Two new salts of the 4-(4-nitrophenyl)piperazin-1-ium cation have been prepared by co-crystallization with aromatic carboxylic acids. The supramolecular assembly in the benzoate salt, which crystallizes as a mono-hydrate, is two dimensional, while that in the 2-carboxy-4,6-dinitrophenolate salt is three dimensional.
Keywords: piperazine, synthesis, crystal structure, molecular structure, hydrogen bonding, supramolecular assembly
Abstract
Crystal structures are reported for two molecular salts containing the 4-(4-nitrophenyl)piperazin-1-ium cation. Co-crystallization from methanol/ethyl acetate solution of N-(4-nitrophenyl)piperazine with benzoic acid gives the benzoate salt, which crystallizes as a monohydrate, C10H14N3O2·C7H5O2·H2O, (I), and similar co-crystallization with 3,5-dinitrosalicylic acid yields the 2-carboxy-4,6-dinitrophenolate salt, C10H14N3O2·C7H3N2O7, (II). In the structure of (I), a combination of O—H⋯O, N—H⋯O and C—H⋯O hydrogen bonds links the components into sheets, while in the structure of (II), the supramolecular assembly, generated by hydrogen bonds of the same types as in (I), is three dimensional. Comparisons are made with the structures of some related compounds.
1. Chemical context
Piperazines and substituted piperazines are important pharmacophores, which can be found in many biologically active compounds (Berkheij, 2005 ▸) such as antifungal (Upadhayaya et al., 2004 ▸), anti-bacterial, anti-malarial and anti-psychotic agents (Chaudhary et al., 2006 ▸). Both the general pharmacological and specific antimicrobial activities of piperazine derivatives have been reviewed in recent years (Elliott, 2011 ▸; Kharb et al., 2012 ▸). Among specific examples of piperazine derivatives, N-(4-nitrophenyl)piperazine has found use in the control of potassium channels (Lu, 2007 ▸). The crystal structures of a number of 4-(4-nitrophenyl)piperazin-1-ium salts have been reported (Lu, 2007 ▸; Mahesha et al., 2022 ▸), and here we report the molecular and supramolecular structures of two further representatives of this family of salts, namely 4-(4-nitrophenyl)piperazin-1-ium benzoate monohydrate, C10H14N3O2·C7H5O2·H2O, (I), and 4-(4-nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate, C10H14N3O2·C7H3N2O7, (II).
2. Structural commentary
In each of compounds (I) and (II) (Figs. 1 ▸ and 2 ▸), the piperazine ring adopts a chair conformation, with the ring-puckering angle θ (Cremer & Pople, 1975 ▸) calculated for the atom sequence (N11/C12/C13/N14/C15/C16) close to the ideal value of zero (Boeyens, 1978 ▸): θ = 6.42 (11) for (I) and 8.75 (11)° for (II). However, in (I), the nitrophenyl substituent occupies an equatorial site, whereas in (II) this substituent occupies an axial site. In each compound, the N-nitrophenyl unit shows the pattern of distances typical of 4-nitroaniline derivatives, namely both C—N distances are short for their types (Allen et al., 1987 ▸), while the nitro N—O distances are long for their type. In addition, the distances C141—C142 and C141—C146 lie in the range 1.4049 (16) to 1.4132 (15) Å whereas the remaining C—C distances for this ring are smaller, falling in the range 1.3764 (17) to 1.3881 (15) Å. These variations are most simply interpreted in terms of some 1,4-quinonoid type bond fixation, moderated by the high electronegativity of the nitro group, generally regarded as similar to that of a fluoro substituent (Huheey, 1966 ▸; Mullay, 1985 ▸).
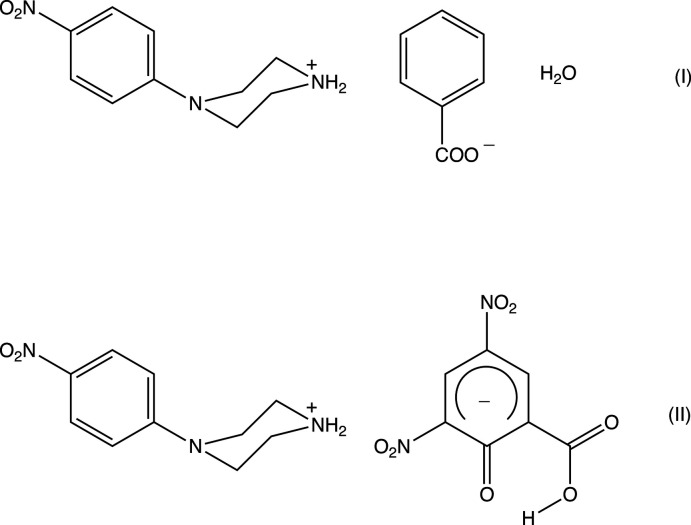
Figure 1.
The molecular structure of (I), showing hydrogen bonds (drawn as dashed lines) within the selected asymmetric unit. Displacement ellipsoids are drawn at the 50% probability level.
Figure 2.
The molecular structure of (II), showing hydrogen bonds (drawn as dashed lines) within the selected asymmetric unit. Displacement ellipsoids are drawn at the 50% probability level.
In the anion of compound (II), the C21—O21 distance, 1.2788 (13) Å is more typical of those in ketones than those in phenols (Allen et al., 1987 ▸); the distances C21—C22 and C21—C26, 1.4394 (15) and 1.4340 (15) Å are longer than the remaining C—C distances in the ring, which are in the range 1.3747 (15) to 1.3869 (15). These observations, taken together, indicate that the negative charge in this anion is delocalized over atoms C22–C26 rather than being localized on atom O21 (see Scheme).
3. Supramolecular features
In each of compounds (I) and (II), the supramolecular assembly involves a combination of O—H⋯O, N—H⋯O and C—H⋯O hydrogen bonds, augmented in the case of (I) by a single C—H⋯π(arene) hydrogen bond: however, aromatic π–π stacking interactions are absent from both structures.
The supramolecular assembly in (I) is di-periodic and the formation of the sheet structure is readily analysed in terms of two mono-periodic sub-structures (Ferguson et al., 1998a
▸,b
▸; Gregson et al., 2000 ▸). Within the selected asymmetric unit for (I) (Fig. 1 ▸), the ionic components are linked by an asymmetric bifurcated (three-centre) N—H⋯(O,O) hydrogen bond (Table 1 ▸), while the water molecule is linked to the anion by an O—H⋯O hydrogen bond. In one of the two sub-structures, a combination of one two-centre N—H⋯O hydrogen bond and a second O—H⋯O hydrogen bond links these three-component aggregates (Fig. 1 ▸) into a chain of rings running parallel to the [100] direction (Fig. 3 ▸) in which there are two different types of
 (12) ring (Bernstein et al., 1995 ▸), centred at (n, 0.5, 0.5) and (n + 0.5, 0.5, 0.5), respectively, where n represents an integer in each case. The second sub-structure, which includes the C—H⋯O hydrogen bond (Table 1 ▸, Fig. 4 ▸), takes the form of another chain of rings in which
(12) ring (Bernstein et al., 1995 ▸), centred at (n, 0.5, 0.5) and (n + 0.5, 0.5, 0.5), respectively, where n represents an integer in each case. The second sub-structure, which includes the C—H⋯O hydrogen bond (Table 1 ▸, Fig. 4 ▸), takes the form of another chain of rings in which
 (12) rings centred at (n + 0.5, n + 0.5, 0.5) alternate with
(12) rings centred at (n + 0.5, n + 0.5, 0.5) alternate with
 (10) rings centred at (n, n, 0.5), where n again represents an integer, so forming a chain of rings running parallel to the [110] direction (Fig. 4 ▸). The combination of chains along [100] and [110] generates a sheet structure lying parallel to (001). The single C—H⋯π(arene) hydrogen bond (Table 1 ▸) lies within this sheet, and so has no influence on the dimensionality of the assembly.
