Abstract
The IncI1 plasmid R64 produces two kinds of sex pili: a thin pilus and a thick pilus. The thin pilus, which belongs to the type IV family, is required only for liquid matings. Fourteen genes, pilI to -V, were found in the DNA region responsible for the biogenesis of the R64 thin pilus (S.-R. Kim and T. Komano, J. Bacteriol. 179:3594–3603, 1997). In this study, we introduced frameshift mutations into each of the 14 pil genes to test their requirement for R64 thin pilus biogenesis. From the analyses of extracellular secretion of thin pili and transfer frequency in liquid matings, we found that 12 genes, pilK to -V, are required for the formation of the thin pilus. Complementation experiments excluded the possible polar effects of each mutation on the expression of downstream genes. Two genes, traBC, were previously shown to be required for the expression of the pil genes. In addition, the rci gene is responsible for modulating the structure and function of the R64 thin pilus via the DNA rearrangement of the shufflon. Altogether, 15 genes, traBC, pilK through pilV, and rci, are essential for R64 thin pilus formation and function.
Type IV pili are long surface filaments produced by gram-negative bacteria such as Pseudomonas aeruginosa, Neisseria gonorrhoeae, Moraxella bovis, Myxococcus xanthus, Vibrio cholerae, and enteropathogenic and enterotoxigenic Escherichia coli (for reviews, see references 22, 25, 26, and 31). Many type IV pili are used to attach bacterial pathogens to epithelial cells of the eukaryotic host. Type IV pili are also associated with the twitching motility of various bacteria and with the social motility of myxobacteria (22, 38).
Type IV pili are composed of small pilin subunits, which are derived from type IV prepilin through cleavage of the N-terminal prepeptide (26, 31). Type IV prepilins share common features within their amino acid sequences. Prepilin peptidase cleaves between the glycine residue at the −1 position and the phenylalanine residue at the +1 position. The fifth amino acid residue of mature pilin is invariantly glutamic acid. The N-terminal 20-amino-acid region is hydrophobic and highly conserved. Type IV pilin has been classified into two groups. Group A, which includes pilins from P. aeruginosa, N. gonorrhoeae, M. bovis, and M. xanthus, contains prepilins whose sequences are conserved among each other (26, 31). Group A prepeptides are usually 6 to 7 amino acids in length. The conserved N-terminal phenylalanine of group A mature pilin is N methylated. Group B, which includes pilins from V. cholerae and enteropathogenic and enterotoxigenic E. coli, contains long prepeptides (24, 29, 30, 34). The amino acid sequences of group B prepilins exhibit far more diversity than those of group A.
The self-transmissible IncI1 plasmids, including R64 and ColIb-P9, produce two kinds of pili: a thick rigid pilus and a thin flexible pilus (4, 5, 10). The thick pilus is essential for transfer of the IncI1 plasmid both in liquid and on surfaces, while the thin pilus is required only for liquid matings (15, 16). One-third of the 54-kb R64 transfer region is responsible for the formation of thin pili (15, 16). E. coli cells harboring pKK641A′, which carries a 21.88-kb segment of R64, were demonstrated to produce thin pili and to be sensitive to IncI1-specific phages Iα and PR64FS (15). DNA sequence analysis of the tra-pil region revealed that 18 genes, traA to -D and pilI to -V, are located in this region (13, 14). Mutational analysis indicated that the traBC genes are required for both liquid and surface matings, suggesting that the traBC genes encode positive regulators of R64 transfer gene expression (13). Some of the R64 pil genes encode proteins which are closely related to type IV pilus biogenesis (14) (see Discussion), indicating that the R64 thin pilus belongs to the type IV pilus family.
Thin pili detached from cells were purified from culture medium in which E. coli cells harboring R64- or ColIb-P9-derived plasmids had been grown and then were characterized (39). In negatively stained thin pilus samples, long rods with diameters of 6 nm, characteristic of type IV pili, were observed under an electron microscope. R64 and ColIb-P9 thin pili are composed of a major 19-kDa subunit, the product of pilS, and a minor 45-kDa subunit, the product of pilVA′. The pilS product was first synthesized as a 22-kDa protein and subsequently processed to a 19-kDa protein by the function of the pilU product. Furthermore, the N-terminal tryptophan of the 19-kDa protein was modified.
