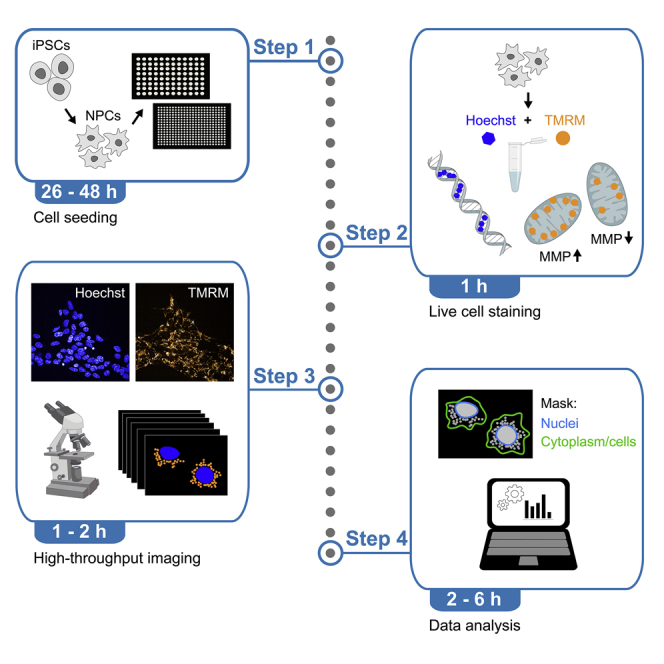
Summary
We present a high-content screening (HCS) protocol for quantifying mitochondrial activity in live neural cells from human induced pluripotent stem cells (iPSCs). The assessment is based on mitochondrial membrane potential, which is influenced by the efficiency of mitochondrial bioenergetics. We describe how to perform the analysis using both an HCS platform and the open-source software CellProfiler. The protocol can identify the mitochondrial fitness of human neurons and may be used to carry out high-throughput compound screenings in patient-derived neural cells.
For complete details on the use and execution of this protocol, please refer to Lorenz et al. (2017) and Zink et al. (2020).
Subject areas: Cell Biology, Cell-based Assays, Metabolism, Microscopy, Neuroscience, Stem Cells
Graphical abstract
Highlights
-
•
High-content screening (HCS) of live neural cells derived from human iPSCs
-
•
Assessment of mitochondrial activity based on mitochondrial membrane potential (MMP)
-
•
Quantification using automated microscopic image analysis
-
•
Data analysis using both an HCS system and the open-source software CellProfiler
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
We present a high-content screening (HCS) protocol for quantifying mitochondrial activity in live neural cells from human induced pluripotent stem cells (iPSCs). The assessment is based on mitochondrial membrane potential, which is influenced by the efficiency of mitochondrial bioenergetics. We describe how to perform the analysis using both an HCS platform and the open-source software CellProfiler. The protocol can identify the mitochondrial fitness of human neurons and may be used to carry out high-throughput compound screenings in patient-derived neural cells.
Before you begin
The protocol described here can be applied to various neural cell types differentiated from human induced pluripotent stem cells (iPSCs). Before starting differentiation, it is important to carefully monitor the genetic integrity of the iPSC lines to be used. This aspect is crucial to ensure that no unwanted abnormalities might have occurred during iPSC culture (Rossi et al., 2022). Proper health of iPSC lines should be ensured by growing cells with appropriate medium and coating material. In our hands, it is helpful to grow iPSCs in incubators with low oxygen concentration (5%). It is also crucial to always monitor the absence of mycoplasma contamination.
With respect to differentiated neural cells, we focus here on neural progenitor cells (NPCs). In fact, NPCs are easier to expand than differentiated neurons, and at the same time already rely on mitochondrial respiration (Lorenz et al., 2017). Homogenous NPC cultures can be obtained from iPSCs in about 4–6 weeks using previously published protocols (Reinhardt et al., 2013; Zink et al., 2021). NPCs generated using this protocol express typical NPC markers including NESTIN, DACH1, SOX2, and PAX6, and the proliferation marker Ki-67. These NPCs retain their multipotent identity over multiple passages (>30), have the potential to form both neural tube and neural crest lineages, and can be effectively differentiated into motor neurons and dopaminergic neurons (Reinhardt et al., 2013). Beside NPCs, it is possible to apply the protocol described here also to other types of neural cells derived from human iPSCs. For example, we successfully employed the HCS protocol to screen for drugs affecting mitochondrial polarization in dopaminergic neurons from iPSCs derived from controls and patients (Zink et al., 2020). For this protocol we used NPCs generated from a published human iPSC line (Wang et al., 2018). This protocol can be used in combination with the quantification of neuronal morphology described in Lickfett et al., 2022.
Institutional permissions
Studies with control iPSCs were performed in accordance with the approval by the Ethic Committee of the Medical Faculty of Heinrich Heine University (study number: 2020-967_2).
Preparation of cell culture materials and buffers
Timing: 1 h
CRITICAL: Adhere to the necessary precautions when working with human samples. They should be handled in a biological safety cabinet. When experimenting with human iPSC lines, comply with relevant local institutional and government regulations.
Prepare cell culture media according to the information provided in the tables below or use media following other standard protocols.
NPC basal medium (sm−)
| Medium | Component | Amount | Stock con. | Final conc. |
|---|---|---|---|---|
| Basal medium for NPCs (sm-) | DMEM/F12 | 240 mL | 1 × | 0.5 × |
| Neurobasal | 240 mL | 1 × | 0.5 × | |
| N2 | 2.5 mL | 100 × | 0.5 × | |
| B27 w/o Vitamin A | 5 mL | 50 × | 0.5 × | |
| Glutamine | 5 mL | 200 mM | 2 mM | |
| MycoZap | 1 mL | 500 × | 1 × | |
| [Pen/Strep∗] | [5 mL] | [100 ×] | [1 ×] |
Note: sm- basal medium can be stored for two weeks at 2°C–8°C.
NPC growth medium (sm+)
| Medium | Component | Amount | Stock con. | Final conc. |
|---|---|---|---|---|
| Growth medium for NPCs (sm+) | sm- | 10 mL | 1 × | 1 × |
| CHIR99021 (Glycogen synthase kinase (GSK)-3 inhibitor) | 5 μL | 6 mM | 3 μM | |
| Purmorphamine | 7.69 μL | 0.65 mM | 500 nM | |
| Vitamin C | 7.5 μL | 200 mM | 150 μM |
Note: sm+ growth medium can be stored for one week at 2°C–8°C.
NPC basal medium (sm−) for live-cell imaging
| Medium | Component | Amount | Stock concentration | Final concentration |
|---|---|---|---|---|
| Phenol red-free basal medium for live-cell imaging of NPCs | DMEM/F12 without phenol red | 240 mL | 1 × | 0.5 × |
| Neurobasal without phenol red | 240 mL | 1 × | 0.5 × | |
| N2 | 2.5 mL | 100 × | 0.5 × | |
| B27 w/o Vitamin A | 5 mL | 50 × | 0.5 × | |
| Glutamine | 5 mL | 200 mM | 2 mM | |
| MycoZap | 1 mL | 500 × | 1 × | |
| [Pen/Strep∗] | [5 mL] | [100 ×] | [1 ×] |
Note: Phenol red-free basal medium can be stored for two weeks at 2°C–8°C.
