Hannaford et al., identify that the interaction between Kinesin-1 and Pericentrin is essential for the direct transport of centrioles on the interphase microtubule network. This transport is important for the correct segregation of centrioles during asymmetric cell division.
Abstract
Centrosome positioning is essential for their function. Typically, centrosomes are transported to various cellular locations through the interaction of centrosomal microtubules (MTs) with motor proteins anchored at the cortex or the nuclear surface. However, it remains unknown how centrioles migrate in cellular contexts in which they do not nucleate MTs. Here, we demonstrate that during interphase, inactive centrioles move directly along the interphase MT network as Kinesin-1 cargo. We identify Pericentrin-Like-Protein (PLP) as a novel Kinesin-1 interacting molecule essential for centriole motility. In vitro assays show that PLP directly interacts with the cargo binding domain of Kinesin-1, allowing PLP to migrate on MTs. Binding assays using purified proteins revealed that relief of Kinesin-1 autoinhibition is critical for its interaction with PLP. Finally, our studies of neural stem cell asymmetric divisions in the Drosophila brain show that the PLP–Kinesin-1 interaction is essential for the timely separation of centrioles, the asymmetry of centrosome activity, and the age-dependent centrosome inheritance.
Introduction
Centrosomes are organelles comprised of two centrioles surrounded by a matrix of proteins termed the pericentriolar material (PCM). The PCM recruits gamma-tubulin ring complexes to centrosomes in a cell cycle-dependent manner to create a microtubule (MT) organizing center (MTOC; Azimzadeh and Bornens, 2007). Centrosomes are important for organizing cilia and mitotic spindles, both of which rely on proper centrosome positioning (Tang and Marshall, 2012).
Most research on centrosome movement has focused on the separation of centrosomes during late G2 before mitosis (Tanenbaum and Medema, 2010; Agircan et al., 2014). Separation is coordinated by motor proteins exerting pushing and pulling forces on centrosomal MTs. The key motor proteins involved in prophase centrosome separation is Kinesin-5/Eg5, which acts on antiparallel MTs to slide them in opposing directions (Kapitein et al., 2005), and dynein located at the cell cortex and the nuclear envelope, which exerts pulling forces on the MTs (Dujardin and Vallee, 2002). Together these motor and position centrosomes for bipolar spindle formation. Incomplete centrosome separation before nuclear envelope breakdown (NEB) can result in the formation of merotelic kinetochore-MT attachments (Silkworth et al., 2012). Merotelic attachments lead to lagging chromosomes and chromosome instability. In severe cases, failure to separate centrosomes can result in supernumerary centrosomes in the following cell cycle. This can lead to spindle abnormalities that further promote chromosome instability (Ganem et al., 2009), a common feature among cancer cells (Bakhoum and Cantley, 2018).
Importantly, centrioles are also motile during interphase, and this motility is poorly understood. In multi-ciliated cells, hundreds of centrioles are produced which must then migrate to the cell cortex to function as basal bodies (Spassky and Meunier, 2017; Jord et al., 2019; Ching et al., 2022). If centriole migration is impaired, then cilia formation is affected, which can result in a range of disorders termed ciliopathies (Reiter and Leroux, 2017). Interestingly, despite the importance of basal body positioning at the cell cortex, little is known about how they reach their destination (Dawe et al., 2006). Centrioles have also been observed to be highly motile before cytokinesis, with a proposed function at the midbody to regulate abscission (Piel et al., 2001; Krishnan et al., 2022).
One of the most documented instances of interphase centriole motility occurs in Drosophila neuroblasts (NBs). In NBs the centrioles are highly asymmetric at PCM levels. At the mitotic exit, the daughter centriole recruits the protein Centrobin which is simultaneously shed by the mother centriole (Januschke et al., 2011, 2013; Gallaud et al., 2020). The localization of Centrobin to the daughter centriole precedes PCM recruitment and MTOC formation. It is through its MT-nucleation capacity that the daughter centriole is stably localized to the apical side of the interphase NB resulting in a polarized MT network (Rebollo et al., 2007; Rusan and Peifer, 2007; Januschke and Gonzalez, 2010). Meanwhile, the mother centriole sheds PCM (Conduit and Raff, 2010; Ramdas Nair et al., 2016), resulting in a centriole incapable of MTOC activity; it then moves from the apical to the basal side of the cell throughout the interphase. As the NB enters the next prophase, the mother centriole once again recruits PCM in preparation for bipolar spindle formation (Rusan and Peifer, 2007; Rebollo et al., 2007). Analysis of mutant NBs has revealed that the centriolar proteins Pericentrin-like-protein (PLP; Lerit and Rusan, 2013) and Bld10/Cep135 (Singh et al., 2014), the motor protein Kinesin-1, and the Kinesin-1 activator Ensconsin/Map7 (Gallaud et al., 2014; Métivier et al., 2019) are involved in centriole separation. However, the underlying mechanism regulating centriole movement remains unclear.
In this study, we take advantage of Drosophila to examine the mechanism of centriole motility. We show that centrioles are transported along MTs by Kinesin-1 via direct interaction of the kinesin heavy chain (KHC) with PLP. We propose that Kinesin-1 and PLP work together to ensure mother centriole motility in NBs. The consequence of failed Kinesin–PLP driven motility is defective centrosome asymmetry and errors in age-dependent centriole segregation.
Results
Inactive centrioles are highly motile in interphase
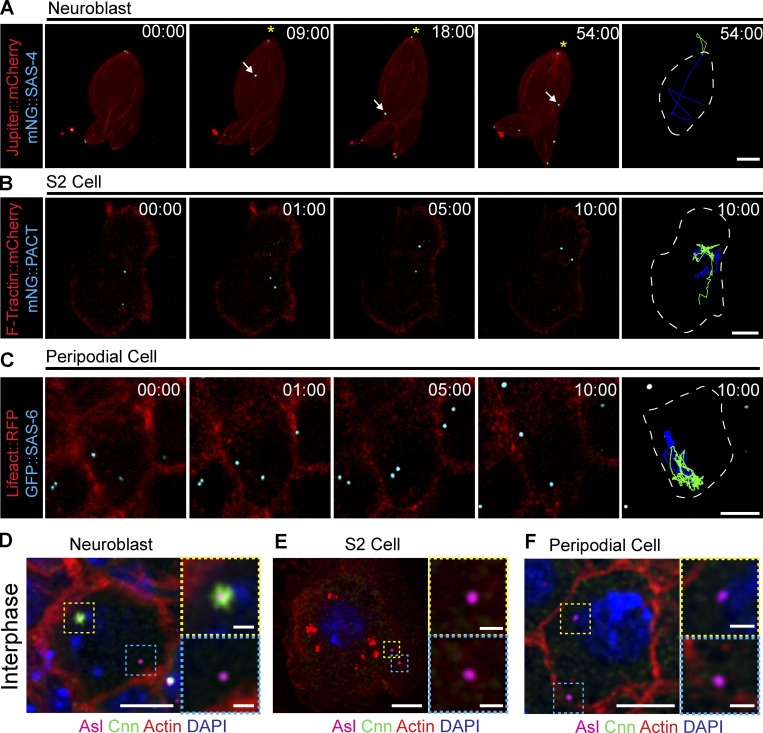
During interphase, Drosophila centrosomes shed their PCM, leaving behind a centriole that does not nucleate MTs; we will refer to these centrioles as inactive. Inactive centriole dynamics are best characterized in Drosophila NBs where the mother centriole is inactivated in the late stages of mitosis and during interphase, migrates from the apical side of the cell toward the basal side (Fig. 1, A and D; and Video 1). The high temporal resolution imaging required for accurate quantitative analysis of centriole movement is challenging in NBs due to the rapid speed of centrioles and the three-dimensional nature of motility. Therefore, we sought to identify non-NB cell populations with inactive, motile centrioles that would be suitable for our study. Inactive centrioles are found in other cell populations (Rogers et al., 2008), and we found that both Drosophila S2 cells (Fig. 1, B and E; and Video 2) and the squamous epithelial cells that constitute the peripodial membrane of the imaginal disks (McClure and Schubiger, 2005), hereon referred to as peripodial cells (PCs; Fig. 1, C and F; and Video 3), contained inactive and highly motile centrioles. The flat nature of the PC layer allowed for tracking multiple centrioles simultaneously with high temporal resolution. We confirmed that the motile centrioles in NBs, S2 cells, and PCs did not recruit the PCM component Centrosomin (Cnn), indicating they are indeed inactive during interphase (Fig. 1, D–F).
Figure 1.
Drosophila centrioles are motile in interphase cells. (A) Z-stack projection of an interphase NB expressing Jupiter::mCherry (red) and mNG::SAS-4 (cyan). The mother centriole (asterisk) remains closely associated with the apical cell cortex; the daughter centriole (arrow) moves throughout the cell. (B) Cultured S2 cell transfected with F-tractin::mCherry (red) to visualize the cell and mNG::PACT (cyan) to label centrioles. Both centrioles are highly motile through the acquisition. (C) Peripodial cell expressing Lifeact::RFP (red) to visualize the cell and GFP::SAS-6 (cyan) labelling the centrioles. Both centrioles are highly motile within the cell. Last columns in A, B, and C show cell outline (white line) and 10-min time projections of centriole movement (green and blue lines). (D) Fixed NB showing that Cnn (green) is restricted to one of the two centrioles (magenta). (E and F) Fixed S2 cell (E) and peripodial cell (F) showing no Cnn (green) present on the centrioles (magenta). Scale bars: 5 µm; inset scale bars: 1 µm. Time stamp: mm:ss.