(10) rings centred at (n, n, 0.5), where n again represents an integer, so forming a chain of rings running parallel to the [110] direction (Fig. 4 ▸). The combination of chains along [100] and [110] generates a sheet structure lying parallel to (001). The single C—H⋯π(arene) hydrogen bond (Table 1 ▸) lies within this sheet, and so has no influence on the dimensionality of the assembly.
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
Cg1 is the centroid of the C21–C26 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N11—H11⋯O271 | 0.926 (14) | 2.564 (14) | 3.1009 (13) | 117.4 (10) |
| N11—H11⋯O272 | 0.926 (14) | 1.857 (14) | 2.7781 (12) | 172.9 (13) |
| N11—H12⋯O31i | 0.920 (15) | 1.884 (15) | 2.7965 (14) | 171.0 (12) |
| O31—H31⋯O271 | 0.892 (18) | 1.757 (18) | 2.6486 (13) | 179 (3) |
| O31—H32⋯O272ii | 0.908 (17) | 1.862 (17) | 2.7581 (12) | 168.8 (16) |
| C12—H12B⋯O272iii | 0.99 | 2.45 | 3.3751 (15) | 156 |
| C146—H146⋯Cg1iv | 0.95 | 2.67 | 3.4363 (13) | 138 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 .
.
Figure 3.
Part of the crystal structure of compound (I) showing the formation of a chain of hydrogen-bonded rings running parallel to the [100] direction. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have all been omitted.
Figure 4.
Part of the crystal structure of compound (I) showing the formation of a chain of hydrogen-bonded rings running parallel to the [110] direction. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to those C atoms that are not involved in the motif shown have been omitted.
The supramolecular assembly for compound (II), by contrast, is tri-periodic (three dimensional) and, as for (I), the formation of the framework is readily analysed in terms of simple sub-structures. Within the selected asymmetric unit (Fig. 2 ▸), there is an intramolecular O—H⋯O hydrogen bond in the anion, and the hydroxyl H atom plays no part in the supramolecular assembly. The two independent components are linked by a very asymmetric bifurcated N—H⋯(O,O) hydrogen bond (Table 2 ▸), and a two-centre N—H⋯O hydrogen bond links these ion pairs into a chain of rings running parallel to the [010] direction (Fig. 5 ▸). There are four C—H⋯O hydrogen bonds in the structure of (II) and that involving atom C145 (Table 2 ▸) links the ion pairs into a second chain, this time running parallel to the [101] direction (Fig. 6 ▸). The two C—H⋯O hydrogen bonds involving atoms C12 and C16 link inversion-related pairs of cations into a centrosymmetric motif containing
 (8) rings (Fig. 7 ▸), and the aggregates of this type are further linked by the final C—H⋯O hydrogen bond, that involves atom C146, to form a complex chain of rings running parallel to the [001] direction (Fig. 8 ▸). The combination of hydrogen-bonded chains parallel to [010], [001] and [101] generates a three-dimensional network. We also note a fairly short nitro–nitro contact, 2.823 (4) Å, between atom O142 at (x, y, z) and atom N24 at (1 + x, y, 1 + z): this probably represents a dipolar attraction between negatively charged O and positively charged N atoms.
(8) rings (Fig. 7 ▸), and the aggregates of this type are further linked by the final C—H⋯O hydrogen bond, that involves atom C146, to form a complex chain of rings running parallel to the [001] direction (Fig. 8 ▸). The combination of hydrogen-bonded chains parallel to [010], [001] and [101] generates a three-dimensional network. We also note a fairly short nitro–nitro contact, 2.823 (4) Å, between atom O142 at (x, y, z) and atom N24 at (1 + x, y, 1 + z): this probably represents a dipolar attraction between negatively charged O and positively charged N atoms.
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N11—H11⋯O21 | 0.894 (14) | 1.869 (14) | 2.7356 (12) | 162.8 (13) |
| N11—H11⋯O262 | 0.894 (14) | 2.396 (14) | 2.8937 (13) | 115.4 (11) |
| N11—H12⋯O271i | 0.910 (14) | 1.874 (14) | 2.7668 (12) | 166.2 (12) |
| O272—H272⋯O21 | 1.000 (17) | 1.549 (17) | 2.5020 (12) | 157.4 (15) |
| C12—H12B⋯O142ii | 0.99 | 2.41 | 3.3921 (14) | 173 |
| C16—H16A⋯O141ii | 0.99 | 2.54 | 3.4906 (14) | 161 |
| C145—H145⋯O242iii | 0.95 | 2.46 | 3.3927 (15) | 168 |
| C146—H146⋯O241iv | 0.95 | 2.55 | 3.4227 (15) | 153 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 .
.
Figure 5.
Part of the crystal structure of compound (II), showing the formation of a hydrogen-bonded chain of rings running parallel to [010]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have all been omitted.
Figure 6.
Part of the crystal structure of compound (II) showing the formation of a chain of hydrogen-bonded rings running parallel to the [101] direction. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to those C atoms that are not involved in the motif shown have been omitted.
Figure 7.
Part of the crystal structure of compound (II) showing the linkage of an inversion-related pair of cations by two independent C—H⋯O hydrogen bonds, drawn as dashed lines. For the sake of clarity, the anions, the H atoms bonded to those C atoms that are not involved in the motif shown, and the unit-cell outline have been omitted. The atoms marked with an asterisk (*) are at the symmetry position (1 − x, 1 − y, 2 − z).
Figure 8.

Part of the crystal structure of compound (II) showing the formation of a chain of hydrogen-bonded rings running parallel to the [001] direction. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to those C atoms that are not involved in the motif shown have been omitted.
4. Database survey
The first structure report on a salt of N-(4-nitrophenyl)piperazine concerned the chloride salt, which crystallizes as a monohydrate (Lu, 2007 ▸); despite the presence of hydrogen bonds of N—H⋯O, N—H⋯Cl and O—H⋯Cl types, the supramolecular assembly is only mono-periodic. The structures of six salts of N-(4-nitrophenyl)piperazine with aromatic carboxylic acids have recently been reported (Mahesha et al., 2022 ▸): in all but one of these, the supramolecular assembly is mono-periodic, although it is di-periodic in the 4-ethoxybenzoate salt. This may be contrasted with the triperiodic assembly found here for compound (II).
In addition, we note that structures have been reported for a wide variety of salts derived from N-(4-fluorophenyl)piperazine (Harish Chinthal, Yathirajan, Archana et al., 2020 ▸; Harish Chinthal, Yathirajan, Kavitha et al., 2020 ▸), and from N-(4-methoxyphenyl)piperazine (Kiran Kumar et al., 2019 ▸, 2020 ▸). Finally, the structure of 4-(2-methoxyphenyl)piperazin-1-ium 3,5-dinitrosalicylate has been reported, but without any description of discussion of the geometry of the anion (Subha et al., 2022 ▸).
5. Synthesis and crystallization
For the preparation of compounds (I) and (II), a solution of N-(4-nitrophenyl)piperazine (100 mg, 0.483 mmol) in methanol (10 ml) was mixed with a solution of either benzoic acid (59 mg, 0.483 mmol) for (I) or 3,5-dinitrosalicylic acid (110 mg, 0.483 mmol) for (II) in methanol/ethyl acetate (1:1 v/v, 20 ml). The solutions of the base and the corresponding acid were mixed, stirred at ambient temperature for 15 min, and then set aside to crystallize at ambient temperature and in the presence of air. After one week, crystals suitable for single-crystal X-ray diffraction were collected by filtration and dried in air: compound (I), pale yellow, m.p. 410–413 K; compound (II), orange, m.p. 446–448 K.
