Abstract
A key to improving vaccine design and vaccination strategy is to understand the mechanism behind the variation of vaccine response with host factors. Glycosylation, a critical modulator of immunity, has no clear role in determining vaccine responses. To gain insight into the association between glycosylation and vaccine-induced antibody levels, we profiled the pre- and postvaccination serum protein glycomes of 160 Caucasian adults receiving the FLUZONE influenza vaccine during the 2019–2020 influenza season using lectin microarray technology. We found that prevaccination levels of Lewis A antigen (Lea) are significantly higher in nonresponders than responders. Glycoproteomic analysis showed that Lea-bearing proteins are enriched in complement activation pathways, suggesting a potential role of glycosylation in tuning the activities of complement proteins, which may be implicated in mounting vaccine responses. In addition, we observed a postvaccination increase in sialyl Lewis X antigen (sLex) and a decrease in high mannose glycans among high responders, which were not observed in nonresponders. These data suggest that the immune system may actively modulate glycosylation as part of its effort to establish effective protection postvaccination.
Keywords: vaccine, influenza, immunity, antibody response, glycosylation, glycome, lectin microarray
Introduction
Vaccination is a powerful means to prevent infection and the development of severe disease. However, the response to vaccination varies across populations in ways that we have yet to understand.1 Vaccines against influenza, a major respiratory illness causing millions of hospitalizations and hundreds of thousand deaths worldwide each year, are perhaps the most widely studied, in part because vaccinations are administered annually.2−4 In the past decade, research has identified several potential biomarkers associated with influenza vaccine response. These include hormones, cytokines, pre-existing antibodies, and pre-existing T cell populations.5−7
Glycosylation is an underexplored modulator of immune response to influenza vaccines, in spite of the multifaceted roles glycans have been found to play in immunity.8,9 For example, antibody glycosylation has been found to alter the ability of antibodies to activate the immune system and changes their circulation time in sera.10−12 Two pioneering studies conducted in influenza-vaccinated cohorts revealed changes in N-glycan sialylation, galactosylation, and fucosylation on influenza-specific IgG postvaccination.13,14 However, the level of viral binding antibodies produced (antibody response) in association with glycosylation prevaccination was not examined. Sera contain a host of other proteins involved in immunity beyond IgG, including innate immune lectins, complement factors, and other glycoproteins. Comprehensive profiling of the serum glycome may provide valuable insights into the mechanisms underlying vaccine response and identify new glycan-based markers.
Here we analyze pre- and postvaccination serum glycomes and associated antibody responses in a cohort of 160 Caucasian adults vaccinated with the widely used FLUZONE influenza vaccine. Glycomic analysis was performed using our dual-color lectin microarray technology, which has been used in a wide variety of studies including analysis of exosomes, host-response to influenza, and identification of cancer drivers in human tissues.15−21 We found that individuals who lacked response to vaccination had high levels of the blood group epitope Lewis A antigen (Lea). Glycoproteomic analysis identified multiple complement components and other immune-related proteins as marked with this epitope, which was enriched in nonresponders. We also observed postvaccination upregulation of sialyl Lewis X antigen (sLex) and downregulation of high mannose glycans in high responders, neither of which was observed in nonresponders. Our data suggest that glycosylation may help predict immune response to vaccine and that specific glycoforms of immune proteins may be important in tuning vaccination response.
Methods
Cohort Recruitment
160 Caucasian adults were enrolled at the University of Georgia Clinical and Translational Research Unit (Athens, GA, USA) from September 2019 to February 2020. All volunteers were enrolled with written, informed consent. Participants were excluded if they already received the seasonal influenza vaccine. Other exclusion criteria included acute or chronic conditions that would put the participant at risk for an adverse reaction to the blood draw or the flu vaccine (e.g., Guillain–Barre syndrome or allergies to egg products), or conditions that could skew the analysis (e.g., recent flu symptoms or steroid injections/medications). All participants received a FLUZONE (Sanofi Pasteur, Lyon, France) seasonal inactivated influenza vaccine. Most received a quadrivalent, standard dose formulation made up of 15 μg HA per strain of A/H1N1 (A/Brisbane/02/2018), A/H3N2 (A/Kansas/14/2017), B/Yamagata (B/Phuket/3073/2013), and B/Victoria (B/Colorado/6/2017-like strain).
Hemagglutination Inhibition Assays
Blood samples were collected from participants prior to vaccination (d0) and again postvaccination at d28 (22–35 days). Hemagglutinin inhibition (HAI) assays were performed with serum from each participant at d0 and d28. Sera was used at a starting concentration of 1:10 following treatment with a receptor-destroying enzyme (RDE) (Denka Seiken, Tokyo, Japan) to inactivate nonspecific inhibitors. RDE-treated sera (25 μL), including positive and negative controls, were serially diluted in PBS (2.67 mM KCl, 1.47 mM KH2PO4, 8.10 mM Na2HPO4, 138 mM NaCl, pH = 7.4, same hereinafter) 2-fold across 96-well V-bottom microtiter plates. An equal volume of influenza virus (25 μL), which was adjusted beforehand via hemagglutination (HA) assay to a concentration of 8 hemagglutination units (HAU) per 50 μL, was added to each well and incubated at room temperature for 20 min. Finally, 0.8% turkey red blood cells (Lampire Biologicals, Pipersville, PA, USA) in PBS were added, and plates were mixed by agitation and then incubated at room temperature for 30 min. The HAI titer was determined by the reciprocal dilution of the last well that contained nonagglutinated RBCs.
Definition of Antibody Responses
Response scores for each strain of influenza were calculated on the basis of the fold changes of antibody titers (d28 titer/d0 titer). For each strain, antibody response was scored in the following steps: (i) calculate the initial score by taking the logarithmic (base 2) value of the titer fold change; (ii) change the score to zero if the d28 antibody titer is lower or equal to 20, a conventional cutoff for effective protection;22,23 (iii) change the score to 4 if the initial score is greater than 4 (i.e., an over 16-fold increase in titer). This is to prevent the total response score (see below) from being biased toward one single strain. This strain-specific score was used to categorize the participants into three response groups: high responders (score ≥ 2), low/moderate responders (1 ≤ score < 2), and nonresponders (score < 1) for each strain. The total response score is the sum of the four strain-specific scores. Similarly, total response scores were used to define overall high responders (score ≥ 8), overall low responders (4 ≤ score < 8), and overall nonresponders (score < 4). Note: People with low/moderate overall responses may be classified as high or nonresponders in a strain-specific manner dependent on their response.
We also categorized antibody response using the metric of Wu et al., which takes into account BMI, age, and gender.24 For our comparison, we ranked participants by the modified response score from high to low. The upper third was considered high responders and the lower third nonresponders.
Fluorescent Labeling of Serum Proteins
Total protein concentrations of serum samples were measured with a DC protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). Each volunteer serum sample was fluorescently labeled with Alexa Fluor 555 NHS ester (Thermo Fisher Scientific, Waltham, MA, USA). First, 10 μg of total protein was diluted in PBS to 27 μL. The pH of the solution was adjusted with 3 μL of 1 M sodium bicarbonate. Then 0.21 μL of a stock solution (10 mg/mL) of Alexa Fluor 555 NHS ester was added to the mixture. The reaction lasted for 1 h in the dark at room temperature. Unconjugated dye molecules were then removed by Zeba Dye and Biotin Removal Filter Plates (Thermo Fisher Scientific, Waltham, MA, USA). The reference material, NIST human serum 909c (Millipore Sigma, Darmstadt, Germany), was fluorescently labeled with Alexa Fluor 647 NHS ester (Thermo Fisher Scientific, Waltham, MA, USA) similarly. The amounts of reagents were scaled linearly to the starting protein amount (4 mg). Finally, each Alexa Fluor 555-labeled sample (10 μg of total protein) was mixed with a proper volume of Alexa Fluor 647-labeled reference material containing the same amount of protein, and the final volume was adjusted to 50 μL with PBS.