Video 1.
Centriole motility in Drosophila neuroblasts. Spinning disk confocal timelapse of an isolated neuroblast expressing SAS-4::Neon (cyan) and mCherry::Jupiter (red). Note that one centriole migrates away from the apical side of the cell through interphase. Apical is up. Time stamp: hh:mm. Scale bar: 5 µm.
Video 2.
Centrioles are motile in interphase S2 cells. Spinning disk confocal time-lapse of an Interphase Drosophila S2 cell expressing F-Tractin::RFP (red) and GFP::PACT (centrioles, cyan). Both centrioles are highly motile. Time stamp: mm:ss. Scale bar: 5 µm.
Video 3.
Centrioles are motile in interphase peripodial cells. Spinning disk confocal timelapse of an interphase peripodial cell expressing UAS-Lifeact::RFP (red) by AGIR-GAL4. Centrioles are labelled with ubi-GFP::SAS-6 (cyan). Time stamp: mm:ss. Scale bar: 5 µm.
Inactive centrioles are MT cargo
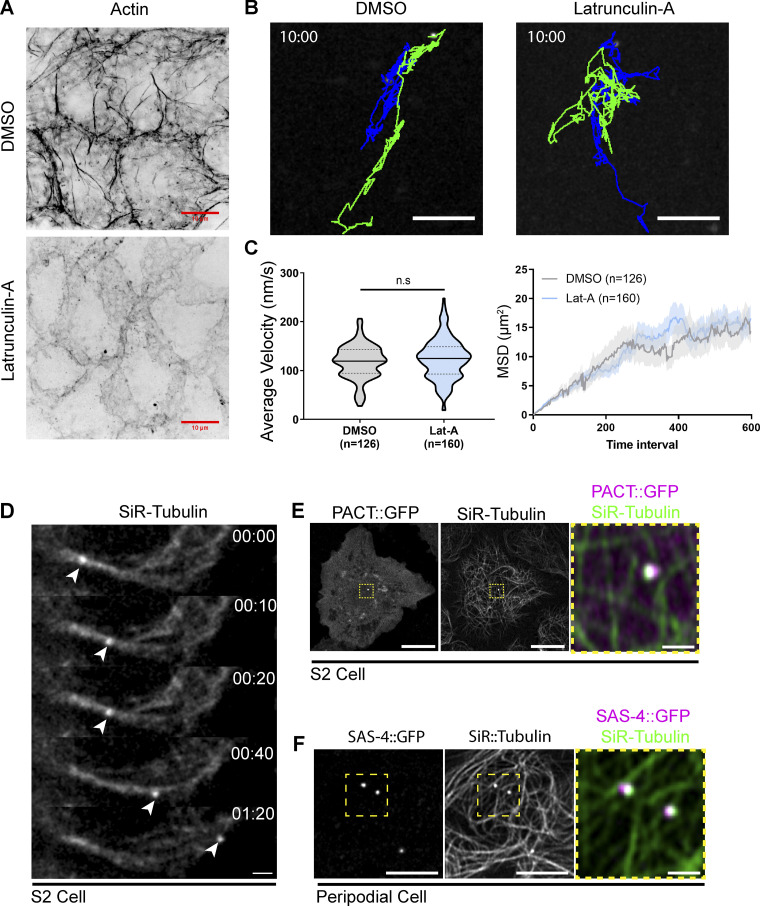
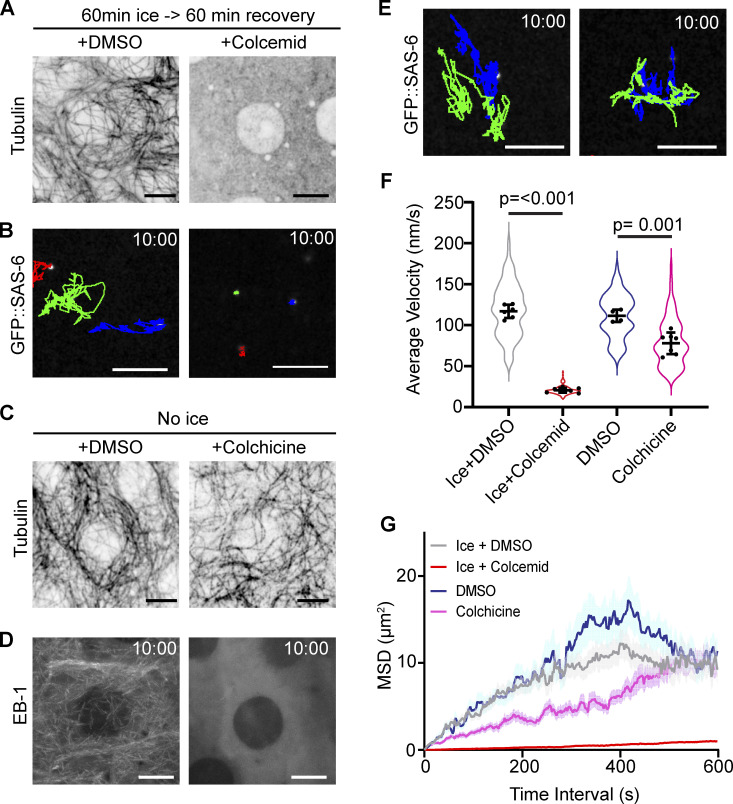
The precise mechanism of centriole motility is unknown, but both the MT and actin cytoskeleton have been implicated in centriole movement and positioning (Piel et al., 2000; Burakov et al., 2003). Using Latrunculin-A to depolymerize the F-Actin network in PCs, we found that an intact actin network was not required for centriole movement (Fig. S1 and Video 4). To investigate the role of the MT cytoskeleton, we depleted MTs by incubating wing discs on ice and then allowing them to recover in Colcemid-containing media (Fig. 2 A and Video 5). Tracking centrioles in MT-depleted PCs revealed a near complete loss of motility with average instantaneous velocity and mean square displacement approaching zero (Fig. 2, B, F, and G; and Video 5). We conclude that the MT network is essential for centriole motility. To test if dynamic MTs were required for motile centrioles, we treated wing discs with Colchicine in the absence of ice, which did not disassemble MTs (Fig. 2 C), but did block MT polymerization (growth) as revealed by the lack of EB-1 localization (Fig. 2 D). This treatment resulted in a 30% decrease in average instantaneous velocity but did not ablate centriole movement (Fig. 2, E–G and Video 5). Therefore, MTs are necessary for centriole movement, but their nucleation and dynamics are not.
Figure S1.
Centriole motility is independent of the Actin network and centrioles associate with the interphase MT cytoskeleton. (A) Z-stack projection of example PC’s labelled with Phalloidin showing that 10 µM Latrunculin-A treatment destroys the Actin network. Scale bars: 10 μm. (B) Tracks showing the movement of centrioles over a 10-min period. Latrunculin-A treatment does not inhibit centriole motility in peripodial cells. Scale bars: 5 μm. (C) Quantification of average velocity. (DMSO: 117.6 nm/s ± 34, n = 126, Lat-A: 123.5 nm/s ± 41, n = 140. Data = mean ± SD. Unpaired, two tailed, t test: P = 0.19). (D) Mean squared displacement is not affected by Latrunculin A treatment. Data = Mean ± SD (D) Timelapse series of an S2 cell labelled with SiR-Tubulin. Arrowhead denotes brighter spot corresponding to the centriole moving along the MT network. Scale bar: 2 μm. (E) Projection of a live S2 cell transfected with PACT::GFP to label the centriole (magenta). Centriolar signal is coincident with bright accumulation of SiR-Tubulin (green). Scale bar: 10 μm, inset: 1 μm. (F) Live PC expressing SAS-4::GFP (centriole, magenta) and labelled with SiR tubulin (green). SiR tubulin accumulation corresponds to SAS-4 positive centrioles. Scale bar: 5 μm; inset: 1 μm.
Video 4.
Centriole movement is independent of the actin cytoskeleton. Spinning disk confocal timelapse of peripodial cells expressing ubi-GFP::SAS-6 to label centrioles. Wing discs were treated with DMSO (left) or Latrunculin-A (right). Note: in both cases, centrioles are highly motile. Time stamp: mm:ss. Scale bar: 5 µm.
Figure 2.
