of the regulation and activation of the phoD promoter in B. subtilis showed that phoD transcription was phosphate starvation inducible and regulated by the two-component signal transduction system PhoP and PhoR (3). To determine where transcription initiated at this promoter, we extracted RNA from strain MH5441 grown under low-phosphate conditions and performed primer extension using primers FMH 231 and FMH 232. The transcription initiation site was the same using both primers (Fig. 1) and was situated 26 bp upstream from the putative translation initiation codon.
FIG. 1.
Primer extension analysis of phoD. The end-labeled primer was annealed to RNA from phosphate-depleted vegetative cells grown in LPDM (lane +1) and extended with reverse transcriptase. Lanes C, G, A, and T are a sequencing ladder made by annealing the same end-labeled primer to a plasmid containing the 5′ end of phoD and extending it with Sequenase (United States Biochemical). The +1 indicates the base (shown in bold print) to which the primer extension product maps.
PhoP binds to the phoD promoter.
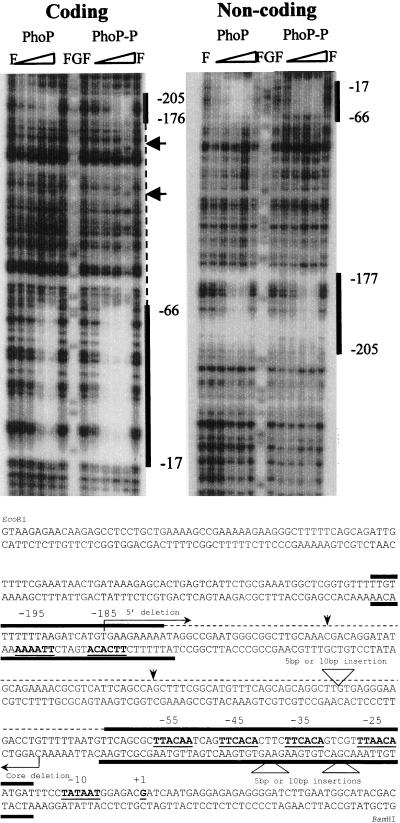
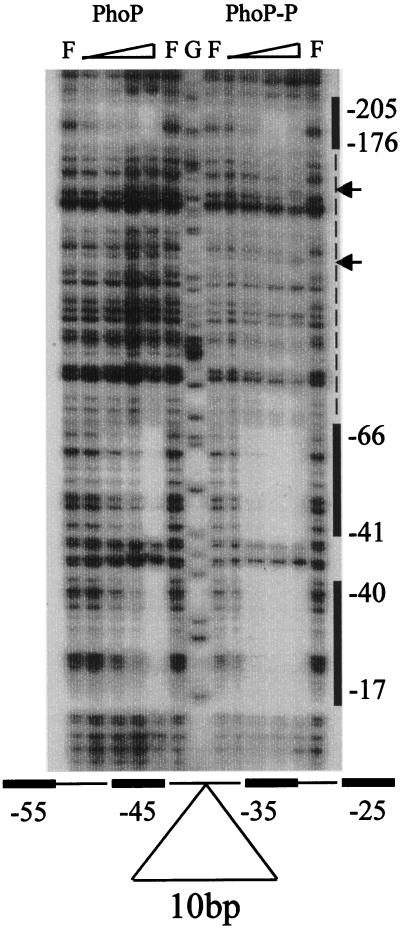
In a previous study showing that phoD was regulated by PhoP and PhoR (3), we used a 365-bp fragment containing the region of the phoD promoter extending from −321 (the numbering is based on the transcriptional start site determined above) to +44. This same promoter fragment was used as a probe to perform DNase I footprinting experiments. Both PhoP and PhoP∼P were able to bind to the phoD promoter on two main regions (Fig. 2A and B). The first region, termed the core binding region, extended from −17 to −66 (coding strand), and the second region (the 5′ binding region) extended from −176 to −205. The core binding region was almost fully protected by unphosphorylated PhoP at a concentration of 220 nM, whereas PhoP∼P was able to fully protect it at a concentration of 55 nM. The 5′ secondary binding region, which contained two TT(A/T)ACA-like repeats, was fully protected by unphosphorylated PhoP at 220 nM but required a higher concentration of PhoP∼P than the core for full protection. It was not fully protected by PhoP∼P until a concentration of 110 nM was used. Similar results were seen in the noncoding strand, both in the length of the binding sites and the concentrations of unphosphorylated PhoP and PhoP∼P necessary for protection. On the coding strand, intermittent protection was observed between the two main binding regions when a concentration of 220 nM PhoP∼P was used in the reaction. Extension of the footprinted regions, however, was not seen on the noncoding strand at these concentrations.