Dual-Color Lectin Microarray
Lectin microarray slides were fabricated as previously described.25 A list of probes printed on the array is in the Supporting Information, Table S1. The print was quality controlled as previously described.25 Prior to hybridization, each dual-color mixture was diluted with 50 μL 0.2% PBST (PBS with 0.2% Tween-20, v/v). Each mixture was then allowed to hybridize with the arrays for 1 h in the dark at room temperature. Arrays were washed twice with 0.005% PBST for 5 min and once with PBS for 5 min. The slides were briefly rinsed with ultrapure water and dried. Fluorescence signals were obtained with Genepix 4400A fluorescence slide scanner (Molecular Devices, San Jose, CA, USA) in the 532 nm channel and the 635 nm channel that correspond to the excitation/emission profiles of Alexa Fluor 555 and Alexa Fluor 647, respectively. Raw fluorescence signal and background signal of each spot were generated by the Genepix Pro 7 software (Molecular Devices, San Jose, CA, USA), which were further processed and analyzed with a custom script as previously described.16 Heatmaps and volcano plots were generated with R (R version 4.0.1, r-project.org). Lectin annotation was done using data from Bojar et al.26 In general, epitopes are annotated when unambiguous (e.g., multiple related binders trending together). Lectin microarray data are available at Synapse.org (DOI: 10.7303/syn26956958).
Lectin/Antibody Affinity Pulldown
80 μg of BambL (expressed in-house) or anti-Lea (Abcam, Cambridge, United Kingdom) was immobilized on columns using AminoLink Plus Micro Immobilization Kit (Thermo Fisher Scientific, Waltham, MA, USA) as per the manufacturer’s protocol. Coupling was carried out at 4 °C overnight with gentle agitation. For the beads-only controls, PBS was added to the columns instead of BambL/anti-Lea in the coupling step.
For protein identification by proteomics, a serum pool was prepared by combining 10 μL of each day 0 serum sample. The pooled serum was incubated at 54 °C for 1 h to inactivate proteases prior to the pulldown experiments. For BambL pulldown, 10 μL of pooled serum was diluted in PBS to 200 μL and incubated on the column for 1 h at room temperature with gentle agitation. The column was washed with 300 μL PBS three times (5 min per wash with gentle agitation). The column was eluted with 200 μL 50 mM methyl α-l-fucopyranoside (TCI America, Portland, OR, USA) in PBS. For anti-Lea pulldown, 50 μL of pooled serum was diluted in PBS to 400 μL and incubated on the column for 1 h at room temperature with gentle agitation. The column was washed with 300 μL PBS three times (5 min per wash with gentle agitation) before being eluted with 100 μL 0.1 M glycine (pH = 2.8). The eluate was immediately neutralized with 30 μL 0.5 M Tris (pH = 8.5). The protocol for preparing the six BambL-pulldown samples for Western blotting is the same as the protocol for glycoproteomics, except that the columns were incubated with 200 μg serum protein diluted in 100 μL PBS, washed with 100 μL PBS, and eluted with 80 μL elution buffer.
Peptide Preparation and LC-MS/MS Analysis
The enriched samples were incubated at 95 °C for 10 min. One μg/μL of sequencing grade-modified trypsin (Promega, Madison, WI, USA) was added to samples and overnight at 37 °C with gentle agitation. Digestion was quenched by pH < 4.0 using 2.5% trifluoroacetic acid (TFA). Samples were subsequently desalted using Pierce C18 spin tips (Thermo Fisher Scientific, Waltham, MA, USA) as per manufacturer’s protocol. The peptides were eluted using aqueous buffer with 60% acetonitrile (ACN) and 0.1% formic acid (FA). The samples were dried, and peptides were resuspended in 10 μL of buffer (0.1% FA in 5% ACN).
Each sample (∼3 μL) was loaded onto Acclaim PepMap 100 trap column (75 μm × 2 cm) nanoViper, attached to an EASY-spray analytical column (PepMap RSLC C18, 2 μm, 100 Å, 75 μm ID × 50 cm) in an EASY nano-LC 1000 liquid chromatography instrument (Thermo Scientific). Chromatography solvent A consisted of LC-MS grade water with 0.1% FA, and solvent B of 80% acetonitrile with 0.1% FA. The 155 min gradient consisted of the following: 2–5% of solvent B for 5 min, 5–25% for 110 min, 25–40% for 25 min, 40–80% for 5 min, 80–95% for 5 min, followed by 95–5% for 5 min. Mass spectrometry data were collected in data dependent mode on an Orbitrap Eclipse mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The MS1 spectra were recorded with a resolution of 240 000, AGC target of 1 × 106, with maximum injection time of 50 ms, and a scan range of 400 to 1500 m/z. The MS2 spectra were collected using quadrupole isolation mode, AGC target of 2 × 104, maximum injection time of 18 ms, one microscan, 0.7 m/z isolation window, collision energy of 27%, excluding ions of charge state < +2 and > +7.
Spectra were searched against the Uniprot human fasta sequence database (UP000005640, downloaded on July 24, 2020) using the MaxQuant software (version 1.5.5.1) with default settings, including 2 missed cleavages, first search with peptide tolerance of 20 ppm and for the main search with peptide tolerance of 4.5 ppm. Carbamidomethylation of Cysteine was set as a static modification. The false discovery rates for peptide and protein identifications were both set to 0.01. Oxidation of Met and acetylation of the protein N terminus were the allowed variable modifications, and proteins were quantified using the Label Free Quantification (LFQ) option.
A protein was identified as a positive binder if the enriched sample (E) and the corresponding control sample (C) satisfied the following: (i) the sum of log10-transformed LFQ intensities of this protein in E and C was >3; and (ii) the difference of log2-transformed LFQ intensities of this protein between E and C was >2 (E – C). The remaining proteins were searched in GlyGen,27 a database that compiles the experimental evidence for glycosylation of proteins. Proteins without any experimental evidence for glycosylation or solely with experimental evidence for O-GlcNAcylation were removed, as they are not the targets of interest of the pulldown experiments.
Pathway Enrichment Analysis
Gene Ontology (GO) enrichment analysis was performed with PANTHER Overrepresentation Test (Release 20210224).28,29 The input analyzed lists are the lists of glycoproteins identified in BambL or anti-Lea pulldowns (Supporting Information, Table S4 and Table S5). A full list of plasma proteins was used as the reference list.28 “GO biological process complete”, “Fisher’s Exact”, and “Calculate False Discovery Rate” were selected as the annotation data set, test type, and correction method, respectively.
Western Blotting
All steps were performed at room temperature. Twenty μg of pulldown samples or input serum samples were resolved by 4–20% SDS/PAGE and transferred to a nitrocellulose membrane. Total protein was stained with Revert 700 Total Protein Stain Kit (LI-COR, Lincoln, Nebraska, USA) as per the manufacturer’s instruction. After the total protein stain was erased, the membrane was blocked with blocking buffer [PBS with 3% (w/v) BSA and 0.05% (v/v) Tween-20] for 1 h. Then the membrane was incubated with primary antibody working solution [rabbit anti-C4BPA (Abcam, Cambridge, United Kingdom), diluted to 0.5 μg/mL in blocking buffer] for 1 h, washed with PBST three times for 5 min per wash, incubated with secondary antibody working solution [goat antirabbit IgG CF640R conjugate (Millipore Sigma, Darmstadt, Germany), diluted to 0.1 μg/mL in blocking buffer] for 15 min, and washed with PBST three times for 5 min per wash before imaging.
Results and Discussion
Lectin Microarray Analysis Shows Clear Glycomic Differences between High and Nonresponders Prior to Influenza Vaccination
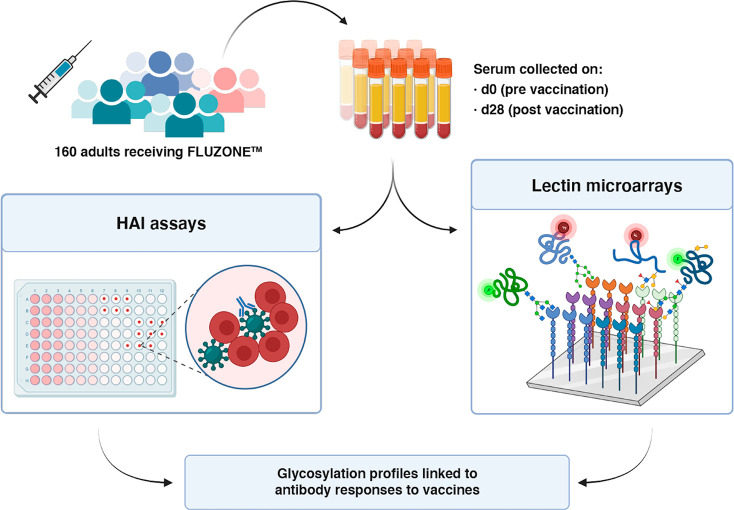
To evaluate whether the glycome varies in individuals with differing vaccine response, we characterized the serum glycomes of 160 Caucasian adults who received FLUZONE quadrivalent vaccines in the 2019–2020 flu season in a medical facility in Georgia, USA. The cohort is described in Table 1. The FLUZONE vaccine is composed of four inactivated strains of influenza virus, including two type A strains (A/Brisbane/02/2018 (subtype H1N1), A/Kansas/14/2017 (subtype H3N2)) and two type B strains (B/Phuket/3073/2013 (Yamagata lineage) and B/Colorado/6/2017-like (Victoria lineage)). Prevaccination sera of participants were collected on the day of vaccination (d0). Postvaccination sera were collected approximately 4 weeks post vaccination (d28). Antibody titers were determined via serum hemagglutination inhibition (HAI) assays, and the resulting log2 fold change (d28/d0) was used to calculate a response score for each strain (Scheme 1 and Methods). To assess the overall antibody response across all four strains, we calculated a total response score by summing the individual response scores. Participants were categorized into high responders (total score ≥ 8, N = 67), low/moderate responders (4 ≤ total score < 8, N = 39), and nonresponders (total score < 4, N = 54).
