Abstract
Severe shrimp disease outbreaks have a destructive impact on shrimp aquaculture and its associated downstream food processing industries. Thus, it is essential to develop proper methods for shrimp disease control, which emphasizes the importance of food safety. In this study, we performed biochemical tests and gut microbiome analysis using uninfected control and Vp AHPND‐infected Penaeus monodon samples. Biochemical tests were performed to assess the phenoloxidase (PO) activity, respiratory Burst (RB) activity, nitrite concentration, superoxide dismutase (SOD) activity, total hemocyte count (THC), and total protein concentrations. Overall, upregulations were detected in these biochemical tests, which showed the activation of the immune response in P. monodon during acute hepatopancreatic necrosis disease (AHPND) infection, especially at 6 hpi and 12 hpi. Besides that, shrimp gut samples were collected and pooled (n = 3), followed by DNA extraction, PCR amplification targeting the V3/V4 16S ribosomal RNA (rRNA) region, next‐generation sequencing (NGS), and bioinformatics analysis. Proteobacteria was the most abundant phylum in both samples. The Rhodobacteraceae family and Maritimibacter genus were proposed to be vital forshrimp health maintenance. Vp AHPND bacterial colonization and secondary Vibrio infections were postulated to have occurred based on the higher abundances of Vibrionaceae family and Vibrio genus in the Vp AHPND‐infected sample. Firmicutes phylum together with Photobacterium and Aliiroseovarius genera were inferred to be pathogenic or related factors of AHPND infections. In conclusion, physiology (immune response activation) and gut microbiome changes of disease tolerant P. monodon during AHPND infection were identified. Both biochemical tests and 16S rRNA analysis are proposed as a combined strategy for shrimp health diagnosis for ensuring shrimp health maintenance, disease control, and food safety.
Keywords: 16S rRNA analysis, acute hepatopancreatic necrosis disease, biochemical tests, food safety, Penaeus monodon
Severe shrimp disease outbreaks have a destructive impact on shrimp aquaculture and its associated downstream food processing industries. Thus, it is essential to develop proper methods for disease control, which emphasizes on the importance of food safety. In this study, we successfully performed biochemical tests and 16S gut microbiome analysis using uninfected control and Vp AHPND‐infected P. monodon samples.
1. INTRODUCTION
The global shrimp aquaculture industry is a crucial source of aquatic food and protein, in which aquaculture shrimp production constituted more than half of the world's shrimp supplies since the year 2015 (Anderson et al., 2016). In Asia, shrimp production successfully achieved 3.75 million metric tons in the year 2018, with an estimated production of 4.00 million metric tons in the year 2021 (Anderson et al., 2019). However, shrimp diseases remain a major challenge for shrimp farmers (Anderson et al., 2019). Some recent important shrimp diseases are acute hepatopancreatic necrosis disease (AHPND), hepatopancreatic microsporidiosis, white spot disease, yellow head disease, hepatopancreatic haplosporidiosis, covert mortality disease, aggregated transformed microvilli, and infectious myonecrosis (Thitamadee et al., 2016).
Acute hepatopancreatic necrosis disease disease is a shrimp bacterial disease that can cause 40% to 100% mortality within the early 35 days of shrimp stocking (Hong et al., 2016). This disease is caused by a pathogenic strain of Gram‐negative Vibrio parahaemolyticus bacteria called Vp AHPND (Lee et al., 2015; Tran et al., 2013). These Vp AHPND bacteria are capable of producing Photorhabdus insect‐related (Pir) toxins, PirA and PirB, encoded by corresponding genes located within a 70‐kbp plasmid (pVA1) (Lee et al., 2015). Since its first emergence in China in the year 2009, AHPND had spread to Southeast Asia and Mexico regions, which caused significant reductions in shrimp aquaculture production (Fao, 2013; Nunan et al., 2014). The common gross clinical signs of AHPND infection are slow growth, empty gut, empty stomach, and pale or atrophied hepatopancreas (Hong et al., 2016). The shrimp species susceptible to AHPND infection include Penaeus monodon, Litopenaeus vannamei, and Fenneropenaeus chinensis (Zorriehzahra & Banaederakhshan, 2015).
Despite being one of the seafood that cause human illnesses globally (Bondad‐Reantaso et al., 2008), there had been insufficient studies on shrimp diseases from the perspective of food safety. The majority of shrimp‐related food safety studies were focused on dietary supplementation (Li et al., 2007) or the food processing (Kaur et al., 2013). Seafood‐borne illnesses are majorly caused by Vibrio species, especially V. parahaemolyticus, V. cholerae, and V. vulnificus (Bondad‐Reantaso et al., 2008; Gopal et al., 2005). The bacterial infections that are potentially pathogenic to humans may have occurred in diseased aquatic animals, particularly those caused by Vibrios (Austin, 2010). In a previous study, Gopal et al. (2005) showed that even though not all isolated Vibrio species possessed pathogenicity or toxicity traits, a significant percentage composition of these Vibrio species found and isolated from the farmed shrimp samples clearly exhibited hidden risks associated with food safety in shrimp consumption. Such hidden risks can be eliminated by ensuring proper shrimp health detection and shrimp gut microbiome monitoring.
The correct and accurate determination of shrimp physiological changes is vital for shrimp health detection and disease prevention. Physiological changes in shrimps in response to stress or diseases are usually detected through various biochemical tests. Common biochemical tests applied are for the estimation of phenoloxidase (PO) activity (Hsieh et al., 2008; Lin et al., 2011), respiratory burst (RB) activity (Lin et al., 2011), superoxide dismutase (SOD) activity (Hsieh et al., 2008; Lin et al., 2011), nitrite concentration (Zokaeifar et al., 2014), total protein (Santhoshkumar et al., 2017; Zokaeifar et al., 2014), and total hemocyte count (THC) (Hsieh et al., 2008; Lin et al., 2011).
Other than that, another essential strategy for shrimp health diagnosis and disease prevention would be shrimp gut microbiome analysis. This is because of the crucial roles played by gut microbiota in the various physiological processes, including metabolism (Tremaroli & Bäckhed, 2012), immune regulation (Maynard et al., 2012), endocrine function (Clarke et al., 2014), and pathogen elimination (Endt et al., 2010). Ever since the widescale application of high throughput next‐generation sequencing (NGS) technology, there had been increasing demands of NGS application in gut microbiome analysis especially involving 16S rRNA diversity due to cost and effectiveness concerns (Caporaso et al., 2011). 16S rRNA studies are usually conducted for the identification of microbial diversity related to host or environmental parameters (Marzinelli et al., 2015) and pattern of gene content (Konstantinidis & Tiedje, 2005). The V3/V4 hypervariable region of the 16S rRNA gene is commonly targeted for 16S rRNA sequencing analysis, as demonstrated in some previous works (Fan et al., 2019; Porchas‐Cornejo et al., 2017; Zoqratt et al., 2018). An advantage of 16S rRNA sequencing analysis would be its less reliance on the quality of extracted DNA samples (Rintala et al., 2017). Although in fewer frequencies, there were also some publications of 16S rRNA analysis involving healthy and diseased shrimps (Cornejo‐Granados et al., 2017; Liang et al., 2020; Zhou et al., 2019).
Therefore, this study aims to identify the physiology and gut microbiome changes of P. monodon during AHPND infection. The biochemical tests and 16S rRNA sequencing technique are also proposed as a combined enhanced strategy for the diagnosis of shrimp disease and the regulation of shrimp health, from the perspective of food safety and nutrition.
2. MATERIALS AND METHODS
2.1. Shrimp pathogenic challenge and sample collection
Juvenile disease tolerant crossbred P. monodon shrimps (5th generation Malaysian strain crossed with 13th generation Madagascar strain) with an average body length of 15–20 cm were collected from a local commercial farm. A modified immersion method (Tran et al., 2013) based AHPND bacterial challenge experiment was conducted as described previously (Devadas, 2019; Soo et al., 2019). Initial isolation, validation, incubation, and selective enrichment of the Vp AHPND bacteria local strain, KS17.S5‐1 was done (Devadas et al., 2018). The incubation and selective enrichment of the Vp AHPND bacteria were done using tryptic soy broth (TSB+) (2% sodium chloride supplemented) (Merck), thiosulfate citrate bile salt (TCBS) agar (Merck), and tryptic soy agar (2% sodium chloride supplemented) (Merck). The incubation condition utilized was 28°C for 18 h at 120 rpm. The obtained Vp AHPND bacteria was verified using AP3 polymerase chain reaction (PCR) method (Sirikharin et al., 2015).
For the AHPND experimental challenge, Vp AHPND bacteria (KS17.S5‐1 strain) at a concentration of 2 × 106 cfu/ml was used for the Vp AHPND‐infected treatment group, whereas sterile TSB + broth was used for the uninfected control treatment group. 27 shrimps were placed in each tank filled with aerated artificial seawater (30 ppt) at 28 ± 1.0°C under an aseptic setup. Shrimp acclimatization was performed for 7 days before the challenge experiment. Three shrimp samples from each treatment group were collected at 0, 3, 6, 12, 24, 36, and 48 h post‐infection (hpi) and their vital organs (hepatopancreas, gut, muscle, and haemolymph) were stored at −80°C. The AHPND infection was validated by the observation of gross clinical symptoms (pale white hepatopancreas, empty stomach, and empty gut) and the AP3 PCR detection method (Sirikharin et al., 2015). The Ethical approval for this work was granted by the University of Malaya (Ethical Application Ref: S/31012019/26112018‐05/R).
2.2. Biochemical test validation
Hepatopancreas from the collected P. monodon samples were homogenized in 1X phosphate‐buffered saline (PBS) at a ratio of 1:9 and centrifuged at 13,300 g for 20 min at 4°C. The supernatant was collected from homogenized samples, diluted based on downstream applications, and stored at −20°C. The collected shrimp muscle and hemolymph samples were also utilized for biochemical analysis. The biochemical experiments were performed in a Greiner 96‐well U bottom microplate (Greiner Bio‐One).
All biochemical assay experiments were conducted with three biological replicates and three technical replicates for each. Statistical analysis involving one‐way analysis of variance (ANOVA) accompanied by Duncan's post hoc test was performed. The statistical significance value was set at p < .05. The raw data for the biochemical assay experiments was provided in Data S1.
2.2.1. Bradford protein assay
Hepatopancreas and muscle samples of P. monodon were homogenized (mixed with 1X PBS at the ratio of 1:9 in ice water) and centrifuged (580 g for 10 min), and the supernatant was collection (diluted as and when needed) and utilized for the Bradford protein assay experiment. The total protein concentrations were identified by the Bradford Assay method (Bradford, 1976) with slight modifications. For the modified Bradford protein estimation assay, 5 µl of homogenized sample supernatant was mixed with 250 µl of 1X Bradford reagent (Coomassie brilliant blue G‐250 dissolved in methanol, phosphoric acid, and water) and was subsequently incubated for 50 min at room temperature. Then, the absorbance of the solution was measured at 595 nm, using a Tecan M200 Infinite Pro Microplate Reader (Tecan Group). The positive control used was bovine serum albumin (BSA) solution (for standard curve plotting), whereas the negative control used was 1X PBS.