NPC growth medium (sm+) for live-cell imaging
| Medium | Component | Amount | Stock concentration | Final concentration |
|---|---|---|---|---|
| Phenol red-free growth medium for live-cell imaging of NPCs | Sm- without phenol red | 10 mL | 1 × | 1 × |
| CHIR99021 | 5 μL | 6 mM | 3 μM | |
| Purmorphamine | 7.69 μL | 0.65 mM | 500 nM | |
| Vitamin C | 7.5 μL | 200 mM | 150 μM |
Note: Phenol red-free growth medium can be stored for one week at 2°C–8°C.
Note: All media should be sterile-filtered and prepared under sterile conditions. Before usage, pre-warm the media to room temperature (20°C–25°C).
∗Note: We supplement our cell culture media with MycoZap Plus-CL antibiotics to minimize the loss of valuable cells due to contamination (including mycoplasma). The use of MycoZap-Plus-CL replaces Penicillin and Streptomycin (Pen/Strep) but can still be used in parallel. At the same time, it is essential to always work under clean and sterile conditions, and to perform routine tests to monitor the absence of mycoplasma in the cultures. For mycoplasma testing, we use PCR analysis of cell supernatants or the test kit Venor®GeM OneStep from MB Minerva Biolabs (Cat. No. 11-8100).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Accutase™ Cell Dissociation Reagent | Gibco | A1110501 |
| Antimycin A, 25 mM | Sigma-Aldrich | A8674 |
| B27 w/o Vitamin A | Gibco | 12587010 |
| CHIR99021 | Sigma-Aldrich | SML1046 |
| DMEM/F-12 | Gibco | 31330038 |
| FCCP, 25 mM | Sigma-Aldrich | C2920 |
| Geltrex | Gibco | A1413201 |
| Glutamine | Gibco | 25030149 |
| Hoechst 33342 | Life Technologies | H3570 |
| Matrigel, Growth Factor Reduced | BD Biosciences | 356231 |
| MycoZap | Lonza | VZA-2012 |
| N2 | Gibco | 17502-048 |
| Neurobasal | Gibco | 21103-049 |
| Purmorphamine | Sigma-Aldrich | 540220 |
| TMRE | Invitrogen/Molecular Probes | T-669 |
| TMRM | Invitrogen/Molecular Probes | T-668 |
| Vitamin C | Sigma-Aldrich | A4034 |
| Software and algorithms | ||
| CellProfiler 4.2.1 | Broad Institute | www.cellprofiler.org |
| Columbus 2.9.0 | PerkinElmer | n/a |
| GraphPad Prism 5.01 | GraphPad Software | https://www.graphpad.com/ |
| Critical commercial assays | ||
| Venor®GeM OneStep | MB Minerva Biolabs | 11-8100 |
| Other | ||
| 384-well Cell Culture Microplate, PS, F-Bottom, black TC, μCLEAR, 384-well black, clear bottom |
Greiner Bio-One | 781091 |
| 96-well Cell Culture Microplate, PS, F-Bottom, black TC, μCLEAR, 96-well black, clear bottom |
Greiner Bio-One | 655090 |
| Operetta CLS™ | PerkinElmer | https://www.perkinelmer.com/de/category/operetta-cls-high-content-analysis-system |
| PhenoPlate 384-well, black, optically clear flat-bottom, tissue-culture treated, lids | PerkinElmer | 6057302 |
| Neubauer hemocytometer | VWR | 6310696 |
| Deposited data | ||
| Pipeline and raw images | GitHub | https://github.com/StemCellMetab/Mitochondrial-membrane-potential (https://doi.org/10.5281/zenodo.6794537) |
| Experimental models: Cell lines | ||
| Human: NPCs from iPSC line HMGUi001-A | Wang et al. | https://hpscreg.eu/cell-line/HMGUi001-A |
Materials and equipment
High-content imaging system
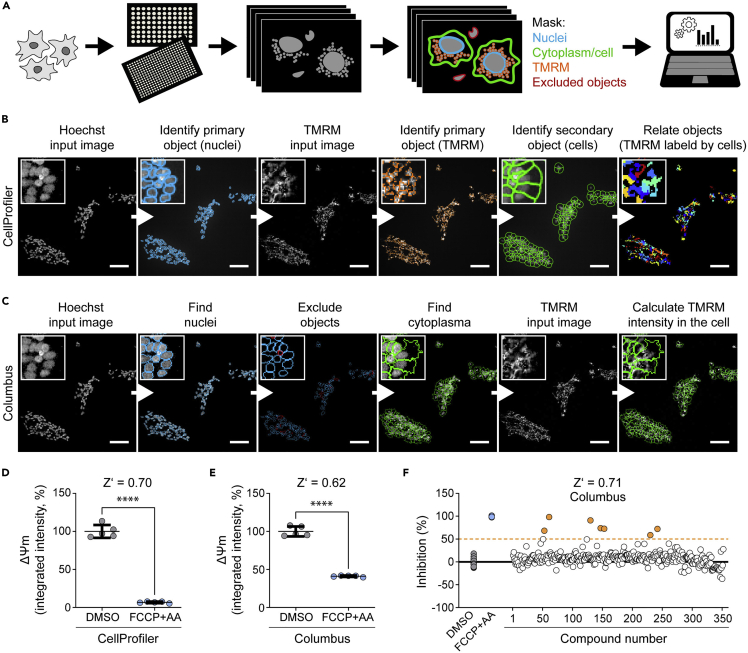
For HCS, we here describe the use of the Operetta CLS High-Content Analysis System (Manufacturer PerkinElmer, https://www.perkinelmer.com/de/Product/operetta-cls-system-hh16000000) controlled by the software Harmony™ (PerkinElmer). Following image acquisition, data analysis can be performed using different approaches. Here, we describe two different pipelines. The first one is based on the open-source software CellProfiler (www.cellprofiler.org). The use of such external software may be ideal for laboratories that do not have access to expensive HCS software and are faced with limited computational possibilities. The second pipeline is based on the image data storage and analysis system Columbus™ (PerkinElmer). This system is more user-friendly, and therefore the establishment of new analysis routines requires less time investment. In order to compare the two pipelines, we show here results obtained by running the same datasets using the two approaches (Figure 1). The comparable results confirm the reproducibility of the HCS protocol, and demonstrate how the protocol outcome should not be influenced by the type of software used for data analysis.
Automated imaging of mitochondrial membrane potential
| Cellular target | Channel | Dye/antibody |
|---|---|---|
| Nucleus | 1 | Hoechst 33342 (Invitrogen) |
| Mitochondrial membrane potential (ΔΨm) | 2 | TMRM (Invitrogen/Molecular Probes T-668) |
Figure 1.
High-content screening (HCS) of mitochondrial membrane potential (ΔΨm) in iPSC-derived neural progenitor cells (NPCs)
(A) Schematic of the HCS protocol steps, including the mask established for the detection of ΔΨm in live-cells.
(B) Flow of imaging analysis using CellProfiler software. Scale bar: 100 µm.
(C) Flow of imaging analysis using Columbus software. Scale bar: 100 µm.