To test whether all of the 14 pil genes are required for the biogenesis of the R64 thin pilus, we have introduced frameshift mutations into all the pil genes. The 12 genes, pilK to -V, were shown to be indispensable for the biogenesis of thin pili.
MATERIALS AND METHODS
Bacterial strains, phages, and plasmids.
The E. coli K-12 strains used in this study were JM83 [Δ(lac-proAB) rpsL thi ara φ80 dlacZΔM15] (35) and TN102 [Nalr] (15).
Bacteriophages Iα (6) and PR64FS (7) were used to test the formation of thin pilus.
Plasmid vectors pUC118, pUC119 (35), and pCL1920 (20) were used for cloning. pKK641A′ contained a 21.88-kb HindIII fragment of R64 (drd-11) bearing the rep and tra-pil region, together with a 1.3-kb DNA fragment for kanamycin resistance from Tn5 (15). pUC7Tc was described previously (13).
Media.
Luria-Bertani medium was prepared as described previously (28). Solid media contained 1.5% agar. Antibiotics were added to liquid or solid media at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 12.5 μg/ml.
Construction of plasmids.
The preparation of plasmid DNA, construction of plasmids, transformation, and other methods of DNA manipulation were performed as previously described (28).
Frameshift mutations were introduced into the pilI to -V genes as well as the noncoding sequence of pKK641A′ as described previously (13). pKK641A′ DNA was partially digested with AluI, HaeIII, HpaI, NaeI, NruI, or SspI and ligated with a tetracycline resistance cassette. The DNA cassette was removed by BamHI digestion. This 22-bp sequence, AATTCCCCGGATCCGGGGAATT, remaining at each restriction site, gave rise to a frameshift mutation. pKK641A′ DNA was partially digested with NspI, treated with the Klenow fragment of E. coli DNA polymerase I, and ligated to give frameshift mutations with a 4-bp deletion. The locations of mutations were determined by restriction enzyme analysis or DNA sequencing.
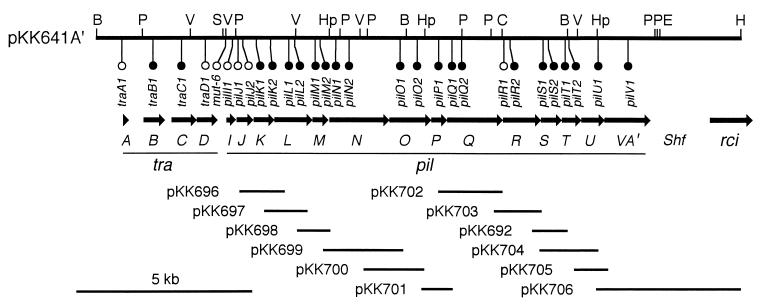
For the complementation experiments, each pil gene was separately cloned into pUC118, pUC119, or pCL1920 as shown in Fig. 1. Thus, the following plasmids were constructed: pKK696 (carrying pilK, nucleotides [nt] 4255 to 6199 in the DNA sequence of GenBank accession no. D88588 [14]), pKK697 (pilL, nt 4881 to 6395), pKK698 (pilM, nt 6194 to 6993), pKK699 (pilN, nt 6672 to 9426), pKK700 (pilO, nt 7769 to 9764), pKK701 (pilP, nt 9427 to 10482), pKK702 (pilQ, nt 9993 to 11997), pKK703 (pilR, nt 11652 to 12889), pKK692 (pilS, nt 12890 to 13514), pKK704 (pilT, nt 12890 to 14380), pKK705 (pilU, nt 13826 to 15184), and pKK706 (pilV, nt 12381 to 18523). Plasmids in which the pil genes are expressed under the control of the lac promoter of the vector, pUC118 or pUC119, are indicated by the suffix “a” (for example, pKK696a), while pUC118- or pUC119-derived plasmids containing the pil genes in the opposite orientation are indicated by the suffix “b”. Plasmids cloned in low-copy-number vector pCL1920 are indicated by the suffix “c”.
FIG. 1.
Gene organization of the traA to -D and pilI to -V regions of pKK641-A′. The top horizontal line represents a restriction map. B, BglII; C, ClaI; E, EcoRI; H, HindIII; Hp, HpaI; P, PstI; S, SmaI; V, PvuII. Locations of insertion or deletion mutations and their transfer capacity are indicated (open circles, transfer proficient; solid circles, transfer deficient). Below the map, open reading frames are represented by arrows. tra, transfer; pil, formation of thin pilus; Shf, shufflon; rci, shufflon-specific recombinase. The solid lines on the bottom indicate DNA portions present in complementing plasmids.