2.2.2. Phenoloxidase activity assay
Phenoloxidase activity assay was done by a previously described modified method (Hong et al., 2019; Park et al., 2019). The assay involved the transformation of L‐3, 4‐dihydroxyphenylalanine (L‐DOPA) to dopachrome. 50 µl of the homogenized hepatopancreas sample supernatant was mixed with 50 µl of 1X PBS and 50 µl of L‐DOPA (3 mg/ml) (dissolved in PBS). The dopachrome formation was then measured at 490 nm for every one min for a total time of 10 min using the Tecan M200 Infinite Pro Microplate Reader (Tecan Group). The PO activity was measured as the maximum change of A490 nm/min × 103 per mg protein. One unit was defined as the increase of 0.001 A490 nm/min. The result normalization was done using the protein concentrations previously identified through the Bradford Assay method. 1X PBS solution was used instead of sample for the negative control.
2.2.3. Respiratory burst activity assay
Respiratory burst activity assay was carried out based on a previously described method (Huynh et al., 2011) with slight modifications. The 96‐well microplate was coated with 100 µl of poly‐L‐lysine solution (0.2% v/v) (diluted with deionized water) 24 h before use for increased cell adhesion. 50 µl of diluted hemolymph samples were mixed with 100 µl of Zymosan A (1 mg/ml) in modified complete Hank's balanced salt solution (MCHBSS) and the mixture was left to react for 30 min at room temperature. 100 µl of nitro blue tetrazolium chloride (NBT) solution (0.3% w/v) was then added to the mixture and it was incubated for 30 min at room temperature. Then, 50 µl of 100% methanol was added to stop the reaction. The mixture was discarded, and the microplate was washed thrice with 100 µl of 70% methanol and air‐dried for 30 min. 120 µl of potassium hydroxide (KOH) (2 M) (dissolved in deionized water) and 140 µl of dimethyl sulfoxide (DMSO) were added for dissolving the insoluble formazan crystals formed from NBT reduction. The RB activity was measured at 630 nm using the Tecan M200 Infinite Pro Microplate Reader (Tecan Group) as the measurement of superoxide anion generation. Zymosan A solution in MCHBSS was used as the positive control, whereas 1X PBS was used as the negative control.
2.2.4. Relative superoxide dismutase activity assay
The relative SOD activity was measured based on a method described by Perera et al. (2017) with slight modifications. Initially, the reaction mixture containing 20 µl of homogenized hepatopancreas sample supernatant or 1X PBS (negative control), 160 µl of glycine‐NaOH buffer (pH 9; 0.1 M) (dissolved in deionized water), and 6.75 µl of each: ethylenediaminetetraacetic acid (EDTA) (3 mM) (dissolved in deionized water), 0.15% BSA, xanthine (3 mM) (dissolved in deionized water), and NBT (0.75 mM) (dissolved in deionized water) was prepared. The reaction mixture was incubated for 10 min at room temperature. The reaction was then initiated by adding 6 mU of xanthine oxidase (dissolved in deionized water) and allowed to run for 20 min at room temperature. The SOD activity was measured at 560 nm every two mins for a total period of 20 min, using the Tecan M200 Infinite Pro Microplate Reader (Tecan Group). The relative SOD activity was calculated as the percentage of respective SOD enzyme activity from the highest SOD enzyme activity (100%).
2.2.5. Nitrite concentration assay
The nitrite concentration measurement was estimated by a modified method based on previous publications (Al‐Amin et al., 2015, 2016; Tracey et al., 1995). 6 µl of homogenized hepatopancreas sample supernatant was mixed with 44 µl of deionized water and 20 µl of phosphate buffer (pH 7.5; 0.31 M) (dissolved in deionized water). The sample mixture was placed in the dark for one hour at room temperature. Subsequently, 200 µl of Griess reagent (1:1 mixture of sulfanilamide solution [1% sulfanilamide in 5% phosphoric acid] and NED solution [0.1% w/v N‐1‐naphthylethylenediamine dihydrochloride dissolved in deionized water]) was added. The assay was incubated in the dark at room temperature for another 10 min before being measured at 540 nm using the Tecan M200 Infinite Pro Microplate Reader (Tecan Group). A serially diluted 100 µM nitrite solution (sodium nitrite dissolved in deionized water) was used for the plotting of a nitrite standard reference curve. The A540 nm measurements of the samples were then converted to nitrite concentrations by matching the nitrite standard reference curve plotted. The negative control used was 1X PBS.
2.2.6. Total hemocyte count
The THC was obtained using a modified standard method as described previously (Huynh et al., 2018). Hemolymph samples were mixed with 0.5% trypan blue solution (diluted with deionized water) at a dilution ratio of 1:1. The Neubauer improved hemocytometer (Marienfeld) and its coverslip were wiped with 70% ethanol before usage. The coverslip was set onto the hemocytometer, and 10 µl of diluted hemolymph mixture was filled into one of the chambers of the hemocytometer. The cells were counted under 40× magnification using Leica DM750 upright microscope (Leica Microsystems).
2.3. Gut microbiome analysis
2.3.1. DNA extraction and 16S rRNA sequencing
DNA samples were extracted from the pooled P. monodon gut samples (n = 3 for each group) using the phenol‐chloroform DNA extraction method (Barker, 1998; PacBio, 2012) with modifications. The DNA extraction involved the initial cutting of 50–100 mg of sample into small pieces. The next step was the addition of 300 µl of lysis buffer (10 mM Tris‐HCl [pH 8], 25 mM EDTA [Merck], 100 mM NaCl, and 2% sodium dodecyl sulfate) (OIE, 2019), 18 µl of RNase A solution (10 mg/ml) (Bio Basic Inc.), and 18 µl of Proteinase K solution (20 mg/ml) (Bioteke Corporation), and then the solution was mixed by inversion and incubation in heat at 65°C for 2 h. 300 µl of phenol‐chloroform‐isoamyl alcohol (25:24:1) was then added, mixed, and centrifuged at 16,000 g for 5 min at 27°C to achieve 3 layers separation. 150 µl of the upper aqueous layer was removed and mixed with 300 µl of chloroform‐isoamyl alcohol (24:1), and then separated under the same previous centrifugation conditions. Cold precipitation was conducted at −80°C for 1 h using the newly separated 150 µl of the upper aqueous layer with the addition of 0.5 × volume of ammonium acetate and 2.5 × volume of 100% ethanol. This was followed by pellet formation (30 min centrifugation at 16,000 g and 4°C), and the pellet formed was washed twice with 150 µl of 70% ethanol (15 min centrifugation at 16,000 g and 4°C) and air dried (5–10 min), and the final DNA elution done using 50 µl of TE buffer. The supernatant was discarded for the steps before the final elution. The concentrations and qualities of the extracted DNA samples were checked using a NanoDrop™ 2000/2000c Spectrophotometer (Thermo Fisher Scientific).
The targeted V3/V4 region of the 16S rRNA gene was amplified through PCR using the condition of initial denaturation of 95°C for 2 min, followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and final extension of 72°C for 5 min. The PCR mixtures used involved 4 μl of 5 × FastPfu Buffer, 2 μl of 2.5 mM dNTPs, 0.8 μl of each primer (5 μM), 0.4 μl of FastPfu polymerase, and template DNA. The PCR products were then verified with 2% agarose gel electrophoresis, purified using AxyPrep DNA Gel Extraction Kit (Axygen Biosciences) according to the manufacturer's instructions, and quantified using QuantiFluor™ ‐ST (Promega). The library was then constructed that involved “Y” adapter linkage, bead‐based adapter dimer removal, PCR amplification for library concentration, and generation of single‐stranded DNA fragments using sodium hydroxide. Finally, the sample libraries were pooled in equimolar concentration and were paired‐end sequenced (2 × 250/300 bp) with the help of the Illumina MiSeq platform based on the standard protocol.
2.3.2. Bioinformatics analysis and statistical validation
The quality of the raw data sequences was checked using FastQC software (Andrews, 2010) and Usearch (Edgar, 2010) software. The sequences were then merged and trimmed using Vsearch software (Rognes et al., 2016) and Trimmomatic software (Bolger et al., 2014) (Average quality score of 20, 50 bp minimum length, 50 bp sliding window). The trimmed sequences obtained were clustered into operational taxonomic units (OTUs) using UPARSE software (Edgar, 2013) with a 97% similarity cut‐off. The chimera sequences were removed using Usearch software (Edgar, 2010). The 16S rRNA OTUs obtained were taxonomically classified using RDP Classifier (Wang et al., 2007) by matching against Silva (SSU123) 16S rRNA database (confidence threshold of 0.7). The microbial communities of the sequenced samples were plotted in bar charts at phylum, family, and genus levels. Analysis with the Linear discriminant analysis effect size (LEfSe) tool was carried out using OTUs obtained to identify the differentially abundant bacterial taxa up to the genus level (LDA cutoff value: 5.0 or higher) (Segata et al., 2011). The alpha diversity parameters, including Good's coverage, Chao 1 estimator, Shannon index, and Simpson index, were calculated using Mothur software (Schloss et al., 2009). The alpha diversity parameters (Chao 1 estimator, Shannon index, and Simpson index) were further plotted as box plot diagrams using R software (Team RC, 2013). Rarefaction and Shannon rarefaction curves were determined and plotted using Mothur software (Schloss et al., 2009) and R software (Team RC, 2013). The beta diversity parameters, including Bray‐Curtis dissimilarity, Euclidean distance, Jaccard coefficient, and Manhattan distance were also calculated and plotted as PCOA diagrams using Usearch software (Edgar, 2010) and R software (Team RC, 2013). The raw data sequences (healthy and infected samples) of previous studies (Foysal et al., 2021; Hossain et al., 2021) were retrieved from NCBI database (BioProject ID: PRJNA662111; PRJNA662500) and used in the beta diversity analysis for cross comparison purpose. A Venn diagram was plotted using Mothur software (Schloss et al., 2009) and R software (Team RC, 2013).
2.3.3. Data availability
The 16S rRNA analysis raw data is available on the NCBI Sequence Read Archive (SRA) database (SRA Accession Number: SRR12199297; SRR12199298) (BioProject ID: PRJNA645513) (BioSample ID: SAMN15507524; SAMN15507525).
3. RESULTS
3.1. Detection of activated immune response through biochemical tests
Similar to previous work (Pan et al., 2008), the activated immune response of disease tolerant P. monodon during AHPND infection can be detected through suitable key immune parameters such as biochemical‐related ones. In this study, several biochemical tests including, PO activity assay, RB activity assay, nitrite concentration assay, relative SOD activity assay, and THC, were carried out and they successfully showed the differential biochemical activities associated with immune response activation of P. monodon during AHPND infection. The biochemical test results are presented in Table 1 and Figures [Link], [Link], [Link], [Link], [Link], [Link]. Statistical validations were also conducted through one‐way ANOVA (p < .05) and post hoc Duncan's test as shown in Tables [Link], [Link], [Link], [Link], [Link]. The Bradford protein assay was also performed for validation or normalization purposes. The Bradford protein assay results and subsequent statistical validation using one‐way ANOVA (p < .05) and post hoc Duncan's test are provided in Figures [Link], [Link], [Link] and Tables S6 and S7. The overall pathogenic and toxin flow of AHPND infection was also shown in Figure 1.
TABLE 1.