FIG. 2.
(A) DNase I footprint analysis of the phoD promoter bound by PhoP and PhoP∼P. A 365-bp phoD promoter fragment (−321 to +44) was used as the probe. F represents free lanes where no PhoP was used; G represents the G-sequencing reaction lane used as a reference. The reactions for both the coding and noncoding strands contained 0.6 μg (0.6 μM) of ∗PhoR. The amounts of PhoP in each of the reactions from the left to right are as follows: 20 ng (27.5 nM), 40 ng (55 nM), 80 ng (110 nM), and 160 ng (220 nM). When PhoP∼P was needed, a final concentration of 4 mM ATP was added. The vertical solid lines mark the regions on the promoter which were bound by both PhoP and PhoP∼P. The dashed line represents the area on the coding strand where extension of the footprint by PhoP∼P occurred at intermittent places. The hypersensitive sites are indicated by arrows. (B) The phoD promoter sequence showing the PhoP and PhoP∼P binding sites. The coding and noncoding sequence of the 365-bp fragment is shown. The binding sites for both PhoP and PhoP∼P are represented by bold lines either above (coding) or below (noncoding) the sequence. A dashed line indicates the area between the two main binding sites which was intermittently protected when the highest concentration of PhoP∼P (220 nM) was used. The arrows indicate the hypersensitive sites. The 6-bp TT(A/T)ACA-like consensus repeats (shown in bold) are underlined on both strands, and their locations in the promoter are indicated above the coding strand. The −10 and the transcriptional start site are shown in bold, underlined, and labeled. The locations of the various deletion or insertion mutations are marked for reference. The numbering is based on the transcriptional start site established by primer extension.
Point mutations in the PhoP core binding region of the phoD promoter and their effects on promoter activity.
In studies of other Pho regulon promoters, TT(A/T)ACA-like repeats were protected by unphosphorylated PhoP or PhoP∼P (14–17) and two sets of these TT(A/T)ACA-like repeats were defined to be the PhoP core binding unit (16). However, no mutational analysis of these repeats or the intervening sequences between them has been done to establish which bases are necessary for proper PhoP-DNA interaction in any Pho regulon promoters. Using site-directed mutagenesis, we made single-base-pair substitutions in the 3′ half of the proposed core binding region of phoD. This region contains a pair of TT(A/T)ACA-like repeats, with one TT(A/T)ACA repeat centered at bp −25 and the other centered at bp −35 (the numbering is based on the transcription initiation site) with an intervening sequence between them. The sequence, TTCACAGTCGTTTAACA, is found on the coding strand of the promoter. Each base was transitionally mutated. At each position, purines were mutated to the other purine, and pyrimidines were mutated to the other pyrimidine. The strains containing these mutated phoD promoter-lacZ fusions in single copy on the chromosome were grown in LPDM, and β-galactosidase activity was assayed. Our results clearly suggest that mutations of the TT(A/T)ACA-like repeats were deleterious to promoter strength (Table 1 and Fig. 3a). Most of the single point mutations in these repeats reduced promoter activity to less than 40% of wild-type levels. However, while the majority of point mutations in the TT(A/T)ACA-like repeats were deleterious, there appeared to be some flexibility within the recognition sequence, as certain mutations within these repeats were not as detrimental as others. Changing a cytosine to a thymine seemed to have the least effect of any of the point mutations, especially in the third position of the −35 repeat in which TTCACA was changed to TTTACA, bringing the wild-type sequence closer to the previously proposed consensus (16). Similar results were seen in the fifth position of the −35 and the fifth position of the −25 repeats.
To compare the effect of point mutations in the TT(A/T)ACA sequences with those in the intervening sequence, we mutated the intervening sequence in a similar way. Unlike the consensus repeats, however, point mutations in the intervening sequence had little or no effect on promoter strength, with the most deleterious mutation reducing promoter activity by 23%. Closer examination of all Pho regulon promoters verified that the intervening sequences show very little preference for a specific sequence of bases.