Conjugal transfer and phage sensitivity.
Liquid mating was performed as described previously (15). E. coli JM83 and TN102 cells were used as donor and recipient cells, respectively. A culture of log-phase donor cells was mixed with an overnight culture of recipient cells. The mixture was incubated for 90 min at 37°C. Sensitivity to phages Iα and PR64FS was determined as described previously (15).
Thin pilus fraction.
The thin pilus fraction was prepared as previously described (39). E. coli cells harboring pKK641A′ with various mutations were grown overnight with shaking at 37°C. The culture was centrifuged twice at 9,200 × g for 10 min to remove the bacterial cells. The supernatant was again centrifuged at 140,000 × g for 1 h. The pellet was used as a crude thin pilus fraction.
Western blot analysis.
Thin pilus fractions or lysates of E. coli cells harboring various plasmids were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (39). Proteins were transferred to a polyvinylidene difluoride membrane with a model AE-6675 semidry transfer apparatus (Atto Corp.) and detected with antipilin antiserum by using antirabbit immunoglobulin G conjugated to horseradish peroxidase with chromogenic substrates.
RESULTS
Introduction of frameshift mutations into 14 pil genes of R64.
To reveal the roles of the pilI to -V genes in the biogenesis of the R64 thin pilus, insertion or deletion mutations were introduced into the pilI to -V genes as well as the intergenic noncoding sequence. All mutations were designed to result in a frameshift of coding sequences. Furthermore, insertions and deletions were restricted to a small number of nucleotides to minimize the effects of mutations on the transcription of the downstream genes. Twenty-five insertion or deletion derivatives of pKK641A′ were constructed (Fig. 1). Twenty-four mutations were located within the pil genes, while the mut-6 mutation was in the noncoding sequence. At least one mutation was introduced into each of the pil genes. The locations of four tra mutations previously described (13) are also indicated in Fig. 1.
Effects of pil mutations on expression and processing of the pilS product.
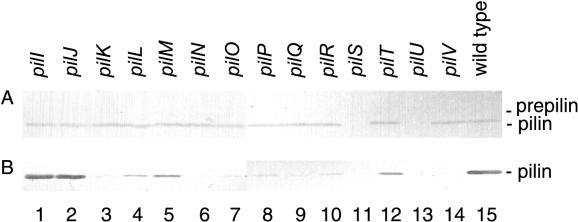
The R64 thin pilus comprises a major pilin subunit, the pilS product, and a minor subunit, the pilV product (39). The pilS product was shown to be synthesized as a 22-kDa prepilin and then processed to a 19-kDa protein via the function of the pilU product. The expression and processing of the pilS product in various pil mutants were examined. Whole-cell extracts from E. coli JM83 harboring pKK641A′ containing various pil mutations were separated by SDS-PAGE and subjected to Western blot analysis with antipilin antiserum (Fig. 2A). E. coli cells harboring pKK641A′ derivatives with mutations in all 14 pil genes except pilS and pilU produced similar amounts of 19-kDa pilin as cells harboring wild-type pKK641A′. E. coli cells harboring pKK641A′ (pilS1) produced neither pilin nor prepilin, confirming that pilin is the pilS product. The prepilin production by E. coli cells harboring pKK641A′ (pilU1) was not detected under these conditions.
FIG. 2.
(A) Effects of various pil mutations on the expression and processing of the pilS product. Whole extracts of E. coli cells harboring pKK641A′ with the indicated pil mutations were separated by SDS-PAGE and subjected to Western blot analysis with antipilin antiserum. (B) Effects of pil mutations on the formation of thin pili. A thin pilus fraction was prepared from the culture media of E. coli cells harboring pKK641A′ with the indicated pil mutations. Proteins were separated by SDS-PAGE and subjected to Western blot analysis with antipilin antiserum. The positions of prepilin (22 kDa) and pilin (19 kDa) are indicated on the right.
Effects of pil mutations on the biogenesis of the R64 thin pilus.