Differential biochemical activities detected at different post‐infection time points of Vp AHPND‐infected Penaeus monodon shrimps
| Hours post‐infection (hpi) |
PO activity (U/total mg protein) |
RB activity (OD 630 nm) |
Average relative SOD activity (%) | Nitrite concentration (nmol/ml) | THC (×105 ml−1) |
|---|---|---|---|---|---|
| Control | 2.069 ± 0.0963 | 0.0483 ± 0.0144 | 68.83 ± 8.04 | 42.944 ± 9.316 | 4.778 ± 0.755 |
| 0 | 4.375 ± 1.142 | 0.0585 ± 0.0148 | 72.98 ± 25.47 | 33.278 ± 11.505 | 5.250 ± 1.710 |
| 3 | 3.483 ± 0.806 | 0.0498 ± 0.0106 | 81.85 ± 16.80 | 34.148 ± 14.492 | 3.878 ± 1.922 |
| 6 | 4.004 ± 0.869 | 0.0449 ± 0.0101 | 100.00 ± 27.26 | 55.185 ± 12.086 | 3.867 ± 0.404 |
| 12 | 14.664 ± 3.177 | 0.0781 ± 0.00535 | 38.50 ± 13.58 | 88.389 ± 13.455 | 4.161 ± 0.857 |
| 24 | 11.807 ± 3.016 | 0.0763 ± 0.00990 | 26.26 ± 16.21 | 54.796 ± 10.278 | 1.961 ± 0.351 |
| 36 | 6.237 ± 2.063 | 0.0397 ± 0.0188 | 47.00 ± 11.46 | 43.296 ± 14.500 | 1.300 ± 0.304 |
| 48 | 6.448 ± 3.886 | 0.0288 ± 0.00496 | 35.03 ± 18.87 | 48.000 ± 7.581 | 2.272 ± 0.300 |
| PTC | — | 0.0661 ± 0.00552 | — | — | — |
The standard deviations (SD) were presented as ±SD values. All measured biochemical test results were statistically significant (p < .05).
Abbreviations: Control, uninfected control; PO, phenoloxidase; PTC, positive control; RB, respiratory burst; SOD, superoxide dismutase; THC, total hemocyte count.
FIGURE 1.
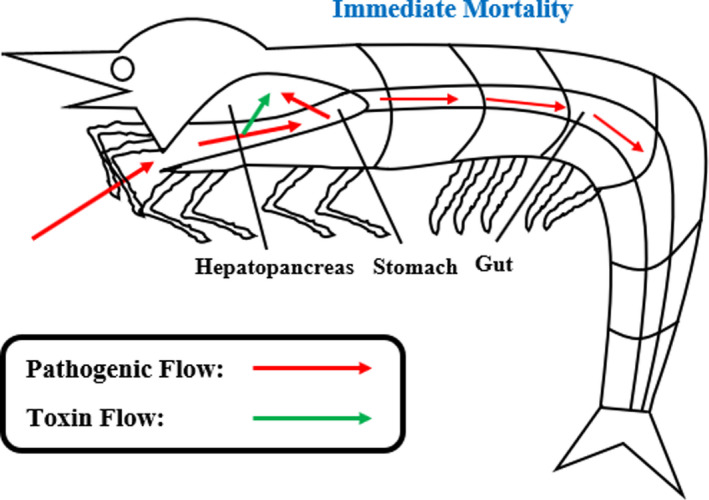
Overall pathogenic and toxin flow of AHPND infection in Penaeus monodon
Phenoloxidase activity was generally upregulated during AHPND infection with the highest peak identified at 12 hpi. The sharp upregulation of RB activity was detected at 12 hpi and 24 hpi by which the highest point was found at 12 hpi. The RB activity was then downregulated at 36 hpi and 48 hpi. In addition, SOD activity was stably upregulated until its highest peak at 6 hpi. However, it was then drastically downregulated at 12 hpi. Nitrite concentrations showed initial increment until the highest point at 12 hpi and were downregulated from 24 hpi to 48 hpi. The THC results were generally reduced by which the lowest point was detected at 36 hpi. Compared to the stable total protein concentration of Vp AHPND‐infected P. monodon hepatopancreas samples with no significant changes, the total protein concentration of Vp AHPND‐infected P. monodon muscle samples showed a significant decrease from 0 hpi to 48 hpi with the lowest point identified at 36 hpi. Overall, based on the biochemical tests results, the immune response activation of disease tolerant P. monodon was mainly detected at early AHPND infection period, particularly at 6 hpi and 12 hpi.
3.2. General analysis and quality assessment of 16S rRNA sequencing
From the 16S rRNA sequencing analysis conducted, a total of 102,455 raw reads and 95,556 clean reads were obtained followed by 99.95% of operational taxonomic units (OTUs) determined to possess sequence lengths of 401–450 bp. The 16S rRNA sequencing data quality was further validated through several alpha diversity parameters as demonstrated in Figure 2, supported by Figures S10 and S11 together with Table S8. The Good's coverage values were 99.99% (0.9999) for both samples, which signified sufficient sequencing depth. In addition, the Vp AHPND‐infected sample demonstrated higher values in Chao 1, Shannon, and Simpson indexes compared to the uninfected control sample, which suggested greater levels of bacterial richness and diversity. Furthermore, beta diversity analysis was conducted as shown in Figure 3. The healthy and infected sample groups were clustered accordingly with greater distance observed between uninfected control and Vp AHPND‐infected sample groups compared to other sample groups in Euclidean distance and Manhattan distance of the beta diversity analysis (Figure 3). The sample groups of the current study were clustered apart from those of previous studies (Figure 3). Besides that, a Venn diagram showing the OTU distribution between the uninfected control and Vp AHPND‐infected samples was plotted, as shown in Figure S12. From the total OTUs (60), the number of unique OTUs was higher in the Vp AHPND‐infected sample (26) compared to the uninfected control sample, (16) whereas the number of overlapping OTUs was 18.
FIGURE 2.
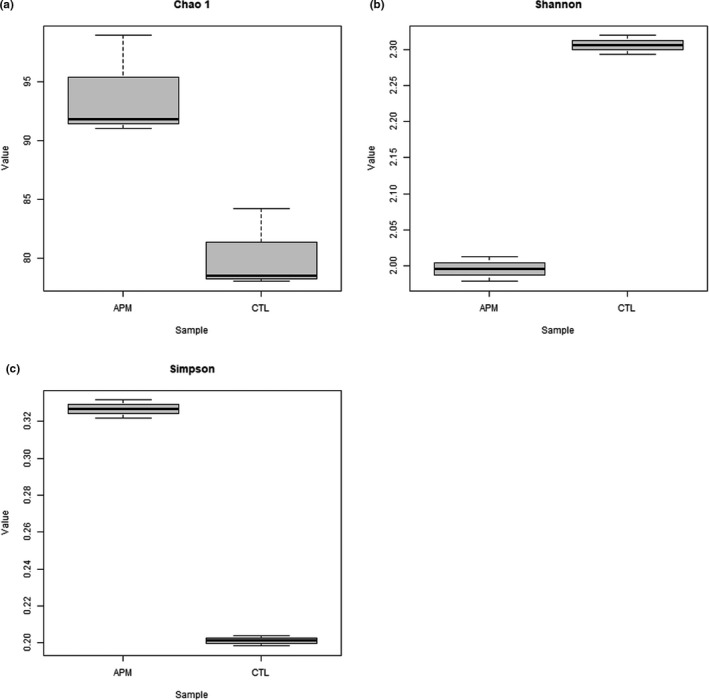
The alpha diversity parameters of 16S rRNA sequencing data: (a) Chao 1, (b) Shannon, (c) Simpson. APM: Vp AHPND‐infected; CTL: uninfected control
FIGURE 3.
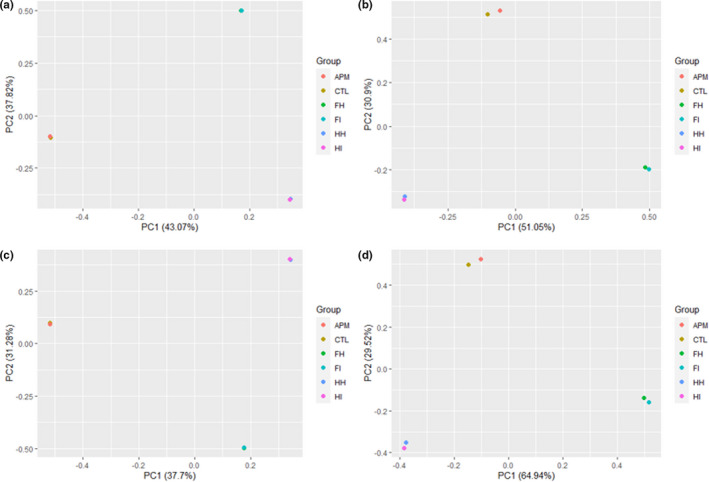
The beta diversity parameters of 16S rRNA sequencing data: (a) Bray–Curtis Dissimilarity, (b) Euclidean distance, (c) Jaccard coefficient, (d) Manhattan distance. APM: Vp AHPND‐infected; CTL: uninfected control; FH: Foysal et al. (2021) healthy; FI: Foysal et al. (2021) infected; HH: Hossain et al. (2021) healthy; HI: Hossain et al. (2021) infected
3.3. Overall comparison of gut microbiota diversity (relative abundance) of uninfected control and Vp AHPND‐infected shrimps
The microbiota diversity or relative abundance of OTUs (phylum, family, and genus levels) between uninfected control and Vp AHPND‐infected P. monodon gut samples were determined by 16S rRNA sequencing analysis as shown in Figure 4a‐c. There were 8 phyla in the uninfected control sample and 9 phyla in the Vp AHPND‐infected sample. The most abundant phylum for both groups in this study was Proteobacteria by which the percentages were 99.37% and 97.87% for uninfected control and Vp AHPND‐infected samples respectively. Intriguingly, Firmicutes phylum showed a near 28‐fold relative abundance increment from the uninfected control sample (0.046%) to the Vp AHPND‐infected sample.
FIGURE 4.