(D) ΔΨm intensity evaluated using the CellProfiler image analysis pipeline. Robustness of the HCS protocol was calculated based on Z′ score (for effective high-throughput assays, values should be between 0.5 and 1). Mean ± standard deviation (SD), unpaired Student’s t-test, ∗∗∗∗p<0.0001.
(E) ΔΨm intensity evaluated using the Columbus image analysis pipeline. Mean ± SD, unpaired Student’s t-test, ∗∗∗∗p < 0.0001.
(F) Compound screening performed in NPCs differentiated from human iPSCs based on the HCS protocol described here. Blue dots indicate the positive control samples treated with FCCP and antimycin A (AA). These drugs are known to decrease ΔΨm and their values are considered as 100% inhibition of ΔΨm. Orange dots indicate screened compounds that can lead to 50%–100% inhibition of ΔΨm.
Step-by-step method details
Cell seeding on the assay plate for HCS
Timing: 26–48 h
In this step, the seeding of the cells on the assay plate is described.
-
1.Preparation of HCS plates (∼2–24 h).
-
a.Coat a black wall, clear F-bottom 96- or 384-well plate with appropriate coating solution. We recommend using Geltrex (1:100 dilution).
-
a.
CRITICAL: The use of transparent clear bottom microplates is essential for this assay. To determine the appropriate microplate type to be used, it is helpful to check beforehand which microplates are already calibrated with the correct dimensions in the instrument. Further helpful information on specific requirements and plate considerations can be found here: https://www.perkinelmer.com/de/lab-products-and-services/application-support-knowledgebase/microplates/high-content-screening-plates.html.
Alternatives: Other coating reagents may also be used. Instead of Geltrex, it is also possible to use Matrigel (200 μg/mL). However, Matrigel is not a defined material, so it may be possible to observe batch-related variability. We obtained comparable results also using poly-l-ornithine (50 μg/mL on glass; 20 μg/mL on polystyrene) plus laminin (10 μg/mL on glass; 5 μg/mL on polystyrene).
Note: Follow manufacturer’s guidelines for coating with Matrigel (https://www.corning.com/worldwide/en/products/life-sciences/products/surfaces/matrigel-matrix.html).
-
2.Seeding of iPSC-derived neural cells (∼ 24 h).
-
a.Split the cells using 0.5 mL Accutase for 6-well plates for 5 min at room temperature (RT) (20°C–25°C).
-
b.Dilute the Accutase to reduce its activity by adding at least double amount of culture media.
-
c.Transfer the cells into a 15 mL falcon tube.
-
d.Centrifuge for 3 min at 120 × g, remove the supernatant, and resuspend the cells in culture media.
-
e.Count the cells using a hemocytometer or cell counter.
-
f.Pipette 100 μL cell suspension containing 30,000–50,000 cells per well of a 96-well plate.
-
g.Place the plate in a tissue culture incubator for 24 h.
-
a.
CRITICAL: Optimal seeding density might need to be adapted according to the cell type used. Very dense cultures make HCS image analysis difficult, as the individual cell identification becomes more challenging. When using NPCs, we recommend using 30,000–50,000 cells per well of 96-well plates (90,000–150,000 cells∗cm-2) and 13,000–16,000 cells per well for 384-well plates (130,000–150,000 cells∗cm-2) (Zink et al., 2020).
Note: The described NPCs retain their multipotent identity over multiple passages and can therefore be used in a wide passage range. We generally use NPCs in the range of 10–25 passages.
Staining procedure for monitoring mitochondrial membrane potential
Timing: 1 h
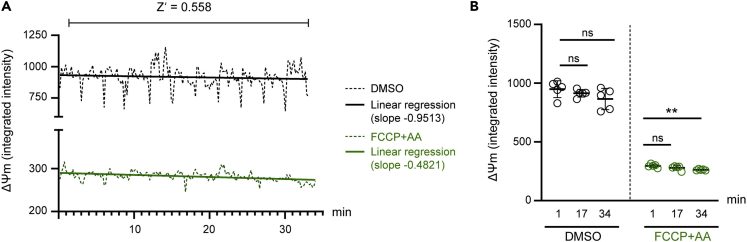
To detect changes in ΔΨm in live cells, iPSC-derived neural cells are stained with the potentiometric fluorescent dye tetramethylrhodamine methyl ester (TMRM). This dye is a lipophilic cation that accumulates across plasma membranes and within the mitochondrial matrix in an inverse proportion to the mitochondrial membrane potential according to the Nernst equation. TMRM accumulation in the mitochondria is dependent on both the plasma membrane potential and the mitochondrial membrane potential. Based on the surface-to-volume ratio, equilibration of TMRM across the mitochondrial membrane occurs more rapidly than across the plasma membrane (Nicholls, 2006). At high concentration, TMRM forms non-fluorescent aggregates within the mitochondrial matrix. Therefore, high concentration of TMRM should be avoided, and the dye should be used in non-quenching mode, which is considered to be the case for concentrations below 30 nM (Ward et al., 2007). The incorporation of the dye indicates active mitochondria and does not affect mitochondrial function. Several papers describe the use of TMRM to analyze ΔΨm with slight differences in the staining method (Zimmermann et al., 2008; Perry et al., 2011; Nicholls, 2018). In our protocol, iPSC-derived NPCs are stained with 10 nM TMRM for 30 min, and then washed before imaging. In our hands, this is the most robust approach, as during imaging the ΔΨm signal remains relatively stable for 30 min (Figure 2). For imaging experiments exceeding 30 min, we recommend to not wash out TMRM before the analysis but instead to dilute the TMRM concentration by adding more media to each well. This approach may allow the TMRM dye to remain in equilibrium for a longer time window (Iannetti et al., 2016). Lastly, given the influence of plasma membrane potential on TMRM accumulation, we advise to not use TMRM in case there might be indications that the plasma membrane potential is changing during the specific experimental set-up. In these cases, alternative strategies to TMRM should be applied (Nicholls, 2018).
-
3.Prepare TMRM stocks.
-
a.Prepare a stock solution of TMRM at a concentration of 25 mM in dimethylsulfoxide (DMSO).
-
a.
Alternatives: Instead of TMRM, it is also possible to use TMRE (Invitrogen/Molecular Probes T-669).
Note: Protect TMRM solution from light, and store sealed at −20°C. Prepare further dilutions in media fresh before each experiment.
-
4.Prepare staining solution.
-
a.Add 10 nM TMRM and 1 μg/mL Hoechst 33342 dye to the culture media.
-
a.
CRITICAL: The concentration of TMRM should be adjusted depending on the neural cell type used. For iPSC-derived NPCs, we carried out dose-dependent experiments and determined 10 nM as the lowest concentration of TMRM showing a reliable signal for HCS.
-
5.For control wells, prepare the staining solution containing ΔΨm modulators: FCCP and antimycin A (collectively defined as F+AA).
-
a.Add 10 nM TMRM, 1 μg/mL Hoechst 33342 dye, 10 μM FCCP and 10 μM antimycin A to the culture media.
-
a.
Note: FCCP and antimycin A lead to ΔΨm depolarization and therefore serve as internal controls for the HCS assay. FCCP and antimycin A are dissolved in DMSO as 25 mM stock solutions. Adjust FCCP and antimycin A concentrations depending on the neural cell type used if needed.