Detached thin pili were recovered by ultracentrifugation of the culture medium in which E. coli cells harboring pKK641A′ had been grown (thin pilus fraction) (39). The production of extracellular thin pilus in various pil mutants was examined by Western blot analysis of the amounts of pilin in the crude thin pilus fractions from E. coli cells harboring pKK641A′ with various pil mutations (Fig. 2B). E. coli cells harboring pKK641A′ (pilI1) and pKK641A′ (pilJ1) produced amounts of extracellular pilin similar to those of cells harboring wild-type pKK641A′. Very small amounts of extracellular pilin were produced by E. coli cells harboring pKK641A′ (pilL1), pKK641A′ (pilM1), and pKK641A′ (pilT1), and trace amounts of extracellular pilin were produced by cells harboring pKK641A′ (pilK1), pKK641A′ (pilN1), pKK641A′ (pilO1), pKK641A′ (pilP1), pKK641A′ (pilQ1), pKK641A′ (pilR2), and pKK641A′ (pilV1). No extracellular pilin was detected for pKK641A′ (pilS1) and pKK641A′ (pilU1).
Effects of pil mutations on liquid matings and phage sensitivity.
R64 liquid mating requires the presence of thin pilus (15). Hence, the formation of a functional thin pilus in various pil mutants was assessed by the transfer frequency in liquid matings. For this purpose, we used the pKK641-pKK661 system, in which the transfer frequency in R64 liquid matings can be accurately estimated, since a lack of the rci gene prevents the DNA rearrangement of the shufflon (15). E. coli donor cells harboring pKK641A′ and pKK661 transmitted pKK661 carrying the oriT sequence into the recipient cells by conjugation at a frequency of 1.2% (Table 1). When pKK641A′ (mut-6) carrying a mutation outside the coding sequences was used instead of pKK641A′, the transfer frequency was equivalent to that of pKK641A′, confirming that the mut-6 mutation has no effect on the expression of the other pil genes. E. coli cells harboring three different pKK641A′ derivatives with mutations in the pilI and pilJ genes were also transfer proficient (Table 1). In contrast, cells harboring pKK661 and pKK641A′ derivatives with mutations in the 12 genes pilK through pilV were transfer deficient, with the exception of pKK641A′ (pilR1). pKK641A′ (pilR1) which lacks 4 nt, including the ATG initiation codon of the pilR gene, was transfer proficient. In this plasmid, pilR translation may restart from the next methionine codon at the fourth codon of the pilR gene. For the pilT1 and pilT2 mutants, residual transfer activities were observed (Table 1). These results indicate that 12 genes, pilK, pilL, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilT, pilU, and pilV, are required for R64 liquid mating.
TABLE 1.
Effects of pil mutations on transfer frequency and sensitivity to phages
| Plasmid | Mutationa
|
Transfer frequency (%)b | Sensitivity to phagec:
|
||
|---|---|---|---|---|---|
| Location | Length (bp) | Iα | PR64FS | ||
| pKK641A′ | 1.2 | ||||
| pKK641A′ (mut-6) | 3713 | +22 | 1.0 | + | + |
| pKK641A′ (pilI1) | 3893 | +22 | 1.3 | + | + |
| pKK641A′ (pilJ1) | 4075 | +22 | 1.1 | + | + |
| pKK641A′ (pilJ2) | 4189 | +22 | 1.3 | + | + |
| pKK641A′ (pilK1) | 4566 | +22 | 0.005 | − | − |
| pKK641A′ (pilK2) | 5000 | −4 | 0.002 | − | − |
| pKK641A′ (pilL1) | 5510 | +22 | 0.002 | − | − |
| pKK641A′ (pilL2) | 5682 | +22 | 0.002 | − | − |
| pKK641A′ (pilM1) | 6266 | +22 | 0.003 | − | − |
| pKK641A′ (pilM2) | 6413 | +22 | 0.003 | − | − |
| pKK641A′ (pilN1) | 6854 | +22 | 0.004 | − | − |
| pKK641A′ (pilN2) | 7238 | +22 | 0.002 | − | − |
| pKK641A′ (pilO1) | 8719 | +22 | 0.002 | − | − |
| pKK641A′ (pilO2) | 9189 | −4 | 0.003 | − | − |
| pKK641A′ (pilP1) | 9798 | +22 | 0.002 | − | − |
| pKK641A′ (pilQ1) | 10201 | +22 | 0.002 | − | − |
| pKK641A′ (pilQ2) | 10467 | +22 | 0.001 | − | − |
| pKK641A′ (pilR1) | 11676 | −4 | 0.9 | + | + |
| pKK641A′ (pilR2) | 11873 | +22 | 0.002 | − | − |
| pKK641A′ (pilS1) | 12807 | +22 | 0.002 | − | − |
| pKK641A′ (pilS2) | 12945 | +22 | 0.003 | − | − |
| pKK641A′ (pilT1) | 13433 | +22 | 0.013 | + | + |
| pKK641A′ (pilT2) | 13551 | +22 | 0.018 | + | + |
| pKK641A′ (pilU1) | 14381 | +22 | 0.002 | − | − |
| pKK641A′ (pilV1) | 15254 | +22 | 0.003 | − | − |
Locations (nucleotide number in the DNA sequence of GenBank accession no. D88588) and lengths (+, insertion; −, deletion) of the mutations are indicated.