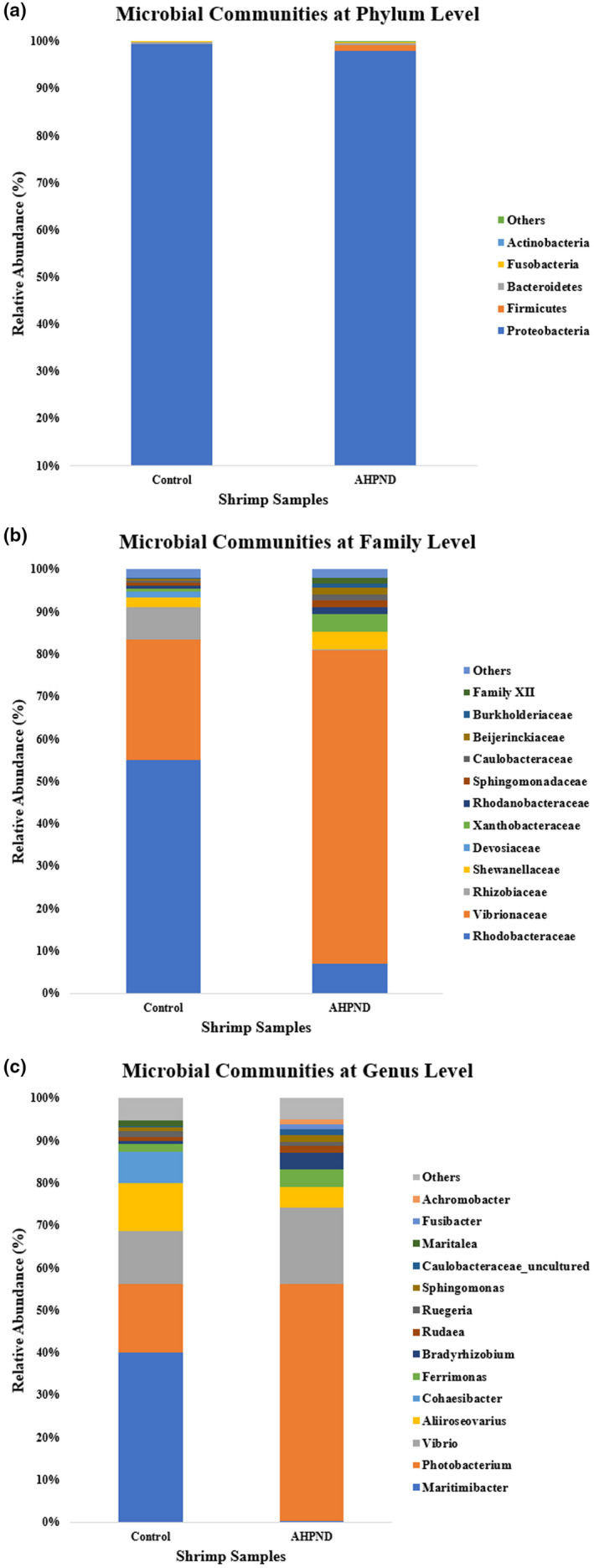
Microbial communities of uninfected control (Control) and Vp AHPND‐infected (AHPND) Penaeus monodon gut samples at (a) Phylum, (b) Family, and (c) Genus levels
At family and genus levels, Rhodobacteraceae (55.0%), Vibrionaceae (28.6%), and Rhizobiaceae (7.5%) families together with Maritimibacter (40.1%), Photobacterium (16.1%), and Vibrio (12.4%) genera had the highest abundances in the uninfected control sample. On the other hand, Vibrionaceae (74.0%), Rhodobacteraceae (7.0%), and Shewanellaceae (4.1%) families together with Photobacterium (56.0%), Vibrio (17.9%), and Aliiroseovarius (4.8%) genera had the highest abundances in Vp AHPND‐infected sample. Vibrionaceae demonstrated a significant increase of relative abundance from the uninfected control sample (28.6%) to the Vp AHPND‐infected sample (74.0%), whereas Rhodobacteraceae showed a significant decrease of relative abundance from the uninfected control sample (55.0%) to the Vp AHPND‐infected sample (7.0%). At the genus level, the relative abundances of Maritimibacter, Aliiroseovarius, and Cohaesibacter in the Vp AHPND‐infected sample (0.21%; 4.8%; 0.10%) were significantly reduced compared to the uninfected control sample (40.1%; 11.3%; 7.4%). There was also a significant increment of the relative abundances of Photobacterium, Vibrio, and Bradyrhizobium from the uninfected control sample (16.1%; 12.4%; 0.69%) to the Vp AHPND‐infected sample (56.0%; 17.9%; 3.9%). Besides that, based on the linear discriminant analysis Effect Size (LEfSe) analysis results (Figure 5), Proteobacteria and Alphaproteobacteria were differentially abundant in uninfected control group whereas Vibrionales, Vibrionaceae, and Vibrio were differentially abundant in Vp AHPND‐infected group.
FIGURE 5.
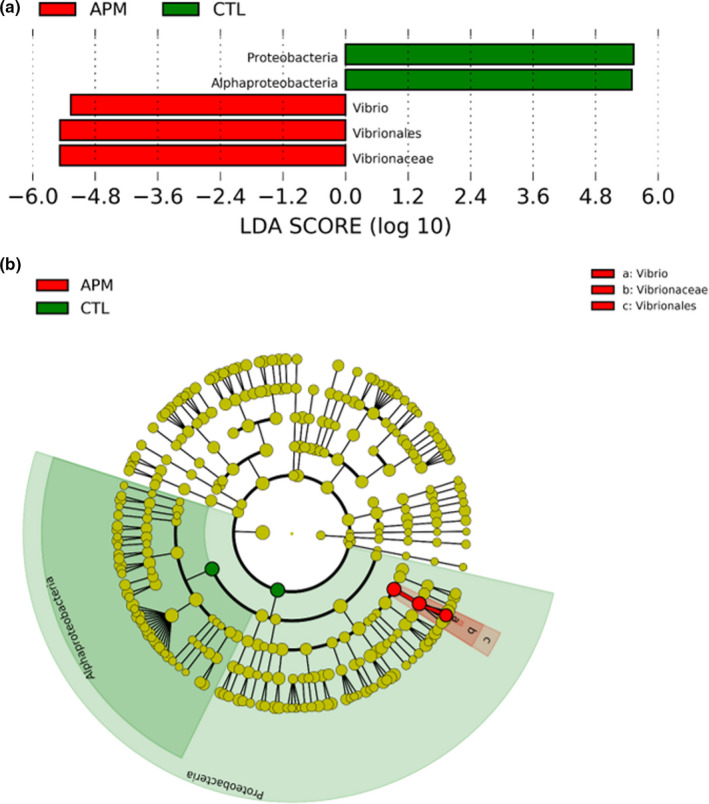
Linear discriminant analysis effect size (LEfSe) analysis conducted using OTUs obtained: (a) Differentially abundant taxa among compared sample groups based on computed Linear discriminant analysis (LDA) scores (LDA cutoff value: 5.0 or higher), (B) Circular cladogram plotted using LEfSe analysis (the dots represent operational taxonomic units [OTUs] at different taxonomic levels from center phylum level to outer circle genus level). APM: Vp AHPND‐infected; CTL: uninfected control
4. DISCUSSION
4.1. Immune response activation during AHPND infection
In this study, a series of biochemical tests (Table 1) using hepatopancreas, muscle, or haemolymph samples of uninfected control and Vp AHPND‐infected P. monodon shrimps was conducted. These biochemical tests successfully validated the immune response activation during AHPND infection. The immune response activation was shown by the upregulated PO activity, relative SOD activity, nitrite concentration, and RB activity during early post‐AHPND infection time points.
The upregulation of PO activity led to a stronger melanization response as melanization is controlled by the phenoloxidase enzyme activity in the prophenoloxidase (proPO) activation system (Alvarez & Chung, 2013; Amparyup et al., 2013; Hong et al., 2019; Park et al., 2019). There was a similar case of PO activity increment in M. rosenbergii after bacterial infection (Sung et al., 2004). The upregulation of RB and SOD activities inferred an elevated level of superoxide anion that caused increased antioxidation activity during AHPND infection. The elevation of nitrite concentration can trigger the activation of metabolic and immune reactions. In addition, nitrite formation in the redox reaction between nitric oxide and reactive oxygen species is vital for the elimination of pathogen through phagocytosis (Wink et al., 2011). However, excessively high nitrite concentration could cause adverse effects, including suppressed immune response, increased cytotoxic level, elevated susceptibility to bacterial infection, and increased superoxide anion level (Tseng & Chen, 2004).
The occurrence of hemocyte depletion due to apoptotic activity was also inferred through the decrease of THC levels. A similar scenario would be the occurrence of hemocyte depletion in Drosophila due to apoptotic activity that caused pro‐inflammatory state formation and alteration of the immune effector pathway (Arefin et al., 2015). Besides that, the general reduction of total protein concentrations in Vp AHPND‐infected P. monodon muscle samples, especially at 36 hpi, inferred protein degradation in shrimp muscle and probable muscle disturbance or degradation during AHPND infection. The inference was done based on a similar previous study that showed reduced total protein concentrations in both muscle and hepatopancreas of WSSV‐infected Penaeus indicus (Yoganandhan et al., 2003).
Furthermore, the importance of PO, SOD, and RB activities together with THC in the detection of shrimp immune response activation was highlighted in previous studies for either activated (Tayag et al., 2010) or suppressed (Li et al., 2008) immunity. As shown in a previous Litopenaeus vannamei challenge using Vibrio harveyi (Huang et al., 2013), despite the common reduction of SOD activity, THC, and PO activity during early infection time points, increased levels of SOD activity, THC, and PO activity were detected in disease‐resistant shrimps compared to non‐resistant shrimps. The elevated biochemical measurements were also accompanied by a faster recovery rate (Huang et al., 2013). Other than that, the immune humoral parameters involved, such as THC, PO, and NBT reduction, had been previously proposed as good potential stress indicators for aquatic animal health status (Verghese et al., 2007). Hence, this suggests the suitability of the biochemical parameters tested in this study as indicators of shrimp health status and immune response activation during pathogenic infection.
4.2. Gut microbiome changes during AHPND infection
Based on the alpha diversity parameters obtained (Figure 2 and Table S8), Good's coverage values of more than 0.99 (99%) identified for both uninfected control and Vp AHPND‐infected samples suggested satisfactory sequencing depth (Shin et al., 2019) and thus able to represent all bacterial communities. The higher alpha diversity parameter values determined in the Vp AHPND‐infected sample compared to the uninfected control sample showed stronger bacterial richness and diversity caused by AHPND infection. These alpha diversity parameters included Chao 1 richness estimation (Chao, 1984; Colwell & Coddington, 1994; Gotelli & Colwell, 2011) together with Shannon (Shannon, 1948) and Simpson (Simpson, 1949) diversity indices. The plateau stage was reached as the number of reads increased in the rarefaction (Figure S10) and Shannon rarefaction (Figure S11) curves plotted, which showed good sequencing depth and coverage (Liu, Yang, et al., 2020; Zhang et al., 2020). In general, the sequencing depth, coverage, richness, and diversity of the 16S data were successfully validated through all the alpha diversity parameters calculated.
From the microbiome relative abundances compared between uninfected control and Vp AHPND‐infected gut samples (Figure 4), the Proteobacteria phylum was most abundantly found in both samples. Similar relative abundances of Proteobacteria phylum were previously reported that involved cultured healthy (69.3%) and cultured Vp AHPND‐infected (70.2%) L. vannamei shrimps (Cornejo‐Granados et al., 2017). Notably, the Firmicutes phylum, which had a near 28‐fold relative abundance increment from the uninfected control sample to Vp AHPND‐infected sample, can be suggested as a potential determinant factor in the colonization or infection process of Vp AHPND bacteria. Firmicutes bacteria can survive in different environments, including extreme conditions, and produce endospores (Galperin, 2016). Firmicutes bacteria was previously reported to have elevated gut relative abundance in V. alginolyticus‐infected crab (Shi et al., 2019) and a positive correlation of its gut relative abundance with immune gene expressions in dietary supplemented Fenneropenaeus merguiensis (Liu, Zhou, et al., 2020).
The Rhodobacteraceae family possessed the highest relative abundance in the uninfected control sample. Rhodobacteraceae family are marine bacteria that possess functional importance in sulfur and carbon biogeochemical cycling together with microorganism or macroorganism symbiosis (Pujalte et al., 2014). The significantly decreased Rhodobacteraceae family relative abundance in the Vp AHPND‐infected sample was inferred to be the effect of Vp AHPND bacterial colonization and subsequently disrupted microbiome symbiosis during AHPND infection. At the genus level, for the uninfected control sample, Maritimibacter had the highest relative abundance and thus may play an essential role in the upkeep of normal healthy P. monodon gut microbiome balance. However, there are insufficient studies on this genus by which only two species were found at the current moment (Lee et al., 2007; Zhong et al., 2015).
In addition, the Vibrionaceae family and Vibrio genus showed higher abundances in the Vp AHPND‐infected sample compared to the uninfected control sample. These higher abundances were postulated to be caused by Vp AHPND bacterial colonization and related to secondary Vibrio infections. The inhabitation of Vibrio spp. bacteria in the shrimp intestine was due to its chitin‐rich environment identical to other crustaceans (Sugita & Ito, 2006). The secondary bacterial infections that occurred were mentioned previously (Santos et al., 2020). The Photobacterium genus bacteria had the highest relative abundance in the Vp AHPND‐infected sample that may be correlated to secondary luminous bacterial infections (Prayitno & Latchford, 1995). Aliiroseovarius genus can be proposed as pathogenic bacteria related to Vibrio bacteria based on a previous decrement of relative abundances for Vibrio and Aliiroseovarius genera bacteria in L. vannamei treated with beneficial seaweed feeding (Elizondo‐González et al., 2020).