Table 1. Characteristics of Study Participants (N = 160).
| high responders (N = 66) | low/moderate responders (N = 39) | nonresponders (N = 54) | |
|---|---|---|---|
| male/female | 22/44 | 15/24 | 24/30 |
| median age in years (IQR) | 46.5 (31.0–58.8) | 56 (39.9–67.0) | 56.5 (42.3–70.0) |
| median body-mass index (IQR) | 28.7 (25.2–32.8) | 30.8 (27.4–34.1) | 26.9 (24.4–31.0) |
Scheme 1. Workflow of Integrated Analysis.
Hemagglutination inhibition assays (HAI) and lectin microarray assays were run on sera collected pre- (day 0, d0) and post- (day 28, d28) vaccination.
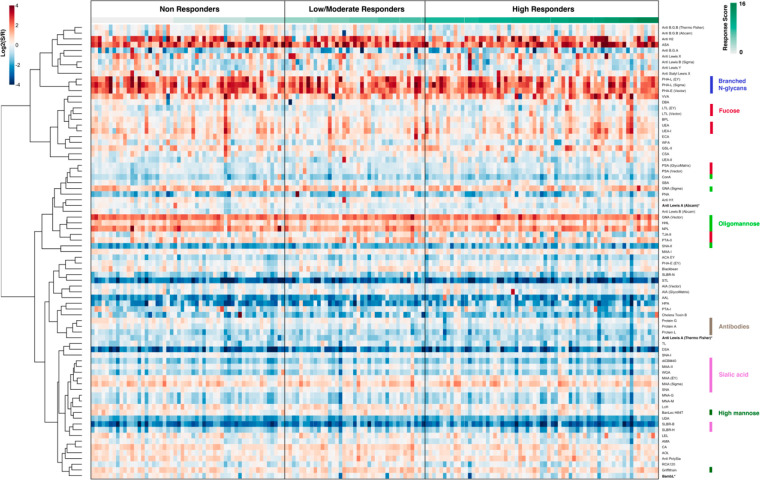
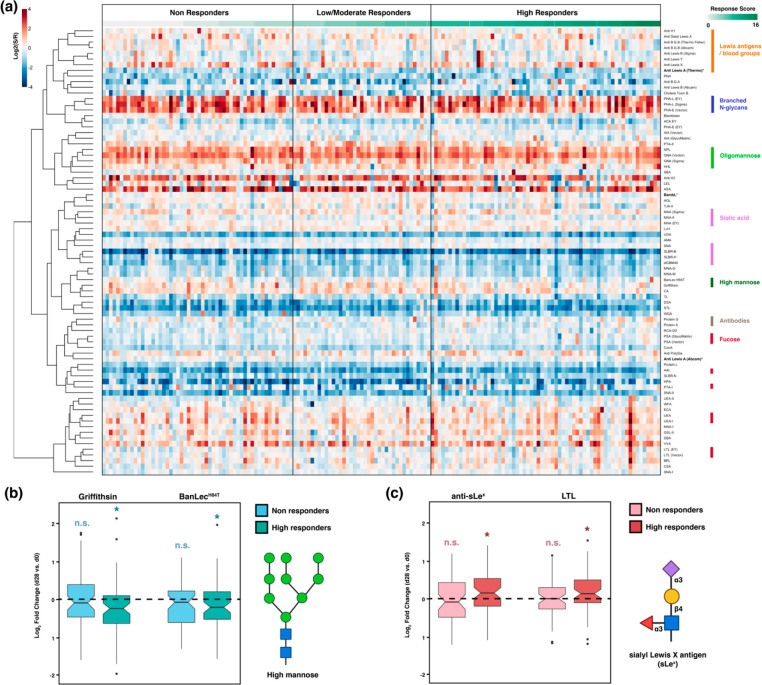
To analyze the glycome, we used our dual-color lectin microarray technology (Scheme 1).15 Lectin microarrays, which utilize well-characterized glycan probes to identify glycomic changes at the substructure level, have been used to identify glycans driving cancer progression and metastasis,19−21 involved in exosome biogenesis,16,30 and associated with influenza severity.17,18 For this study, the probes included 68 lectins and 14 carbohydrate-binding antibodies. In addition, we printed protein A, protein G, and protein L to assess serum immunoglobulin levels. In brief, each sample was labeled with Alexa Fluor 555. A serum standard (human serum 909c, NIST) was labeled with the orthogonal dye, Alexa Fluor 647, and used as the biological reference. Equal amounts (10 μg) of sample and reference were incubated on each array. Data were analyzed as previously described.16 An annotated heatmap of the prevaccination glycomic profiles with response scores is shown in Figure 1.
Figure 1.
Heatmap of lectin microarray data for d0 serum samples. Columns represent the participants and rows represent the probes. Color of cells represent the normalized log2 ratios (Sample signal (S)/Reference signal (R)). Total response scores are annotated with a green-white sliding scale bar on the top of the heatmap.
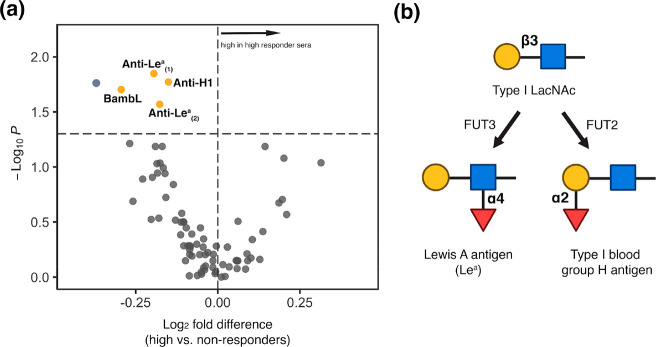
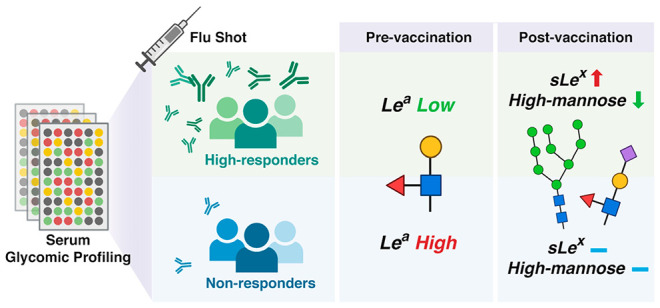
To clearly identify glycan epitopes that might be predictive of vaccine response, we compared the glycomes at d0 of high responders to nonresponders (∼76% of the cohort). We observed a clear pattern of glycan motifs associated with a lack of response to the vaccine. In comparison to high responders, people with poor antibody response exhibited significantly higher binding to fucosylated Type I LacNAc antigens (Figure 2; Supporting Information, Figure S1, probes: BambL, anti-Lea, anti-H1). The specificity of the anti-Lea antibody overlaps with the Burkholderia ambifaria lectin (BambL), a pan-Lewis antigen binder that binds Lea.31 Upon examination of the low/moderate response cohort, a variable group that contains people who had high responses to a single strain and/or moderate responses across strains also showed lower levels of BambL binding when compared with the high responders (Supporting Information, Figure S2). No statistical association of baseline serum Lea with age, gender, or BMI was identified (Supporting Information, Figure S3). Consistent with this, regrouping the samples using a modified response score that takes into account age, gender, and BMI found the same pattern when comparing the high responders to low/nonresponders (Supporting Information, Figure S4).24
Figure 2.