Transfer frequency of pKK661 from donor cells harboring the indicated plasmids and pKK661 in liquid mating is indicated as a percentage of transconjugants relative to donor cells.
Sensitivity to phages Iα and PR64FS of E. coli cells harboring the indicated plasmid (+, sensitive; −, resistant).
The sensitivity of E. coli cells harboring pKK641A′ derivatives to phages Iα and PR64FS was also tested (Table 1). Phages Iα and PR64FS specifically adsorb to the shaft and tip, respectively, of thin pili produced by the IncI1 plasmid, and the infected cells subsequently produce progeny phage (6, 7). Therefore, the sensitivity of cells to phages Iα and PR64FS can be used as an indication of thin pilus formation. The results shown in Table 1 indicate that 11 genes, pilK, pilL, pilM, pilN, pilO, pilP, pilQ, pilR, pilS, pilU, and pilV, are required for the formation of thin pili as receptors for phages Iα and PR64FS.
Complementation of pil mutations by pil+ plasmids.
To test complementation of pil mutations, various plasmids carrying each pil gene were constructed (Fig. 1). The pKK696a series of plasmids contained pil genes in the orientation in which they were expressed under the control of the lac promoter of the vector, while the pKK696b series of plasmids contained the pil genes in the opposite orientation. E. coli cells harboring pKK661 and pKK641A′ with each mutation were transformed by plasmids carrying the corresponding pil gene (Table 2). E. coli cells harboring pKK706a and pKK641A′ (pilV1) did not grow well, while those harboring pKK706b and pKK641A′ (pilV1) grew normally. Similarly, E. coli cells harboring pKK706a together with wild-type pKK641A′ did not grow well. These results suggest that overproduction of the pilV gene in E. coli cells forming R64 thin pili is harmful for cell growth. In the other combinations, defects in liquid mating of mutants containing the pilK1, pilL1, pilM1, pilN1, pilO1, pilP1, pilQ1, pilR2, pilS1, pilU1, and pilV1 mutations were complemented by the corresponding pil+ plasmids, although the defect of the pilT1 mutant could not be complemented (Table 2). The defect of the pilV1 mutant was complemented by low-copy-number plasmid pKK706c. Defects in phage sensitivity for all of the pil mutants were also complemented by the corresponding pil+ plasmids (data not shown). These results indicate that the pil mutants used in this study did not exhibit polar effects on the expression of downstream genes.
TABLE 2.
Complementation of pil mutations by pil+ plasmids
| Plasmid | Complementation with plasmid froma:
|
|
|---|---|---|
| pKK696a series | pKK696b series | |
| pKK641A′ | ||
| pKK641A′ (pilK1) | + (pKK696a) | + (pKK696b) |
| pKK641A′ (pilL1) | + (pKK697a) | + (pKK697b) |
| pKK641A′ (pilM1) | + (pKK698a) | + (pKK698b) |
| pKK641A′ (pilN1) | + (pKK699a) | + (pKK699b) |
| pKK641A′ (pilO1) | + (pKK700a) | + (pKK700b) |
| pKK641A′ (pilP1) | + (pKK701a) | + (pKK701b) |
| pKK641A′ (pilQ1) | + (pKK702a) | + (pKK702b) |
| pKK641A′ (pilR2) | + (pKK703a) | + (pKK703b) |
| pKK641A′ (pilS1) | + (pKK692a) | + (pKK692b) |
| pKK641A′ (pilT1) | − (pKK704a) | − (pKK704b) |
| pKK641A′ (pilU1) | + (pKK705a) | + (pKK705b) |
| pKK641A′ (pilV1) | Unstable (pKK706a) | + (pKK706b) |
Transfer frequency of pKK661 from donor cells harboring pKK661 and the indicated plasmids with complementing plasmids (indicated in parentheses) in liquid matings was estimated as in Table 1. +, with complementation activity (transfer frequencies recovered to a level similar to that of wild-type pKK641A′); −, without complementation activity.