In the process of AHPND infection, Vp AHPND bacteria started to colonize the shrimp's stomach after entry through the oral route. The bacteria then released PirA and PirB toxins, which led to damaging of the shrimp hepatopancreas. There was also the identification of both Vp AHPND bacteria and its toxins in the shrimp hepatopancreas during later post‐infection time points (Lai et al., 2015). Additionally, the occurrence of Vp AHPND bacterial colonization and hemocytic infiltration at the shrimp anterior midgut were detected during post‐AHPND infection time points (Soonthornchai et al., 2016). The Vp AHPND bacterial colonization locations were also shown by the common use of shrimp digestive organs such as stomach, hepatopancreas, midgut, and hindgut in AHPND diagnosis (Zorriehzahra & Banaederakhshan, 2015).
Gut microbiota is vital in the gut immune response such that disrupted microbiota will lead to immune dysregulation (Round & Mazmanian, 2009). A balanced gut microbiome composition is crucial in disease control (Biesebeke, 2018; Buttó & Haller, 2016). The shrimp gut microbiota dysbiosis is correlated to disease severity, which also involves environmental stress factors (Xiong et al., 2015). A balanced composition of shrimp gut microbial communities can be maintained or enhanced using different methods, such as shrimp feed composition (Huang et al., 2016; Jescovitch et al., 2018; Landsman et al., 2019; Li et al., 2018; Ringø et al., 2016), microbiota supplementation (prebiotics and probiotics) (Butt et al., 2021; Holt et al., 2020; Vargas‐Albores et al., 2017), and water quality assessment (Bentzon‐Tilia et al., 2016). 16S rRNA sequencing technique had been previously applied in the selection of beneficial indigenous (natural marine environment) microbial communities (Vargas‐Albores et al., 2017) and microbes with disease inhibitory potential (Wanka et al., 2018) for probiotic supplementation. In general, the important P. monodon gut microbiome changes due to AHPND infection were identified through the 16S rRNA sequencing technique in this study.
4.3. Diagnostic and food safety applications
In this study, biochemical tests and 16S rRNA analysis are suggested as suitable diagnostic tools for the determination of shrimp health status and gut microbiome changes, especially between healthy and diseased shrimps. Such applications are vital for ensuring food safety in downstream consumption. Some of the commonly investigated biochemical aspects of food safety include biochemical lesions, enzyme inhibition, and congenital metabolic disorders (Walker, 1980). Biochemical tests were utilized in food safety studies, such as bacteria biochemical tests (ALatawi et al., 2015) and immunological biochemical tests (Sun et al., 2011). The common applications of immunological biochemical tests in food safety‐related studies would be probiotics or dietary supplementation works (Gupta et al., 2014; Kumar et al., 2013; Sun et al., 2011), challenged shrimp works (Vaseeharan et al., 2013), and combined works (Citarasu et al., 2006; Gholamhosseini et al., 2020). However, there had been a lack of attention and effort in connecting immunological biochemical tests of shrimp challenge works to food safety‐oriented applications.
On the other hand, due to the time and labor constraints of traditional food microbiology detection methods (Rodríguez‐Lázaro et al., 2007), PCR had risen to become the standard rapid detection method for food microbes (Hameed et al., 2018). 16S rRNA analysis either through conventional PCR (ALatawi et al., 2015) or real‐time qPCR (Wolffs et al., 2004) had been previously utilized in food safety studies. Furthermore, the importance of cultured environment bacterial composition and associated shrimp gut microbiota changes was successfully highlighted from the beneficial changes in growth, immune response, survival, and gut microbiome of probiotics‐supplemented P. indicus cultured under biofloc system (Panigrahi et al., 2020). Such importance suggests the necessity of 16S rRNA analysis to be applied in the diagnosis of shrimp health and detection of shrimp disease outbreaks. The shrimp health diagnosis can be achieved through the close comparison of shrimp gut relative abundances, particularly focusing on potentially pathogenic or disease‐related microbes such as Vibrio genera.
5. CONCLUSION
In conclusion, the biochemical tests performed in this study, including the estimation of PO activity, RB activity, SOD activity, nitrite concentration, and THC, successfully demonstrated P. monodon's immune response activation at 6 hpi and 12 hpi of the post‐AHPND infection time points. In addition, from the 16S rRNA analysis, microbial communities of the Rhodobacteraceae family and Maritimibacter genus were postulated to be important for shrimp health maintenance. On the other hand, potential AHPND‐related pathogenic factors determined would be the Firmicutes phylum, Vibrionaceae family, and Photobacterium, Vibrio, and Aliiroseovarius genera. The occurrence of secondary Vibrio infections associated with Vp AHPND bacterial colonization was also suggested. Overall, the physiology and gut microbiota changes of P. monodon in response to AHPND infection were successfully determined using biochemical tests and the 16S rRNA sequencing technique. Hence, both biochemical tests and the 16S rRNA sequencing technique involved in this study are proposed as a combined strategy to be applied in ensuring shrimp health status diagnosis and disease control. The successful application of such strategies can then lead to stronger food safety and nutrition starting from the beginning of the food processing chain. Shrimp diseases are potentially accompanied by pathogens that are harmful to humans. Healthy shrimps not only are safer for consumption but also possess higher nutrition values compared to diseased shrimps.
Based on the results obtained, enhanced biochemical tests can be developed to achieve cost‐effective and efficient detection of shrimp health status changes, followed by the establishment of a biochemical‐based profiling system. The important beneficiary shrimp gut microbial communities identified in this study can assist in the development of enhanced shrimp supplements to achieve better shrimp health conditions and stronger disease resistance or tolerance. Other than that, an associated shrimp gut microbial profiling system can also be established for shrimp health diagnosis, disease detection, disease severity estimation (Dai et al., 2020; Xiong et al., 2015), and identification of potential polymicrobial infections (Dai et al., 2018).
CONFLICT OF INTEREST
No conflict of interest is declared.
Supporting information
Data S1
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Figure S8
Figure S9
Figure S10
Figure S11
Figure S12
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
ACKNOWLEDGEMENTS
This work was supported by the Technofund grant from the Ministry of Science and Technology, Malaysia (PV019‐2016) and the Centre for Research in Biotechnology for Agriculture (CEBAR) grant (RU004G‐2020). The authors acknowledge the prawn/shrimp farmers that helped them in providing the shrimps and fellow AGAGEL lab members.
Soo, T. C. C. , & Bhassu, S. (2022). Biochemical indexes and gut microbiota testing as diagnostic methods for Penaeus monodon health and physiological changes during AHPND infection with food safety concerns. Food Science & Nutrition, 10, 2694–2709. 10.1002/fsn3.2873
REFERENCES
- Al‐Amin, M. M. , Rahman, M. M. , Khan, F. R. , Zaman, F. , & Reza, H. M. (2015). Astaxanthin improves behavioral disorder and oxidative stress in prenatal valproic acid‐induced mice model of autism. Behavioural Brain Research, 286, 112–121. 10.1016/j.bbr.2015.02.041 [DOI] [PubMed] [Google Scholar]
- Al‐Amin, M. M. , Reza, H. M. , Saadi, H. M. , Mahmud, W. , Ibrahim, A. A. , Alam, M. M. , Kabir, N. , Saifullah, A. , Tropa, S. T. , & Quddus, A. H. M. R. (2016). Astaxanthin ameliorates aluminum chloride‐induced spatial memory impairment and neuronal oxidative stress in mice. European Journal of Pharmacology, 777, 60–69. 10.1016/j.ejphar.2016.02.062 [DOI] [PubMed] [Google Scholar]
- ALatawi, A. R. A. , Susilowati, A. , & Hailu, H. (2015). Biochemical and molecular characterization of food contaminating bacteria isolates from food stall vegetables. Microbiology Research Journal International, 5, 405–411. 10.9734/BMRJ/2015/13792 [DOI] [Google Scholar]
- Alvarez, J. V. , & Chung, J. S. (2013). Cloning of prophenoloxidase from hemocytes of the blue crab, Callinectes sapidus and its expression and enzyme activity during the molt cycle. Fish & Shellfish Immunology, 35, 1349–1358. 10.1016/j.fsi.2013.07.041 [DOI] [PubMed] [Google Scholar]
- Amparyup, P. , Charoensapsri, W. , & Tassanakajon, A. (2013). Prophenoloxidase system and its role in shrimp immune responses against major pathogens. Fish & Shellfish Immunology, 34, 990–1001. 10.1016/j.fsi.2012.08.019 [DOI] [PubMed] [Google Scholar]
- Anderson, J. L. , Valderrama, D. , & Jory, D. (2016). Shrimp production review. Global Aquaculture Alliance: Presentation Global Aquaculture Production Data and Analysis, 1–50. Retrieved from https://www.aquaculturealliance.org/wp‐content/uploads/2018/01/Global‐Shrimp‐Production‐Data‐Analysis‐Dr.‐James‐Anderson‐GOAL‐2017.pdf [Google Scholar]
- Anderson, J. L. , Valderrama, D. , & Jory, D. E. (2019). GOAL 2019: Global shrimp production review. Global Aquaculture Advocate, 1–5. Retrieved from https://www.aquaculturealliance.org/advocate/goal‐2019‐global‐shrimp‐production‐review/ [Google Scholar]
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Retrieved from https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Arefin, B. , Kucerova, L. , Krautz, R. , Kranenburg, H. , Parvin, F. , & Theopold, U. (2015). Apoptosis in hemocytes induces a shift in effector mechanisms in the Drosophila immune system and leads to a pro‐inflammatory state. PLoS One, 10(8), 1–19. 10.1371/journal.pone.0136593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, B. (2010). Vibrios as causal agents of zoonoses. Veterinary Microbiology, 140, 310–317. 10.1016/j.vetmic.2009.03.015 [DOI] [PubMed] [Google Scholar]
- Barker, K. (1998). At the bench: A laboratory navigator. Cold Spring Laboratory. [Google Scholar]