Alternatives: In addition to F+AA, other mitochondrial inhibitors and modulators may be included to support ΔΨm quantification (Nicholls, 2018). For example, rotenone can be used to lower ΔΨm by inhibiting mitochondrial complex I, oligomycin to increase ΔΨm and prevent reversal of ATP synthase that may lead to unwanted usage of cytosolic ATP, and cyclosporine A to mitigate the opening of the mitochondrial permeability transition pore (mPTP).
-
6.
Include DMSO controls accordingly.
CRITICAL: Wear protective clothing, gloves, and eye/face protection when handling DMSO or solutions containing DMSO. DMSO irritates the skin and eyes. Dispose DMSO in accordance with applicable regional, national, and local laws and regulations.
Optional: For high-throughput compound screenings, we recommend to add the compounds to the iPSC-derived neural cells prior to the staining, and then incubate the drugs for 1–24 h. The incubation time depends on the respective compounds and may need to be experimentally validated and adjusted accordingly. Following appropriate incubation, neural cells can be stained by adding 2 × TMRM and Hoechst (in medium) directly to the cells (without aspiration of the old medium) to obtain a final TMRM concentration of 10 nM and 1 μg/mL Hoechst 33342. After addition of the staining solution, continue with step 9.
-
7.
Aspirate the medium and wash once with PBS without MgCl2/CaCl2.
-
8.
Incubate the iPSC-derived neural cells with 50 μL live-cell staining solution for 96-well plates and 30 μL for 384-well plates (with and without ΔΨm modulators) and incubate for 30 min at 37°C, 5% CO2.
-
9.
Turn on the HCS imaging platform. If possible, set the temperature to 37°C and the CO2 level to 5%. If CO2 control is not available, ensure that the imaging media contains an alternative pH buffer such as HEPES.
-
10.
After the incubation time, remove the live-cell staining solution and gently wash 1–2 times with pre-warmed (RT) PBS without MgCl2/CaCl2.
-
11.
Add 100 μL for 96-well plates and 30 μL for 384-well plates of phenol red-free cell culture media appropriate for your cell type of use (e.g., for NPCs see media list above).
CRITICAL: In our hands, the TMRM signal is relatively stable for 30 min when washed out and kept without temperature control (Figure 2). In case of time-dependent variability or lack of comparable and robust TMRM signal intensity across the plate, we recommend to perform image acquisition without complete washing of TMRM and to dilute the concentration of the dye by adding imaging medium (Iannetti et al., 2016) (see also troubleshooting problem 4).
-
12.
Proceed to step 13 (HCS imaging) immediately for HCS.
Figure 2.
Kinetic assessment of ΔΨm
(A) Continuous monitoring of the ΔΨm over time and assessment of the robustness of the assay (Z′ score) in NPCs treated with DMSO or with FCCP and antimycin A (AA). iPSC-derived NPCs were stained with 10 nM TMRM (in non-quenching mode) for 30 min followed by a washing step and HCS-based quantification of ΔΨm.
(B) Quantification of ΔΨm intensity based on the TMRM signal in iPSC-derived NPCs treated with DMSO or with FCCP and antimycin A (AA) at dedicated time points. Mean ± SD, unpaired Student’s t-test, ∗∗p < 0.01.
HCS imaging
Timing: 1–2 h (depending on image acquisition settings)
This step describes the image acquisition using the Operetta CLS High Content Analysis System (PerkinElmer).
-
13.
Place the plate in the HCS imaging platform.
-
14.Define the plate type (e.g., 96- or 384-well plate based on step 1.a).
-
a.Check manufacturer comments on plate usage.
-
a.
-
15.
Select the appropriate light source and filter: for instance, detection at wavelength ranges from 365–465 nm for Hoechst 33342 and 550–610 nm for Alexa Fluor 568 / TMRM.
Alternatives: Instead of Alexa 568, comparable results can be obtained using Cy3.
-
16.
Choose operational mode non-confocal (wide-field fluorescence).
-
17.Select the objective.
-
a.See the notes section for advice on which objective to use.
-
a.
Note: For live-cell imaging, always apply the control of temperature and CO2 if possible.
Alternatives: It is possible to select confocal as operational mode. For measuring TMRM intensity, the non-confocal wide-field fluorescence mode is sufficient. However, for other HCS experiments (for example to assess detailed mitochondrial structure) confocal imaging may be crucial.
Note: For image acquisition of an entire 96- or 384-well plate, we recommend using the 20 × objective, as it reduces the image acquisition time. In our hands, the 20 × objective (air or water immersion) provides most robust results.
-
18.Determine microscope settings, exposure time, and Z-set-off for additional Z-positions. We suggest binning of 2 × 2, which reduces noise and the image size and increases acquisition speed, even if it lowers the digital resolution.
-
a.Select a well containing iPSC-derived neural cells and take a snapshot. Find focus height using the Z-stack. The Z-stack heights may need to be optimized. Start at negative values and go up to positive until cells are in focus. Set the appropriate focus height for each channel individually.
-
b.Evaluate brightness and set exposure times for each channel individually.
-
a.
CRITICAL: Set up the image acquisition based on a well containing control cells. However, check the settings also in the wells treated with ΔΨm modulators (or test compounds) in order to cover the dynamic range of the experiments and avoid over- or underexposure. Different drugs may cause big changes in ΔΨm and TMRM fluorescence. Hence, imaging should be tuned to avoid signal saturation and take into account the expected dynamic range of the experimental set up.
-
19.
Choose a different well to confirm or adjust settings and exclude plate-tilting.
Note: Accidental tilting of the plate could lead to out-of-focus images on one side of the plate. Ensure that the plate is placed correctly in the plate holder.
-
20.Define the plate layout.
-
a.Select scan area and number of fields.
-
b.Determine the appropriate number of fields needed to represent the well (e.g., 8 fields in a single well of a 96-well plate).
-
a.
-
21.
Start the run.
Note: Maximal excitation for TMRM may vary depending on the concentration of the dye in the mitochondria and the solvents used.
Note: Choosing “all fields” in plate layout definition will prolong the time of measurement.
Note: Avoid using the outer rows and outer columns of any given HCS plate due to edge effects (e.g., thermal evaporation and differences near the plate edge). The outer rows and outer columns should not be left blank but should be filled with buffer solution or PBS (at least the same volume as the wells containing cells, e.g., 100 μL for 96-well plates).
HCS analysis of the mitochondrial membrane potential using open-source software (CellProfiler)
Timing: ∼2–6 h (depending on the computer capacity and the number of images to be processed)
This step describes the HCS image analysis for the quantification of ΔΨm in iPSC-derived neural cells based on the open-source software CellProfiler (Figures 1A, 1B and 1D), which is also suitable to quantify images from high-throughput experiments. The image processing time is based on the number of images acquired per field and the computing capacity. In our hands, 4–8 images (fields of view) per well are sufficient to generate robust data. When using the Operetta CLS, 16-bit images are generated with a pixel resolution of 2160 × 2160 resulting in 8–10 GB imaging data per 96-well plate (no outer row image acquisition).
The pipelines and examples images that can be used to try out our HCS protocol can all be downloaded here: https://github.com/StemCellMetab/Mitochondrial-membrane-potential. We also included a video showing all the steps of the CellProfiler analysis pipeline.
-
22.