Interphase motility is dependent on intact MT networks. (A) Peripodial cells following 1 h ice treatment followed by recovery in DMSO or 50 µM Colcemid. Note: no visible MTs remaining in the Colcemid treated wing disc. (B) 10-min time projection of centriole movement (colored tracks). Centrioles are not motile following ice treatment and Colcemid recovery. (C and D) Colchicine treatment does not destroy the pre-existing MT network (C) but does block MT dynamics revealed by EB-1 localization (D). (E) 10-min time projections of centriole movement (colored tracks). Centrioles remain highly motile following Colchicine treatment. (F) Quantification of instantaneous velocity in the indicated conditions (ice + DMSO: 117 ± 8.1 n = 6 wing discs, 95 centrioles. ice + Colcemid: 20.7 ± 2.8, n = 7 wing discs, 158 centrioles. DMSO: 115.5 ± 7.4, n = 4 wing discs, 78 centrioles. Colchicine: 77.9 ± 13.2, n = 7 wing discs, 109 centrioles). Data = mean ± SD, P values derived from unpaired t test. (G) Average mean squared displacement of centrioles. Scale bars: 5 µm. Time stamp: mm:ss.
Video 5.
Centriole movement is dependent upon the MT network but not MT dynamics. Spinning disk confocal timelapse of interphase peripodial cells expressing ubi-GFP::SAS-6 to label centrioles. Top panels: Centrioles are highly motile after recovering in the presence of DMSO following exposure to ice (left). Recovery from ice in the presence of Colcemid blocked centriole movement (right). Bottom panels: Treatment of wing discs with colchicine to inhibit MT dynamics does not block centriole movement (right) compared to those treated with DMSO (left). Time stamp: mm:ss. Scale bar: 5 µm.
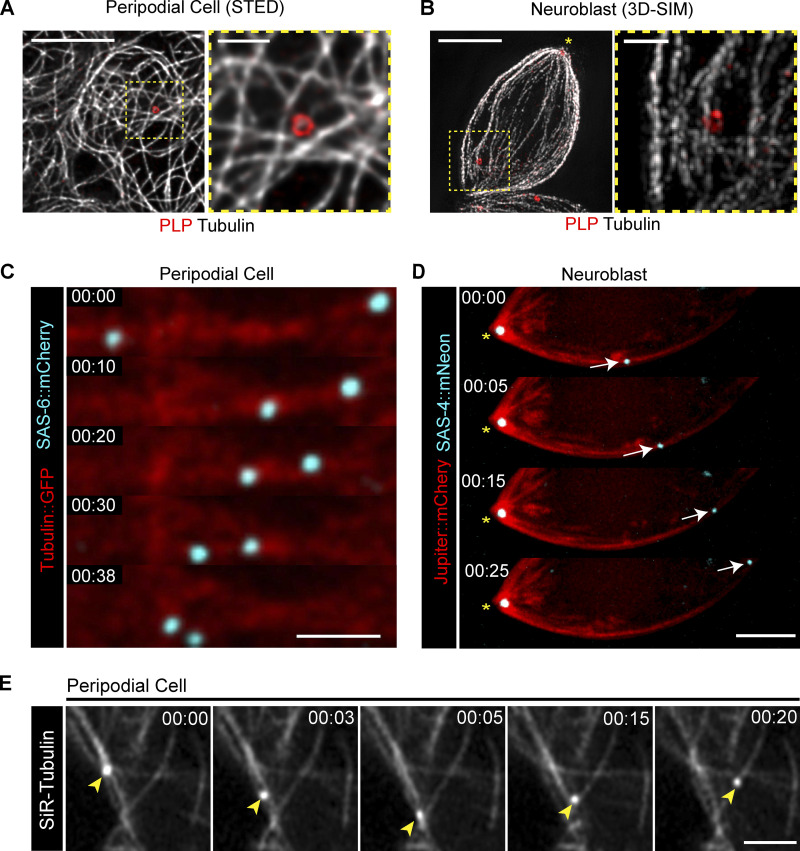
To further explore the relationship between centrioles and the MT network, we performed super resolution microscopy. Fixed imaging of MTs and centrioles in PCs and NBs revealed a close positional relationship between centrioles and MTs (Fig. 3, A and B), while live imaging revealed centrioles moving along MTs (Fig. 3, C and D; and Videos 6 and 7). To robustly and easily label both centrioles and MTs in live PCs, we used the MT dye SIR-Tubulin (Fig. S1, D–F; Lukinavičius et al., 2014), which revealed centrioles moving on MTs, even changing directions by switching their MT tracks (Fig. 3 E and Video 8). These data show that centrioles likely travel on MTs as cargo.
Figure 3.
Centrioles are MT cargo. (A and B) Super-resolution imaging reveals a close relationship between centrioles (PLP; red) and MTs (gray) in PCs (A) and NBs (B). Scale bars: 5 µm; inset 1 µm. (C and D) Centrioles (cyan) move on the MTs (red) in PCs (C) and NBs (D). Scale bar: 2 µm. Fluorescence transgenes are as indicated on left. (E) PCs treated with SiR-Tubulin showing centrioles (arrowheads) moving along MTs and switching tracks. Scale bar: 2 µm. Time stamp: mm:ss.
Video 6.
Centrioles move on the MT network in peripodial cells. Aireyscan microscopy timelapse showing centrioles (RFP::Pact, cyan) moving along MTs (GFP::Tubulin, red). Time stamp: mm:ss. Scale bar: 2 µm.
Video 7.
Centrioles move on the MT network in neuroblasts. Spinning disk confocal timelapse showing a mother centriole (SAS-4::Neon, cyan, yellow arrowhead) moving away from the daughter centriole (yellow asterisk) along the MT network (mCherry::Jupiter, red). Time stamp: mm:ss. Scale bar: 2 µm.
Video 8.
Centrioles switch between MTs at junctions. Aireyscan timelapse of a peripodial cell labelled with SIR- Tubulin. The centriole (yellow arrowhead) moves along multiple MTs, changing tracks at MT-MT junctions. Time stamp: mm:ss. Scale bar: 2 µm.
Kinesin-1 is required for efficient centriole transport
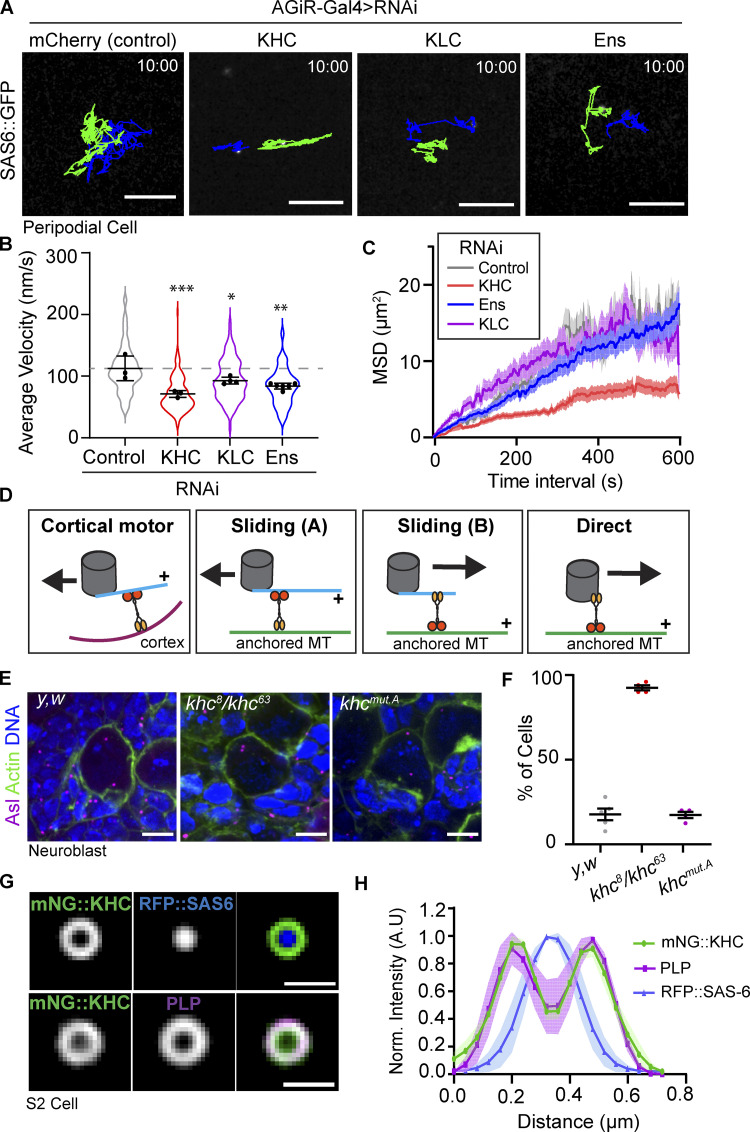
To investigate the MT transport of centrioles, we aimed to identify which motor proteins were required for their motility. Previously, the heavy chain of Drosophila Kinesin-1 (KHC) and its activator MT Associated Protein 7/Ensconsin (Map7, Ens) were shown to be important for centrosome separation at prophase during asymmetric NB division (Gallaud et al., 2014; Métivier et al., 2019). Therefore, we tested if Kinesin-1 was involved in MT-mediated centriole transport by knocking down KHC, Kinesin Light Chain (KLC), or Ens in PCs using the PC-specific driver AGIR-GAL4 (Gibson et al., 2002). Control centrioles moved rapidly around the PCs as expected, while knockdown of KHC, KLC, or Ens significantly reduced centriole motility (Fig. 4, A–C and Video 9). Thus, Kinesin-1 is important for interphase centriole motility.