- Bentzon‐Tilia, M. , Sonnenschein, E. C. , & Gram, L. (2016). Monitoring and managing microbes in aquaculture–Towards a sustainable industry. Microbial Biotechnology, 9, 576–584. 10.1111/1751-7915.12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesebeke, R. (2018). Balancing microbial ecosystems within humans and animals to prevent medical conditions. Journal of Nutrition and Food Technology, 1, 40. 10.30881/jnfrt.00009 [DOI] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondad‐Reantaso, M. G. , Arthur, J. R. , & Subasinghe, R. P. (Eds.) (2008). Understanding and applying risk analysis in aquaculture. Food and Agriculture Organization of the United Nations. [Google Scholar]
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Analytical Biochemistry, 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Butt, U. D. , Lin, N. , Akhter, N. , Siddiqui, T. , Li, S. , & Wu, B. (2021). Overview of the latest developments in the role of probiotics, prebiotics and synbiotics in shrimp aquaculture. Fish & Shellfish Immunology, 114, 263–281. 10.1016/j.fsi.2021.05.003 [DOI] [PubMed] [Google Scholar]
- Buttó, L. F. , & Haller, D. (2016). Dysbiosis in intestinal inflammation: Cause or consequence. International Journal of Medical Microbiology, 306, 302–309. 10.1016/j.ijmm.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Caporaso, J. G. , Lauber, C. L. , Walters, W. A. , Berg‐Lyons, D. , Lozupone, C. A. , Turnbaugh, P. J. , Fierer, N. , & Knight, R. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America, 108, 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scandinavian Journal of Statistics, 11, 265–270. [Google Scholar]
- Citarasu, T. , Sivaram, V. , Immanuel, G. , Rout, N. , & Murugan, V. (2006). Influence of selected Indian immunostimulant herbs against white spot syndrome virus (WSSV) infection in black tiger shrimp, Penaeus monodon with reference to haematological, biochemical and immunological changes. Fish & Shellfish Immunology, 21, 372–384. 10.1016/j.fsi.2006.01.002 [DOI] [PubMed] [Google Scholar]
- Clarke, G. , Stilling, R. M. , Kennedy, P. J. , Stanton, C. , Cryan, J. F. , & Dinan, T. G. (2014). Minireview: Gut microbiota: The neglected endocrine organ. Molecular Endocrinology, 28, 1221–1238. 10.1210/me.2014-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell, R. K. , & Coddington, J. A. (1994). Estimating terrestrial biodiversity through extrapolation. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 345, 101–118. 10.1098/rstb.1994.0091 [DOI] [PubMed] [Google Scholar]
- Cornejo‐Granados, F. , Lopez‐Zavala, A. A. , Gallardo‐Becerra, L. , Mendoza‐Vargas, A. , Sánchez, F. , Vichido, R. , Brieba, L. G. , Viana, M. T. , Sotelo‐Mundo, R. R. , & Ochoa‐Leyva, A. (2017). Microbiome of Pacific Whiteleg shrimp reveals differential bacterial community composition between Wild, Aquacultured and AHPND/EMS outbreak conditions. Scientific Reports, 7(1), 1–15. 10.1038/s41598-017-11805-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W. , Sheng, Z. , Chen, J. , & Xiong, J. (2020). Shrimp disease progression increases the gut bacterial network complexity and abundances of keystone taxa. Aquaculture, 517, 1–8. 10.1016/j.aquaculture.2019.734802 [DOI] [Google Scholar]
- Dai, W. , Yu, W. , Xuan, L. , Tao, Z. , & Xiong, J. (2018). Integrating molecular and ecological approaches to identify potential polymicrobial pathogens over a shrimp disease progression. Applied Microbiology and Biotechnology, 102, 3755–3764. 10.1007/s00253-018-8891-y [DOI] [PubMed] [Google Scholar]
- Devadas, S. (2019). Acute hepatopancreatic necrosis disease in cultured shrimp in Malaysia (Master's thesis). University Putra Malaysia. [Google Scholar]
- Devadas, S. , Bhassu, S. , Christie Soo, T. C. , Mohamed Iqbal, S. N. , Yusoff, F. M. , & Shariff, M. (2018). Draft genome sequence of a Vibrio parahaemolyticus strain, KS17. S5–1, with multiple antibiotic resistance genes, which causes acute hepatopancreatic necrosis disease in Penaeus monodon in the West Coast of Peninsular Malaysia. Microbiology Resource Announcements, 7(2), 1–2. 10.1128/MRA.00829-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 10, 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Elizondo‐González, R. , Quiroz‐Guzmán, E. , Howe, A. , Yang, F. , Flater, J. , Gemin, M. , Palacios, E. , & Peña‐Rodríguez, A. (2020). Changes on the intestinal bacterial community of white shrimp Penaeus vannamei fed with green seaweeds. Journal of Applied Phycology, 32, 2061–2070. 10.1007/s10811-020-02072-w [DOI] [Google Scholar]
- Endt, K. , Stecher, B. , Chaffron, S. , Slack, E. , Tchitchek, N. , Benecke, A. , Van Maele, L. , Sirard, J. C. , Mueller, A. J. , Heikenwalder, M. , Macpherson, A. J. , Strugnell, R. , von Mering, C. , & Hardt, W. D. (2010). The microbiota mediates pathogen clearance from the gut lumen after non‐typhoidal Salmonella diarrhea. PLoS Path, 6(9), 1–18. 10.1371/journal.ppat.1001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, L. , Wang, Z. , Chen, M. , Qu, Y. , Li, J. , Zhou, A. , Xie, S. , Zeng, F. , & Zou, J. (2019). Microbiota comparison of Pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Science of the Total Environment, 657, 1194–1204. 10.1016/j.scitotenv.2018.12.069 [DOI] [PubMed] [Google Scholar]
- FAO . (2013). Report of the FAO/MARD technical workshop on early mortality syndrome (EMS) or acute hepatopancreatic necrosis syndrome (AHPNS) of cultured shrimp (UNDER TCP/VIE/3304). FAO Fisheries and Aquaculture Report No. 1053. Retrieved from http://www.fao.org/3/i3422e/i3422e00.htm [Google Scholar]
- Foysal, M. J. , Momtaz, F. , Kawser, A. Q. M. R. , Ali, M. H. , Raihan, T. , Siddik, M. A. B. , Rahman, M. M. , & Tay, A. (2021). Amplicon sequencing reveals significantly increased Vibrio abundance and associated gene functions in vibriosis‐infected black tiger shrimp (Penaeus monodon). Journal of Fish Diseases, 44(5), 591–599. 10.1111/jfd.13304 [DOI] [PubMed] [Google Scholar]
- Galperin, M. Y. (2016). Genome diversity of spore‐forming Firmicutes. In Driks A., & Eichenberger P. (Eds.), The bacterial spore: From molecules to systems (pp. 1–18). ASM Press. [Google Scholar]
- Gholamhosseini, A. , Kheirandish, M. R. , Shiry, N. , Akhlaghi, M. , Soltanian, S. , Roshanpour, H. , & Banaee, M. (2020). Use of a methanolic olive leaf extract (Olea europaea) against white spot virus syndrome in Penaeus vannamei: Comparing the biochemical, hematological and immunological changes. Aquaculture, 528, 1–10. 10.1016/j.aquaculture.2020.735556 [DOI] [Google Scholar]
- Gopal, S. , Otta, S. K. , Kumar, S. , Karunasagar, I. , Nishibuchi, M. , & Karunasagar, I. (2005). The occurrence of Vibrio species in tropical shrimp culture environments; implications for food safety. International Journal of Food Microbiology, 102, 151–159. 10.1016/j.ijfoodmicro.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Gotelli, N. J. , & Colwell, R. K. (2011). Estimating species richness. Biological Diversity: Frontiers in Measurement and Assessment, 12, 39–54. [Google Scholar]
- Gupta, A. , Gupta, P. , & Dhawan, A. (2014). Dietary supplementation of probiotics affects growth, immune response and disease resistance of Cyprinus carpio fry. Fish & Shellfish Immunology, 41, 113–119. 10.1016/j.fsi.2014.08.023 [DOI] [PubMed] [Google Scholar]
- Hameed, S. , Xie, L. , & Ying, Y. (2018). Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends in Food Science & Technology, 81, 61–73. 10.1016/j.tifs.2018.05.020 [DOI] [Google Scholar]
- Holt, C. C. , Bass, D. , Stentiford, G. D. , & van der Giezen, M. (2020). Understanding the role of the shrimp gut microbiome in health and disease. Journal of Invertebrate Pathology, 186, 1–14. 10.1016/j.jip.2020.107387 [DOI] [PubMed] [Google Scholar]
- Hong, X. , Lu, L. , & Xu, D. (2016). Progress in research on acute hepatopancreatic necrosis disease (AHPND). Aquaculture International, 24, 577–593. 10.1007/s10499-015-9948-x [DOI] [Google Scholar]
- Hong, Y. , Huang, Y. , Yan, G. , Pan, C. , & Zhang, J. (2019). Antioxidative status, immunological responses, and heat shock protein expression in hepatopancreas of Chinese mitten crab, Eriocheir sinensis under the exposure of glyphosate. Fish & Shellfish Immunology, 86, 840–845. 10.1016/j.fsi.2018.12.020 [DOI] [PubMed] [Google Scholar]
- Hossain, M. S. , Dai, J. , & Qiu, D. (2021). Dysbiosis of the shrimp (Penaeus monodon) gut microbiome with AHPND outbreaks revealed by 16S rRNA metagenomics analysis. Aquaculture Research, 52(7), 3336–3349. 10.1111/are.15178 [DOI] [Google Scholar]
- Hsieh, S. L. , Ruan, Y. H. , Li, Y. C. , Hsieh, P. S. , Hu, C. H. , & Kuo, C. M. (2008). Immune and physiological responses in Pacific white shrimp (Penaeus vannamei) to Vibrio alginolyticus . Aquaculture, 275, 335–341. 10.1016/j.aquaculture.2007.12.019 [DOI] [Google Scholar]
- Huang, H. H. , Liu, X. L. , Xiang, J. H. , & Wang, P. (2013). Immune response of Litopenaeus vannamei after infection with Vibrio harveyi . Aquaculture, 406, 115–120. 10.1016/j.aquaculture.2013.05.010 [DOI] [Google Scholar]
- Huang, Z. , Li, X. , Wang, L. , & Shao, Z. (2016). Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei . Aquaculture Research, 47, 1737–1746. 10.1111/are.12628 [DOI] [Google Scholar]
- Huynh, T. G. , Cheng, A. C. , Chi, C. C. , Chiu, K. H. , & Liu, C. H. (2018). A synbiotic improves the immunity of white shrimp, Litopenaeus vannamei: Metabolomic analysis reveal compelling evidence. Fish & Shellfish Immunology, 79, 284–293. 10.1016/j.fsi.2018.05.031 [DOI] [PubMed] [Google Scholar]
- Huynh, T. G. , Yeh, S. T. , Lin, Y. C. , Shyu, J. F. , Chen, L. L. , & Chen, J. C. (2011). White shrimp Litopenaeus vannamei immersed in seawater containing Sargassum hemiphyllum var. chinense powder and its extract showed increased immunity and resistance against Vibrio alginolyticus and white spot syndrome virus. Fish & Shellfish Immunology, 31, 286–293. 10.1016/j.fsi.2011.05.014 [DOI] [PubMed] [Google Scholar]
- Jescovitch, L. N. , Ullman, C. , Rhodes, M. , & Davis, D. A. (2018). Effects of different feed management treatments on water quality for Pacific white shrimp Litopenaeus vannamei . Aquaculture Research, 49, 526–531. 10.1111/are.13483 [DOI] [Google Scholar]
- Kaur, B. P. , Kaushik, N. , Rao, P. S. , & Chauhan, O. P. (2013). Effect of high‐pressure processing on physical, biochemical, and microbiological characteristics of black tiger shrimp (Penaeus monodon). Food and Bioprocess Technology, 6, 1390–1400. 10.1007/s11947-012-0870-1 [DOI] [Google Scholar]