Install CellProfiler (https://cellprofiler.org/releases) and download the analysis pipeline (CellProfiler-Pipeline_MMP_20×.cpproj), and pipelines for illumination correction (CorrectIlluminationCalculate.cpproj, ApplyIllumCorr_DNA.cppoj, AplyIllumCorr_TMRM.cpproj) from GitHub (https://github.com/StemCellMetab/Mitochondrial-membrane-potential).
-
23.
Open the “CorrectIlluminationCalculate” pipeline.
Note: For measuring intensity, correction of the illumination variation can be important. Due to the optics of the microscope, or imperfection in the slide or sensor bias, the intensity within a fluorescence image can vary by more than two-fold across the field of view. By applying the “CorrectIlluminationCalculate” and “CorrectIlluminationApply” modules, it is possible to compensate for non-uniformities in illumination. If correction of the illumination is not needed, move on to step 39.
-
24.
Add the images to be analyzed by dragging and dropping them into CellProfiler.
-
25.Go to “Metadata”.
-
a.Enter the extraction method and metadata source. To set an expression to enable CellProfiler to interpret metadata from a file name or folder click on the magnifying glass. Adjust the regular expression (“Regex”). For further help on defining the regular expression syntax click on the “?” next to the magnifying glass.
-
b.Click “Submit”.
-
c.Click “Update” to extract and display the metadata in the CellProfiler window.
-
a.
-
26.Go to “NamesAndTypes”.
-
a.Type in a name to assign to the images.
-
a.
-
27.Go to “CorrectIlluminationCalculate” and adjust the following steps.
-
a.Select the input name (based on the name given in “NamesAndTypes”).
-
b.Name the output image (e.g., IllumNameOfTheExperiment).
-
c.Adjust the smoothing filter size if necessary.
-
i.In the example pipeline, the filter method is set to ‘automatic’. Should this setting not lead to sufficient illumination correction, try the method ‘Object size’ and enter the approximate diameter of a typical object in pixels. Use the ‘start test mode’ to step through the pipeline and visually check the outcome, and increase or decrease the diameter if necessary.
-
i.
-
a.
Note: For more help on the “CorrectIlluminationPipeline” use the “?” button for detailed explanations or visit the CellProfiler homepage (https://cellprofiler.org/previous-examples#illumination-correction).
-
28.Go to “SaveImages”.
-
a.Select the image to save (from “CorrectIlluminationCalculate”).
-
b.Enter the single file name.
-
c.Define the output file location.
-
a.
-
29.Click “Start Test Mode” to check for errors in the pipeline.
-
a.Errors are marked with a red “x”.
-
b.Click “Analyze Images” to start the pre-processing of the images using the “CorrectIlluminationCalculate” module.
-
c.The representative image is stored at the destination set in step 28c.
-
a.
Note: The module “CorrectIlluminationCalculate” is used to create the illumination function, which is an image that is representative of the overall pattern of uneven illumination in the image set.
-
30.
Open the “ApplyIllumCalculate_DNA” pipeline (https://github.com/StemCellMetab/Mitochondrial-membrane-potential).
-
31.
Add the images to be analyzed by dragging and dropping them into CellProfiler.
-
32.Go to “Metadata”.
-
a.Enter the extraction method and metadata source as under step 25.
-
b.Click “Submit”.
-
c.Click “Update” to extract and display the metadata in the CellProfiler window.
-
a.
-
33.Go to “NamesAndTypes”.
-
a.Type in a name to assign to the images.
-
b.Select the single image location.
-
c.Choose the single image to add to all image sets. This image has been generated in the “CorrectIlluminationCalculate” step.
-
a.
-
34.Go to “CorrectIlluminationApply” and adjust the following steps.
-
a.Select the input image.
-
i.Image to be corrected (from “NamesAndTypes”).
-
i.
-
b.Name the output image (e.g., CorrNameOfTheExperiment).
-
c.Select the Illumination correction function.
-
i.Image that will be used to perform the correction and has been generated in the “CorrectIlluminationCalculate” step.
-
i.
-
a.
-
35.Go to “SaveImages”.
-
a.Set the type of image to save to “image”.
-
b.Select the image to save (entry done for “CorrectIlluminationApply”).
-
c.Select the image name for file prefix.
-
d.Enter the text to append to the image name.
-
e.Define the output file location.
-
a.
-
36.
Click “Start Test Mode” to check for errors in the pipeline.
-
37.Click “Analyze Images” to start the pre-processing of the images using the “CorrectIlluminationApply” module.
-
a.The images generated are saved in the defined file location and are ready to be analyzed using the CellProfiler analysis pipeline.
-
a.
-
38.
Repeat steps 30–37 using the “ApplyIllumCalculate_TMRM” pipeline (https://github.com/StemCellMetab/Mitochondrial-membrane-potential).
Note: The illumination pattern may differ between experiments. For example, different staining reagents for a batch of images or changes in the settings in the optical path of the microscope will change the illumination pattern. This means that the illumination function has to be performed for each set of imaging or sample preparation condition separately.
Note: For small image sets, the workflow for creating and applying the illumination correction can be integrated into the CellProfiler analysis pipeline. For large image sets, it is advisable to first create and save the illumination functions using one CellProfiler pipeline, and then retrieve and apply the illumination functions using the CellProfiler analysis pipeline as described in this protocol.
-
39.
For image analysis of ΔΨm using CellProfiler, open the CellProfiler analysis pipeline (CellProfiler-Pipeline_MMP_20x.cpproj: https://github.com/StemCellMetab/Mitochondrial-membrane-potential).
-
40.
Add the pre-processed images (or raw images if illumination correction has not been applied) generated in step 37 by dragging and dropping them into CellProfiler.
-
41.Go to “Metadata”.
-
a.Enter the extraction method and metadata source as under step 25.
-
a.
-
42.Go to “NameAndTypes” and adjust the settings.
-
a.In the default settings of the downloaded pipeline channel 1 is set as DNA (Hoechst staining for nuclei) and channel 2 is set as TMRM (TMRM staining for ΔΨm).
-
a.
-
43.Set the default input and output folders.
-
a.Select the “File” dropdown menu and select “Preferences”. Select “Browse” next to the “Default Output Folder” and “Default Input Folder”.
-
b.Enter a filename for saving.
-
a.
-
44.Go to the “ExportToSpreadsheet” module.
-
a.Enter a name at “filename prefix”.
-
a.
-
45.Testing and adjusting the pipeline.
-
a.Click on “Start Test Mode”.
-
b.A green checkmark will display next to the modules that have no errors. A red “x” will display next to the modules that have an error. The “ExportToSpreadsheet” module always has a red “x” during the test mode.
-
c.Use the “Step” button to step through each module. Visually check the images and adjust each module according to your cells and image intensity. The applied analysis algorithm and a short description of the building blocks can be found in the following table (see also Figure 1B for a graphical visualization of the pipeline):
-
a.