Figure 4.
Kinesin-1 is required for efficient centriole motility. (A) 10-min time projections of centriole movement (colored tracks) in the indicated knockdown conditions in PCs. Scale bars: 5 µm. (B) Average velocity is significantly reduced following the knockdown of Kinesin-1 components (control: 112 ± 20, n = 3 wing discs, 100 centrioles. KHCRNAi: 71 ± 5, n = 3 wing discs, 110 centrioles. KLCRNAi: 92 ± 5, n = 4 wing discs, 97 centrioles. EnsRNAi: 83 ± 8, n = 6 wing discs, 84 centrioles; data = mean ± SD. ANOVA P = 0.001, Dunnett’s pairwise comparisons: Ctrl vs. KHCRNAi P = 0.0005, ***, Ctrl vs KLCRNAi P = 0.043, *, Ctrl vs. EnsRNAi P = 0.0027, **). (C) Mean squared displacement is reduced following KHC knockdown in PCs. (D) Diagram summarizing the models by which Kinesin-1 could move centrioles in cells. Kinesin-1 cargo domain is shown in yellow; motor domain (orange) always walks toward the indicated + sign, and the black arrow indicates the movement direction of the centriole. (E) Z-stack projections of fixed NBs sowing centriole positioning. Note: in khc8/khc63 NBs, the centrioles are adjacent to the apical side of the cell. (F) Quantification of the percentage of neuroblasts with adjacent apical centrioles. y,w: 17.8% ± 7, n = 5 brains; khc8/khc63: 92.5% ± 3, n = 4 brains, khcmut.A: 17.4% ± 3.5, n = 4 brains. Data = mean ± SD. (G) Averaged OMX-SIM micrograph showing mNG::KHC localizes to the outer centriole edge. Scale bar: 500 nm. (H) Quantification of rotational averaged centrioles showing the distribution of mNG::KHC relative to PLP and RFP::SAS-6 (n = 4).
Video 9.
Centriole movement requires Kinesin-1. Spinning disk confocal timelapse showing centriole (ubi-GFP::SAS-6) movement following indicated knockdowns. RNAi was expressed under UAS promoter with the AGIR-GAL4. Time stamp: mm:ss. Scale bar: 5 µm.
Based on these data, we hypothesized that Kinesin could move centrioles via three possible mechanisms. First, by indirect motor transport whereby the centriole nucleates a small number of MTs, that in-turn contact anchored Kinesin motor at the cortex (Fig. 4 D). We do not favor this model as, unlike Dynein, Kinesin-1 is not known to act in this manner. In addition, this model requires dynamic MTs that search the cortex for motors; however, our Colchicine treatment showed that MT dynamics are not required for inactive centriole motility (Fig. 2 E). We also note that previous studies on inactive centrioles did not identify the presence of centriolar MTs in interphase Drosophila cells (Rusan and Peifer, 2007; Rebollo et al., 2007; Rogers et al., 2008).
The second possible mechanism of centriole motility is via the MT “sliding” activity of Kinesin-1 (Winding et al., 2016; Lu et al., 2016; Fig. 4 D, Sliding A and B). KHC slides MTs by anchoring its C-terminus to one MT via a binding site within the cargo binding domain and walking along a second MT with its motor domain. MT sliding activity can be reduced by 50% using a khc allele in which C-terminal MT binding site is mutated (khcmut.A: R914A, K915A, R916A, and Q918A; Lu et al., 2016; Winding et al., 2016). To test whether sliding was essential for centriole motility, we compared centriole positioning between control, khc8/khc63 (hypomorphic), and khcmut.A NBs (Fig. 4 E). As expected, the khc8/khc63 NBs exhibited a failure to separate their centrioles (Gallaud et al., 2014). However, reducing the sliding activity of Kinesin-1 had no effect on centriole separation compared to controls (Fig. 4 F). Additionally, either sliding model (Fig. 4 D, Sliding A and B) would require that both the centriole-attached MTs and the anchored MT be precisely polarized relative to each other and relative to the apical-basal cell axis to generate the required directional motility in NB. We believe this would be extremely difficult to achieve, and thus do not favor a sliding model.
Finally, the third possible mechanism involves Kinesin-1 directly binding the centriole surface and moving it as cargo. For this model to be plausible, Kinesin-1 would have to localize to centrioles. Kinesin-1 has previously been shown to localize to centrosomes in vertebrate cultured cells (Neighbors et al., 1988), but its localization to centrosomes in Drosophila has not been investigated. To examine Kinesin-1 localization we expressed and imaged N-terminal KHC fusion proteins. Importantly, KHC localized to centrioles in PCs, NBs, and S2 cells (Fig. S3, D–F). Structured illumination microscopy of S2 cells revealed that mNeon::KHC localized as a ring around RFP::SAS-6 (core centriole marker), while occupying the same spatial position as Pericentrin-like-protein (PLP; Fig. 4, G and H). These data indicate that KHC is positioned on the outer layer of the centriole in a region occupied by proteins that bridge the centriole and PCM in mitosis. Collectively, our data thus far suggest a model whereby Kinesin-1 moves centrioles as cargo by directly binding the centriole surface.
Figure S3.
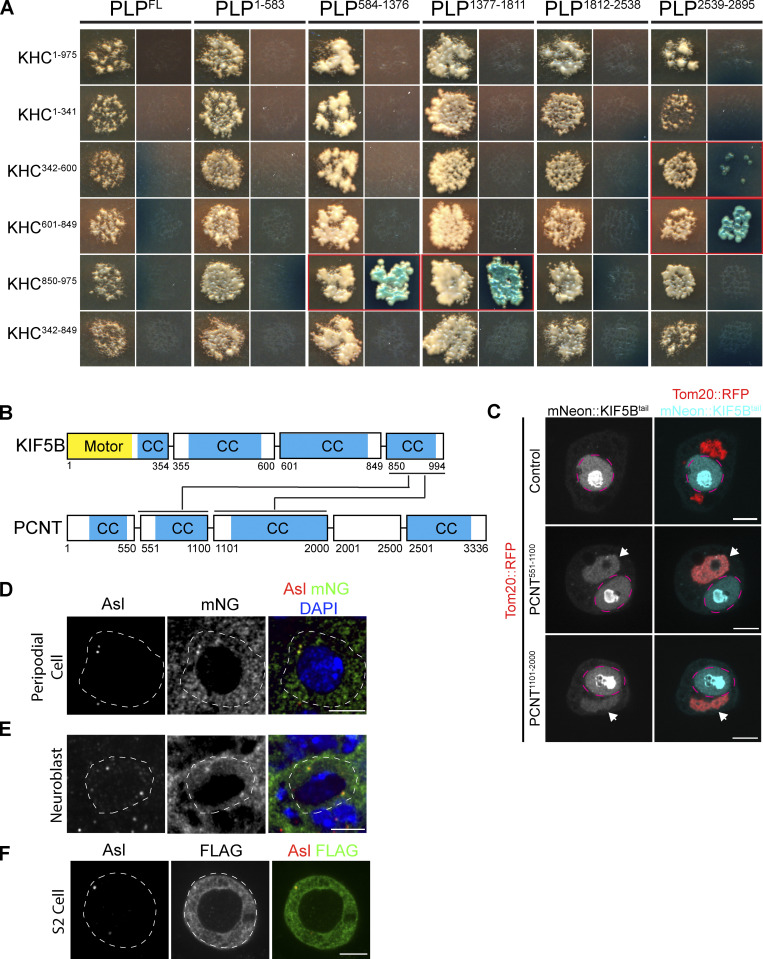
PLP–Kinesin-1 interaction is conserved. (A) Images of mated yeast clones. For each PLP fragment, the left column is DDO plates to select for both bait and prey. Right column is QDOXA plates to select for interaction. Red outlined clones indicate a positive interaction, identified by the growth of blue yeast colonies. (B) Diagram illustrating interactions identified between human KIF5B and PCNT fragments, predicted coiled coils are highlighted in blue (CC). (C) Example images of S2 cells in which fragments of PCNT (Red) have been targeted to the mitochondria by a Tom20 mitochondrial targeting sequence. The KIF5B tail (aa850-994) was tagged with mNeonGreen. Note that KIF5B is only recruited to mitochondria in the presence of PCNT fragments (arrow heads) indicative of interaction. mNeon::KIF5B also accumulated in the nucleus (magenta dashed line). (D) Representative image of a peripodial cell expressing mNG::KHC. Note mNG signal (green) coincident with the centriole marker Asterless (red). (E) Representative image of an interphase NB expressing mNG::KHC. mNG::KHC localizes to both the apical and the basal centriole. (F) Representative image of an S2 cell expressing FLAG::KHC stained for FLAG and Asterless. Scale bar: 5 μm.