(a) Volcano plot comparing lectin microarray data for high responders (N = 65) and nonresponders (N = 54) prevaccination. Mann–Whitney U test was used to determine p-values. Probes with p < 0.05 are colored in yellow. BambL: Burkholderia ambifaria lectin; Anti-Lea(1): anti-Lewis A, Invitrogen; Anti-Lea(2): anti-Lewis A, Abcam. Anti-H1: antitype I blood group H (O), Invitrogen. (b) Partial biosynthetic routes of Lewis A antigen and type I blood group H (O) antigen. Glycans are shown in the Symbolic Nomenclature for Glycomics (SNFG). Symbols are defined as follows: galactose (yellow ●), N-acetylglucosamine (blue ■), fucose (red ▲). FUT2: galactoside alpha-(1,2)-fucosyltransferase 2; FUT3: 3-galactosyl-N-acetylglucosaminide 4-alpha-l-fucosyltransferase.
Immune response to influenza has been found to be strain-dependent.32,33 The FLUZONE vaccine contains four different strains of influenza (two type A and two type B). To assess the impact of strain on the glycomic association with antibody response, we compared the high-responders and nonresponders for each strain (high response: score ≥ 2, nonresponse: score < 1; Supporting Information, Figure S5–S8). The strain-specific scores are separate from the overall score and have different subsets of participants (Supporting Information, Table S2). In line with our previous analysis, we observed higher binding to Lea probes (anti-Lea, BambL) in nonresponders for three of the four strains. Only the B/Victoria (B/Colorado/6/2017-like) strain did not show this association. In aggregate, we observed an association between high baseline serum Lea and nonresponsiveness to influenza vaccination in both the overall and strain-specific analysis.
Lea lacks well-characterized roles in immunity. In a study among children vaccinated against rotavirus, higher levels of Lea correlated with low seroconversion rates.34 High levels of this epitope are observed in and often used as a marker for nonsecretors, a subset of the population who have a functional defect in galactoside alpha-(1,2)-fucosyltransferase 2 (FUT2, Figure 2b).35 A significant proportion of the human population is genetically FUT2 deficient (∼20%), although we do not have genetic data for our cohort.36,37 Immunological consequences of FUT2 deficiency have emerged, many of which have focused on microbial colonization and infection.35,38 For instance, nonsecretors have lower microbial diversity, especially in bifidobacteria species.39,40 Higher levels of microbial diversity have been correlated with better response to influenza vaccination in porcine models.41 In addition, treatment of mice with 2′-fucosyllactose, which mimics secreted FUT2 epitopes, enhances influenza vaccine efficacy.42 Together with our data, this suggests a potential role for FUT2 in controlling vaccine response. Whether differential levels of the enzyme in secretors also alter response, precise mechanisms, and whether Lea antigens correlate with FUT2 levels in secretors will require further study.
Glycoproteomic Identification of Serum Glycoproteins Marked by Lea
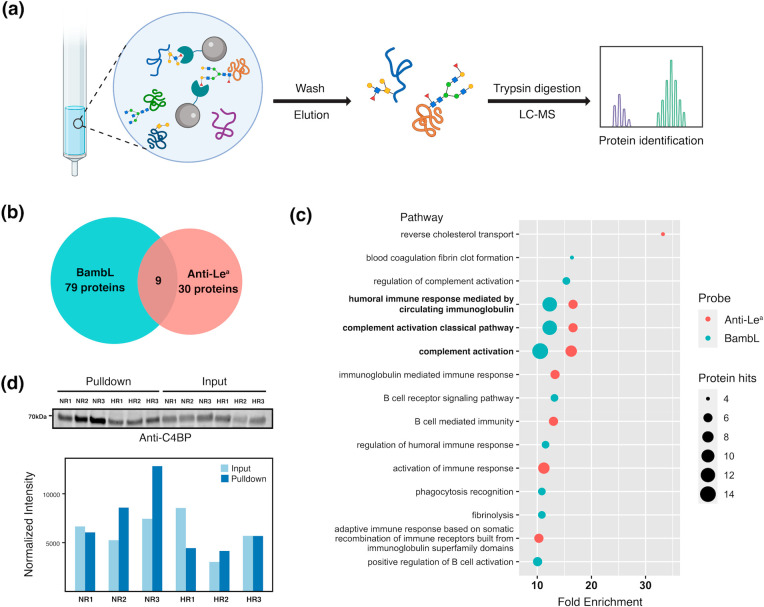
To gain more insight into the association between Lea and lack of vaccine efficacy, we performed glycoproteomic analysis using the anti-Lea antibody and BambL. In brief, we pooled the serum of all participants and performed pulldowns with either BambL or anti-Lea antibody. We then analyzed the isolated proteins by mass spectrometry (Figure 3a). After removal of all nonglycosylated proteins and those that bound to control beads, we identified 79 glycoproteins in the BambL pulldown sample and 30 for the anti-Lea pulldown. Glycoproteins enriched by the two probes are listed in Supporting Information, Table S4 and Table S5. Major glycoproteins enriched by BambL included immunoglobulins, complement proteins, cell adhesion molecules, protease inhibitors, and proteins in the blood coagulation pathways. As expected, the spectrum of proteins enriched by anti-Lea is narrower since BambL has a broader specificity than anti-Lea (Figure 3b). Gene ontology enrichment analysis showed enrichment for complement activation and humoral immunity in both samples. Among the pathways with more than 10-fold enrichment, the complement-related pathways had the highest number of protein hits (Figure 3c and Table 2).
Figure 3.
(a) Scheme of the experimental approach of glycoproteomic analysis. (b) Number of glycoproteins identified in BambL/anti-Lea pulldown experiments. (c) Gene ontology pathway enrichment analysis for glycoproteins enriched with BambL/anti-Lea. The false discovery rates (FDRs) of the enriched pathways shown are all <0.05. (d) Differential C4BP glycosylation. Western blot analysis for C4BP of BambL pulldown samples and corresponding input for three high responders (HR1, HR2, and HR3) and three nonresponders (NR1, NR2, and NR3) are shown. Signal intensities of the bands (normalized to total protein stain) are depicted in the bar plot.
Table 2. Complement-Related Serum Glycoproteins Enriched by BambL and/or Anti-Lea.
| Swiss-Prot accession number | entry name | protein name | BambL | Anti-Lea |
|---|---|---|---|
| P04003 | C4BPA_HUMAN | C4b-binding protein alpha chain | + | + |
| Q9NZP8 | C1RL_HUMAN | Complement C1r subcomponent-like protein | + | |
| P09871 | C1S_HUMAN | Complement C 1s subcomponent | + | |
| P01024 | CO3_HUMAN | Complement C3 | + | |
| P07357 | CO8A_HUMAN | Complement component C8 alpha chain | + | |
| P02748 | CO9_HUMAN | Complement component C9 | + | |
| P00751 | CFAB_HUMAN | Complement factor B | + | |
| P08603 | CFAH_HUMAN | Complement factor H | + | |
| P05156 | CFAI_HUMAN | Complement factor I | + | |
| Q15485 | FCN2_HUMAN | Ficolin-2 | + | |
| P48740 | MASP1_HUMAN | Mannan-binding lectin serine protease 1 | + | |
| O00187 | MASP2_HUMAN | Mannan-binding lectin serine protease 2 | + |
Of the glycoproteins identified in our analysis, we selected C4b-binding protein (C4BP) for validation because it was the most abundant complement protein in our analysis and was pulled down by both BambL and anti-Lea. C4BP has a high serum concentration (∼0.2 mg/mL),43 thus it may have a significant contribution to the differences in BambL binding observed in lectin microarrays. For our analysis, we performed BambL-pulldowns from six individual sera samples: three from nonresponders with high BambL binding and three from high responders with low BambL binding. We then performed Western blot analysis with an anti-C4BP antibody (Figure 3d). As expected, Western blot analysis showed that C4BP was pulled down by BambL in all samples. In nonresponders, we observed an enrichment in C4BP pulled down by BambL that was not due to a significant change in C4BP sera levels, as seen in the input samples. Our results confirm that complement component C4BP is differentially glycosylated in non- versus high responders.