A comparison of the known TT(A/T)ACA-like repeats in the Pho regulon revealed the absence of guanine (16). In an effort to assess the effect of the presence of guanine in the core binding sequence on promoter activity, each of the C and T nucleotides within the 3′ half of the core binding region were individually changed to G. The results from this mutational analysis demonstrated that, in the TT(A/T)ACA-like consensus repeats, guanines were also detrimental to promoter activity even in the positions previously shown to have some variability (Fig. 3a). However, these same mutations in the intervening sequence, again, proved to have very little effect on promoter strength.
Three additional point mutations were made in the fourth position of the −45 repeat (TTCACA) and in the third and fourth positions of the −55 repeat (TTACAA) (Table 1). Each of the bases at these positions was changed to a guanine, and their promoter activities were reduced by at least 70% compared with that of the wild-type strain. In addition, a phoD promoter construct was created with point mutations in the fourth positions of the −25 and −35 repeat simultaneously, changing both to guanine. This construct was devoid of all activity.
Point mutations in the PhoP core binding region of the phoD promoter and their effects on PhoP binding activity.
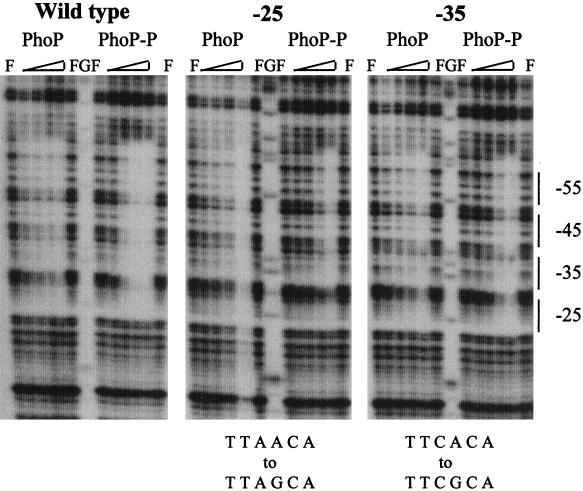
To determine if lower promoter activity was due to a reduction of PhoP’s affinity to the mutant promoters, we compared the footprints of several mutant promoter constructions with the wild-type promoter. Our results show that a single point mutation in the core binding region, whether it was in the −25 or −35 consensus repeat, reduced the affinity of unphosphorylated PhoP and PhoP∼P for the entire core binding region. In Fig. 4, the −25 or −35 repeats were mutated in the fourth position (both from A to G). Full protection of the core binding region in these mutant promoters was not seen until a concentration of 220 nM PhoP∼P was used as opposed to 55 nM PhoP∼P for the wild-type sequence. The 5′ secondary binding region, however, did not appear to be affected (data not shown). In addition to DNase I footprint analysis of the promoters mutated in the TT(A/T)ACA-like consensus repeats, we also assessed changes in PhoP binding to a promoter mutated in the intervening sequence (GTCGT to GGCGT) by comparing it with the wild-type promoter (data not shown). There was a slight decrease in the ability of both unphosphorylated PhoP and PhoP∼P to bind to the mutated promoter. The wild-type promoter was fully protected at a concentration of 55 nM PhoP∼P, whereas the mutated promoter was only partially protected at 55 nM. However, such a difference did not appear to influence promoter activity. Taken together, these data suggest that PhoP∼P must be able to recognize and bind the TT(A/T)ACA-like repeats but not the intervening sequences for full promoter activation.
FIG. 4.
DNase I footprint analysis of the wild-type phoD core binding region versus a −25 or a −35 mutant phoD core binding region using PhoP and PhoP∼P. The mutant phoD promoters in pSE274b (−25, TTAACA to TTAGCA) and pSE292b (−35, TTCACA to TTCGCA) were used as probes, and only the coding strands are shown. The amounts of PhoP, PhoR, and ATP in each reaction were the same as those used for footprinting both the coding and noncoding strands of the wild-type phoD promoter shown in Fig. 2A. The core binding region is marked for reference by a dashed line along with the general location of the TT(A/T)ACA-like repeats.