- Konstantinidis, K. T. , & Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proceedings of the National Academy of Sciences of the United States of America, 102, 2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N. R. , Raman, R. P. , Jadhao, S. B. , Brahmchari, R. K. , Kumar, K. , & Dash, G. (2013). Effect of dietary supplementation of Bacillus licheniformis on gut microbiota, growth and immune response in giant freshwater prawn, Macrobrachium rosenbergii (de Man, 1879). Aquaculture International, 21, 387–403. 10.1007/s10499-012-9567-8 [DOI] [Google Scholar]
- Lai, H. C. , Ng, T. H. , Ando, M. , Lee, C. T. , Chen, I. T. , Chuang, J. C. , Mavichak, R. , Chang, S. H. , Yeh, M. D. , Chiang, Y. A. , Takeyama, H. , Hamaguchi, H. O. , Lo, C. F. , Aoki, T. , & Wang, H. C. (2015). Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish & Shellfish Immunology, 47, 1006–1014. 10.1016/j.fsi.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Landsman, A. , St‐Pierre, B. , Rosales‐Leija, M. , Brown, M. , & Gibbons, W. (2019). Impact of aquaculture practices on intestinal bacterial profiles of pacific whiteleg shrimp Litopenaeus vannamei . Microorganisms, 7(4), 1–14. 10.3390/microorganisms7040093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. T. , Chen, I. T. , Yang, Y. T. , Ko, T. P. , Huang, Y. T. , Huang, J. Y. , Huang, M. F. , Lin, S. J. , Chen, C. Y. , Lin, S. S. , Lightner, D. V. , Wang, H. C. , Wang, A. H. , Wang, H. C. , Hor, L. I. , & Lo, C. F. (2015). The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proceedings of the National Academy of Sciences of the United States of America, 112, 10798–10803. 10.1073/pnas.1503129112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. , Choo, Y. J. , Giovannoni, S. J. , & Cho, J. C. (2007). Maritimibacter alkaliphilus gen. nov., sp. nov., a genome‐sequenced marine bacterium of the Roseobacter clade in the order Rhodobacterales. International Journal of Systematic and Evolutionary Microbiology, 57, 1653–1658. 10.1099/ijs.0.64960-0 [DOI] [PubMed] [Google Scholar]
- Li, C. C. , Yeh, S. T. , & Chen, J. C. (2008). The immune response of white shrimp Litopenaeus vannamei following Vibrio alginolyticus injection. Fish & Shellfish Immunology, 25, 853–860. 10.1016/j.fsi.2008.09.014 [DOI] [PubMed] [Google Scholar]
- Li, E. , Chang, X. , Wang, X. , Wang, S. , Zhao, Q. , Zhang, M. , Qin, J. G. , & Chen, L. (2018). Gut microbiota and its modulation for healthy farming of Pacific white shrimp Litopenaeus vannamei . Reviews in Fisheries Science & Aquaculture, 26, 381–399. 10.1080/23308249.2018.1440530 [DOI] [Google Scholar]
- Li, P. , Burr, G. S. , Gatlin, D. M. III , Hume, M. E. , Patnaik, S. , Castille, F. L. , & Lawrence, A. L. (2007). Dietary supplementation of short‐chain fructooligosaccharides influences gastrointestinal microbiota composition and immunity characteristics of Pacific white shrimp, Litopenaeus vannamei, cultured in a recirculating system. The Journal of Nutrition, 137, 2763–2768. 10.1093/jn/137.12.2763 [DOI] [PubMed] [Google Scholar]
- Liang, Q. , Li, Z. , Ou, M. , Wu, X. , Qiao, X. , Wei, W. , Liu, Y. , Ye, J. M. , & Wang, W. (2020). Hypoimmunity and intestinal bacterial imbalance are closely associated with blue body syndrome in cultured Penaeus vannamei . Aquaculture, 522, 1–8. 10.1016/j.aquaculture.2020.735118 [DOI] [Google Scholar]
- Lin, Y. C. , Yeh, S. T. , Li, C. C. , Chen, L. L. , Cheng, A. C. , & Chen, J. C. (2011). An immersion of Gracilaria tenuistipitata extract improves the immunity and survival of white shrimp Litopenaeus vannamei challenged with white spot syndrome virus. Fish & Shellfish Immunology, 31, 1239–1246. 10.1016/j.fsi.2011.07.021 [DOI] [PubMed] [Google Scholar]
- Liu, W. C. , Zhou, S. H. , Balasubramanian, B. , Zeng, F. Y. , Sun, C. B. , & Pang, H. Y. (2020). Dietary seaweed (Enteromorpha) polysaccharides improves growth performance involved in regulation of immune responses, intestinal morphology and microbial community in banana shrimp Fenneropenaeus merguiensis . Fish & Shellfish Immunology, 104, 202–212. 10.1016/j.fsi.2020.05.079 [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Yang, Y. , Lei, Y. , Yang, L. , Zhang, X. , Yuan, J. , & Lei, Z. (2020). Effects of dihydroartemisinin on the gut microbiome of mice. Molecular Medicine Reports, 22, 707–714. 10.3892/mmr.2020.11165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzinelli, E. M. , Campbell, A. H. , Zozaya Valdes, E. , Vergés, A. , Nielsen, S. , Wernberg, T. , de Bettignies, T. , Bennett, S. , Caporaso, J. G. , Thomas, T. , & Steinberg, P. D. (2015). Continental‐scale variation in seaweed host‐associated bacterial communities is a function of host condition, not geography. Environmental Microbiology, 17, 4078–4088. 10.1111/1462-2920.12972 [DOI] [PubMed] [Google Scholar]
- Maynard, C. L. , Elson, C. O. , Hatton, R. D. , & Weaver, C. T. (2012). Reciprocal interactions of the intestinal microbiota and immune system. Nature, 489, 231–241. 10.1038/nature11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunan, L. , Lightner, D. , Pantoja, C. , & Gomez‐Jimenez, S. (2014). Detection of acute hepatopancreatic necrosis disease (AHPND) in Mexico. Diseases of Aquatic Organisms, 111, 81–86. 10.3354/dao02776 [DOI] [PubMed] [Google Scholar]
- OIE , (2019). Manual of diagnostic tests for aquatic animals‐ infection with white spot syndrome virus. Retrieved from https://www.oie.int/fileadmin/Home/eng/Health_standards/aahm/current/chapitre_wsd.pdf [Google Scholar]
- PacBio . (2012). Extracting DNA using phenol‐chloroform. Retrieved from https://www.pacb.com/wp‐content/uploads/2015/09/SharedProtocol‐Extracting‐DNA‐usinig‐Phenol‐Chloroform.pdf [Google Scholar]
- Pan, Z. C. , He, J. G. , Weng, S. P. , Yin, Z. X. , Fu, X. Z. , & Li, S. D. (2008). Changes in mortality and immunological variables of Litopenaeus vannamei parents and their filial families infected with white spot syndrome under different experimental conditions. Fish & Shellfish Immunology, 25, 459–471. 10.1016/j.fsi.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Panigrahi, A. , Das, R. R. , Sivakumar, M. R. , Saravanan, A. , Saranya, C. , Sudheer, N. S. , Kumaraguru Vasagam, K. P. , Mahalakshmi, P. , Kannappan, S. , & Gopikrishna, G. (2020). Bio‐augmentation of heterotrophic bacteria in biofloc system improves growth, survival, and immunity of Indian white shrimp Penaeus indicus . Fish & Shellfish Immunology, 98, 477–487. 10.1016/j.fsi.2020.01.021 [DOI] [PubMed] [Google Scholar]
- Park, K. , Kim, W. S. , & Kwak, I. S. (2019). Endocrine‐disrupting chemicals impair the innate immune prophenoloxidase system in the intertidal mud crab, Macrophthalmus japonicus . Fish & Shellfish Immunology, 87, 322–332. 10.1016/j.fsi.2019.01.025 [DOI] [PubMed] [Google Scholar]
- Perera, N. C. N. , Godahewa, G. I. , Lee, S. , Kim, M. J. , Hwang, J. Y. , Kwon, M. G. , Hwang, S. D. , & Lee, J. (2017). Manganese‐superoxide dismutase (MnSOD), a role player in seahorse (Hippocampus abdominalis) antioxidant defense system and adaptive immune system. Fish & Shellfish Immunology, 68, 435–442. 10.1016/j.fsi.2017.07.049 [DOI] [PubMed] [Google Scholar]
- Porchas‐Cornejo, M. A. , Martínez‐Porchas, M. , Vargas‐Albores, F. , Gollas‐Galvan, T. , Martínez‐Córdova, L. R. , Vazquez‐Euan, R. , & Peña‐Messina, E. (2017). High‐resolution detection of bacterial profile of ocean water, before and after being used by shrimp farms. Aquaculture International, 25, 1833–1843. 10.1007/s10499-017-0160-z [DOI] [Google Scholar]
- Prayitno, S. B. , & Latchford, J. W. (1995). Experimental infections of crustaceans with luminous bacteria related to Photobacterium and Vibrio. Effect of salinity and pH on infectiosity. Aquaculture, 132, 105–112. 10.1016/0044-8486(94)00374-W [DOI] [Google Scholar]
- Pujalte, M. J. , Lucena, T. , Ruvira, M. A. , Arahal, D. R. , & Macián, M. C. (2014). The family rhodobacteraceae. In Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., & Thompson F. (Eds.), The prokaryotes (pp. 439–512). Springer. [Google Scholar]
- Ringø, E. , Zhou, Z. , Vecino, J. G. , Wadsworth, S. , Romero, J. , Krogdahl, Å. , Olsen, R. E. , Dimitroglou, A. , Foey, A. , Davies, S. , Owen, M. , Lauzon, H. L. , Martinsen, L. L. , De Schryver, P. , Bossier, P. , Sperstad, S. , & Merrifield, D. L. (2016). Effect of dietary components on the gut microbiota of aquatic animals. A never‐ending story? Aquaculture Nutrition, 22, 219–282. 10.1111/anu.12346 [DOI] [Google Scholar]
- Rintala, A. , Pietilä, S. , Munukka, E. , Eerola, E. , Pursiheimo, J. P. , Laiho, A. , Pekkala, S. , & Huovinen, P. (2017). Gut microbiota analysis results are highly dependent on the 16S rRNA gene target region, whereas the impact of DNA extraction is minor. Journal of Biomolecular Techniques: JBT, 28, 19–30. 10.7171/jbt.17-2801-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Lázaro, D. , Lombard, B. , Smith, H. , Rzezutka, A. , d'Agostino, M. , Helmuth, R. , Schroeter, A. , Malorny, B. , Miko, A. , Guerra, B. , Davison, J. , Kobilinsky, A. , Hernández, M. , Bertheau, Y. , & Cook, N. (2007). Trends in analytical methodology in food safety and quality: Monitoring microorganisms and genetically modified organisms. Trends in Food Science & Technology, 18, 306–319. 10.1016/j.tifs.2007.01.009 [DOI] [Google Scholar]
- Rognes, T. , Flouri, T. , Nichols, B. , Quince, C. , & Mahé, F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ, 4, 1–22. 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round, J. L. , & Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology, 9, 313–323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhoshkumar, S. , Sivakumar, S. , Vimal, S. , Abdul Majeed, S. , Taju, G. , Haribabu, P. , Uma, A. , & Sahul Hameed, A. S. (2017). Biochemical changes and tissue distribution of Enterocytozoon hepatopenaei (EHP) in naturally and experimentally EHP‐infected whiteleg shrimp, Litopenaeus vannamei (Boone, 1931), in India. Journal of Fish Diseases, 40, 529–539. 10.1111/jfd.12530 [DOI] [PubMed] [Google Scholar]