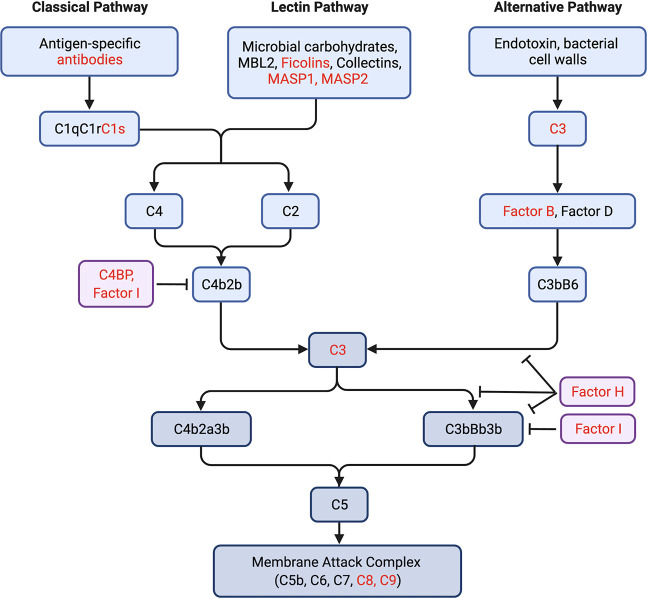
The complement system is an essential aspect of both innate and adaptive immune responses and can be activated by multiple pathways (Figure 4).44,45 The role of complement in vaccine efficacy is unclear, although several studies have found a direct link.46,47 Complement can be triggered both by immune lectins, such as MBL2 and ficolins, or by antibodies. Immunogens that can bind MBL2 and activate complement increase response to immunogens including influenza in mouse models.48,49 In line with this, deletion of the downstream complement component C3 in mice was found to lower antibody response to both live influenza virus and influenza antigen.50 There is also evidence that C2 deficiency may be associated with weak antibody response to pneumococcal vaccines.51 This collective work argues that complement activation is important in antibody response to vaccination. Any role glycosylation might have in the interplay between the complement cascade and antibody response has not yet been explored. However, in a study of complement activation in the oral cavity, it was found that activation was higher in secretors than in nonsecretors.52 In that work, C4 deposition was inhibited in the presence of fucose, implying a direct connection between glycosylation and the complement cascade. Whether glycosylation of complement itself alters the intricate binding networks required in this cascade, and whether interventions such as 2′-fucosyllactose can alter glycan composition of complement will be important points for future exploration. Our work suggests that the glycoform of complement proteins might play a role in mediating antibody induction in response to vaccination and as a result may predict immune response to vaccines.
Figure 4.
Multiple components of the complement activation pathways are glycosylated with Lea. For simplicity, only select components in the pathways are shown. Glycoproteins enriched by BambL or anti-Lea are colored in red.
Serum High-Mannose Is Downregulated and Fucosylation Is Upregulated in High Responders Postvaccination
To assess whether changes in sera glycosylation as a function of immunity occur postvaccination, we generated glycomic profiles of the day 28 sera (Figure 5a). In general, the d28 glycomes significantly correlated with the d0 glycomes of the same individuals, indicating stability of the overall serum glycome postvaccination compared to prevaccination (median correlation coefficient = 0.91, Supporting Information, Figure S9). However, vaccine induced changes were observed when we compared paired pre- (d0) and post- (d28) vaccination glycomes. In specific, we identified a loss of high mannose glycans postvaccination (Griffithsin, BanLecH84T, HHL, Supporting Information, Figure S10). This loss is clear in paired high responders but not in nonresponders (Figure 5b; Supporting Information, Figure S11 and Figure S12). In our prevaccination cohort, we also observed higher binding to BanLecH84T in participants categorized as nonresponders to the influenza B strains (Supporting Information, Figure S7 and Figure S8). High mannose is a ligand for the mannose binding lectin (MBL), the first step in the lectin-mediated complement pathway. As previously mentioned, engagement of MBL and complement has recently been shown to increase response to immunogens including influenza in mouse models.48,49 Whether the changes in high mannose observed in high responders are related to this phenomenon is a point for further analysis.
Figure 5.
(a) Heatmap of lectin microarray data for postvaccination (day 28) serum samples. Columns represent the participants and rows represent the probes. Shown is the normalized log2 ratios (Sample signal (S)/Reference signal (R)). Total response scores are annotated with a green-white sliding scale bar on the top of the heatmap. Rough specificities of select lectins are annotated on the right of the heatmap. (b,c) Boxplots of log2 fold-change for paired d28 and d0 samples in non- and high responders. (b) High-mannose binding lectins (Griffithisin and BanLecH84T). (c) Sialyl Lewis X binding probes (Anti-sLex). Paired Mann–Whitney U test was used to determine p-values. n.s.: not statistically significant (no difference is observed d28/d0); (*) p < 0.05. Glycans are shown in SNFG notation at the side of the boxplots. Symbols are defined as follows: galactose (yellow ●), N-acetylglucosamine (blue ■), mannose (green ●), sialic acid (purple ◆), fucose (red ▲).
We also observed overall upregulation of fucosylation (AAL) and of Lewis X antigens (Lex) postvaccination (Supporting Information, Figure S10). These changes are clear in high-responders, where we also observed increases in Lewis B antigen and sialyl-Lewis X (sLex) at d28 (Figure 5c; Supporting Information, Figure S11). No other clear shifts in glycan levels were identified. Interestingly, total serum immunoglobulins seemed to decrease slightly in nonresponders, as protein A, protein G, and protein L all had lower binding to postvaccination sera (Supporting Information, Figure S12).
Direct comparison between the d28 glycomes of high and nonresponders showed few probes with differential binding (Supporting Information, Figure S13). At d28, high responders have lower levels of high mannose (BanLecH84T) consistent with the loss observed upon vaccination. At this time point the difference in Lea is no longer seen. This is most likely due to the overall increase in fucosylation and Lewis antigens observed. Fucosylated glycans are increased in macrophages and other cells, such as intestinal epithelia, upon immune stimulation, consistent with our results.53
Conclusion
Awareness of how vaccine effectiveness varies across populations is currently at an all-time high. The underlying reasons for such differences, however, are still opaque. Until recently, glycosylation, which is a critical modulator of immunity, has been missing from the picture. With the exception of glycosylation on IgG, there is no information on how glycosylation of proteins in the sera impacts or correlates with vaccination. In this exploratory study, we examine the glycosylation of sera glycoproteins pre- and postvaccination with the FLUZONE influenza vaccine in 160 individuals. Our data identified high baseline levels of Lea on sera glycoproteins, a potential indicator of FUT2 levels and secretor status, as a possible marker of unresponsiveness to vaccination. There is emerging literature identifying a role for FUT2 in mediating immunity, with potential pathways for ameliorating the absence of this enzyme.42 Glycoproteomics showed that Lea was enriched in complement components such as C4BP. Multiple studies argue a direct connection between complement activation and vaccine efficacy, although the exact mechanisms are still unclear.46−49 Our data argue that this may partly be due to the importance of glycoforms in mediating complement, which have not previously been considered. Further studies to explore how glycans such as Lea influence the functions of serum glycoproteins are warranted, especially in regard to components of the complement system and the predictive ability of glycoforms to identify nonresponders to specific vaccines.
Acknowledgments
Some graphical contents were created with biorender.com. This project has been funded by the National Institute of Allergy and Infectious Diseases, a component of the NIH, Department of Health and Human Services, under contract 75N93019C00052.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.2c00251.