CellProfiler image analysis pipeline
| Building block | Specification/details |
|---|---|
| Images | The image module is used to compile the images for analysis. |
| Metadata | The Metadata module allows to extract information describing your images (i.e., metadata). |
| NamesAndTypes | The NamesAndTypes module allows to assign a meaningful name to each image by which other modules will refer to it. |
| IdentifyPrimaryObjects | The nuclei are identified based on Hoechst staining. Nuclei at the border of the image are removed as well as objects not matching the size and intensity criteria (threshold). |
| IdentifyPrimaryObjects | The TMRM staining is identified. Objects not matching the size and intensity criteria (threshold) are excluded. |
| IdentifySecondaryObjects | Cell edges are defined based on the previously selected nuclei, here defined as “Cell”. |
| RelateObjects | TMRM objects (child object) are associated with the previously defined Cell objects (parent object). This enables to calculate mean measurement values for TMRM associated to the Cell. |
| MeasureImageAreaOccupied | The area occupied by previously identified objects is measured (DNA, Cells and TMRM). |
| MeasureObjectIntensity | TMRM intensity is calculated in relation to the cell. Additional intensity parameters can be chosen (e.g., intensity of the nuclei). |
| MeasureObjectSizeShape | Additional output parameter for size and shape of the objects can be chosen. |
| ExportToSpreadsheet | Output results and storage location are defined. |
Note: The “IdentifyPrimaryObjects” and “IdentifySecondaryObjects” modules might need to be adjusted according to the cell type and intensity of the dyes. The first parameters to optimize are the typical size of objects limits and the threshold method and values.
-
46.Start analysis.
-
a.Ensure to exit the test mode (click “Exit Test Mode”) and click the “Analyze Images” button.
-
b.CellProfiler will analyze the image sets and will export the results to a spreadsheet on the selected drive and folder. The pipeline will also save a spreadsheet containing the used settings (filename prefixExperiment). For opening in Excel, choose “,” as delimiter, “.” as decimal separator, and “(space)” as thousands separator.
-
a.
Note: The time necessary to complete analysis depends on the computer used. For the analysis of large datasets, a fast computer with sufficient RAM (32 GB recommended) would help to speed up the analysis.
Note: The number of workers can be adjusted in the preferences of CellProfiler (File - Preferences - Maximum number of workers). This allows machines with many computational cores to use their resources efficiently.
Note: Representative graphs for the quantification of ΔΨm can be created in Microsoft Excel or statistics analysis software such as GraphPad Prism.
HCS analysis of the mitochondrial membrane potential using commercial software (Columbus)
Timing: ∼2–6 h (depending on the computer capacity and the number of images to be processed)
This step describes the HCS image analysis for the quantification of ΔΨm in iPSC-derived neural cells based on the custom image data storage and analysis system Columbus™ (PerkinElmer) (Figures 1A, 1C, 1E). The image analysis pipeline and exemplary images for trial analysis can be downloaded here: https://github.com/StemCellMetab/Mitochondrial-membrane-potential.
-
47.For data migration from the Operetta CLS High-Content Analysis System to Columbus export TIF files and image-related metadata from the Harmony software and import data to Columbus.
-
a.Open “Settings” and then “Data Management”, and “Export Data” in the Harmony software.
-
b.Specify “Columbus Export (Index and Images)”.
-
c.Select data and measurements to be exported and specify “Export Path”.
-
d.Start the export from Harmony by clicking “Start”.
-
e.Open the Columbus software and move to the “Import” screen. The “Columbus Helper Required” window will appear. Download the connection file and connect the Columbus Helper App. If the Columbus Helper App is not installed yet follow the instructions for installation.
-
f.Specify the “Import Type: Operetta IDX/TIF” and “Source Folder”. Optional: Specify “Import Mode” and “Screen Name”.
-
g.Start the import to Columbus by clicking “Start”.
-
a.
Note: Alternatively, the “Columbus Transfer” function can be used to reduce time and to automate the data migration process. For “Columbus Transfer”, a network connection between Columbus and the Operetta CLS High-Content Analysis System is required.
-
48.
For image analysis of ΔΨm quantification using Columbus, open the Columbus software.
-
49.Move to “Image Analysis” screen.
-
a.Select the measurement you want to analyze from the Data Tree on the left.
-
b.Select one single image (e.g., well and field) as representative image.
-
c.The selected image is displayed in the Image View section.
-
a.
-
50.Load existing image analysis pipeline for ΔΨm quantification.
-
a.Click “Load Analysis from Disk”.
-
b.“Choose File” and open the ∗.aas file analysis script file (https://github.com/StemCellMetab/Mitochondrial-membrane-potential).
-
c.Click “Open” and “OK”.
-
a.
Note: As an alternative, the image analysis pipeline can be set up by the user using the building blocks implemented in the Columbus software. The applied analysis algorithm and a short description of the building blocks can be found in the following table (see also Figure 1C for a graphical visualization of the pipeline):
Columbus image analysis pipeline
| Building block | Specification/details |
|---|---|
| Input Image | Selected input image is displayed. |
| Find Nuclei | Nuclei are detected based on Hoechst signal. Output population: I |
| Calculate Intensity Properties | Intensities of nuclei are calculated. |
| Calculate Morphology Properties | Area and roundness of nuclei are calculated. |
| Select Population | Nuclei at the border of the image are removed. Output population: II |
| Select Population (2) | Additional nuclei are excluded based on individual area, roundness, and intensity parameters. Output population: III |
| Find Cytoplasm | Cell cytoplasms are defined based on previously selected nuclei (Output population: III). |
| Calculate Intensity Properties (2) | ΔΨm intensity is calculated based on TMRM channel within the previously defined cell regions. |
| Define Results | Output results are defined. “Population III” is used to select “Number of Objects” (nuclei) and the mean “ΔΨm intensity” in the cell as results. |
| Sequence Comment | Comment section for the script. |
-
51.Adjust and test the settings.
-
a.The mask/segmentation of defined objects is displayed in the Image View. The illustration of the mask (Regions, Overlays) can be adjusted as preferred. Intermediate results for the selected building block are displayed in the Table View section.
-
b.If required, adjust the filters and input parameters of the building blocks to optimize the segmentation of objects. Define the final readout parameters of the analysis in the last building block “Define Results”.
-
c.Once you have achieved good results, select a few other image fields to test and optimize the analysis with these images.
-
a.
Note: In particular, the building blocks “Find Nuclei”, “Find Cytoplasm”, and “Select Population” typically require adjustment of the filter methods and parameters according to imaging settings, the cell type and density used, and the intensity of the dyes.
-
52.
Save the analysis to the database or the file system as an ∗.aas file.
-
53.Start Analysis.
-
a.Move to “Batch Analysis” screen.
-
b.Select one or multiple measurements in the Data Tree (right click “Select”).
-
c.Click "..." to load the saved analysis script file. Alternatively, you can select the analysis script directly from the database in the Data Tree via left mouse click.
-
d.Click the green arrow to start the batch analysis. A background job is started to analyze all selected measurements using the selected script file.
-
e.Use the Job Status screen to view the progress of the analysis.
-
f.The results are written to the database.
-
a.
Note: Make sure that the used channels for the selected measurements and the channel names are the same, and correspond to the names defined in the analysis.
-
54.Export of results.
-
a.Move to “Export” screen.
-
b.Connect to Columbus Helper.
-
c.Select your measurement from the Data Tree.
-
a.
-
55.
Specify export options.
-
56.Choose a destination export folder on your server.
-
a.Tick the type of file for the export (for the results choose “Results” as ∗.txt File).
-
b.Click the green arrow to start the export.
-
a.
-
57.
The results can be found as a Text Document in previously defined folder.