The centriole bridge protein PLP is required for centriole motility
PLP is a recruiter and organizer of PCM, and to perform this function it localizes to the outer edge of the centriole (Varadarajan and Rusan, 2018). Importantly, previous mutant analysis has implicated PLP in centrosome positioning in NBs as well as basal body positioning in sensory neurons (Martinez-Campos et al., 2004; Lerit and Rusan, 2013; Galletta et al., 2014; Roque et al., 2018). However, the mechanism by which PLP influences centriole/centrosome motility remains unknown. Based on the localization of PLP to the outer layer of the centriole and PLP’s role in centriole/centrosome positioning, we hypothesized that PLP is required for MT-based motility of centrioles in interphase cells.
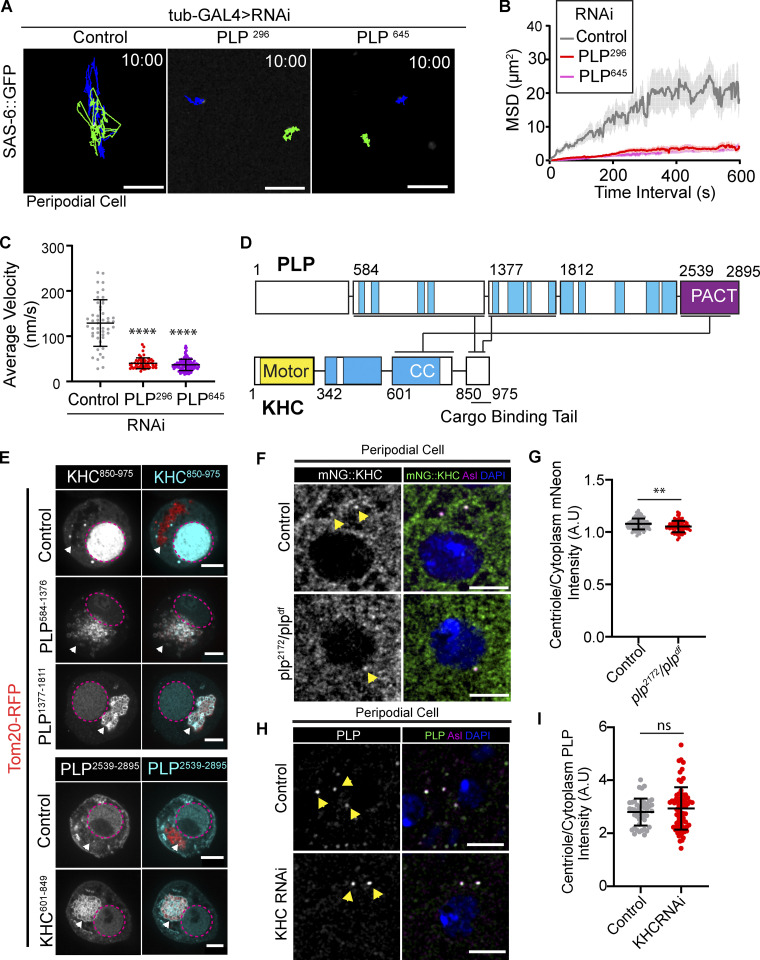
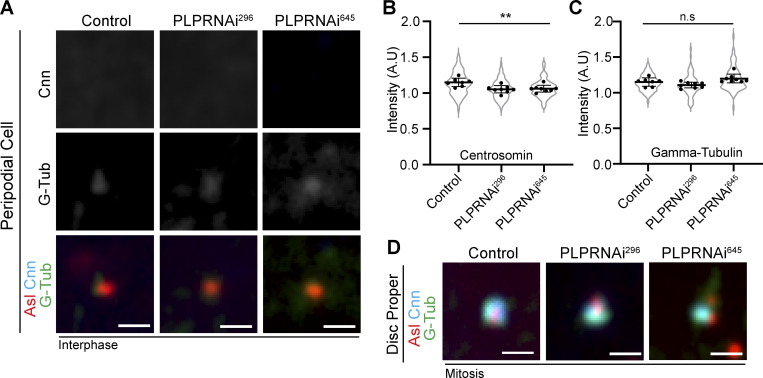
To test our hypothesis, we knocked down PLP in PCs using two independent RNAi lines. In each case, PLP knockdown caused a substantial 70% reduction in centriole motility (Fig. 5, A–C and Video 10). The loss of centriole motility could be explained in two ways. Firstly, it is possible that PLP depletion resulted in ectopic PCM recruitment, similar to what was previously described in NBs (Lerit and Rusan, 2013), leading to the formation of centrosomal MTs that anchor the centrosome to the cortex. However, the lack of PCM on centrioles in PLP RNAi PCs excludes this possibility (Fig. S2). Therefore, we favor a second explanation that PLP is directly involved in centriole motility, potentially functioning as a motor-cargo adaptor molecule.
Figure 5.
PLP is essential for centriole motility and interacts with KHC. (A) 10-min time projections of centriole movement (colored tracks) in PCs following knockdown of PLP with tub-GAL4. Scale bars: 5 µm. (B and C) Quantification of mean squared displacement (B) and average velocity (C) following PLP knockdown (control average velocity = 128.8 nm/s ± 51.48, n = 49; PLPRNAi296 = 40.19 nm/s ±12.04, n = 54; PLPRNAi645 = 36.6 nm/s ±12.48, n = 112, data = mean ± SD, ANOVA: P = <0.0001, Dunnett’s pairwise comparison P = 0.0001 between Ctrl and RNAi conditions, ****). (D) Diagram showing the three independent interactions (black lines) found through yeast two-hybrid screening of PLP and KHC subfragments (Fig. S4). Blue = predicted coiled coils. Yellow = KHC motor domain. The PACT domain of PLP is located in the final C-terminal fragment (purple). (E) Mitochondrial recruitment assay validating interactions found through Y2H. Prey fragments (gray/cyan) accumulate in nucleus (magenta dashed line). In the presence of a co-transfected bait fragments (red, arrowhead) that is targeted to the mitochondria via a Tom20-RFP tag, a positive interacting fragment (gray/cyan) is also recruited to the mitochondria. (F) Examples showing mNG::KHC (green) localization in control or plp− PCs. Yellow arrowheads indicated centriolar localization determined by Asterless staining (magenta). Blue = nucleus. (G) Quantification of mNG::KHC localization determined by the mean intensity at the centriole divided by mean cytoplasmic intensity. Mean ± SD: Control = 1.078 ± 0.05, n = 116 centrioles; plp2172/Df = 1.05 ± 0.06, n = 69 centrioles, unpaired t test: P = 0.003, **. (H) Examples showing endogenous PLP (green) localization in control or KHCRNAi expressing PCs. Yellow arrowheads indicated centriolar localization determined by Asterless staining (magenta). Blue = nucleus. (I) Quantification of PLP localization determined by the mean intensity at the centriole divided by mean cytoplasmic intensity. Mean ± SD: Control = 2.8 ± 0.5, n = 44 centrioles; KHCRNAi = 2.9 ± 0.8, n = 71 centrioles, unpaired t test: P = 0.3, ns.
Video 10.
Centriole movement is dependent upon PLP. Spinning disk confocal timelapse showing centriole (ubi-GFP::SAS-6) movement. Expressing UAS-PLPRNAi with Tub-GAL4 (right) blocks centriole movement compared to controls (left). Time stamp: mm:ss. Scale bar: 5 µm.
Figure S2.
PLP knockdown does not cause precocious centriole activation in PCs. (A) Fixed PCs stained for Asl (red), Cnn (cyan), and Gamma-tubulin (green) shows that PCM does not accumulate in PLPRNAi expressing PCs. (B) Quantification of Centrosomin intensity relative to cytoplasm (Control: 1.16 ± 0.05 n = 7 wing discs, 67 centrioles; PLPRNAi296: 1.1 ± 0.05, n = 8 wing discs, 91 centrioles; PLPRNAi645: 1.1 ± 0.05, n = 8 wing discs, 86 centrioles. Mean ± SD. ANOVA: P = 0.003. Dunnett’s multiple comparison: Control vs PLPRNAi296: P = 0.0035, Control vs PLPRNAi645: P = 0.0065). (C) Quantification of Gamma Tubulin intensity relative to cytoplasm (Control: 1.15 ± 0.05 n = 7 wing discs, 67 centrioles; PLPRNAi296: 1.1 ± 0.04, n = 8 wing discs, 91 centrioles; PLPRNAi645: 1.2 ± 0.06, n = 8 wing discs, 86 centrioles. ANOVA: P = 0.01, multi-comparison test showed no significance between Control and RNAi groups. Mean ± SD). (D) Fixed mitotic wing disc cells stained for Asl (red), Cnn (cyan), and Gamma-tubulin (green). PLP knockdown does not prevent PCM accumulation in mitosis. But PCM can appear disorganized. Scale bars: 1 µm.