6. Refinement
Crystal data, data collection and refinement details are summarized in Table 3 ▸. All H atoms were located in difference maps. The H atoms bonded to C atoms were then treated as riding atoms in geometrically idealized positions with C—H distances of 0.95 Å (aromatic) or 0.99 Å (CH2), and with U iso(H) = 1.2U eq(C). For the H atoms bonded to N or O atoms, the atomic coordinates were refined with U iso(H) = 1.2U eq(N) or 1.5U eq(O), giving the N—H and O—H distances shown in Tables 1 ▸ and 2 ▸.
Table 3. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C10H14N3O2 +·C7H5O2 −·H2O | C10H14N3O2 +·C7H3N2O7 − |
| M r | 347.37 | 435.35 |
| Crystal system, space group | Triclinic, P

|
Triclinic, P

|
| Temperature (K) | 90 | 90 |
| a, b, c (Å) | 6.0768 (3), 7.4427 (4), 18.4737 (9) | 7.9599 (4), 8.5391 (4), 14.2227 (5) |
| α, β, γ (°) | 78.894 (2), 85.870 (3), 83.668 (2) | 90.426 (2), 105.273 (1), 98.538 (2) |
| V (Å3) | 813.77 (7) | 921.15 (7) |
| Z | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.11 | 0.13 |
| Crystal size (mm) | 0.24 × 0.22 × 0.17 | 0.22 × 0.18 × 0.12 |
| Data collection | ||
| Diffractometer | Bruker D8 Venture | Bruker D8 Venture |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.912, 0.971 | 0.919, 0.971 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 27142, 3737, 3164 | 38287, 4212, 3662 |
| R int | 0.066 | 0.043 |
| (sin θ/λ)max (Å−1) | 0.651 | 0.650 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.036, 0.091, 1.04 | 0.029, 0.077, 1.04 |
| No. of reflections | 3737 | 4212 |
| No. of parameters | 238 | 289 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.25, −0.20 | 0.28, −0.18 |
Supplementary Material
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S2056989022007472/hb8030sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022007472/hb8030Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989022007472/hb8030IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989022007472/hb8030Isup4.cml
Supporting information file. DOI: 10.1107/S2056989022007472/hb8030IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
HJS is grateful to the University of Mysore for research facilities.
supplementary crystallographic information
4-(4-Nitrophenyl)piperazin-1-ium benzoate monohydrate (I). Crystal data
| C10H14N3O2+·C7H5O2−·H2O | Z = 2 |
| Mr = 347.37 | F(000) = 368 |
| Triclinic, P1 | Dx = 1.418 Mg m−3 |
| a = 6.0768 (3) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.4427 (4) Å | Cell parameters from 3737 reflections |
| c = 18.4737 (9) Å | θ = 2.3–27.6° |
| α = 78.894 (2)° | µ = 0.11 mm−1 |
| β = 85.870 (3)° | T = 90 K |
| γ = 83.668 (2)° | Block, pale yellow |
| V = 813.77 (7) Å3 | 0.24 × 0.22 × 0.17 mm |
4-(4-Nitrophenyl)piperazin-1-ium benzoate monohydrate (I). Data collection
| Bruker D8 Venture diffractometer | 3737 independent reflections |
| Radiation source: microsource | 3164 reflections with I > 2σ(I) |
| Multilayer mirror monochromator | Rint = 0.066 |
| φ and ω scans | θmax = 27.6°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −7→7 |
| Tmin = 0.912, Tmax = 0.971 | k = −9→9 |
| 27142 measured reflections | l = −23→23 |
4-(4-Nitrophenyl)piperazin-1-ium benzoate monohydrate (I). Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.036 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.091 | w = 1/[σ2(Fo2) + (0.030P)2 + 0.2902P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.001 |
| 3737 reflections | Δρmax = 0.25 e Å−3 |
| 238 parameters | Δρmin = −0.20 e Å−3 |
| 0 restraints |
4-(4-Nitrophenyl)piperazin-1-ium benzoate monohydrate (I). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Nitrophenyl)piperazin-1-ium benzoate monohydrate (I). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N11 | 0.22281 (16) | 0.29238 (14) | 0.44679 (5) | 0.0163 (2) | |
| H11 | 0.172 (2) | 0.2234 (19) | 0.4908 (8) | 0.020* | |
| H12 | 0.255 (2) | 0.403 (2) | 0.4565 (7) | 0.020* | |
| C12 | 0.04640 (18) | 0.33021 (16) | 0.39257 (6) | 0.0175 (2) | |
| H12A | −0.0854 | 0.3983 | 0.4129 | 0.021* | |
| H12B | 0.0020 | 0.2124 | 0.3839 | 0.021* | |
| C13 | 0.12736 (18) | 0.44242 (16) | 0.32002 (6) | 0.0172 (2) | |
| H13A | 0.0112 | 0.4575 | 0.2838 | 0.021* | |
| H13B | 0.1524 | 0.5664 | 0.3278 | 0.021* | |
| N14 | 0.33332 (15) | 0.35614 (13) | 0.28973 (5) | 0.0147 (2) | |
| C15 | 0.50502 (18) | 0.30670 (16) | 0.34411 (6) | 0.0163 (2) | |
| H15A | 0.5561 | 0.4205 | 0.3546 | 0.020* | |
| H15B | 0.6336 | 0.2365 | 0.3230 | 0.020* | |
| C16 | 0.42143 (19) | 0.19258 (16) | 0.41561 (6) | 0.0177 (2) | |
| H16A | 0.3830 | 0.0734 | 0.4063 | 0.021* | |
| H16B | 0.5394 | 0.1671 | 0.4515 | 0.021* | |
| C141 | 0.40488 (18) | 0.44038 (15) | 0.21853 (6) | 0.0149 (2) | |