- Santos, H. M. , Tsai, C. Y. , Maquiling, K. R. A. , Tayo, L. L. , Mariatulqabtiah, A. R. , Lee, C. W. , & Chuang, K. P. (2020). Diagnosis and potential treatments for acute hepatopancreatic necrosis disease (AHPND): A review. Aquaculture International, 28, 169–185. 10.1007/s10499-019-00451-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P. D. , Westcott, S. L. , Ryabin, T. , Hall, J. R. , Hartmann, M. , Hollister, E. B. , Lesniewski, R. A. , Oakley, B. B. , Parks, D. H. , Robinson, C. J. , Sahl, J. W. , Stres, B. , Thallinger, G. G. , Van Horn, D. J. , & Weber, C. F. (2009). Introducing mothur: Open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75, 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata, N. , Izard, J. , Waldron, L. , Gevers, D. , Miropolsky, L. , Garrett, W. S. , & Huttenhower, C. (2011). Metagenomic biomarker discovery and explanation. Genome Biology, 12(6), 1–18. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, C. E. (1948). A mathematical theory of communication. The Bell System Technical Journal, 27, 379–423. 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- Shi, C. , Xia, M. , Li, R. , Mu, C. , Zhang, L. , Liu, L. , Ye, Y. , & Wang, C. (2019). Vibrio alginolyticus infection induces coupled changes of bacterial community and metabolic phenotype in the gut of swimming crab. Aquaculture, 499, 251–259. 10.1016/j.aquaculture.2018.09.031 [DOI] [Google Scholar]
- Shin, J. H. , Park, Y. H. , Sim, M. , Kim, S. A. , Joung, H. , & Shin, D. M. (2019). Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Research in Microbiology, 170, 192–201. 10.1016/j.resmic.2019.03.003 [DOI] [PubMed] [Google Scholar]
- Simpson, E. H. (1949). Measurement of diversity. Nature, 163, 688. 10.1038/163688a0 [DOI] [Google Scholar]
- Sirikharin, R. , Taengchaiyaphum, S. , Sanguanrut, P. , Chi, T. D. , Mavichak, R. , Proespraiwong, P. , Nuangsaeng, B. , Thitamadee, S. , Flegel, T. W. , & Sritunyalucksana, K. (2015). Characterization and PCR detection of binary, Pir‐like toxins from Vibrio parahaemolyticus isolates that cause acute hepatopancreatic necrosis disease (AHPND) in shrimp. PLoS One, 10, 1–16. 10.1371/journal.pone.0126987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo, T. C. C. , Devadas, S. , Din, M. S. M. , & Bhassu, S. (2019). Differential transcriptome analysis of the disease tolerant Madagascar‐Malaysia crossbred black tiger shrimp, Penaeus monodon hepatopancreas in response to acute hepatopancreatic necrosis disease (AHPND) infection: Inference on immune gene response and interaction. Gut Pathogens, 11, 1–13. 10.1186/s13099-019-0319-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonthornchai, W. , Chaiyapechara, S. , Klinbunga, S. , Thongda, W. , Tangphatsornruang, S. , Yoocha, T. , Jarayabhand, P. , & Jiravanichpaisal, P. (2016). Differentially expressed transcripts in stomach of Penaeus monodon in response to AHPND infection. Developmental & Comparative Immunology, 65, 53–63. 10.1016/j.dci.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Sugita, H. , & Ito, Y. (2006). Identification of intestinal bacteria from Japanese flounder (Paralichthys olivaceus) and their ability to digest chitin. Letters in Applied Microbiology, 43, 336–342. 10.1111/j.1472-765X.2006.01943.x [DOI] [PubMed] [Google Scholar]
- Sun, Y. Z. , Yang, H. L. , Ma, R. L. , Zhang, C. X. , & Lin, W. Y. (2011). Effect of dietary administration of Psychrobacter sp. on the growth, feed utilization, digestive enzymes and immune responses of grouper Epinephelus coioides . Aquaculture Nutrition, 17(3), e733–e740. 10.1111/j.1365-2095.2010.00837.x [DOI] [Google Scholar]
- Sung, H. H. , Huang, Y. T. , & Hsiao, L. T. (2004). Phenoloxidase activity of Macrobrachium rosenbergii after challenge with two kinds of pathogens: Lactococcus garvieae and Aeromonas veronii . Fish Pathology, 39, 1–8. 10.3147/jsfp.39.1 [DOI] [Google Scholar]
- Tayag, C. M. , Lin, Y. C. , Li, C. C. , Liou, C. H. , & Chen, J. C. (2010). Administration of the hot‐water extract of Spirulina platensis enhanced the immune response of white shrimp Litopenaeus vannamei and its resistance against Vibrio alginolyticus . Fish & Shellfish Immunology, 28, 764–773. 10.1016/j.fsi.2010.01.023 [DOI] [PubMed] [Google Scholar]
- Team RC . (2013). R: A language and environment for statistical computing. Retrieved from https://www.R‐project.org/ [Google Scholar]
- Thitamadee, S. , Prachumwat, A. , Srisala, J. , Jaroenlak, P. , Salachan, P. V. , Sritunyalucksana, K. , Flegel, T. W. , & Itsathitphaisarn, O. (2016). Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture, 452, 69–87. 10.1016/j.aquaculture.2015.10.028 [DOI] [Google Scholar]
- Tracey, W. R. , Tse, J. , & Carter, G. (1995). Lipopolysaccharide‐induced changes in plasma nitrite and nitrate concentrations in rats and mice: Pharmacological evaluation of nitric oxide synthase inhibitors. Journal of Pharmacology and Experimental Therapeutics, 272, 1011–1015. [PubMed] [Google Scholar]
- Tran, L. , Nunan, L. , Redman, R. M. , Mohney, L. L. , Pantoja, C. R. , Fitzsimmons, K. , & Lightner, D. V. (2013). Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Diseases of Aquatic Organisms, 105, 45–55. 10.3354/dao02621 [DOI] [PubMed] [Google Scholar]
- Tremaroli, V. , & Bäckhed, F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature, 489, 242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- Tseng, I. T. , & Chen, J. C. (2004). The immune response of white shrimp Litopenaeus vannamei and its susceptibility to Vibrio alginolyticus under nitrite stress. Fish & Shellfish Immunology, 17, 325–333. 10.1016/j.fsi.2004.04.010 [DOI] [PubMed] [Google Scholar]
- Vargas‐Albores, F. , Porchas‐Cornejo, M. A. , Martínez‐Porchas, M. , Villalpando‐Canchola, E. , Gollas‐Galván, T. , & Martínez‐Córdova, L. R. (2017). Bacterial biota of shrimp intestine is significantly modified by the use of a probiotic mixture: A high throughput sequencing approach. Helgoland Marine Research, 71, 1–10. 10.1186/s10152-017-0485-z [DOI] [Google Scholar]
- Vaseeharan, B. , Ramasamy, P. , Wesley, S. G. , & Chen, J. C. (2013). Influence of acute salinity changes on biochemical, hematological and immune characteristics of Fenneropenaeus indicus during white spot syndrome virus challenge. Microbiology and Immunology, 57, 463–469. 10.1111/1348-0421.12057 [DOI] [PubMed] [Google Scholar]
- Verghese, B. , Radhakrishnan, E. V. , & Padhi, A. (2007). Effect of environmental parameters on immune response of the Indian spiny lobster, Panulirus homarus (Linnaeus, 1758). Fish & Shellfish Immunology, 23, 928–936. 10.1016/j.fsi.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Walker, R. (1980). Biochemical aspects of food safety. In Birch G. G., & Parker K. J. (Eds.), Food and health: Science and technology (pp. 167–181). Springer. [Google Scholar]
- Wang, Q. , Garrity, G. M. , Tiedje, J. M. , & Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73, 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanka, K. M. , Damerau, T. , Costas, B. , Krueger, A. , Schulz, C. , & Wuertz, S. (2018). Isolation and characterization of native probiotics for fish farming. BMC Microbiology, 18, 1–13. 10.1186/s12866-018-1260-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink, D. A. , Hines, H. B. , Cheng, R. Y. , Switzer, C. H. , Flores‐Santana, W. , Vitek, M. P. , Ridnour, L. A. , & Colton, C. A. (2011). Nitric oxide and redox mechanisms in the immune response. Journal of Leukocyte Biology, 89, 873–891. 10.1189/jlb.1010550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffs, P. , Knutsson, R. , Norling, B. , & Rådström, P. (2004). Rapid quantification of Yersinia enterocolitica in pork samples by a novel sample preparation method, flotation, prior to real‐time PCR. Journal of Clinical Microbiology, 42, 1042–1047. 10.1128/JCM.42.3.1042-1047.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, J. , Wang, K. , Wu, J. , Qiuqian, L. , Yang, K. , Qian, Y. , & Zhang, D. (2015). Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Applied Microbiology and Biotechnology, 99, 6911–6919. 10.1007/s00253-015-6632-z [DOI] [PubMed] [Google Scholar]
- Yoganandhan, K. , Thirupathi, S. , & Hameed, A. S. (2003). Biochemical, physiological and hematological changes in white spot syndrome virus‐infected shrimp, Penaeus indicus . Aquaculture, 221, 1–11. 10.1016/S0044-8486(02)00220-X [DOI] [Google Scholar]
- Zhang, X. L. , Xu, T. W. , Wang, X. G. , Geng, Y. Y. , Liu, H. J. , Hu, L. Y. , Zhao, N. , Kang, S. P. , Zhang, W. M. , & Xu, S. X. (2020). The effect of transitioning between feeding methods on the gut microbiota dynamics of yaks on the Qinghai‐Tibet Plateau. Animals, 10, 1–13. 10.3390/ani10091641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z. P. , Liu, Y. , Zhou, Y. G. , Liu, H. C. , Wang, F. , & Liu, Z. P. (2015). Maritimibacter lacisalsi sp. nov., isolated from a salt lake, and emended description of the genus MaritimibacterLee et al 2007. International Journal of Systematic and Evolutionary Microbiology, 65, 3462–3468. 10.1099/ijsem.0.000437 [DOI] [PubMed] [Google Scholar]
- Zhou, L. , Chen, C. , Xie, J. , Xu, C. , Zhao, Q. , Qin, J. G. , Chen, L. , & Li, E. (2019). Intestinal bacterial signatures of the “cotton shrimp‐like” disease explain the change of growth performance and immune responses in Pacific white shrimp (Litopenaeus vannamei). Fish & Shellfish Immunology, 92, 629–636. 10.1016/j.fsi.2019.06.054 [DOI] [PubMed] [Google Scholar]
- Zokaeifar, H. , Babaei, N. , Saad, C. R. , Kamarudin, M. S. , Sijam, K. , & Balcazar, J. L. (2014). Administration of Bacillus subtilis strains in the rearing water enhances the water quality, growth performance, immune response, and resistance against Vibrio harveyi infection in juvenile white shrimp, Litopenaeus vannamei . Fish & Shellfish Immunology, 36, 68–74. 10.1016/j.fsi.2013.10.007 [DOI] [PubMed] [Google Scholar]
- Zoqratt, M. Z. H. M. , Eng, W. W. H. , Thai, B. T. , Austin, C. M. , & Gan, H. M. (2018). Microbiome analysis of Pacific white shrimp gut and rearing water from Malaysia and Vietnam: Implications for aquaculture research and management. PeerJ, 6, 1–22. 10.7717/peerj.5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorriehzahra, M. J. , & Banaederakhshan, R. (2015). Early mortality syndrome (EMS) as new emerging threat in shrimp industry. Advances in Animal and Veterinary Sciences, 3, 64–72. 10.14737/journal.aavs/2015/3.2s.64.72 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Figure S8
Figure S9
Figure S10
Figure S11
Figure S12
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Data Availability Statement
The 16S rRNA analysis raw data is available on the NCBI Sequence Read Archive (SRA) database (SRA Accession Number: SRR12199297; SRR12199298) (BioProject ID: PRJNA645513) (BioSample ID: SAMN15507524; SAMN15507525).