Effect of base pair insertions in the core binding region on promoter activity and PhoP binding activity.
To determine how changes in spacing between the TT(A/T)ACA-like sequences in the core binding region would affect PhoP binding affinity and promoter activation, we inserted 5 and 10 random base pairs in the intervening sequences between the −25 and −35 repeats or the −35 and −45 repeats, respectively (Fig. 2B). DNase I footprinting analysis of the promoters with the insertions between the −25 and −35 repeats revealed that both mutations were detrimental to PhoP binding affinity. Full protection of the entire core binding region was not observed with the 5-bp insertion mutant until a concentration of 220 nM PhoP∼P was used (data not shown), approximately fourfold more than is required for full protection of the core binding region in the wild-type promoter. Full protection of the core binding region with the 10-bp insertion was nearly achieved at a concentration of 110 nM PhoP∼P (data not shown), more than a twofold difference in binding affinity compared to the wild-type promoter. Unlike the 5-bp insertion sequence, the 10-bp insertion sequence was not protected by PhoP∼P even at the highest concentrations.
The 5-bp insertion between the −35 and −45 repeats had the same effect on PhoP∼P binding affinity as the 5-bp insertion between the −25 and −35 repeats. PhoP∼P was not able to bind the entire core binding region until a concentration of 220 nM was used (data not shown). In contrast, the core binding region was fully protected by 55 nM PhoP∼P in the promoter with the 10-bp insertion between the −35 and −45 repeats (again, the 10-bp insertion sequence was not protected), the same concentration of PhoP∼P needed to protect the core binding region in the wild-type promoter (Fig. 5). These results indicate that the spacing within the core binding region is very important for PhoP to bind efficiently. It also suggests that binding occurs more efficiently when the TT(A/T)ACA-like repeats are on the same face of the helix.
FIG. 5.
DNase I footprint analysis of the phoD promoter with a 10-bp insertion between the −35 and −45 consensus repeats of the core binding region using PhoP and PhoP∼P. The mutant phoD promoter in pSE344b was used as a probe. Only the coding strand was labeled. The amounts of PhoP, PhoR, and ATP in each reaction were the same as those used for footprinting both the coding and noncoding strands of the wild-type phoD promoter shown in Fig. 2A. The labels are also the same as those in Fig. 2A.
To correlate these data with promoter activity in vivo, we used promoter-lacZ fusions. Both 5-bp insertions eliminated all promoter activity. This was consistent with the significantly larger concentrations of PhoP∼P required to bind to these constructions. The 10-bp insertion between the −25 and −35 repeats reduced promoter activity by 97%. This, again, is consistent with PhoP∼P’s reduced binding affinity. In contrast, the 10-bp insertion between the −35 and the −45 repeats reduced promoter activity by 86%, a result which is inconsistent with PhoP∼P binding affinity, suggesting that the positioning of PhoP within the core binding region is important for promoter activity regardless of how well it binds.
The 5′ secondary binding region is necessary for promoter activation.
All other Pho regulon promoters studied to date have either no secondary binding site (15, 16) or a secondary binding site in the coding region (17). The phoD promoter, however, has a secondary binding site 5′ of the core binding region. To determine whether this 5′ secondary binding site is necessary for full promoter activation, we made a deletion mutation, removing this site while leaving the entire sequence between it and the core binding region intact (Fig. 2B). The resulting promoter activity was only 3% of wild-type activity determined by β-galactosidase assays (Table 1), indicating that the additional PhoP binding site was necessary for full promoter activity. Interestingly, the TT(A/T)ACA-like repeats in this binding site are inverted compared with those in the core binding site (Fig. 2B).
To investigate whether PhoP bound to the 5′ binding site must be on the correct face of the helix in order to activate transcription, we inserted five random base pairs 5′ of the core binding region, effectively placing the 5′ PhoP binding site on the opposite face of the helix. As a control, we also inserted 10 bp, returning the repeats to the correct face of the helix but moving them further away from the core. The promoter activity from these two constructs was compared to that of the wild-type phoD promoter. By moving these consensus repeats to the opposite face of the helix, we reduced promoter activity by 83%, while bringing them back around to the proper face of the helix brought the activity back up to 52% of wild-type levels. In addition, we also inverted the 5′ binding sequence through oligo-directed mutagenesis. Promoter activity in this construct was reduced to 2% of wild-type levels. The results (Table 1) indicate that there is some flexibility within the system but that the location of the 5′ binding region, in terms of both the face of the helix and the distance from the core binding region, is important for full promoter activity.