PLP directly binds KHC
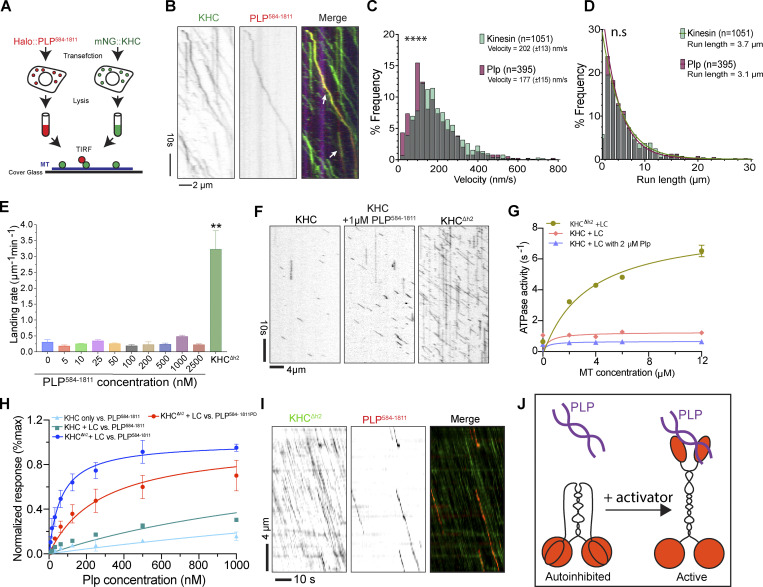
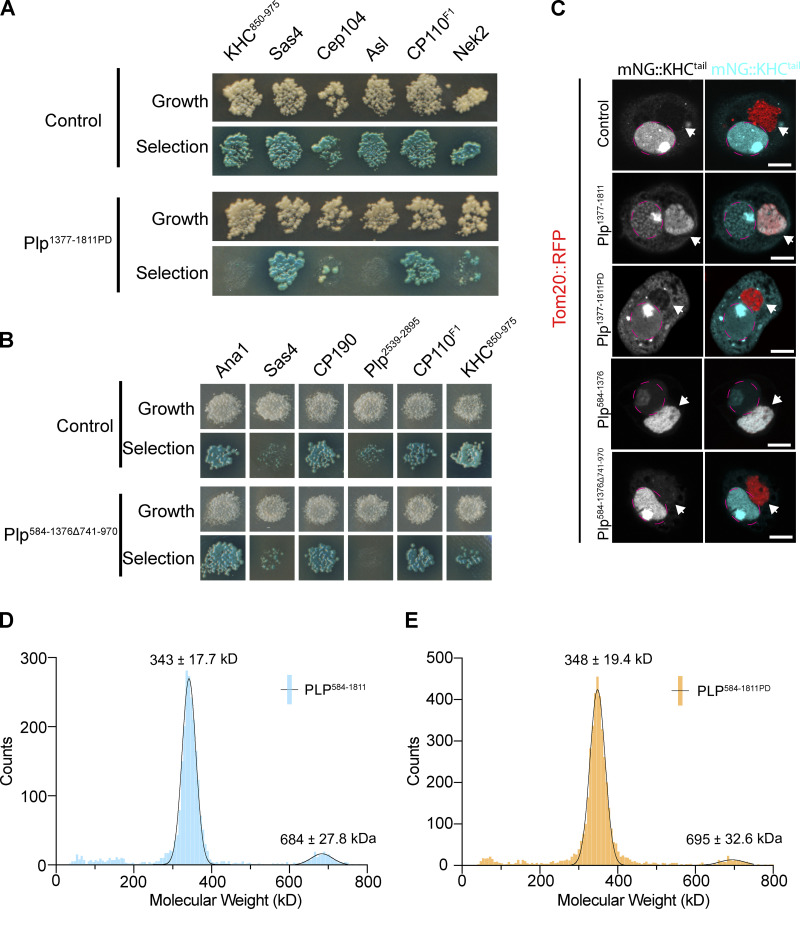
Given that both PLP and Kinesin-1 are required for centriole motility, and that they colocalize on the centriole, we hypothesized that PLP and Kinesin-1 directly interact. To test for direct protein–protein interactions (PPIs), we performed a yeast two-hybrid (Y2H) assay using subdivided fragments of PLP and KHC (Fig. 5 D). The Y2H identified three interactions: PLP584-1376-KHC850-975, PLP1377-1811-KHC850-975, and Plp2539-2895-KHC601-849 (Fig. 5 D and Fig. S3 A); the former two interactions are with the KHC cargo binding tail domain. We confirmed all three interactions in vivo using a mitochondria-targeting assay that utilizes colocalization to test PPIs (Fig. 5 E; Galletta et al., 2014; Schoborg et al., 2015). This same assay confirmed that the PLP–KHC interaction is conserved between human Pericentrin (PCNT) and the cargo binding domain of the human Kinesin-1 heavy chain (KIF5B; Fig. S3, B and C).
Because PLP physically interacts with the cargo binding domain of KHC (KHC850-975), we hypothesized that PLP serves as an adaptor required to anchor Kinesin-1 to the centriole. We therefore compared the localization of mNG::KHC in control and plp mutant PCs and found an extremely small (1.9%) decrease in Kinesin levels. Therefore, PLP is not essential for KHC localization to the centriole (Fig. 5, F and G). Importantly, analysis of PLP localization in PCs expressing KHC RNAi demonstrated that KHC was also not important for PLP localization in the interphase (Fig. 5, H and I).
If not an adaptor, we hypothesized that PLP could function as an activator, or enhancer of Kinesin-1 processive motility, possibly by relieving the Kinesin-1 autoinhibited state that occurs by the interaction between the motor domain and cargo binding tail (Verhey and Hammond, 2009). To test this hypothesis, we used an in vitro cell extract approach (Kelliher et al., 2018) where Drosophila S2 cells were transfected with full-length mNG::KHC and another with HALO::PLP584-1811. We used PLP584-1811 in this assay as it encompassed both central fragments of PLP that bind to the cargo domain of KHC (Fig. 5 D). Whole-cell lysates were applied to polymerized MTs and imaged using single molecule TIRF microscopy, which revealed PLP584-1811 and KHC comigrating along MTs (Fig. 6, A and B; and Video 11). Interestingly, our data showed that the dynamics of Kinesin-1 with and without PLP584-1811 were similar, suggesting that PLP584-1811 was not sufficient to activate the Kinesin-1 motor (Fig. 6, C and D; and Video 12). However, the use of lysate could confound our assay through other interactors with the KHC-PLP complex, as well as other proteins on the MTs.
Figure 6.
In vitro analysis of KHC motility. (A) Diagram of in vitro motility experiment. mNG::KHC (red) and Halo::PLP584-1811 (green) were transfected into S2 cells. Cleared lysate was then flowed onto MTs (blue) for TIRF analysis. (B) Co-migration of mNG-KHC (green) with Halo-PLP584-1811 (red) on HiLyte-647 MTs (blue). (C) Velocities of KHC motors and PLP584-1811 cargos on MTs. Kinesin mean velocity = 202 (± 113, SD) nm/s (n = 1,051). PLP584-1811 mean velocity = 177 (± 115, SD) nm/s (n = 395). Mann-Whitney test P value = <0.0001, ****. (D) Characteristic run lengths of KHC motors and PLP584-1811 cargo. PLP584-1811 run length = 3.1 μm, (n = 1,051). KHC run length = 3.7 μm (n = 395). Mann-Whitney test P value = 0.08, ns. (E) Quantification of the landing rates of 25 nM KHC + LC with PLP584-1811 at indicated concentrations in comparison with that of 25 nM KHCΔh2 + LC. Error bars represent the SD of landing rates determined from three or four independent movies. For each condition, several thousand landing events were quantified on a total MT length of several millimeters. Unpaired, two tailed t test P value = <0.001 (**), KHCΔh2 + LC vs. KHC + LC + PLP (at varying concentrations). (F) Kymographs showing landing and movement of KHC on MTs in the absence and presence of PLP and compared with the activated KHCΔh2. (G) Steady-state ATPase activities of different Kinesin-1 constructs in the presence or absence of 2 μM PLP584-1811 as a function of MT concentration. Data were presented as mean ± SD and fitted with Michaelis-Menten kinetics. N = 3 independent titrations. (H) Steady-state analysis of Kinesin-1 to PLP binding affinity using biolayer interferometry (BLI). KHC, KHC + LC, or KHCΔh2 + LC were loaded onto the biosensors and exposed to WT PLP584-1811 or PLP584-1811PD mutant at the indicated concentrations. BLI signals at equilibrium were plotted against PLP concentrations, and dissociation constant (Kd) were determined by fitting the data with nonlinear regression single-site binding: WT PLP584-1811 vs. KHC, 4.4 μM; vs. KHC + LC, 1.7 μM; vs. KHCΔh2 + LC, 65 nM; PLPPD vs. KHCΔh2 + LC, 281 nM. (I) Kymograph analysis showing events of PLP584-1811 comigrating with KHCΔh2 + LC. (J) Diagram highlighting how the interaction between the motor domain and cargo binding tail could inhibit PLP interaction.
Video 11.
KHC and PLP584-1811 comigrate on MTs in vitro. TIRF timelapse showing comigration of PLP584-1811 (red) and KHC (green) on MTs (blue). Arrows indicate events of comigration. PLP molecules appear to precede KHC due to the slower multichannel imaging rate relative to the motility. Time: seconds. Scale bar: 5 µm.
Video 12.
PLP motility. Movie showing movement of PLP584-1811 (red) on MTs in the presence of KHC. A maximum intensity projection of the KHC (blue) channel is used to highlight MT position. Time: seconds. Scale bar: 5 µm.