| C142 | 0.26572 (19) | 0.57118 (16) | 0.17307 (6) | 0.0199 (2) | |
| H142 | 0.1188 | 0.6040 | 0.1906 | 0.024* | |
| C143 | 0.3394 (2) | 0.65311 (16) | 0.10304 (6) | 0.0213 (3) | |
| H143 | 0.2432 | 0.7399 | 0.0725 | 0.026* | |
| C144 | 0.5535 (2) | 0.60750 (16) | 0.07820 (6) | 0.0189 (2) | |
| C145 | 0.6944 (2) | 0.47632 (16) | 0.12051 (6) | 0.0199 (2) | |
| H145 | 0.8406 | 0.4442 | 0.1021 | 0.024* | |
| C146 | 0.61972 (19) | 0.39295 (16) | 0.18972 (6) | 0.0178 (2) | |
| H146 | 0.7152 | 0.3015 | 0.2186 | 0.021* | |
| N144 | 0.63672 (18) | 0.70469 (14) | 0.00703 (6) | 0.0242 (2) | |
| O141 | 0.50758 (17) | 0.81136 (14) | −0.03222 (5) | 0.0352 (2) | |
| O142 | 0.83458 (17) | 0.67621 (15) | −0.01004 (6) | 0.0421 (3) | |
| C21 | 0.13555 (18) | 0.09842 (14) | 0.71032 (6) | 0.0156 (2) | |
| C22 | −0.05414 (19) | 0.01147 (15) | 0.73666 (6) | 0.0170 (2) | |
| H22 | −0.1565 | −0.0072 | 0.7030 | 0.020* | |
| C23 | −0.0937 (2) | −0.04784 (16) | 0.81206 (6) | 0.0207 (2) | |
| H23 | −0.2239 | −0.1058 | 0.8299 | 0.025* | |
| C24 | 0.0564 (2) | −0.02246 (16) | 0.86119 (6) | 0.0229 (3) | |
| H24 | 0.0297 | −0.0636 | 0.9127 | 0.027* | |
| C25 | 0.2458 (2) | 0.06303 (16) | 0.83528 (7) | 0.0236 (3) | |
| H25 | 0.3494 | 0.0793 | 0.8691 | 0.028* | |
| C26 | 0.2842 (2) | 0.12465 (16) | 0.76032 (7) | 0.0203 (2) | |
| H26 | 0.4127 | 0.1852 | 0.7429 | 0.024* | |
| C27 | 0.18134 (19) | 0.16532 (15) | 0.62878 (6) | 0.0179 (2) | |
| O271 | 0.32916 (15) | 0.27141 (13) | 0.61005 (5) | 0.0293 (2) | |
| O272 | 0.06693 (14) | 0.11185 (11) | 0.58360 (4) | 0.02171 (19) | |
| O31 | 0.70609 (15) | 0.35347 (12) | 0.53628 (5) | 0.02152 (19) | |
| H31 | 0.579 (3) | 0.325 (2) | 0.5608 (9) | 0.032* | |
| H32 | 0.813 (3) | 0.268 (2) | 0.5570 (9) | 0.032* |
4-(4-Nitrophenyl)piperazin-1-ium benzoate monohydrate (I). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N11 | 0.0197 (5) | 0.0150 (5) | 0.0136 (5) | −0.0029 (4) | 0.0017 (4) | −0.0017 (4) |
| C12 | 0.0152 (5) | 0.0199 (6) | 0.0168 (5) | −0.0021 (4) | 0.0012 (4) | −0.0027 (4) |
| C13 | 0.0147 (5) | 0.0189 (5) | 0.0165 (5) | 0.0002 (4) | 0.0009 (4) | −0.0011 (4) |
| N14 | 0.0129 (4) | 0.0170 (5) | 0.0133 (4) | −0.0004 (4) | 0.0003 (3) | −0.0016 (3) |
| C15 | 0.0142 (5) | 0.0186 (5) | 0.0153 (5) | 0.0005 (4) | −0.0002 (4) | −0.0023 (4) |
| C16 | 0.0189 (6) | 0.0174 (5) | 0.0154 (5) | 0.0014 (4) | −0.0004 (4) | −0.0015 (4) |
| C141 | 0.0166 (5) | 0.0138 (5) | 0.0149 (5) | −0.0035 (4) | 0.0003 (4) | −0.0038 (4) |
| C142 | 0.0173 (6) | 0.0229 (6) | 0.0178 (6) | 0.0004 (5) | 0.0000 (4) | −0.0011 (4) |
| C143 | 0.0238 (6) | 0.0203 (6) | 0.0181 (6) | 0.0008 (5) | −0.0032 (5) | −0.0002 (4) |
| C144 | 0.0267 (6) | 0.0166 (5) | 0.0138 (5) | −0.0061 (5) | 0.0018 (4) | −0.0023 (4) |
| C145 | 0.0203 (6) | 0.0205 (6) | 0.0186 (6) | −0.0023 (5) | 0.0036 (4) | −0.0048 (4) |
| C146 | 0.0181 (6) | 0.0167 (5) | 0.0174 (5) | 0.0009 (4) | 0.0003 (4) | −0.0020 (4) |
| N144 | 0.0324 (6) | 0.0222 (5) | 0.0170 (5) | −0.0051 (4) | 0.0035 (4) | −0.0018 (4) |
| O141 | 0.0423 (6) | 0.0372 (6) | 0.0203 (5) | −0.0024 (5) | −0.0027 (4) | 0.0089 (4) |
| O142 | 0.0364 (6) | 0.0463 (6) | 0.0329 (5) | 0.0013 (5) | 0.0165 (4) | 0.0092 (5) |
| C21 | 0.0167 (5) | 0.0110 (5) | 0.0180 (5) | 0.0009 (4) | 0.0008 (4) | −0.0019 (4) |
| C22 | 0.0169 (5) | 0.0158 (5) | 0.0186 (5) | −0.0011 (4) | 0.0002 (4) | −0.0043 (4) |
| C23 | 0.0226 (6) | 0.0172 (6) | 0.0209 (6) | −0.0024 (5) | 0.0060 (5) | −0.0026 (4) |
| C24 | 0.0348 (7) | 0.0164 (6) | 0.0158 (5) | 0.0018 (5) | 0.0008 (5) | −0.0021 (4) |
| C25 | 0.0302 (7) | 0.0180 (6) | 0.0236 (6) | 0.0003 (5) | −0.0100 (5) | −0.0047 (5) |
| C26 | 0.0190 (6) | 0.0142 (5) | 0.0271 (6) | −0.0019 (4) | −0.0013 (5) | −0.0024 (4) |
| C27 | 0.0171 (5) | 0.0130 (5) | 0.0211 (6) | 0.0017 (4) | 0.0036 (4) | −0.0004 (4) |
| O271 | 0.0260 (5) | 0.0318 (5) | 0.0282 (5) | −0.0127 (4) | 0.0070 (4) | 0.0012 (4) |
| O272 | 0.0275 (5) | 0.0213 (4) | 0.0157 (4) | −0.0036 (3) | 0.0013 (3) | −0.0020 (3) |
| O31 | 0.0203 (4) | 0.0214 (4) | 0.0228 (4) | −0.0045 (4) | 0.0042 (3) | −0.0044 (3) |
4-(4-Nitrophenyl)piperazin-1-ium benzoate monohydrate (I). Geometric parameters (Å, º)
| N11—C16 | 1.4857 (14) | C144—C145 | 1.3841 (17) |
| N11—C12 | 1.4870 (14) | C144—N144 | 1.4581 (14) |
| N11—H11 | 0.925 (14) | C145—C146 | 1.3780 (16) |
| N11—H12 | 0.921 (15) | C145—H145 | 0.9500 |
| C12—C13 | 1.5167 (15) | C146—H146 | 0.9500 |
| C12—H12A | 0.9900 | N144—O141 | 1.2226 (14) |
| C12—H12B | 0.9900 | N144—O142 | 1.2268 (14) |
| C13—N14 | 1.4671 (14) | C21—C26 | 1.3911 (16) |
| C13—H13A | 0.9900 | C21—C22 | 1.3953 (16) |
| C13—H13B | 0.9900 | C21—C27 | 1.5084 (15) |
| N14—C141 | 1.4044 (14) | C22—C23 | 1.3896 (16) |