The effect of the 5′ secondary PhoP binding site on PhoP binding to the core binding region.
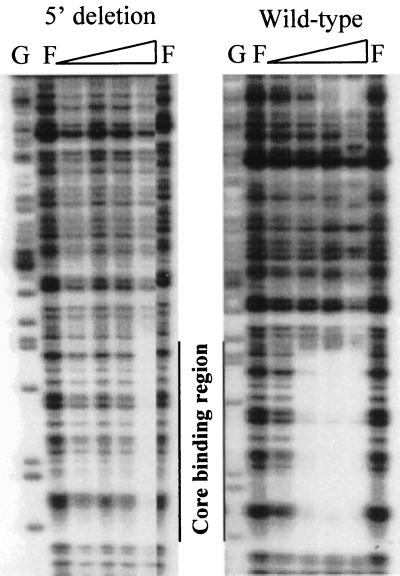
In order to investigate the role of the 5′ binding site in PhoP binding to the core binding region of the promoter, the 5′ deletion mutant promoter was labeled on the coding strand and used as a probe for DNase I footprinting experiments. The core binding region was bound by PhoP∼P but only at a concentration of 220 nM (Fig. 6), nearly four times the amount of PhoP∼P necessary for full protection of the core region in the wild-type promoter. Concurrently, we footprinted the 5′ secondary binding region without the core binding region (Fig. 7). Full protection was achieved at a concentration of 220 nM PhoP∼P, which is approximately double the concentration of PhoP∼P necessary to bind it when the core binding region is present. These results suggest that both regions are required for PhoP to bind efficiently to this promoter.
FIG. 6.
DNase I footprint analysis of the wild-type phoD promoter versus the phoD 5′ binding region deletion mutant promoter using PhoP∼P. The amounts of PhoP, PhoR, and ATP in each reaction were the same as those used for footprinting both the coding and noncoding strands of the wild-type phoD promoter in Fig. 2A. The core binding region is marked for reference.
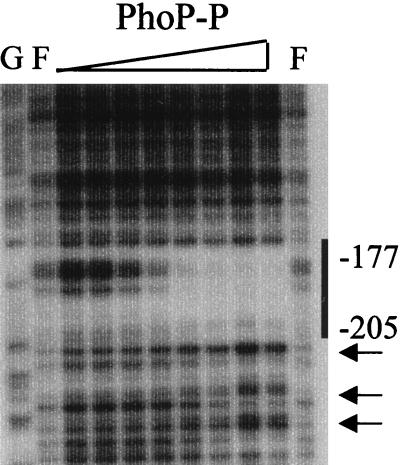
FIG. 7.
DNase I footprint analysis of the core binding region deletion mutant using PhoP∼P. The amounts of PhoR and ATP in each reaction were the same as those used for footprinting both the coding and noncoding strands of the wild-type phoD promoter shown in Fig. 2A. The concentrations of PhoP used in each lane from left to right were 27.5, 55, 110, 165, 220, 275, 330, and 385 nM. Hypersensitive sites are marked with arrows, and the 5′ binding region is marked with a bold line.
DISCUSSION
The 6-bp consensus for PhoP binding derived from Pho promoter sequence alignments or from experimental analysis show good agreement.
Point mutations within the core showed that base pair substitutions within the consensus sequence were deleterious to transcription initiation, generally reducing promoter activity in comparison to the wild-type promoter by more than 60% (Fig. 3a). Similar mutations in the intervening sequence had little or no effect on transcription efficiency. There was some flexibility observed in the consensus repeats, as point mutations at certain positions were not as detrimental as others. A new consensus of T29/34T29/34(A18/34T10/34C6/34)A29/34C27/34A32/34 was apparent (Fig. 3b) upon adding the TT(A/T)ACA-like repeats found in phoD and tagD (14). However, the experimental data and the sequence comparison were not in agreement at the highly conserved cytosine in the fifth position. Promoter function was retained with thymine inserted in that position, a condition found only once among all the Pho promoters studied to date. Further experimental data, showing no enhancement of promoter activity when the phoD TTCACA sequence centered at bp −35 was changed to the previous consensus sequence, TT(A/T)ACA, supports a new consensus sequence, TT(A/T/C)ACA, with the realization that as more Pho regulon genes are discovered, thymine may be interchangeable with cytosine at the fifth position (Fig. 3a). Such findings would support the experimental data and would justify changing the consensus sequence at the fifth position from C to C/T.