Figure S1: Volcano plot comparing lectin microarray data for high responders and nonresponders prevaccination; Figure S2: Volcano plot comparing lectin microarray data for high responders and low/moderate responders prevaccination; Figure S3: Association of multiple factors with microarray data of BambL and anti-Lea data; Figure S4: Volcano plot comparing lectin microarray data for redefined high responders and nonresponders prevaccination; Figure S5: Volcano plot comparing lectin microarray data for high responders and nonresponders prevaccination to the A/H1N1 (A/Brisbane/02/2018) strain; Figure S6: Volcano plot comparing lectin microarray data for high responders and nonresponders prevaccination to the A/H3N2 (A/Kansas/14/2017) strain; Figure S7: Volcano plot comparing lectin microarray data for high responders and nonresponders prevaccination to the B/Yamagata (B/Phuket/3073/2013) strain; Figure S8: Volcano plot comparing lectin microarray data for high responders and nonresponders prevaccination to the B/Victoria (B/Colorado/6/2017-like) strain; Figure S9: Boxplot analysis of the correlation coefficient between pre- and postvaccination lectin microarray data; Figure S10: Volcano plots comparing the paired pre- and postvaccination lectin microarray data of all participants; Figure S11: Volcano plots comparing the paired pre- and postvaccination lectin microarray data of high responders; Figure S12: Volcano plots comparing the paired pre- and postvaccination lectin microarray data of nonresponders; Figure S13: Volcano plot comparing lectin microarray data for high responders and low/moderate-responders postvaccination; Table S1: List of Probes Printed on Lectin Microarrays; Table S3: Serum Glycoproteins Enriched by BambL; Table S4: Serum Glycoproteins Enriched by Anti-Lea; Table S5: Characteristics of Study Participants Grouped by Strain-Specific Response (PDF)
Table S2: Raw LFQ Data from Pulldowns (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Dhakal S.; Klein S. L. Host Factors Impact Vaccine Efficacy: Implications for Seasonal and Universal Influenza Vaccine Programs. J. Virol. 2019, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano A. D.; Roguski K. M.; Chang H. H.; Muscatello D. J.; Palekar R.; Tempia S.; Cohen C.; Gran J. M.; Schanzer D.; Cowling B. J.; Wu P.; Kyncl J.; Ang L. W.; Park M.; Redlberger-Fritz M.; Yu H.; Espenhain L.; Krishnan A.; Emukule G.; van Asten L.; Pereira da Silva S.; Aungkulanon S.; Buchholz U.; Widdowson M. A.; Bresee J. S.; Azziz-Baumgartner E.; Cheng P. Y.; Dawood F.; Foppa I.; Olsen S.; Haber M.; Jeffers C.; MacIntyre C. R.; Newall A. T.; Wood J. G.; Kundi M.; Popow-Kraupp T.; Ahmed M.; Rahman M.; Marinho F.; Sotomayor Proschle C. V.; Vergara Mallegas N.; Luzhao F.; Sa L.; Barbosa-Ramírez J.; Sanchez D. M.; Gomez L. A.; Vargas X. B.; Acosta Herrera a. B.; Llanés M. J.; Fischer T. K.; Krause T. G.; Mølbak K.; Nielsen J.; Trebbien R.; Bruno A.; Ojeda J.; Ramos H.; an der Heiden M.; del Carmen Castillo Signor L.; Serrano C. E.; Bhardwaj R.; Chadha M.; Narayan V.; Kosen S.; Bromberg M.; Glatman-Freedman A.; Kaufman Z.; Arima Y.; Oishi K.; Chaves S.; Nyawanda B.; Al-Jarallah R. A.; Kuri-Morales P. A.; Matus C. R.; Corona M. E. J.; Burmaa A.; Darmaa O.; Obtel M.; Cherkaoui I.; van den Wijngaard C. C.; van der Hoek W.; Baker M.; Bandaranayake D.; Bissielo A.; Huang S.; Lopez L.; Newbern C.; Flem E.; Grøneng G. M.; Hauge S.; de Cosío F. G.; de Moltó Y.; Castillo L. M.; Cabello M. A.; von Horoch M.; Medina Osis J.; Machado A.; Nunes B.; Rodrigues A. P.; Rodrigues E.; Calomfirescu C.; Lupulescu E.; Popescu R.; Popovici O.; Bogdanovic D.; Kostic M.; Lazarevic K.; Milosevic Z.; Tiodorovic B.; Chen M.; Cutter J.; Lee V.; Lin R.; Ma S.; Cohen A. L.; Treurnicht F.; Kim W. J.; Delgado-Sanz C.; de mateo Ontañón S.; Larrauri A.; León I. L.; Vallejo F.; Born R.; Junker C.; Koch D.; Chuang J. H.; Huang W. T.; Kuo H. W.; Tsai Y. C.; Bundhamcharoen K.; Chittaganpitch M.; Green H. K.; Pebody R.; Goñi N.; Chiparelli H.; Brammer L.; Mustaquim D. Estimates of Global Seasonal Influenza-Associated Respiratory Mortality: A Modelling Study. Lancet 2018, 391 (10127), 1285–1300. 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A. M.; Kim I. K.; Reed C.; Thompson M.; Chaves S. S.; Finelli L.; Bresee J. Modeling the Effect of Different Vaccine Effectiveness Estimates on the Number of Vaccine-Prevented Influenza-Associated Hospitalizations in Older Adults. Clin. Infect. Dis. 2014, 59 (3), 406–409. 10.1093/cid/ciu328. [DOI] [PubMed] [Google Scholar]
- Evaluation of Influenza Vaccine Effectiveness: A Guide to the Design and Interpretation of Observational Studies; World Health Organization: Geneva, Switzerland, 2017; pp 1–47. [Google Scholar]
- Potluri T.; Fink A. L.; Sylvia K. E.; Dhakal S.; Vermillion M. S.; vom Steeg L.; Deshpande S.; Narasimhan H.; Klein S. L. Age-Associated Changes in the Impact of Sex Steroids on Influenza Vaccine Responses in Males and Females. npj Vaccines 2019, 4 (1), 29. 10.1038/s41541-019-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J. S.; Dobano C.; VanDamme P.; Moncunill G.; Marchant A.; Othman R. B.; Sadarangani M.; Koff W. C.; Kollmann T. R. Improving Vaccine-Induced Immunity: Can Baseline Predict Outcome?. Trends Immunol. 2020, 41 (6), 457–465. 10.1016/j.it.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J. S.; Schwartzberg P. L.; Kotliarov Y.; Biancotto A.; Xie Z.; Germain R. N.; Wang E.; Olnes M. J.; Narayanan M.; Golding H.; Moir S.; Dickler H. B.; Perl S.; Cheung F.; et al. Global Analyses of Human Immune Variation Reveal Baseline Predictors of Postvaccination Responses. Cell 2014, 157 (2), 499–513. 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Biological Roles of Glycans. Glycobiology 2017, 27 (1), 3–49. 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R.; Mahal L. K. The Host Glycomic Response to Pathogens. Curr. Opin. Struct. Biol. 2021, 68, 149–156. 10.1016/j.sbi.2020.12.011. [DOI] [PubMed] [Google Scholar]
- Irvine E. B.; Alter G. Understanding the Role of Antibody Glycosylation through the Lens of Severe Viral and Bacterial Diseases. Glycobiology 2020, 30 (4), 241–253. 10.1093/glycob/cwaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G.; Ottenhoff T. H. M.; Joosten S. A. Antibody Glycosylation in Inflammation, Disease and Vaccination. Semin. Immunol. 2018, 39 (June), 102–110. 10.1016/j.smim.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura Y.; Katoh T.; Saldova R.; O’Flaherty R.; Izumi T.; Mimura-Kimura Y.; Utsunomiya T.; Mizukami Y.; Yamamoto K.; Matsumoto T.; Rudd P. M. Glycosylation Engineering of Therapeutic IgG Antibodies: Challenges for the Safety, Functionality and Efficacy. Protein Cell 2018, 9 (1), 47–62. 10.1007/s13238-017-0433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman M. H. J.; De Jong S. E.; Soonawala D.; Kroon F. P.; Adegnika A. A.; Deelder A. M.; Hokke C. H.; Yazdanbakhsh M.; Wuhrer M. Changes in Antigen-Specific IgG1 Fc N-Glycosylation upon Influenza and Tetanus Vaccination. Mol. Cell. Proteomics 2012, 11 (4), 1–10. 10.1074/mcp.M111.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. T.; Maamary J.; Tan G. S.; Bournazos S.; Davis C. W.; Krammer F.; Schlesinger S. J.; Palese P.; Ahmed R.; Ravetch J. V. Anti-HA Glycoforms Drive B Cell Affinity Selection and Determine Influenza Vaccine Efficacy. Cell 2015, 162 (1), 160–169. 10.1016/j.cell.