| N14—C15 | 1.4688 (14) | C22—H22 | 0.9500 |
| C15—C16 | 1.5134 (15) | C23—C24 | 1.3840 (18) |
| C15—H15A | 0.9900 | C23—H23 | 0.9500 |
| C15—H15B | 0.9900 | C24—C25 | 1.3869 (18) |
| C16—H16A | 0.9900 | C24—H24 | 0.9500 |
| C16—H16B | 0.9900 | C25—C26 | 1.3838 (17) |
| C141—C142 | 1.4049 (16) | C25—H25 | 0.9500 |
| C141—C146 | 1.4081 (16) | C26—H26 | 0.9500 |
| C142—C143 | 1.3853 (16) | C27—O271 | 1.2476 (14) |
| C142—H142 | 0.9500 | C27—O272 | 1.2692 (14) |
| C143—C144 | 1.3764 (17) | O31—H31 | 0.893 (17) |
| C143—H143 | 0.9500 | O31—H32 | 0.909 (17) |
| C16—N11—C12 | 109.33 (9) | C141—C142—H142 | 119.5 |
| C16—N11—H11 | 110.2 (8) | C144—C143—C142 | 119.36 (11) |
| C12—N11—H11 | 110.3 (8) | C144—C143—H143 | 120.3 |
| C16—N11—H12 | 111.3 (8) | C142—C143—H143 | 120.3 |
| C12—N11—H12 | 108.0 (8) | C143—C144—C145 | 121.48 (11) |
| H11—N11—H12 | 107.6 (12) | C143—C144—N144 | 119.32 (11) |
| N11—C12—C13 | 110.80 (9) | C145—C144—N144 | 119.16 (11) |
| N11—C12—H12A | 109.5 | C146—C145—C144 | 119.05 (11) |
| C13—C12—H12A | 109.5 | C146—C145—H145 | 120.5 |
| N11—C12—H12B | 109.5 | C144—C145—H145 | 120.5 |
| C13—C12—H12B | 109.5 | C145—C146—C141 | 121.42 (10) |
| H12A—C12—H12B | 108.1 | C145—C146—H146 | 119.3 |
| N14—C13—C12 | 112.32 (9) | C141—C146—H146 | 119.3 |
| N14—C13—H13A | 109.1 | O141—N144—O142 | 123.28 (11) |
| C12—C13—H13A | 109.1 | O141—N144—C144 | 118.78 (11) |
| N14—C13—H13B | 109.1 | O142—N144—C144 | 117.94 (10) |
| C12—C13—H13B | 109.1 | C26—C21—C22 | 119.26 (10) |
| H13A—C13—H13B | 107.9 | C26—C21—C27 | 119.54 (10) |
| C141—N14—C13 | 115.67 (9) | C22—C21—C27 | 121.19 (10) |
| C141—N14—C15 | 115.77 (9) | C23—C22—C21 | 120.20 (11) |
| C13—N14—C15 | 112.34 (9) | C23—C22—H22 | 119.9 |
| N14—C15—C16 | 112.12 (9) | C21—C22—H22 | 119.9 |
| N14—C15—H15A | 109.2 | C24—C23—C22 | 119.98 (11) |
| C16—C15—H15A | 109.2 | C24—C23—H23 | 120.0 |
| N14—C15—H15B | 109.2 | C22—C23—H23 | 120.0 |
| C16—C15—H15B | 109.2 | C23—C24—C25 | 120.06 (11) |
| H15A—C15—H15B | 107.9 | C23—C24—H24 | 120.0 |
| N11—C16—C15 | 110.12 (9) | C25—C24—H24 | 120.0 |
| N11—C16—H16A | 109.6 | C26—C25—C24 | 120.10 (11) |
| C15—C16—H16A | 109.6 | C26—C25—H25 | 119.9 |
| N11—C16—H16B | 109.6 | C24—C25—H25 | 119.9 |
| C15—C16—H16B | 109.6 | C25—C26—C21 | 120.38 (11) |
| H16A—C16—H16B | 108.2 | C25—C26—H26 | 119.8 |
| N14—C141—C142 | 121.85 (10) | C21—C26—H26 | 119.8 |
| N14—C141—C146 | 120.53 (10) | O271—C27—O272 | 124.07 (11) |
| C142—C141—C146 | 117.62 (10) | O271—C27—C21 | 117.57 (11) |
| C143—C142—C141 | 121.01 (11) | O272—C27—C21 | 118.35 (10) |
| C143—C142—H142 | 119.5 | H31—O31—H32 | 106.4 (14) |
| C16—N11—C12—C13 | −58.34 (12) | C144—C145—C146—C141 | −0.95 (17) |
| N11—C12—C13—N14 | 54.53 (12) | N14—C141—C146—C145 | −178.85 (10) |
| C12—C13—N14—C141 | 172.83 (9) | C142—C141—C146—C145 | 2.33 (17) |
| C12—C13—N14—C15 | −51.20 (12) | C143—C144—N144—O141 | −7.29 (17) |
| C141—N14—C15—C16 | −171.67 (9) | C145—C144—N144—O141 | 175.18 (11) |
| C13—N14—C15—C16 | 52.40 (12) | C143—C144—N144—O142 | 171.98 (12) |
| C12—N11—C16—C15 | 59.27 (12) | C145—C144—N144—O142 | −5.55 (17) |
| N14—C15—C16—N11 | −56.67 (12) | C26—C21—C22—C23 | 0.09 (16) |
| C13—N14—C141—C142 | −13.70 (15) | C27—C21—C22—C23 | −179.53 (10) |
| C15—N14—C141—C142 | −148.15 (11) | C21—C22—C23—C24 | −0.69 (17) |
| C13—N14—C141—C146 | 167.53 (10) | C22—C23—C24—C25 | 0.35 (18) |
| C15—N14—C141—C146 | 33.07 (14) | C23—C24—C25—C26 | 0.59 (18) |
| N14—C141—C142—C143 | 179.86 (11) | C24—C25—C26—C21 | −1.19 (17) |
| C146—C141—C142—C143 | −1.33 (17) | C22—C21—C26—C25 | 0.85 (17) |
| C141—C142—C143—C144 | −1.02 (18) | C27—C21—C26—C25 | −179.52 (10) |
| C142—C143—C144—C145 | 2.49 (18) | C26—C21—C27—O271 | −12.91 (16) |
| C142—C143—C144—N144 | −174.99 (11) | C22—C21—C27—O271 | 166.71 (11) |
| C143—C144—C145—C146 | −1.51 (18) | C26—C21—C27—O272 | 167.61 (10) |
| N144—C144—C145—C146 | 175.97 (10) | C22—C21—C27—O272 | −12.77 (16) |
4-(4-Nitrophenyl)piperazin-1-ium benzoate monohydrate (I). Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the C21–C26 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H11···O271 | 0.926 (14) | 2.564 (14) | 3.1009 (13) | 117.4 (10) |
| N11—H11···O272 | 0.926 (14) | 1.857 (14) | 2.7781 (12) | 172.9 (13) |
| N11—H12···O31i | 0.920 (15) | 1.884 (15) | 2.7965 (14) | 171.0 (12) |
| O31—H31···O271 | 0.892 (18) | 1.757 (18) | 2.6486 (13) | 179 (3) |
| O31—H32···O272ii | 0.908 (17) | 1.862 (17) | 2.7581 (12) | 168.8 (16) |
| C12—H12B···O272iii | 0.99 | 2.45 | 3.3751 (15) | 156 |
| C146—H146···Cg1iv | 0.95 | 2.67 | 3.4363 (13) | 138 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x+1, y, z; (iii) −x, −y, −z+1; (iv) −x+1, −y, −z+1.