PhoP dimers are proposed to bind to one pair of consensus repeats.
PhoP is a member of the OmpR family of response regulators in which the DNA binding motif is a winged helix-turn-helix (20, 21). Members of this family are known for binding direct repeats (6, 19), and it is presumed that the single recognition helix in each DNA binding region makes contact with a specific sequence of bases in the major groove of the double helix. We have previously demonstrated that PhoP and PhoP∼P are dimers in solution (15) and that both protect sequences of DNA containing direct repeats in multiples of two, suggesting that one dimer of PhoP binds to one pair of TT(A/T/C)ACA-like repeats. In this study, we demonstrated that one set of repeats is sufficient for PhoP-DNA binding (Fig. 7). Therefore, the Pho box consisting of two repeats and the core binding region consisting of a minimum of four repeats are two separate entities, the former being a functional binding unit for PhoP and/or PhoP∼P and the latter being necessary to form the protein complex required for transcriptional activation of Pho regulon genes. Based on these assumptions, the core binding region of the phoD promoter, containing four repeats, should theoretically be bound by two dimers.
Pho dimer binding to the core binding region is cooperative.
In direct correlation with the β-galactosidase-specific activity assays, point mutations in the intervening sequence had little effect on PhoP binding affinity, suggesting that the major PhoP∼P-DNA contacts were retained. In contrast, changes in either of the consensus sequences in the pair centered at −25 and −35 were detrimental to PhoP binding at the mutated binding site and importantly, at the adjacent binding site (−45 and −55) demonstrating a high degree of cooperative binding within the core binding region (Fig. 4). Both unphosphorylated PhoP and PhoP∼P were able to bind the mutated core region at higher concentrations, with partial footprints detected at 220 nM PhoP, a concentration of PhoP∼P similar to that required to bind one pair of consensus repeats in the 5′ secondary binding region (Fig. 7).
Proper spacing of the repeats within the core binding region is necessary for promoter activation but not always required for PhoP to bind efficiently.
In vivo transcription assays consistently showed that both 5- and 10-bp insertions in the core binding region were deleterious to promoter activation whether the insertions were between the repeats within a single dimer binding site (−35 to −25) or between two dimer binding sites (−45 to −35). Either 5-bp insertion eliminated promoter activation, whereas either 10-bp insertion still allowed some promoter activity but at a severely reduced level. As promoter activity suggested, either 5-bp insertion severely reduced PhoP’s affinity for the core region, requiring approximately four times the amount of PhoP∼P to bind, a PhoP∼P concentration similar to that required to bind the core binding region with a point mutation in the −35 or −25 repeats. This is particularly interesting because it again points out that cooperative binding is necessary to bind the core efficiently. These data clearly show that the cooperativity can be disrupted either by mutating a single dimer binding site or by changing the face of the helix of the two dimer binding sites. The binding data for the promoter with the 10-bp insertion between the −25 and −35, separating the consensus repeats within a single binding site, also correlated with promoter activity. However, the correlation between promoter activity and PhoP binding was diminished when a 10-bp insertion (between the −35 and −45) separated the two core dimer binding sites (Fig. 5). Although promoter activity was greatly reduced, PhoP∼P’s affinity for the core binding region was not reduced, suggesting that the two PhoP dimers were still able to make contact and bind cooperatively. This may be due to flexibility in the DNA or perhaps in PhoP itself.
Based on the assumption that two dimers are binding the core binding region, we would expect that a 5- or 10-bp insertion within a single dimer binding site (between the −35 and −25) would be more deleterious to PhoP binding than a 5- or 10-bp insertion between two dimer binding sites (−45 and −35 repeats). This was very apparent when comparing the footprints of the two 10-bp insertion mutations. The 10-bp insertion between the −45 and −35 had no effect on binding except for separation of the two dimer binding sites. It took greater than twice as much PhoP to bind the promoter with the 10-bp insertion between the −35 and −25, suggesting that a single dimer binding site had been disrupted.