DISCUSSION
In the present study, we demonstrated that 12 of 14 pil genes are indispensable for the biogenesis of the R64 thin pilus. We have previously shown that two genes, traBC, are required for the expression of R64 pil genes (13). In addition, the rci gene modulates R64 thin pilus function by DNA rearrangement of the shufflon (16, 18, 19). The R64 shufflon consists of four DNA segments which are flanked by seven 19-bp repeat sequences. The rci product mediates recombinations between any two inverted repeats, resulting in the inversions of the four DNA segments independently or in groups. The shufflon DNA rearrangement selects one of seven possible pilV genes, whereupon the N-terminal region remains constant while the C-terminal region is variable. The seven pilV genes determine recipient specificity in liquid matings (17). High-frequency liquid matings were observed only for proper combinations of recipient bacterial strains and C-terminal segments of the pilV genes in R64 donor cells. For example, donor cells producing thin pili with PilVA′, PilVC, or PilVC′ protein, but not PilVA, PilVB, PilVB′, or PilVD′ protein, were transferred into E. coli K-12 recipient cells at a high frequency in liquid matings. E. coli K-12 cells producing thin pili with PilVA′, PilVC, or PilVC′ protein formed large cell aggregates, which presumably play crucial roles in liquid matings (39).
It has been shown that many gene products are involved in the biogenesis of type IV pili. More than 30 genes are required for the biogenesis and function of type IV pili in P. aeruginosa (2, 22, 37). They are distributed among several loci, forming gene clusters, over the P. aeruginosa chromosome. Several of the pil genes are required for the biogenesis and function of type IV pili, while others regulate type IV pilus gene expression. Several genes homologous to the P. aeruginosa pil genes were identified in N. gonorrhoeae (8, 9). The type IV pilus biogenesis system of N. gonorrhoeae undergoes antigenic variation as well as phase variation (33).
In contrast to the type IVA pilus biogenesis system, the genes encoding type IVB pilus biogenesis appear to form gene clusters. Both the bfp system of enteropathogenic E. coli and the tcp system of V. cholerae are comprised of clusters of 14 genes (24, 29, 30). Proteins involved in general secretory pathways of gram-negative bacteria (26, 27) and DNA uptake systems of gram-positive bacteria (1, 23) share amino acid sequence homology with members of the type IV pilus biogenesis system.
Conserved proteins among the type IV pilus biogenesis systems and related systems are summarized in Table 3. The R64 Pil proteins shown in Table 3 were found to be indispensable for thin pilus formation in the present work. Many proteins of the other systems shown in Table 3 were demonstrated to be essential for type IV pilus biogenesis or a related activity.
TABLE 3.
Conserved proteins in type IV pilus biogenesis and related systems
| Homologuea
|
Characteristic | ||||||
|---|---|---|---|---|---|---|---|
| R64 | EPEC | V. cholerae | P. aeruginosa | N. gonorrhoeae | GSP | Bsu | |
| PilS | BfpA | TcpA | PilA | PilE | Prepilin | ||
| PilU | BfpP | TcpJ | PilD | PilD | PulO | ComC | Prepilin peptidase |
| PilN | BfpB | TcpC | PilQ | PilQ | PulD | Outer membrane protein | |
| PilQ | BfpD | TcpT | PilB | PilF | PulE | ComGA | Nucleotide binding protein |
| PilR | BfpE | TcpE | PilC | PilG | PulF | ComGB | Integral membrane protein |
| PilT | BfpH | Gene X homologue | |||||
| PilV | BfpI, -J, and -K | PilE, -V, -W, and -X; FimT and -U | PulG, -H, -I, and -J | ComGC, -GD, and -GE | Prepilin peptidase cleavage site | ||
The pilS gene is essential, since it encodes the prepilin of R64 thin pilus. Four pilS mutant classes were identified from mutational analyses (12). The products of class I pilS mutants were not processed by prepilin peptidase. The extracellular secretion of the products of class II pilS mutants was inhibited. Class III pilS mutants formed thin pili with reduced activities in liquid matings. Class IV pilS mutants exhibited thin pili with mating activities similar to that of pili formed from the wild-type pilS gene. In addition, various proteins containing prepilin peptidase cleavage sites are essential for the type IV pilus biogenesis systems and related systems (Table 3). As many as six such proteins are involved in type IV pilus biogenesis in P. aeruginosa and are suggested to construct a membrane scaffold (22). Among the other proteins containing type IV prepilin cleavage sites, R64 PilV is the only protein which has been demonstrated to be the minor component of type IV pili (39).