Note: To import or export files, you must connect Columbus to the Columbus Helper App. If the Columbus Helper App is not installed, follow the instructions for installation.
Note: Representative graphs for the quantification of ΔΨm can be created in Microsoft Excel or statistics analysis software such as GraphPad Prism.
Expected outcomes
The protocol described here enables the monitoring and quantification of mitochondrial activity based on ΔΨm in human iPSC-derived neural cells. Previous protocols have been generated for assessing ΔΨm in live cells using HCS (Iannetti et al., 2016). Our protocol is specifically designed for neural cells obtained from human iPSCs. When applied to patient-derived iPSC lines or genetically modified iPSCs, this protocol could lead to identify disease-specific or mutation-specific defects in neural mitochondrial activity. Such defects can be scored as “phenotypes” to perform phenotypic compound screenings using the HCS pipeline that we provide here. We previously applied this approach to identify compounds capable of counteracting ΔΨm defects in neural cells derived from patients affected by Maternally Inherited Leigh Syndrome (MILS) carrying mutations in the mitochondrial gene MT-ATP6 (Lorenz et al., 2017). Our HCS protocol can be used also in combination with another HCS assay that quantifies neurite outgrowth (Lickfett et al., STAR Protocols, 2022). We previously used the combination of these two protocols to screen for modulators of human neurotoxicity that can affect ΔΨm and neurite count in different ways (Zink et al., 2020).
Limitations
This HCS protocol is essentially an endpoint measurement for high-throughput assessment of changes in mitochondrial membrane potential (ΔΨm), which is an important parameter regulating mitochondrial bioenergetics and cellular physiology. For detailed kinetic experiments aimed at addressing dynamic changes of ΔΨm, important modifications should be applied, since the TMRM signal may vary if the cells are assessed for periods longer than 30 min (Figure 2). In these cases, changes in the washing step may be beneficial (see troubleshooting problem 4). Automated dispensing of reagents for example by liquid handling might also be considered to reduce the variability of the assay.
The protocol reliably quantifies changes in the existing baseline rate of dye loss from the mitochondria, but it is not effective in monitoring transient changes of ΔΨm. In order to address this and to increase the identification of modifiers that could lead to increased ΔΨm, we recommend to include positive modulators of ΔΨm (e.g., oligomycin) and to be aware of the strict time sensitivity of the assay (see troubleshooting problem 7).
Since our protocol measures the functional mitochondrial state of live neural cells, it is possible that external conditions may influence the results. For example, changes in environment (temperature, CO2, or humidity), improper coating of the plates, or mechanical forces may induce cellular stress or detachment leading to changes in the ΔΨm signal. This could lead to inconsistent results. In order to avoid this, particular care needs to be applied to the handling of the neural cells and the HCS plates. A reference sample can be included in each plate to serve as internal control in order to prevent low quality data caused by inter-plate variability (Bray and Carpenter, 2004).
Our HCS protocol is based on non-confocal wide-field fluorescence mode, and is therefore not suitable for assessing detailed structures (e.g., mitochondrial morphology). In fact, we only use variations in the Z-axis for optimal cell focus. It is possible to adapt the HCS pipeline to confocal mode and include multiple Z-stacks images for the same field of view. However, this approach generates multiple measurements of the same cells, and this needs to be taken into account during data analysis and processing. In order to correctly assign multiple images to the same cells, it is possible to run a maximal intensity projection on all Z-stacks to generate a pseudo 2D image. This approach should eliminate out-of-focus pixels and could produce acceptable images for analysis.
Lastly, there are challenges associated with data storage. This protocol is based on images that can take up a conspicuous hard drive space, and may require additional computational power for analysis and long-term storage. As an example, we commonly generate 16-bit images with a pixel resolution of 2160 × 2160. For a single 96-well plate, we obtain approximately 8–10 GB of data.
Troubleshooting
Problem 1
iPSC-derived neural cells detach after washing steps (e.g., step 7).
Potential solution
-
•
Try different coating reagents and/or increase the concentration of the coating.
-
•
Reduce number of washing steps. One way to reduce washing steps is to add the staining solution directly to the cells in equal volume as 2 × to obtain a final concentration of 10 nM for TMRM and 1 μg/mL for Hoechst.
-
•
Seed more cells to compensate for cell loss during staining.
Problem 2
Cells are out-of-focus (steps 17–19).
Potential solution
-
•
Try different Z-positions.
-
•
Optimize the seeding conditions. Ideally, cells should grow as single cells.
-
•
Check if the plate type used is suitable for the device and whether the optical quality is sufficient (e.g., evenness).
-
•
Check that the plate is correctly positioned and is not tilted.
-
•
Make sure that the wells contain enough medium so that the liquid level does not affect the autofocus of the instrument.
Problem 3
iPSC-derived neural cells are not correctly visualized and analyzed (step 45).
Potential solution
-
•
Determine the optimal cell density in a pilot experiment. Cells should be dense enough to ensure survival, but also sufficiently sparse for the pipeline to work properly. In general, always analyze more than 1,000 cells per individual well (see also step 2).
Problem 4
The TMRM signal is variable across the plate and within different wells of the same cell line (steps 10–20).
Potential solution
-
•
Instead of washing off the TMRM, add imaging medium in each well in order to dilute the TMRM concentration. This approach has been suggested to maintain the TMRM signal in equilibrium for longer time (Iannetti et al., 2016). Alternatively, it is also possible to not wash the cells, and leave the TMRM concentration as it is during the imaging step (Nicholls, 2018).
-
•
The presence of light-induced oxidative stress may compromise cell viability. Extensive exposure to light may induce reactive oxygen species (ROS), which in turn also influences the TMRM signal. Reduce the images acquired from one well. This will reduce the time of exposure to light. Also, ensure that cell viability is maintained by monitoring the Hoechst signal.
-
•
Check for photobleaching. By reducing the energy of the excitation beam and the irradiation time of the fluorophore, an attempt can be made to minimize the effect of photobleaching.
-
•
Check for quenching effect. Use low concentrations of TMRM (<30 nM is considered for non-quenching mode) in order to avoid auto-quenching of mitochondrial TMRM. In this case, the fluorescence signal of TMRM can be directly correlated to ΔΨm across the inner mitochondrial membrane. A loss of ΔΨm may cause TMRM to leak from mitochondria resulting in a loss of fluorescence intensity. Ensure that the TMRM signal is due to changes in the ΔΨm, e.g., by using a probe specific to the plasma membrane potential such as DiBAC2 (Ward et al., 2007).
Problem 5
The HCS pipeline for ΔΨm quantification does not generate stable results using CellProfiler (steps 39–46).
Potential solution
-
•
The pipeline indicates when analysis for a given position fails. This is usually because primary objects, the Hoechst signal, or the TMRM signal are not properly detected. This can be solved by adjusting the “IdentifyPrimaryObjects” modules. Adjust the typical diameter of the objects (Nuclei and TMRM). To determine the average diameter of the objects, open an exemplary image and measure the distances using the “measure length” tool (“Tools” in the display window menu bar). Use the pixel intensity tool to estimate the pixel intensity and adjust the lower and upper bounds of the threshold accordingly. Adjust the method to distinguish clumped objects (decide between intensity or shape).
-
•
The pipeline might stop with a memory error if the RAM was limiting. Try to restart the analysis running on fewer CPU cores (can be adjusted in CellProfiler settings).