PhoP binding to the secondary binding site and the core binding region is coordinated.
PhoP binds to the core binding region and the 5′ binding site of phoD in a coordinated fashion. Removal of either region reduces PhoP’s affinity to the remaining site (Fig. 6 and 7). It is possible that a DNA loop may form between the two sites to facilitate PhoP binding to the core binding region for activation of the promoter, since deletion of the 5′ binding site nearly eliminates promoter activity (Table 1). DNA loop formation had previously been proposed when analyzing the secondary binding sites in the coding regions of the phoA and pstS promoters, and it seems reasonable that this mechanism could increase the local concentration of PhoP dimers near the core binding region, ultimately strengthening the promoters (17).
The working model, based on the data presented, suggests that three dimers of PhoP∼P bind to the phoD promoter. Two PhoP dimers bind to the core binding region (−77 to −16) in a cooperative manner, and the exact spacing between dimer binding sites is required for promoter function but is not as restricted for PhoP dimer binding as long as the two sites are on the same face of the helix. Another PhoP dimer binds 5′ of the core binding region (−205 to −177). It is hypothesized that all three dimers bind to their respective regions in a coordinated fashion through DNA loop formation to activate transcription. Such binding may change the local DNA conformation and, through an unknown mechanism, the PhoP∼P oligomers may interact with the RNA polymerase to activate gene transcription by forming an open complex. Nothing is known about how PhoP∼P interacts with the RNA polymerase, e.g., what subunit(s) of the RNA polymerase is involved in the interaction with PhoP∼P oligomers. Based on a sequence comparison of the proposed α-loops in the DNA binding domains of several OmpR family members, PhoP may make contact with the sigma factor, as PhoB does with E. coli ς70 (18). Genetic analysis, such as mapping the specific suppressors of PhoP mutants in the RNA polymerase subunits, the crystal structure of PhoP, or an electron micrograph of the PhoP∼P-DNA interaction, may provide more insight into the PhoP∼P activation mechanism.
ACKNOWLEDGMENTS
We thank L. Shi for the use of his *PhoR. We are also grateful to W. Hendrickson and J. Narita for their critical reading of the manuscript.
This work was supported by the Public Health Service research grant GM33471 from the National Institutes of Health to F.M.H.
REFERENCES
- 1.Chesnut R S, Bookstein C, Hulett F M. Separate promoters direct expression of phoAIII, a member of the Bacillus subtilis alkaline phosphatase multigene family, during phosphate starvation and sporulation. Mol Microbiol. 1991;5:2181–2190. doi: 10.1111/j.1365-2958.1991.tb02148.x. [DOI] [PubMed] [Google Scholar]
- 2.Cutting S M, Vander Horn J A. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for bacillus. New York, N.Y: John Wiley; 1990. pp. 27–74. [Google Scholar]
- 3.Eder S, Shi L, Jensen K, Yamane K, Hulett F M. A Bacillus subtilis secreted phosphodiesterase/alkaline phosphatase is the product of a Pho regulon gene, phoD. Microbiology. 1996;142:2041–2047. doi: 10.1099/13500872-142-8-2041. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari E, Howard S, Hoch J A. Effect of stage 0 sporulation mutants on subtilisin expression. J Bacteriol. 1986;166:173–179. doi: 10.1128/jb.166.1.173-179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning II: a practical approach. Washington, D.C: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 6.Harlocker S L, Bergstrom L, Inouye M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 7.Hulett F M. Complex phosphate regulation by sequential switches in Bacillus subtilis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 289–302. [Google Scholar]
- 8.Hulett F M, Bookstein C, Jensen K. Evidence for two structural genes for alkaline phosphatase in Bacillus subtilis. J Bacteriol. 1990;172:735–740. doi: 10.1128/jb.172.2.735-740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulett F M, Kim E E, Bookstein C, Kapp N, Edwards C W, Wycoff H W. Bacillus subtilis alkaline phosphatase III and IV. Cloning, sequencing, and comparisons of deduced amino acid sequence with Escherichia coli alkaline phosphatase three dimensional structure. J Biol Chem. 1991;266:1348–1358. [PubMed] [Google Scholar]