Prepilin peptidases including R64 PilU are required for the processing of type IV prepilins (Table 3). The PilD protein in P. aeruginosa was shown to function as a pilin N-methylase in addition to a prepilin peptidase (32). In R64 thin pili, the N-terminal end is not methylated but modified with an unknown blocking group (39). Outer membrane proteins including R64 PilN are required for type IV pilus biogenesis. Among such outer membrane proteins, R64 PilN, enteropathogenic E. coli BfpB, and V. cholerae TcpC are lipoproteins (14, 24, 29, 30). They are also related to the gene IV proteins of f1-type filamentous phages (11). Nucleotide-binding proteins such as R64 PilQ are conserved among type IV pilus biogenesis and related systems. Similar nucleotide-binding proteins are also found in plasmid transfer systems (RP4 TrbB) (21) and the vir genes of Ti plasmids (Ti VirB11) (36). Additional nucleotide-binding proteins, BfpF and PilT, containing less similarity are also present in enteropathogenic E. coli and P. aeruginosa, respectively (22, 29, 30). Integral membrane proteins such as R64 PilR are also common in the type IV pilus biogenesis and related systems. Gene X homologues PilT and BfpH are present only in R64 and enteropathogenic E. coli. Gene X (gene 19), located in the “leading region” of IncF plasmids, was shown to be required for the efficient transfer of R1 (3).
It was previously shown that E. coli cells harboring a pilS+ multicopy plasmid produced a 22-kDa prepilin and that cells harboring a pilSTU+ multicopy plasmid produced a 19-kDa pilin as well as a 22-kDa prepilin (39). While we had expected that E. coli cells harboring pKK641A′ (pilU1) would produce a 22-kDa prepilin, this was not the case. When a 100-fold amount of cell extract from cells harboring pKK641A′ (pilU1) was electrophoresed in an experiment similar to that in Fig. 2A, a small amount of 22-kDa prepilin was found (data not shown). One possible explanation for this is that unprocessed prepilin is unstable and is degraded rapidly. Alternatively, unprocessed prepilin may negatively regulate the expression of the pilS gene. Whether one or the other mechanism (or both) is responsible remains to be elucidated in future experiments.
ACKNOWLEDGMENTS
We are grateful to N. Furuya for useful discussions and K. Takayama for critical reading of the manuscript.
This work was supported in part by a grant from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Albano M, Breitling R, Dubnau D A. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J Bacteriol. 1989;171:5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm R A, Hallinan J P, Watson A A, Mattick J S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- 3.Bayer M, Eferl R, Zellnig G, Teferle K, Dijkstra A, Koraimann G, Högenauer G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J Bacteriol. 1995;177:4279–4288. doi: 10.1128/jb.177.15.4279-4288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley D E. Derepressed plasmids of incompatibility group I1 determine two different morphological forms of pilus. Plasmid. 1983;9:331–334. doi: 10.1016/0147-619x(83)90011-2. [DOI] [PubMed] [Google Scholar]
- 5.Bradley D E. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K, and Z. J Gen Microbiol. 1984;130:1489–1502. doi: 10.1099/00221287-130-6-1489. [DOI] [PubMed] [Google Scholar]
- 6.Coetzee J N, Bradley D E, Hedges R W. Phages Iα and I2-2: IncI plasmid-dependent bacteriophages. J Gen Microbiol. 1982;128:2797–2804. doi: 10.1099/00221287-128-11-2797. [DOI] [PubMed] [Google Scholar]
- 7.Coetzee J N, Sirgel F A, Lacatsas G. Properties of a filamentous phage which absorbs to pili coded by plasmids of the IncI complex. J Gen Microbiol. 1980;117:547–551. doi: 10.1099/00221287-117-2-547. [DOI] [PubMed] [Google Scholar]
- 8.Drake S L, Sandstedt S A, Koomey M. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol Microbiol. 1997;23:657–668. doi: 10.1046/j.1365-2958.1997.2511618.x. [DOI] [PubMed] [Google Scholar]
- 9.Freitag N E, Seifert H S, Koomey M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 10.Frost L S. Conjugative pili and pilus-specific phages. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Publishing; 1993. pp. 189–212. [Google Scholar]