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilobello K. T.; Slawek D. E.; Mahal L. K. A Ratiometric Lectin Microarray Approach to Analysis of the Dynamic Mammalian Glycome. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (28), 11534–11539. 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista B. S.; Eng W. S.; Pilobello K. T.; Hendricks-Muñoz K. D.; Mahal L. K. Identification of a Conserved Glycan Signature for Microvesicles. J. Proteome Res. 2011, 10 (10), 4624–4633. 10.1021/pr200434y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Kasper B.; Zhang B.; Lashua L. P.; Ross T. M.; Ghedin E.; Mahal L. K. Age-Dependent Glycomic Response to the 2009 Pandemic H1N1 Influenza Virus and Its Association with Disease Severity. J. Proteome Res. 2020, 19 (11), 4486–4495. 10.1021/acs.jproteome.0c00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel D. W.; Koppolu S.; Zhang Y.; Kasper B.; Meche L.; Vaiana C. A.; Bissel S. J.; Carter C. E.; Kelvin A. A.; Elaish M.; Lopez-Orozco J.; Zhang B.; Zhou B.; Chou T.-W.; Lashua L.; Hobman T. C.; Ross T. M.; Ghedin E.; Mahal L. K. Glycomic Analysis of Host Response Reveals High Mannose as a Key Mediator of Influenza Severity. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (43), 26926–26935. 10.1073/pnas.2008203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley D.; Mani V. R.; Mohan N.; Akkad N.; Ochi A.; Heindel D. W.; Lee K. B.; Zambirinis C. P.; Pandian G. S. D. B.; Savadkar S.; Torres-Hernandez A.; Nayak S.; Wang D.; Hundeyin M.; Diskin B.; Aykut B.; Werba G.; Barilla R. M.; Rodriguez R.; Chang S.; Gardner L.; Mahal L. K.; Ueberheide B.; Miller G. Dectin 1 Activation on Macrophages by Galectin 9 Promotes Pancreatic Carcinoma and Peritumoral Immune Tolerance. Nat. Med. 2017, 23 (5), 556–567. 10.1038/nm.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz E.; Chen S.; Vucic E.; Baptiste G.; Loomis C.; Agrawal P.; Hajdu C.; Bar-Sagi D.; Mahal L. K. Integrated Systems Analysis of the Murine and Human Pancreatic Cancer Glycomes Reveals a Tumor-Promoting Role for ST6GAL1. Mol. Cell. Proteomics 2021, 20, 100160. 10.1016/j.mcpro.2021.100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal P.; Fontanals-Cirera B.; Sokolova E.; Jacob S.; Vaiana C. A.; Argibay D.; Davalos V.; McDermott M.; Nayak S.; Darvishian F.; Castillo M.; Ueberheide B.; Osman I.; Fenyö D.; Mahal L. K.; Hernando E. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell 2017, 31 (6), 804–819. 10.1016/j.ccell.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli M. J.; Shaw P. A.; Han A.; Czajkowski L.; Reed S.; Athota R.; Bristol T.; Fargis S.; Risos K.; Powers J. H.; Davey R. T.; Taubenberger J. K. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. MBio 2016, 7 (2), e00417–16. 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.; Nachbagauer R.; Balmaseda A.; Stadlbauer D.; Ojeda S.; Patel M.; Rajabhathor A.; Lopez R.; Guglia A. F.; Sanchez N.; Amanat F.; Gresh L.; Kuan G.; Krammer F.; Gordon A. Novel Correlates of Protection against Pandemic H1N1 Influenza A Virus Infection. Nat. Med. 2019, 25 (6), 962–967. 10.1038/s41591-019-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.; Ross T. M.; Carlock M. A.; Ghedin E.; Choi H.; Vogel C. Evaluation of Determinants of the Serological Response to the Quadrivalent Split-inactivated Influenza Vaccine. Mol. Syst. Biol. 2022, 18 (5), 1–14. 10.15252/msb.202110724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilobello K. T.; Agrawal P.; Rouse R.; Mahal L. K. Advances in Lectin Microarray Technology: Optimized Protocols for Piezoelectric Print Conditions. Curr. Protoc. Chem. Biol. 2013, 5 (1), 1–23. 10.1002/9780470559277.ch120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojar D.; Meche L.; Meng G.; Eng W.; Smith D. F.; Cummings R. D.; Mahal L. K. A Useful Guide to Lectin Binding: Machine-Learning Directed Annotation of 57 Unique Lectin Specificities. ACS Chem. Biol. 2022, 10.1021/acschembio.1c00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York W. S.; Mazumder R.; Ranzinger R.; Edwards N.; Kahsay R.; Aoki-Kinoshita K. F.; Campbell M. P.; Cummings R. D.; Feizi T.; Martin M.; Natale D. A.; Packer N. H.; Woods R. J.; Agarwal G.; Arpinar S.; Bhat S.; Blake J.; Castro L. J. G.; Fochtman B.; Gildersleeve J.; Goldman R.; Holmes X.; Jain V.; Kulkarni S.; Mahadik R.; Mehta A.; Mousavi R.; Nakarakommula S.; Navelkar R.; Pattabiraman N.; Pierce M. J.; Ross K.; Vasudev P.; Vora J.; Williamson T.; Zhang W. GlyGen: Computational and Informatics Resources for Glycoscience. Glycobiology 2020, 30 (2), 72–73. 10.1093/glycob/cwz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S.; Douglass E.; Good B. M.; Unni D. R.; Harris N. L.; Mungall C. J.; Basu S.; Chisholm R. L.; Dodson R. J.; Hartline E.; Fey P.; Thomas P. D.; Albou L. P.; Ebert D.; Kesling M. J.; Mi H.; Muruganujan A.; Huang X.; Mushayahama T.; LaBonte S. A.; Siegele D. A.; Antonazzo G.; Attrill H.; Brown N. H.; Garapati P.; Marygold S. J.; Trovisco V.; dos Santos G.; Falls K.; Tabone C.; Zhou P.; Goodman J. L.; Strelets V. B.; Thurmond J.; Garmiri P.; Ishtiaq R.; Rodríguez-López M.; Acencio M. L.; Kuiper M.; Lægreid A.; Logie C.; Lovering R. C.; Kramarz B.; Saverimuttu S. C. C.; Pinheiro S. M.; Gunn H.; Su R.; Thurlow K. E.; Chibucos M.; Giglio M.; Nadendla S.; Munro J.; Jackson R.; Duesbury M. J.; Del-Toro N.; Meldal B. H. M.; Paneerselvam K.; Perfetto L.; Porras P.; Orchard S.; Shrivastava A.; Chang H. Y.; Finn R. D.; Mitchell A. L.; Rawlings N. D.; Richardson L.; Sangrador-Vegas A.; Blake J. A.; Christie K. R.; Dolan M. E.; Drabkin H. J.; Hill D. P.; Ni L.; Sitnikov D. M.; Harris M. A.; Oliver S. G.; Rutherford K.; Wood V.; Hayles J.; Bähler J.; Bolton E. R.; de Pons J. L.; Dwinell M. R.; Hayman G. T.; Kaldunski M. L.; Kwitek A. E.; Laulederkind S. J. F.; Plasterer C.; Tutaj M. A.; Vedi M.; Wang S. J.; D’Eustachio P.; Matthews L.; Balhoff J. P.; Aleksander S. A.; Alexander M. J.; Cherry J. M.; Engel S. R.; Gondwe F.; Karra K.; Miyasato S. R.; Nash R. S.; Simison M.; Skrzypek M. S.; Weng S.; Wong E. D.; Feuermann M.; Gaudet P.; Morgat A.; Bakker E.; Berardini T. Z.; Reiser L.; Subramaniam S.; Huala E.; Arighi C. N.; Auchincloss A.; Axelsen K.; Argoud-Puy G.; Bateman A.; Blatter M. C.; Boutet E.; Bowler E.; Breuza L.; Bridge A.; Britto R.; Bye-A-Jee H.; Casas C. C.; Coudert E.; Denny P.; Es-Treicher A.; Famiglietti M. L.; Georghiou G.; Gos A. N.; Gruaz-Gumowski N.; Hatton-Ellis E.; Hulo C.; Ignatchenko A.; Jungo F.; Laiho K.; Le Mercier P.; Lieberherr D.; Lock A.; Lussi Y.; MacDougall A.; Ma-Grane M.; Martin M. J.; Masson P.; Natale D. A.; Hyka-Nouspikel N.; Orchard S.; Pedruzzi I.; Pourcel L.; Poux S.; Pundir S.; Rivoire C.; Speretta E.; Sundaram S.; Tyagi N.; Warner K.; Zaru R.; Wu C. H.; Diehl A. D.; Chan J. N.; Grove C.; Lee R. Y. N.; Muller H. M.; Raciti D.; van Auken K.; Sternberg P. W.; Berriman M.; Paulini M.; Howe K.; Gao S.; Wright A.; Stein L.; Howe D. G.; Toro S.; Westerfield M.; Jaiswal P.; Cooper L.; Elser J. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49 (D1), D325–D334. 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H.; Ebert D.; Muruganujan A.; Mills C.; Albou L. P.; Mushayamaha T.; Thomas P. D. PANTHER Version 16: A Revised Family Classification, Tree-Based Classification Tool, Enhancer Regions and Extensive API. Nucleic Acids Res. 2021, 49 (D1), D394–D403. 