- 10.Hulett F M, Lee J, Shi L, Sun G, Chesnut R, Sharkova E. Sequential action of two-component genetic switches regulates the Pho regulon in Bacillus subtilis. J Bacteriol. 1994;176:1348–1358. doi: 10.1128/jb.176.5.1348-1358.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulett F M, Sun G, Liu W. The Pho regulon of Bacillus subtilis is regulated by the sequential action of two genetic switches. In: Torriani-Gorini A, Yogil E, Silver S, editors. Phosphate in microorganisms: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1994. pp. 50–54. [Google Scholar]
- 12.Jensen K, Sharkova E, Duggan M F, Qi Y, Koide A, Hoch J A, Hulett F M. Bacillus subtilis transcription regulator, Spo0A, decreases alkaline phosphatase levels induced by phosphate starvation. J Bacteriol. 1993;175:3749–3756. doi: 10.1128/jb.175.12.3749-3756.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang W K, Glassy K, Archibald A R. Influence of phosphate supply on teichoic acid and teichuronic acid content of Bacillus subtilis cell walls. J Bacteriol. 1982;151:367–375. doi: 10.1128/jb.151.1.367-375.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Eder S, Hulett F M. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP∼P. J Bacteriol. 1998;180:753–758. doi: 10.1128/jb.180.3.753-758.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Hulett F M. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J Bacteriol. 1997;179:6302–6310. doi: 10.1128/jb.179.20.6302-6310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Hulett F M. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology. 1998;144:1443–1450. doi: 10.1099/00221287-144-5-1443. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Qi Y, Hulett F M. Sites internal to the coding regions of phoA and pstS bind PhoP and are required for full promoter activity. Mol Microbiol. 1998;28:119–130. doi: 10.1046/j.1365-2958.1998.00779.x. [DOI] [PubMed] [Google Scholar]
- 18.Makino K, Amemura M, Kawamoto T, Kimura S, Shingawa H, Nakata A, Suzuki M. DNA binding of PhoB and its interaction with RNA polymerase. J Mol Biol. 1996;259:15–26. doi: 10.1006/jmbi.1996.0298. [DOI] [PubMed] [Google Scholar]
- 19.Makino K, Shinagawa H, Amemura M, Kimura S, Nakata A, Ishihama A. Regulation of the phosphate regulon of Escherichia coli, activation of pstS transcription by PhoB protein in vitro. J Mol Biol. 1988;203:85–95. doi: 10.1016/0022-2836(88)90093-9. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure. 1996;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Hackert E, Stock A M. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269:301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 22.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 23.Qi Y, Hulett F M. PhoP∼P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP∼P activator sites within the coding region stimulate transcription in vivo. Mol Microbiol. 1998;28:1187–1197. doi: 10.1046/j.1365-2958.1998.00882.x. [DOI] [PubMed] [Google Scholar]
- 24.Qi Y, Kobayashi Y, Hulett F M. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the Pho regulon. J Bacteriol. 1997;179:2534–2539. doi: 10.1128/jb.179.8.2534-2539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimotsu H, Kuroda M I, Yanofsky C, Henner D J. Novel form of transcription attenuation regulates expression of the Bacillus subtilis tryptophan operon. J Bacteriol. 1986;166:461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive response regulators in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stock J B, Stock A M, Mottenen J M. Signal transduction in bacteria. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 28.Sun G, Birkey S M, Hulett F M. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol Microbiol. 1996;19:941–948. doi: 10.1046/j.1365-2958.1996.422952.x. [DOI] [PubMed] [Google Scholar]
- 29.Yamane K, Maruo B. Alkaline phosphatase possessing alkaline phosphodiesterase activity and other phosphodiesterases in Bacillus subtilis. J Bacteriol. 1978;134:108–114. doi: 10.1128/jb.134.1.108-114.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamane K, Maruo B. Purification and characterization of extracellular soluble and membrane-bound insoluble alkaline phosphatases possessing phosphodiesterase activities in Bacillus subtilis. J Bacteriol. 1978;134:100–107. doi: 10.1128/jb.134.1.100-107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]