- 11.Hill D F, Petersen G B. Nucleotide sequence of bacteriophage f1 DNA. J Virol. 1982;44:32–46. doi: 10.1128/jvi.44.1.32-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horiuchi T, Komano T. Mutational analysis of plasmid R64 thin pilus prepilin: the entire prepilin sequence is required for processing by type IV prepilin peptidase. J Bacteriol. 1998;180:4613–4620. doi: 10.1128/jb.180.17.4613-4620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S-R, Funayama N, Komano T. Nucleotide sequence and characterization of the traABCD region of IncI1 plasmid R64. J Bacteriol. 1993;175:5035–5042. doi: 10.1128/jb.175.16.5035-5042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S-R, Komano T. The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komano T, Funayama N, Kim S-R, Nisioka T. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J Bacteriol. 1990;172:2230–2235. doi: 10.1128/jb.172.5.2230-2235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komano T, Kim S-R, Yoshida T. Mating variation by DNA inversions of shufflon in plasmid R64. Adv Biophys. 1995;31:181–193. doi: 10.1016/0065-227x(95)99391-2. [DOI] [PubMed] [Google Scholar]
- 17.Komano T, Kim S-R, Yoshida T, Nisioka T. DNA rearrangement of the shufflon determines recipient specificity in liquid mating of IncI1 plasmid R64. J Mol Biol. 1994;243:6–9. doi: 10.1006/jmbi.1994.1625. [DOI] [PubMed] [Google Scholar]
- 18.Komano T, Kubo A, Nisioka T. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 1987;15:1165–1172. doi: 10.1093/nar/15.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubo A, Kusukawa A, Komano T. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet. 1988;213:30–35. doi: 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- 20.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capacity. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 22.Mattick J S, Whitchurch C B, Alm R A. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene. 1966;179:147–155. doi: 10.1016/s0378-1119(96)00441-6. [DOI] [PubMed] [Google Scholar]
- 23.Mohan S, Aghion J, Guillen N, Dubnau D. Molecular cloning and characterization of comC, a late competence gene of Bacillus subtilis. J Bacteriol. 1989;171:6043–6051. doi: 10.1128/jb.171.11.6043-6051.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogierman M A, Zabihi S, Mourtzios L, Manning P A. Genetic organization and sequence of the promoter-distal region of the tcp gene cluster of Vibrio cholerae. Gene. 1993;126:51–60. doi: 10.1016/0378-1119(93)90589-u. [DOI] [PubMed] [Google Scholar]
- 25.Ottow J C G. Ecology, physiology, and genetics of fimbriae and pili. Annu Rev Microbiol. 1975;29:79–108. doi: 10.1146/annurev.mi.29.100175.000455. [DOI] [PubMed] [Google Scholar]
- 26.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyss I, Pugsley A P. Five additional genes in the pulC-O of the gram-negative bacterium Klebsiella oxytoca UNF5023 that are required for pullulanase secretion. Mol Gen Genet. 1990;222:176–184. doi: 10.1007/BF00633815. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sohel I, Puente J L, Ramer S W, Bieber D, Wu C-Y, Schoolnik G K. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J Bacteriol. 1996;178:2613–2628. doi: 10.1128/jb.178.9.2613-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone K D, Zhang H-Z, Carlson L K, Donnenberg M S. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol Microbiol. 1996;20:325–337. doi: 10.1111/j.1365-2958.1996.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 31.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 32.Strom M S, Nunn D N, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson J, Koomey J M. Mechanisms for variation of pili and outer membrane protein II in Neisseria gonorrhoeae. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 743–781. [Google Scholar]
- 34.Taniguchi T, Fujino Y, Yamamoto K, Miwatani T, Honda T. Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect Immun. 1995;63:724–728. doi: 10.1128/iai.63.2.724-728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 36.Ward J E, Akiyoshi D E, Regier D, Datta A, Gordon M P, Nester E W. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1988;263:5804–5814. [PubMed] [Google Scholar]
- 37.Whitchurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 38.Wu S S, Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J Bacteriol. 1998;180:2842–2848. doi: 10.1128/jb.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]