Problem 6
The HCS pipeline for ΔΨm quantification does not generate stable results using Columbus (steps 48–57).
Potential solution
-
•
Many nuclei are sorted out that actually look healthy. This is usually because the filter properties for nuclei exclusion do not fit with the cell type used. This can be solved by adjusting these filter properties within the building block “Select Population (2)”. Define the minimal and maximal values for Hoechst intensity (Intensity Nucleus) and for the area (Nucleus area). Also, the Nucleus Roundness filter can be adjusted.
-
•
ΔΨm quantification shows strongly fluctuating values. The reason for this could be an insufficient definition of the cell area. This could be solved by adjusting the methods used under the building block “Find Cytoplasm”. Choose from method A-F and adjust the common/individual thresholds to improve cell segmentation.
Problem 7
iPSC-derived neural cells in the HCS plate treated with FCCP and antimycin A appear unhealthy, or the treatment does not lead to depolarization (i.e., to a decrease of the TMRM signal) (step 6).
Potential solution
-
•
Optimize the FCCP and antimycin A concentration. Perform a dose-response curve to determine the most suitable concentration to induce depolarization, but without inducing cell death.
Optional: Include oligomycin to prevent ATP synthase reversal.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Prof. Dr. Alessandro Prigione (alessandro.prigione@hhu.de).
Materials availability
This study did not generate new unique materials.
Acknowledgments
A.P. acknowledges support from the Deutsche Forschungsgemeinschaft (DFG) (#PR1527/5-1 and PR1527/6-1), the European Joint Programme for Rare Diseases (EJPRD) and German Federal Ministry of Education and Research (BMBF) (#01GM2002A), the United Mitochondrial Disease Foundation (UMDF), People Against Leigh Syndrome (PALS), Fondation Maladies Rares and Association AMMi, and the University Hospital Düsseldorf (Forschungskommission UKD). O.P. acknowledges funding from the European Joint Programme for Rare Diseases (EJPRD). M.B. acknowledges support from the Deutsche Forschungsgemeinschaft (DFG) (INST 208/760-1FUGG).
Author contributions
Conceptualization, A.Z. and A.P.; methodology, A.Z., U.H., A.W., O.P., and M.B.; investigation, A.Z., U.H., and A.W.; writing – original draft, A.Z.; writing – review and editing, A.Z., U.H., A.W., O.P., and A.P.; funding acquisition, A.P., O.P., and M.B.; resources, M.B., O.P., and A.P.; supervision, A.P. and O.P.
Declaration of interests
The authors declare no competing interests.
Data and code availability
The pipelines for ΔΨm analysis and example images have been deposited to GitHub: https://github.com/StemCellMetab/Mitochondrial-membrane-potential [https://doi.org/10.5281/zenodo.6794537].
References
- Bray M.-A., Carpenter A., Imaging Platform, Broad Institute of MIT and Harvard . In: Assay Guidance Manual. Markossian S., Grossman A., Brimacombe K., Arkin M., Auld D., Austin C.P., Baell J., Chung T.D.Y., Coussens N.P., Dahlin J.L., et al., editors. Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004. Advanced assay development guidelines for image-based high content screening and analysis.http://www.ncbi.nlm.nih.gov/books/NBK126174/ [PubMed] [Google Scholar]
- Iannetti E.F., Smeitink J.A.M., Beyrath J., Willems P.H.G.M., Koopman W.J.H. Multiplexed high-content analysis of mitochondrial morphofunction using live-cell microscopy. Nat. Protoc. 2016;11:1693–1710. doi: 10.1038/nprot.2016.094. [DOI] [PubMed] [Google Scholar]
- Lickfett S., Menacho A., Zink A., Telugu N.S., Beller M., Diecke S., Cambridge S., Prigione A. High-content analysis of neuronal morphology in human iPSC-derived neurons. STAR Protocols. 2022 doi: 10.1016/j.xpro.2022.101567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C., Lesimple P., Bukowiecki R., Zink A., Inak G., Mlody B., Singh M., Semtner M., Mah N., Auré K., et al. Human iPSC-derived neural progenitors are an effective drug discovery model for neurological mtDNA disorders. Cell Stem Cell. 2017;20:659–674.e9. doi: 10.1016/j.stem.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Nicholls D.G. Simultaneous monitoring of ionophore- and inhibitor-mediated plasma and mitochondrial membrane potential changes in cultured neurons. J. Biol. Chem. 2006;281:14864–14874. doi: 10.1074/jbc.M510916200. [DOI] [PubMed] [Google Scholar]
- Nicholls D.G. In: Mitochondrial Bioenergetics: Methods and Protocols Methods in Molecular Biology. Palmeira C.M., Moreno A.J., editors. Springer; 2018. Fluorescence measurement of mitochondrial membrane potential changes in cultured cells; pp. 121–135. [DOI] [PubMed] [Google Scholar]
- Perry S.W., Norman J.P., Barbieri J., Brown E.B., Gelbard H.A. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques. 2011;50:98–115. doi: 10.2144/000113610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C.S., Höing S., Moritz S., Parga J.A., Wagner L., Bruder J.M., et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One. 2013;8:e59252. doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Lickfett S., Martins S., Prigione A. A call for consensus guidelines on monitoring the integrity of nuclear and mitochondrial genomes in human pluripotent stem cells. Stem Cell Rep. 2022;17:707–710. doi: 10.1016/j.stemcr.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Sterr M., Burtscher I., Chen S., Hieronimus A., Machicao F., Staiger H., Häring H.U., Lederer G., Meitinger T., et al. Genome-wide analysis of PDX1 target genes in human pancreatic progenitors. Mol. Metab. 2018;9:57–68. doi: 10.1016/j.molmet.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M.W., Huber H.J., Weisová P., Düssmann H., Nicholls D.G., Prehn J.H.M. Mitochondrial and plasma membrane potential of cultured cerebellar neurons during glutamate-induced necrosis, apoptosis, and tolerance. J. Neurosci. 2007;27:8238–8249. doi: 10.1523/JNEUROSCI.1984-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S., Zarse K., Schulz T.J., Siems K., Müller-Kuhrt L., Birringer M., Ristow M. A cell-based high-throughput assay system reveals modulation of oxidative and nonoxidative glucose metabolism due to commonly used organic solvents. Horm. Metab. Res. 2008;40:29–37. doi: 10.1055/s-2007-1004542. [DOI] [PubMed] [Google Scholar]
- Zink A., Conrad J., Telugu N.S., Diecke S., Heinz A., Wanker E., Priller J., Prigione A. Assessment of ethanol-induced toxicity on iPSC-derived human neurons using a novel high-throughput mitochondrial neuronal health (MNH) assay. Front. Cell Dev. Biol. 2020;8:590540. doi: 10.3389/fcell.2020.590540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink A., Lisowski P., Prigione A. Generation of human iPSC-derived neural progenitor cells (NPCs) as drug discovery model for neurological and mitochondrial disorders. Bio. Protoc. 2021;11:e3939. doi: 10.21769/BioProtoc.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The pipelines for ΔΨm analysis and example images have been deposited to GitHub: https://github.com/StemCellMetab/Mitochondrial-membrane-potential [https://doi.org/10.5281/zenodo.6794537].