To more precisely investigate the potential function of PLP as a Kinesin-1 activator, we purified PLP584-1811, KHC, and KLC, as well as the activated KHCΔh2 (a mutant KHC in which the tail is unable to interact with the motor; Friedman and Vale, 1999; Kelliher et al., 2018). Adding increasing concentrations of PLP584-1811 to KHC did not significantly alter the landing rate of the wild-type motor, especially when compared to the significantly higher landing rate of active KHCΔh2 (Fig. 6, E and F; and Video 13). We then measured motor activity through an ATPase assay and, in agreement with the landing rate, the ATPase activity of KHC was also not increased upon addition of PLP584-1811, while the ATPase activity of KHCΔh2 was significantly higher (Fig. 6 G). These results demonstrate that PLP584-1811 is not sufficient to activate Kinesin-1 in vitro.
Video 13.
PLP584-1811 does not enhance KHC landing rate. TIRF timelapse showing GFP::KHC (green) landing on MTs (red). Left panel shows 25 nM GFP::KHC. The center panel shows 25 nM GFP::KHC in the presence of 1 µM PLP584-1811. Right panel shows 25 nM GFP::KHCΔh2. Scale bar: 10 µm.
We next hypothesized that Kinesin-1 activation is required for KHC interaction with PLP. Interestingly, through Y2H, we were unable to observe the interaction between PLP fragments and full-length KHC (Fig. S3 A). This could be due to the autoinhibited conformation masking the PLP binding site. To quantitatively measure PLP–KHC interaction, we performed a biolayer interferometry in vitro binding assay to compare the interaction of PLP584-1811 with either wild-type KHC or KHCΔh2 in the presence or absence of KLC. The complex of KHCΔh2 + LC showed the highest affinity to PLP584-1811, indicating that kinesin activation and the light chain association are both critical for PLP binding (Fig. 6 H). Utilizing KHCΔh2 + KLC we were then able to visualize comigration of Kinesin-1 and PLP584-1811 on MTs in vitro, indicating that these proteins can move together on the MT network through direct interaction (Fig. 6 I).
These data suggest that a multimeric protein complex is required to not only relieve the autoinhibition of Kinesin-1 but also to facilitate the interaction of PLP with the cargo-binding tail of the motor (Fig. 6 J).
PLP–KHC interaction is required for centriole motility
We next sought to determine whether the interaction between PLP and Kinesin-1 was required for centriole motility. To test this, we aimed to specifically block the PLP–KHC interaction while maintaining PLP binding to its other centrosome binding partners (Galletta et al., 2016).
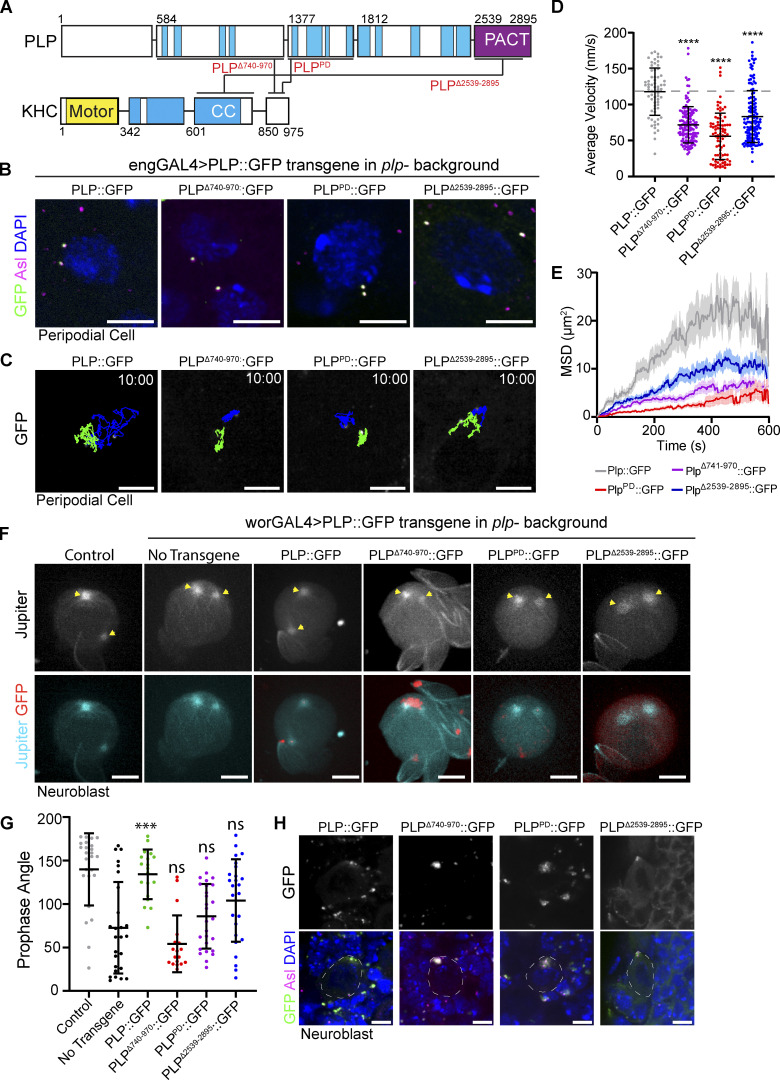
To identify plp alleles that block Kinesin interaction, we took a multifaceted approach (Fig. 7 A). First, to test the significance of the PLP1377-1811–KHC850-975 interaction, we performed random mutagenesis on PLP1377-1811 and screened to identify clones that resulted in the loss of Kinesin interaction (Galletta and Rusan, 2015). Screening over 1,000 clones yielded one harboring two amino acid substitutions—L1663P and G1699D (plpPD; Fig. S4, A and C)—that disrupted PLP’s interaction with KHC and Asl while maintaining the integrity of all four other known interactions (Galletta et al., 2016). Importantly, we were able to purify PLP584-1811 carrying the L1663P and G1699D mutations and found that these mutations significantly weakened the direct interaction with KHC (Fig. 6 H), while preserving the ability of PLP584-1811 to dimerize demonstrating that the major structural properties of the proteins remain intact (Fig. S4, D and E). Second, to test the significance of the PLP584-1376–KHC850-975 interaction, we used a series of previously generated deletions and truncations within PLP584-1376 (Lerit et al., 2015) because our random mutagenesis approach did not yield a useful allele. We found that deletion of amino acids 740–971 (plpΔ740-971) was sufficient to ablate PLP584-1376 interaction with KHC (Fig. S4, B and C). Finally, we were unable to use random mutagenesis or small deletions to test the PLP2539-2895–KHC601-849 interaction, so we completely truncated amino acids 2,539–2,895 (plpΔ2539-2895).
Figure 7.
PLP–KHC interaction mutants show reduced centriole motility. (A) Schematic showing the interactions between PLP and KHC and the corresponding interaction mutations (red text). (B) Example projections of PCs showing PLP transgenes (green) localizing centrioles (Asterless; magenta). DNA (blue) shows cells are in interphase. (C) 10-min time projections of centriole movement (colored tracks) in PCs expressing the indicated PLP transgenes in a mutant background (plp2172/Df(3L)Brd15). (D) Rescue with the mutant transgenes results in a significantly slower instantaneous velocity compared to full length rescue (PLP::GFP: 117 ± 32, n = 66. PLPΔ740-971::GFP: 71 ± 25, n = 83. PLPPD::GFP: 55 ± 32, n = 138, PLPΔ2539-2895::GFP: 83 ± 36 n = 139, ANOVA: ****P = <0.0001, Dunnett’s pairwise comparison between PLP::GFP and all other conditions). Data = mean ± SD. (E) Mean squared displacement shows motility is most effected by mutations (PLPPD and PLPΔ740-971) that interfere with interaction between PLP and the cargo binding tail of KHC. (F) Maximum intensity projections from live NBs showing the position of the MTOCs (yellow arrows; Jupiter::RFP) in prophase. Cells are expressing the indicated PLP transgene in a PLP mutant background (plp2172/Df(3L)Brd15). (G) The angle of the centrosomes relative to the nucleus in prophase is not significantly rescued by the transgenes that block PLP–KHC interaction (Control: 139° ± 41.4, n = 25. No Transgene: 72° ± 52, n = 26. PLP::GFP: 134° ± 28, n = 19, P = <0.0001. PLPΔ740-971::GFP: 54° ± 32.7, n = 18, P = 0.69. PLPPD::GFP: 85.8° ± 37, n = 27, P = 0.86. PLPΔ2539-2895::GFP: 104 ± 47.5, n = 25, P = 0.08. P values determined by Tukey pairwise comparison between no transgene and PLP rescue conditions). Data = mean ± SD. (H) Maximum projections showing the localization of the indicated transgenes in interphase NBs. Scale bars: 5 µm. Time stamp: mm:ss.
Figure S4.