4-(4-Nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate (II). Crystal data
| C10H14N3O2+·C7H3N2O7− | Z = 2 |
| Mr = 435.35 | F(000) = 452 |
| Triclinic, P1 | Dx = 1.570 Mg m−3 |
| a = 7.9599 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.5391 (4) Å | Cell parameters from 4212 reflections |
| c = 14.2227 (5) Å | θ = 2.4–27.5° |
| α = 90.426 (2)° | µ = 0.13 mm−1 |
| β = 105.273 (1)° | T = 90 K |
| γ = 98.538 (2)° | Block, orange |
| V = 921.15 (7) Å3 | 0.22 × 0.18 × 0.12 mm |
4-(4-Nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate (II). Data collection
| Bruker D8 Venture diffractometer | 4212 independent reflections |
| Radiation source: microsource | 3662 reflections with I > 2σ(I) |
| Multilayer mirror monochromator | Rint = 0.043 |
| φ and ω scans | θmax = 27.5°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −10→10 |
| Tmin = 0.919, Tmax = 0.971 | k = −11→11 |
| 38287 measured reflections | l = −18→17 |
4-(4-Nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate (II). Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.029 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.077 | w = 1/[σ2(Fo2) + (0.029P)2 + 0.3527P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.001 |
| 4212 reflections | Δρmax = 0.28 e Å−3 |
| 289 parameters | Δρmin = −0.18 e Å−3 |
| 0 restraints |
4-(4-Nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate (II). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate (II). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N11 | 0.30883 (12) | 0.10386 (11) | 0.64405 (7) | 0.01693 (19) | |
| H11 | 0.2967 (17) | 0.1840 (17) | 0.6040 (10) | 0.020* | |
| H12 | 0.2796 (17) | 0.0129 (17) | 0.6057 (10) | 0.020* | |
| C12 | 0.17915 (14) | 0.10314 (13) | 0.70340 (8) | 0.0188 (2) | |
| H12A | 0.0579 | 0.0733 | 0.6608 | 0.023* | |
| H12B | 0.1890 | 0.2107 | 0.7328 | 0.023* | |
| C13 | 0.21403 (14) | −0.01389 (13) | 0.78350 (8) | 0.0187 (2) | |
| H13A | 0.1321 | −0.0092 | 0.8248 | 0.022* | |
| H13B | 0.1931 | −0.1229 | 0.7541 | 0.022* | |
| N14 | 0.39625 (12) | 0.02405 (11) | 0.84356 (6) | 0.01699 (19) | |
| C15 | 0.51561 (14) | 0.00483 (13) | 0.78325 (8) | 0.0170 (2) | |
| H15A | 0.4884 | −0.1043 | 0.7536 | 0.020* | |
| H15B | 0.6388 | 0.0215 | 0.8245 | 0.020* | |
| C16 | 0.49616 (14) | 0.12299 (13) | 0.70345 (8) | 0.0172 (2) | |
| H16A | 0.5334 | 0.2322 | 0.7330 | 0.021* | |
| H16B | 0.5729 | 0.1055 | 0.6610 | 0.021* | |
| C141 | 0.44287 (14) | 0.13710 (12) | 0.91966 (7) | 0.0156 (2) | |
| C142 | 0.31639 (14) | 0.18318 (13) | 0.96323 (8) | 0.0185 (2) | |
| H142 | 0.1955 | 0.1406 | 0.9370 | 0.022* | |
| C143 | 0.36498 (14) | 0.28859 (13) | 1.04300 (8) | 0.0182 (2) | |
| H143 | 0.2783 | 0.3179 | 1.0717 | 0.022* | |
| C144 | 0.54137 (14) | 0.35176 (12) | 1.08126 (7) | 0.0161 (2) | |
| C145 | 0.66886 (14) | 0.31362 (13) | 1.03852 (8) | 0.0173 (2) | |
| H145 | 0.7887 | 0.3601 | 1.0640 | 0.021* | |
| C146 | 0.62025 (14) | 0.20794 (13) | 0.95897 (8) | 0.0174 (2) | |
| H146 | 0.7078 | 0.1821 | 0.9299 | 0.021* | |
| N144 | 0.59245 (12) | 0.46290 (11) | 1.16455 (6) | 0.01728 (19) | |
| O141 | 0.47859 (11) | 0.49209 (10) | 1.20379 (6) | 0.02406 (19) | |
| O142 | 0.74805 (11) | 0.52457 (10) | 1.19410 (6) | 0.02329 (18) | |
| C21 | 0.24532 (14) | 0.43648 (13) | 0.46951 (8) | 0.0171 (2) | |
| C22 | 0.20094 (14) | 0.59329 (12) | 0.45355 (8) | 0.0168 (2) | |
| C23 | 0.15472 (14) | 0.65011 (13) | 0.36161 (8) | 0.0163 (2) | |
| H23 | 0.1308 | 0.7557 | 0.3537 | 0.020* | |
| C24 | 0.14290 (14) | 0.55395 (13) | 0.28038 (8) | 0.0164 (2) | |
| C25 | 0.17807 (14) | 0.40041 (13) | 0.28998 (8) | 0.0165 (2) | |
| H25 | 0.1656 | 0.3340 | 0.2339 | 0.020* | |
| C26 | 0.23134 (14) | 0.34521 (12) | 0.38199 (8) | 0.0162 (2) | |
| O21 | 0.28898 (12) | 0.38660 (9) | 0.55580 (6) | 0.02378 (19) | |
| C27 | 0.20933 (15) | 0.70160 (13) | 0.53734 (8) | 0.0199 (2) | |
| O271 | 0.16416 (12) | 0.83252 (9) | 0.52573 (6) | 0.02424 (19) | |
| O272 | 0.26860 (13) | 0.65093 (10) | 0.62536 (6) | 0.0294 (2) | |
| H272 | 0.288 (2) | 0.540 (2) | 0.6144 (12) | 0.044* | |
| N24 | 0.09824 (12) | 0.61730 (11) | 0.18421 (7) | 0.01767 (19) | |
| O241 | 0.08439 (11) | 0.75980 (9) | 0.17920 (6) | 0.02152 (18) | |
| O242 | 0.07873 (11) | 0.52884 (10) | 0.11242 (6) | 0.02219 (18) | |
| N26 | 0.27842 (12) | 0.18676 (11) | 0.38689 (7) | 0.01824 (19) | |
| O261 | 0.20718 (11) | 0.09218 (9) | 0.31673 (6) | 0.02451 (19) | |
| O262 | 0.38998 (12) | 0.15381 (10) | 0.45879 (6) | 0.02596 (19) |
4-(4-Nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate (II). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N11 | 0.0230 (5) | 0.0128 (4) | 0.0146 (4) | 0.0040 (4) | 0.0037 (4) | −0.0003 (4) |
| C12 | 0.0177 (5) | 0.0181 (5) | 0.0198 (5) | 0.0037 (4) | 0.0032 (4) | −0.0033 (4) |
| C13 | 0.0188 (5) | 0.0191 (5) | 0.0171 (5) | −0.0008 (4) | 0.0051 (4) | −0.0030 (4) |
| N14 | 0.0184 (4) | 0.0179 (4) | 0.0142 (4) | 0.0015 (4) | 0.0045 (3) | −0.0018 (3) |
| C15 | 0.0195 (5) | 0.0162 (5) | 0.0159 (5) | 0.0045 (4) | 0.0048 (4) | −0.0012 (4) |
| C16 | 0.0194 (5) | 0.0157 (5) | 0.0175 (5) | 0.0030 (4) | 0.0066 (4) | −0.0006 (4) |
| C141 | 0.0210 (5) | 0.0132 (5) | 0.0128 (5) | 0.0032 (4) | 0.0044 (4) | 0.0028 (4) |
| C142 | 0.0163 (5) | 0.0207 (5) | 0.0176 (5) | 0.0029 (4) | 0.0030 (4) | 0.0004 (4) |
| C143 | 0.0192 (5) | 0.0199 (5) | 0.0173 (5) | 0.0064 (4) | 0.0063 (4) | 0.0020 (4) |
| C144 | 0.0218 (5) | 0.0130 (5) | 0.0133 (5) | 0.0031 (4) | 0.0039 (4) | 0.0005 (4) |