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y.; Eng W. S.; Colquhoun D. R.; Dinglasan R. R.; Graham D. R.; Mahal L. K. Complex N-Linked Glycans Serve as a Determinant for Exosome/Microvesicle Cargo Recruitment. J. Biol. Chem. 2014, 289 (47), 32526–32537. 10.1074/jbc.M114.606269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audfray A.; Claudinon J.; Abounit S.; Ruvoën-Clouet N.; Larson G.; Smith D. F.; Wimmerová M.; Le Pendu J.; Römer W.; Varrot A.; Imberty A. Fucose-Binding Lectin from Opportunistic Pathogen Burkholderia Ambifaria Binds to Both Plant and Human Oligosaccharidic Epitopes. J. Biol. Chem. 2012, 287 (6), 4335–4347. 10.1074/jbc.M111.314831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. A.; Slack G. S.; Marriott A. C.; Kane J. A.; Whittaker C. J.; Silman N. J.; Carroll M. W.; Gooch K. E. Cellular Immune Response to Human Influenza Viruses Differs between H1N1 and H3N2 Subtypes in the Ferret Lung. PLoS One 2018, 13 (9), e0202675 10.1371/journal.pone.0202675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Huang Y.; Xu K.; Liu Y.; Li X.; Xu Y.; Zhong W.; Hao P. Differential Responses of Innate Immunity Triggered by Different Subtypes of Influenza a Viruses in Human and Avian Hosts. BMC Med. Genomics 2017, 10 (Suppl 4), 41–54. 10.1186/s12920-017-0304-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucardo F.; Nordgren J.; Reyes Y.; Gonzalez F.; Sharma S.; Svensson L. The Lewis A Phenotype Is a Restriction Factor for Rotateq and Rotarix Vaccine-Take in Nicaraguan Children. Sci. Rep. 2018, 8 (1), 1–8. 10.1038/s41598-018-19718-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooling L. Blood Groups in Infection and Host Susceptibility. Clin. Microbiol. Rev. 2015, 28 (3), 801–870. 10.1128/CMR.00109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampaoli O.; Conta G.; Calvani R.; Miccheli A. Can the FUT2 Non-Secretor Phenotype Associated With Gut Microbiota Increase the Children Susceptibility for Type 1 Diabetes? A Mini Review. Front. Nutr. 2020, 7, 1–9. 10.3389/fnut.2020.606171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M.; Luo G.; Lu R.; Shi W.; Cheng H.; Lu Y.; Jin K.; Yang C.; Wang Z.; Long J.; Xu J.; Ni Q.; Liu C.; Yu X. Distribution of Lewis and Secretor Polymorphisms and Corresponding CA19–9 Antigen Expression in a Chinese Population. FEBS Open Bio 2017, 7 (11), 1660–1671. 10.1002/2211-5463.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L. C.; Brewer-Jensen P. D.; Mallory M. L.; Jensen K.; Yount B. L.; Costantini V.; Collins M. H.; Edwards C. E.; Sheahan T. P.; Vinjé J.; Baric R. S. Virus–Host Interactions Between Nonsecretors and Human Norovirus. Cell. Mol. Gastroenterol. Hepatol. 2020, 10 (2), 245–267. 10.1016/j.jcmgh.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacklin P.; Mäkivuokko H.; Alakulppi N.; Nikkilä J.; Tenkanen H.; Räbinä J.; Partanen J.; Aranko K.; Mättö J. Secretor Genotype (FUT2 Gene) Is Strongly Associated with the Composition of Bifidobacteria in the Human Intestine. PLoS One 2011, 6 (5), e20113. 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacklin P.; Tuimala J.; Nikkilä J.; Tims S.; Mäkivuokko H.; Alakulppi N.; Laine P.; Rajilic-Stojanovic M.; Paulin L.; De Vos W. M.; Mättö J. Faecal Microbiota Composition in Adults Is Associated with the FUT2 Gene Determining the Secretor Status. PLoS One 2014, 9 (4), e94863 10.1371/journal.pone.0094863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borey M.; Blanc F.; Lemonnier G.; Leplat J. J.; Jardet D.; Rossignol M. N.; Ravon L.; Billon Y.; Bernard M.; Estellé J.; Rogel-Gaillard C. Links between Fecal Microbiota and the Response to Vaccination against Influenza A Virus in Pigs. npj Vaccines 2021, 6 (1), 92. 10.1038/s41541-021-00351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L.; Leusink-Muis T.; Kettelarij N.; van Ark I.; Blijenberg B.; Hesen N. A.; Stahl B.; Overbeek S. A.; Garssen J.; Folkerts G.; van’t Land B. Human Milk Oligosaccharide 2’-Fucosyllactose Improves Innate and Adaptive Immunity in an Influenza-Specific Murine Vaccination Model. Front. Immunol. 2018, 9 (MAR), 1–16. 10.3389/fimmu.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.; Gottsäter A.; Nilsson P. M.; Mollnes T. E.; Lindblad B.; Blom A. M. Complement Activation and Plasma Levels of C4b-Binding Protein in Critical Limb Ischemia Patients. J. Vasc. Surg. 2009, 50 (1), 100–106. 10.1016/j.jvs.2008.12.033. [DOI] [PubMed] [Google Scholar]
- Merle N. S.; Church S. E.; Fremeaux-Bacchi V.; Roumenina L. T. Complement System Part I - Molecular Mechanisms of Activation and Regulation. Front. Immunol. 2015, 6 (JUN), 1–30. 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle N. S.; Noe R.; Halbwachs-Mecarelli L.; Fremeaux-Bacchi V.; Roumenina L. T. Complement System Part II: Role in Immunity. Front. Immunol. 2015, 6 (MAY), 1–26. 10.3389/fimmu.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehen N.; Stover C. The Role of Complement in the Success of Vaccination with Conjugated vs. Unconjugated Polysaccharide Antigen. Vaccine 2008, 26 (4), 451–459. 10.1016/j.vaccine.2007.11.049. [DOI] [PubMed] [Google Scholar]
- Mellors J.; Tipton T.; Longet S.; Carroll M. Viral Evasion of the Complement System and Its Importance for Vaccines and Therapeutics. Front. Immunol. 2020, 11 (July), 1–20. 10.3389/fimmu.2020.01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read B. J.; Won L.; Kraft J. C.; Sappington I.; Aung A.; Wu S.; Bals J.; Chen C.; Lee K. K.; Lingwood D.; King N. P.; Irvine D. J. Mannose-Binding Lectin and Complement Mediate Follicular Localization and Enhanced Immunogenicity of Diverse Protein Nanoparticle Immunogens. Cell Rep. 2022, 38 (2), 110217. 10.1016/j.celrep.2021.110217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokatlian T.; Read B. J.; Jones C. A.; Kulp D. W.; Menis S.; Chang J. Y. H.; Steichen J. M.; Kumari S.; Allen J. D.; Dane E. L.; Liguori A.; Sangesland M.; Lingwood D.; Crispin M.; Schief W. R.; Irvine D. J. Innate Immune Recognition of Glycans Targets HIV Nanoparticle Immunogens to Germinal Centers. Science (80-.). 2019, 363 (6427), 649–654. 10.1126/science.aat9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-J.; Kim K.-H.; Ko E.-J.; Kim M.-C.; Lee Y.-N.; Jung Y.-J.; Lee Y.-T.; Kwon Y.-M.; Song J.-M.; Kang S.-M. Complement C3 Plays a Key Role in Inducing Humoral and Cellular Immune Responses to Influenza Virus Strain-Specific Hemagglutinin-Based or Cross-Protective M2 Extracellular Domain-Based Vaccination. J. Virol. 2018, 92 (20), 1–14. 10.1128/JVI.00969-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingels H.; Schejbel L.; Lundstedt A. C.; Jensen L.; Laursen I. A.; Ryder L. P.; Heegaard N. H. H.; Konradsen H.; Christensen J. J.; Heilmann C.; Marquart H. V. Immunodeficiency among Children with Recurrent Invasive Pneumococcal Disease. Pediatr. Infect. Dis. J. 2015, 34 (6), 644–651. 10.1097/INF.0000000000000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunput S. T. G.; Ligtenberg A. J. M.; Terlouw B.; Brouwer M.; Veerman E. C. I.; Wouters D. Complement Activation by Salivary Agglutinin Is Secretor Status Dependent. Biol. Chem. 2015, 396 (1), 35–43. 10.1515/hsz-2014-0200. [DOI] [PubMed] [Google Scholar]
- Li J.; Hsu H. C.; Mountz J. D.; Allen J. G. Unmasking Fucosylation: From Cell Adhesion to Immune System Regulation and Diseases. Cell Chem. Biol. 2018, 25 (5), 499–512. 10.1016/j.chembiol.2018.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.