| C145 | 0.0175 (5) | 0.0168 (5) | 0.0170 (5) | 0.0012 (4) | 0.0045 (4) | 0.0015 (4) |
| C146 | 0.0190 (5) | 0.0176 (5) | 0.0169 (5) | 0.0031 (4) | 0.0071 (4) | 0.0011 (4) |
| N144 | 0.0211 (5) | 0.0150 (4) | 0.0161 (4) | 0.0033 (4) | 0.0054 (4) | 0.0013 (3) |
| O141 | 0.0240 (4) | 0.0273 (4) | 0.0229 (4) | 0.0058 (3) | 0.0092 (3) | −0.0061 (3) |
| O142 | 0.0213 (4) | 0.0234 (4) | 0.0230 (4) | −0.0029 (3) | 0.0058 (3) | −0.0057 (3) |
| C21 | 0.0188 (5) | 0.0154 (5) | 0.0162 (5) | 0.0013 (4) | 0.0039 (4) | −0.0003 (4) |
| C22 | 0.0183 (5) | 0.0142 (5) | 0.0172 (5) | 0.0014 (4) | 0.0042 (4) | −0.0017 (4) |
| C23 | 0.0156 (5) | 0.0146 (5) | 0.0187 (5) | 0.0024 (4) | 0.0043 (4) | −0.0001 (4) |
| C24 | 0.0151 (5) | 0.0189 (5) | 0.0146 (5) | 0.0018 (4) | 0.0034 (4) | 0.0011 (4) |
| C25 | 0.0143 (5) | 0.0176 (5) | 0.0174 (5) | −0.0001 (4) | 0.0054 (4) | −0.0042 (4) |
| C26 | 0.0164 (5) | 0.0131 (5) | 0.0196 (5) | 0.0024 (4) | 0.0059 (4) | −0.0013 (4) |
| O21 | 0.0394 (5) | 0.0160 (4) | 0.0151 (4) | 0.0071 (3) | 0.0044 (3) | 0.0009 (3) |
| C27 | 0.0252 (6) | 0.0158 (5) | 0.0178 (5) | 0.0013 (4) | 0.0051 (4) | −0.0015 (4) |
| O271 | 0.0347 (5) | 0.0153 (4) | 0.0218 (4) | 0.0066 (3) | 0.0046 (3) | −0.0032 (3) |
| O272 | 0.0541 (6) | 0.0178 (4) | 0.0151 (4) | 0.0096 (4) | 0.0051 (4) | −0.0019 (3) |
| N24 | 0.0141 (4) | 0.0220 (5) | 0.0168 (4) | 0.0022 (4) | 0.0043 (3) | 0.0004 (4) |
| O241 | 0.0228 (4) | 0.0206 (4) | 0.0216 (4) | 0.0054 (3) | 0.0056 (3) | 0.0050 (3) |
| O242 | 0.0224 (4) | 0.0284 (4) | 0.0151 (4) | 0.0022 (3) | 0.0049 (3) | −0.0038 (3) |
| N26 | 0.0207 (5) | 0.0153 (4) | 0.0209 (5) | 0.0031 (4) | 0.0093 (4) | −0.0007 (4) |
| O261 | 0.0267 (4) | 0.0178 (4) | 0.0285 (4) | 0.0023 (3) | 0.0074 (3) | −0.0087 (3) |
| O262 | 0.0329 (5) | 0.0245 (4) | 0.0226 (4) | 0.0132 (4) | 0.0061 (3) | 0.0031 (3) |
4-(4-Nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate (II). Geometric parameters (Å, º)
| N11—C16 | 1.4919 (14) | C145—C146 | 1.3765 (15) |
| N11—C12 | 1.4951 (14) | C145—H145 | 0.9500 |
| N11—H11 | 0.894 (14) | C146—H146 | 0.9500 |
| N11—H12 | 0.910 (14) | N144—O142 | 1.2315 (12) |
| C12—C13 | 1.5187 (15) | N144—O141 | 1.2355 (12) |
| C12—H12A | 0.9900 | C21—O21 | 1.2788 (13) |
| C12—H12B | 0.9900 | C21—C26 | 1.4340 (15) |
| C13—N14 | 1.4635 (14) | C21—C22 | 1.4394 (15) |
| C13—H13A | 0.9900 | C22—C23 | 1.3747 (15) |
| C13—H13B | 0.9900 | C22—C27 | 1.4844 (15) |
| N14—C141 | 1.3812 (13) | C23—C24 | 1.3869 (15) |
| N14—C15 | 1.4625 (14) | C23—H23 | 0.9500 |
| C15—C16 | 1.5169 (15) | C24—C25 | 1.3811 (15) |
| C15—H15A | 0.9900 | C24—N24 | 1.4498 (13) |
| C15—H15B | 0.9900 | C25—C26 | 1.3750 (15) |
| C16—H16A | 0.9900 | C25—H25 | 0.9500 |
| C16—H16B | 0.9900 | C26—N26 | 1.4540 (14) |
| C141—C142 | 1.4125 (15) | C27—O271 | 1.2236 (14) |
| C141—C146 | 1.4133 (15) | C27—O272 | 1.3171 (14) |
| C142—C143 | 1.3774 (15) | O272—H272 | 1.003 (18) |
| C142—H142 | 0.9500 | N24—O242 | 1.2277 (12) |
| C143—C144 | 1.3881 (15) | N24—O241 | 1.2389 (12) |
| C143—H143 | 0.9500 | N26—O262 | 1.2303 (12) |
| C144—C145 | 1.3875 (15) | N26—O261 | 1.2343 (12) |
| C144—N144 | 1.4437 (13) | ||
| C16—N11—C12 | 113.90 (8) | C144—C143—H143 | 120.2 |
| C16—N11—H11 | 108.4 (9) | C145—C144—C143 | 120.87 (10) |
| C12—N11—H11 | 108.3 (9) | C145—C144—N144 | 119.43 (10) |
| C16—N11—H12 | 111.0 (8) | C143—C144—N144 | 119.67 (10) |
| C12—N11—H12 | 108.3 (9) | C146—C145—C144 | 119.48 (10) |
| H11—N11—H12 | 106.8 (12) | C146—C145—H145 | 120.3 |
| N11—C12—C13 | 110.16 (9) | C144—C145—H145 | 120.3 |
| N11—C12—H12A | 109.6 | C145—C146—C141 | 121.42 (10) |
| C13—C12—H12A | 109.6 | C145—C146—H146 | 119.3 |
| N11—C12—H12B | 109.6 | C141—C146—H146 | 119.3 |
| C13—C12—H12B | 109.6 | O142—N144—O141 | 122.45 (9) |
| H12A—C12—H12B | 108.1 | O142—N144—C144 | 118.77 (9) |
| N14—C13—C12 | 109.99 (9) | O141—N144—C144 | 118.78 (9) |
| N14—C13—H13A | 109.7 | O21—C21—C26 | 124.86 (10) |
| C12—C13—H13A | 109.7 | O21—C21—C22 | 120.75 (9) |
| N14—C13—H13B | 109.7 | C26—C21—C22 | 114.38 (9) |
| C12—C13—H13B | 109.7 | C23—C22—C21 | 121.89 (10) |
| H13A—C13—H13B | 108.2 | C23—C22—C27 | 117.59 (10) |
| C141—N14—C15 | 121.02 (9) | C21—C22—C27 | 120.49 (10) |
| C141—N14—C13 | 121.10 (9) | C22—C23—C24 | 120.26 (10) |
| C15—N14—C13 | 109.06 (8) | C22—C23—H23 | 119.9 |
| N14—C15—C16 | 110.28 (9) | C24—C23—H23 | 119.9 |
| N14—C15—H15A | 109.6 | C25—C24—C23 | 120.99 (10) |
| C16—C15—H15A | 109.6 | C25—C24—N24 | 119.55 (9) |
| N14—C15—H15B | 109.6 | C23—C24—N24 | 119.43 (10) |
| C16—C15—H15B | 109.6 | C26—C25—C24 | 118.95 (10) |
| H15A—C15—H15B | 108.1 | C26—C25—H25 | 120.5 |
| N11—C16—C15 | 109.87 (9) | C24—C25—H25 | 120.5 |
| N11—C16—H16A | 109.7 | C25—C26—C21 | 123.45 (10) |
| C15—C16—H16A | 109.7 | C25—C26—N26 | 115.98 (9) |
| N11—C16—H16B | 109.7 | C21—C26—N26 | 120.55 (9) |
| C15—C16—H16B | 109.7 | O271—C27—O272 | 121.14 (10) |
| H16A—C16—H16B | 108.2 | O271—C27—C22 | 121.85 (10) |
| N14—C141—C142 | 121.47 (10) | O272—C27—C22 | 117.01 (10) |
| N14—C141—C146 | 121.20 (10) | C27—O272—H272 | 105.0 (10) |
| C142—C141—C146 | 117.30 (10) | O242—N24—O241 | 123.40 (9) |
| C143—C142—C141 | 121.27 (10) | O242—N24—C24 | 118.92 (9) |
| C143—C142—H142 | 119.4 | O241—N24—C24 | 117.67 (9) |
| C141—C142—H142 | 119.4 | O262—N26—O261 | 122.85 (9) |
| C142—C143—C144 | 119.59 (10) | O262—N26—C26 | 119.17 (9) |
| C142—C143—H143 | 120.2 | O261—N26—C26 | 117.95 (9) |
| C16—N11—C12—C13 | −50.40 (12) | C26—C21—C22—C23 | −1.93 (15) |
| N11—C12—C13—N14 | 56.08 (11) | O21—C21—C22—C27 | 1.51 (16) |
| C12—C13—N14—C141 | 84.07 (12) | C26—C21—C22—C27 | −179.98 (10) |
| C12—C13—N14—C15 | −63.61 (11) | C21—C22—C23—C24 | 2.55 (16) |
| C141—N14—C15—C16 | −83.73 (12) | C27—C22—C23—C24 | −179.35 (10) |
| C13—N14—C15—C16 | 63.98 (11) | C22—C23—C24—C25 | −0.39 (16) |
| C12—N11—C16—C15 | 50.41 (11) | C22—C23—C24—N24 | −178.48 (9) |
| N14—C15—C16—N11 | −56.47 (11) | C23—C24—C25—C26 | −2.25 (16) |
| C15—N14—C141—C142 | 164.67 (10) | N24—C24—C25—C26 | 175.84 (9) |
| C13—N14—C141—C142 | 20.80 (15) | C24—C25—C26—C21 | 2.85 (16) |
| C15—N14—C141—C146 | −17.23 (15) | C24—C25—C26—N26 | −175.67 (9) |
| C13—N14—C141—C146 | −161.10 (10) | O21—C21—C26—C25 | 177.64 (10) |
| N14—C141—C142—C143 | 175.90 (10) | C22—C21—C26—C25 | −0.79 (16) |
| C146—C141—C142—C143 | −2.27 (16) | O21—C21—C26—N26 | −3.90 (17) |
| C141—C142—C143—C144 | 0.36 (16) | C22—C21—C26—N26 | 177.67 (9) |
| C142—C143—C144—C145 | 1.84 (16) | C23—C22—C27—O271 | 5.49 (17) |
| C142—C143—C144—N144 | 179.67 (10) | C21—C22—C27—O271 | −176.38 (10) |
| C143—C144—C145—C146 | −2.02 (16) | C23—C22—C27—O272 | −174.21 (10) |
| N144—C144—C145—C146 | −179.86 (10) | C21—C22—C27—O272 | 3.93 (16) |
| C144—C145—C146—C141 | 0.00 (16) | C25—C24—N24—O242 | 5.79 (14) |
| N14—C141—C146—C145 | −176.08 (10) | C23—C24—N24—O242 | −176.09 (9) |
| C142—C141—C146—C145 | 2.09 (16) | C25—C24—N24—O241 | −173.14 (9) |
| C145—C144—N144—O142 | 1.91 (15) | C23—C24—N24—O241 | 4.98 (14) |
| C143—C144—N144—O142 | −175.95 (10) | C25—C26—N26—O262 | 150.18 (10) |
| C145—C144—N144—O141 | −177.77 (10) | C21—C26—N26—O262 | −28.38 (15) |
| C143—C144—N144—O141 | 4.37 (15) | C25—C26—N26—O261 | −27.86 (14) |
| O21—C21—C22—C23 | 179.56 (10) | C21—C26—N26—O261 | 153.57 (10) |
4-(4-Nitrophenyl)piperazin-1-ium 2-carboxy-4,6-dinitrophenolate (II). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H11···O21 | 0.894 (14) | 1.869 (14) | 2.7356 (12) | 162.8 (13) |
| N11—H11···O262 | 0.894 (14) | 2.396 (14) | 2.8937 (13) | 115.4 (11) |
| N11—H12···O271i | 0.910 (14) | 1.874 (14) | 2.7668 (12) | 166.2 (12) |
| O272—H272···O21 | 1.000 (17) | 1.549 (17) | 2.5020 (12) | 157.4 (15) |
| C12—H12B···O142ii | 0.99 | 2.41 | 3.3921 (14) | 173 |
| C16—H16A···O141ii | 0.99 | 2.54 | 3.4906 (14) | 161 |
| C145—H145···O242iii | 0.95 | 2.46 | 3.3927 (15) | 168 |
| C146—H146···O241iv | 0.95 | 2.55 | 3.4227 (15) | 153 |
Symmetry codes: (i) x, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) x+1, y, z+1; (iv) −x+1, −y+1, −z+1.
Funding Statement
HSY thanks the UGC for a BSR Faculty fellowship for three years.
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–S19.
- Berkheij, M., van der Sluis, L., Sewing, C., den Boer, D. J., Terpstra, J. W., Hiemstra, H., Iwema Bakker, W. I., van den Hoogenband, A. & van Maarseveen, J. H. (2005). Tetrahedron Lett. 46, 2369–2371.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Boeyens, J. C. A. (1978). J. Cryst. Mol. Struct. 8, 317–320.
- Bruker (2016). APEX3. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chaudhary, P., Kumar, R., Verma, K., Singh, D., Yadav, V., Chhillar, A. K., Sharma, G. L. & Chandra, R. (2006). Bioorg. Med. Chem. 14, 1819–1826. [DOI] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Elliott, S. (2011). Drug Test. Anal. 3, 430–438. [DOI] [PubMed]
- Ferguson, G., Glidewell, C., Gregson, R. M. & Meehan, P. R. (1998a). Acta Cryst. B54, 129–138.
- Ferguson, G., Glidewell, C., Gregson, R. M. & Meehan, P. R. (1998b). Acta Cryst. B54, 139–150.
- Gregson, R. M., Glidewell, C., Ferguson, G. & Lough, A. J. (2000). Acta Cryst. B56, 39–57. [DOI] [PubMed]
- Harish Chinthal, C., Yathirajan, H. S., Archana, S. D., Foro, S. & Glidewell, C. (2020). Acta Cryst. E76, 841–847. [DOI] [PMC free article] [PubMed]
- Harish Chinthal, C., Yathirajan, H. S., Kavitha, C. N., Foro, S. & Glidewell, C. (2020). Acta Cryst. E76, 1179–1186. [DOI] [PMC free article] [PubMed]
- Huheey, J. E. (1966). J. Phys. Chem. 70, 2086–2092.
- Kharb, R., Bansal, K. & Sharma, A. K. (2012). Der Pharma Chem. 4, 2470–2488.
- Kiran Kumar, H., Yathirajan, H. S., Foro, S. & Glidewell, C. (2019). Acta Cryst. E75, 1494–1506. [DOI] [PMC free article] [PubMed]
- Kiran Kumar, H., Yathirajan, H. S., Harish Chinthal, C., Foro, S. & Glidewell, C. (2020). Acta Cryst. E76, 488–495. [DOI] [PMC free article] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Lu, Y.-X. (2007). Acta Cryst. E63, o3611.
- Mahesha, N., Kiran Kumar, H., Yathirajan, H. S., Foro, S., Abdelbaky, M. S. M. & Garcia-Granda, S. (2022). Acta Cryst. E78, 510–518. [DOI] [PMC free article] [PubMed]
- Mullay, J. (1985). J. Am. Chem. Soc. 107, 7271–7275.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Subha, V., Seethalakshmi, T., Balakrishnan, T., Percino, M. J. & Venkatesan, P. (2022). Acta Cryst. E78, 774–778. [DOI] [PMC free article] [PubMed]
- Upadhayaya, P. S., Sinha, N., Jain, S., Kishore, N., Chandra, R. & Arora, S. K. (2004). Bioorg. Med. Chem. 12, 2225–2238. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S2056989022007472/hb8030sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022007472/hb8030Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989022007472/hb8030IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989022007472/hb8030Isup4.cml
Supporting information file. DOI: 10.1107/S2056989022007472/hb8030IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report