Disrupting the PLP–Kinesin-1 interaction. (A) Yeast clones showing the PLP interactions disrupted by the PLPPD mutation. Note that Asl interaction is also disrupted. Interaction was determined by the growth of blue clones on the selection plate (QDOXA). (B) Yeast clones showing the interactions disrupted by deletion of PLP aa741-970. The interaction with KHC850-975 appears weaker due to decreased growth. Note interaction with PLP2539-2895 is also disrupted. (C) Validation of PLP–KHC interaction mutants by mitochondrial targeting assay. PLP fragments were targeted to the mitochondria using the Tom20 mitochondrial localization sequence (red). The cargo binding tail of KHC (KHC850-975) was tagged with mNeon. Interaction was determined by recruitment of mNeon::KHC850-975 to the mitochondria. (Arrowheads point to mitochondria; magenta dashed line labels the nuclei.) (D and E) Histograms of single particle mass values determined by mass photometry for HALO::PLP584-1811: WT and PD mutant, respectively. Lines are the Gaussian fit to the data yielding the molecular weights, consistent with a predominant dimer (346 kD) and a minor tetramer (692 kD) species of PLP. Scale bars: 5 µm.
To test the physiological relevance of PLP–KHC interaction, we generated transgenic flies expressing each of the three PLP mutant alleles (plpΔ740-971, plpPD, plpΔ2539-2895). Expressing each of these PLP transgenes in the plp− null mutant background (plp2172/Df [Martinez-Campos et al., 2004; Galletta et al., 2020]) did not fully rescue centriole motility in PCs, despite the transgene encoded protein localizing to the centriole (Fig. 7, B–E and Video 14). We next examined the role of PLP–KHC interaction in centriole positioning within NBs. Previously, the angle between the two centrosomes in prophase has been used as a proxy for interphase centriole motility (Lerit and Rusan, 2013; Gallaud et al., 2014). None of the mutant transgenes (plpΔ740-971, plpPD, and plpΔ2539-2895) were able to rescue prophase centrosome positioning in NBs (Fig. 7, F and G). The most severe defects were observed in conditions where the interaction of PLP with the cargo binding tail of Kinesin-1 was disrupted (plpΔ740-971and plpPD). We, therefore, concluded that PLP interaction with Kinesin-1 is critical for proper centriole motility in interphase cells.
Video 14.
PLP–KHC interaction mutants fail to rescue centriole motility. Spinning disk confocal timelapse of peripodial cells expressing indicated rescue constructs under UAS control by Engrailed-GAL4 in a plp− mutant background (plp2172/Df(3L)Brd15). Time stamp: mm:ss. Scale bar: 5 µm.
PLP and Kinesin-1 are required for PCM asymmetry and proper centriole inheritance
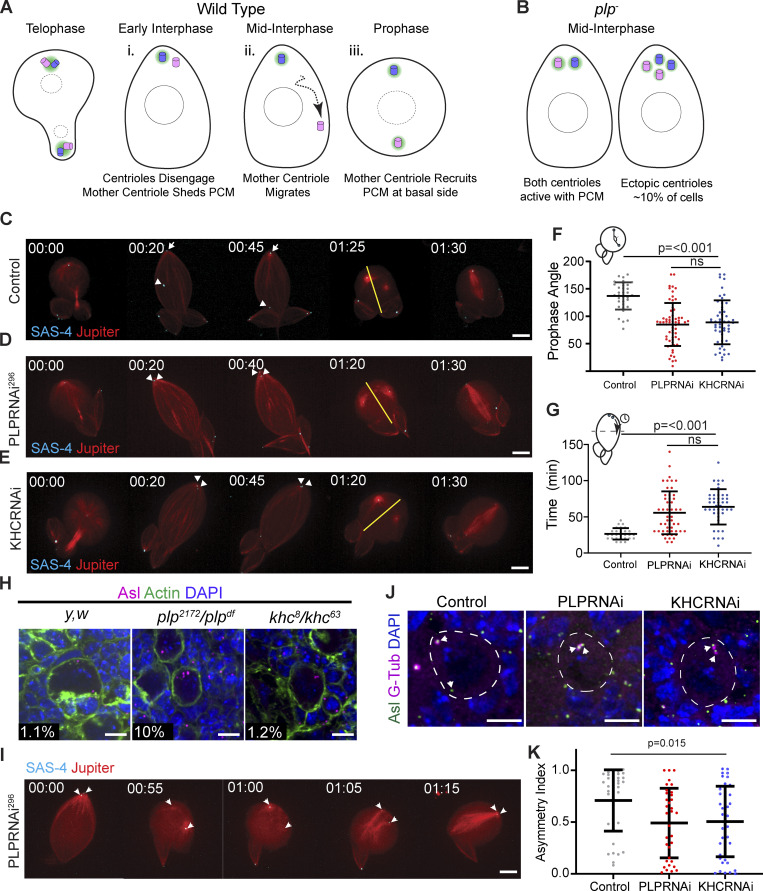
PLP is involved in three processes in NB asymmetric division: the timely migration of the mother centrosome away from the apical cortex (Fig. 8 A ii), maintaining the correct number of centrioles in NBs, and correct PCM asymmetry between the mother and daughter centrosomes (Fig. 8 B; Lerit and Rusan, 2013). While our transgenes that inhibit PLP–KHC interaction result in failed mother centriole migration (Fig. 8, G and H), the severe mutants (plpΔ740-971 and plpPD) formed PLP aggregates in NBs, which could interfere with centriole behavior (Fig. 7 H). We, therefore, augmented our analysis by performing a detailed comparison between PLP and KHC loss-of-function to gain additional insight into the role of PLP–KHC dependent centriole motility in asymmetric cell division.
Figure 8.
Comparison of PLP and KHC loss of function in NBs. (A i–iii) Schematic of centrosome asymmetry in a wild-type NB. (i) Mother centriole (pink) sheds PCM (green). (ii) Mother centriole migrates to the basal side. (iii) Mother centriole recruits PCM in the following prophase. (B) Schematic showing the main phenotypes in plp− mutants: Centrioles do not migrate away from the apical domain, mother centriole retains PCM, some NBs inherit supernumerary centrioles. (C–E) Live imaging of NBs expressing the centriole marker mNG::SAS-4 (cyan, arrows) and the MT marker Jupiter::mCherry (red). The apical daughter centriole is indicated in control NB (arrow), metaphase spindle axis is indicated by yellow line. Unlike controls (C), centrioles do not migrate away from the apical cortex following PLP (D) or KHC (E) knockdown. (F) The angle between the two centrosomes is significantly reduced at prophase following PLP or KHC knockdown (Control angle: 137 ± 25, n = 34. PLPRNAi angle: 84.98 ± 39, n = 62. KHC RNAi angle: 88.94 ± 40, n = 52; data = mean ± SD; ANOVA P = <0.0001, Tukey multiple comparison: P = <0.0001 between Control and RNAi conditions), ns = not significant. (G) The time taken for one centriole to cross the cell midline (equator) is significantly increased following PLP or KHC knockdown compared to controls (Control separation time: 26.43 ± 8, n = 28. PLPRNAi separation time: 55.65 ± 29.7, n = 54. KHCRNAi separation time: 63.84 ± 24.4, n = 43; data = mean ± SD; ANOVA P = <0.0001, Tukey multiple comparison: P = 0.001 between Control and RNAi conditions). (H) Representative NBs showing supernumerary centrioles are present in plp− (plp2171/df(3L) Brd15, 10%, n = 258 NBs, four brains) mutants but not in control (y,w, 1.1%, n = 229 NBs, four brains) or khc− (khc8/khc63, 1.15%, n = 195 NBs, four brains) mutants. Numbers represent percentage of cells carrying >2 centrioles as determined by Asl puncta (magenta) (I) Time series showing supernumerary centrioles following plp− loss of function arise from a failure of centrosomes to separate in prophase. (J) Fixed neuroblasts showing Gamma-Tubulin (magenta) associates with both centrioles (green) following PLP or KHC knockdown. (K) Quantification of the asymmetric index showing a significant reduction in gamma tubulin asymmetry in PLP or KHC knockdown (Control ASI: 0.7 ± 0.29, n = 37. PLPRNAi ASI: 0.49 ± 0.33, n = 34. KHCRNAi ASI: 0.5 ± 0.37, n = 46; ANOVA: P = <0.0001, Tukey pairwise comparison: P = <0.0001 between Control and all other conditions). Data = mean ± SD. Scale bars: 5 µm.
Firstly, we confirmed the rescue experiment results (Fig. 7, F and G) by comparing the centriole separation phenotype following knockdown of PLP or KHC by live imaging. Analyzing the time required for one of the two centrioles to cross the cells’ equator in interphase, as well as the angle between the two centrosomes at prophase revealed no significant difference between PLP and KHC knockdown, but a significant difference to controls (Fig. 8, C–G and Video 15). These results indicate that both PLP and KHC are important for mother centriole motility.
Video 15.
Centriole separation in neuroblasts. Spinning disk confocal timelapses showing maximum projection of isolated neuroblasts expressing SAS-4::Neon (cyan) and mCherry::Jupiter (red). Note centrioles (cyan) do not migrate away from the apical side of the cell following PLP or KHC knockdown. Apical is up. Time stamp: hh:mm. Scale bar: 5 µm.
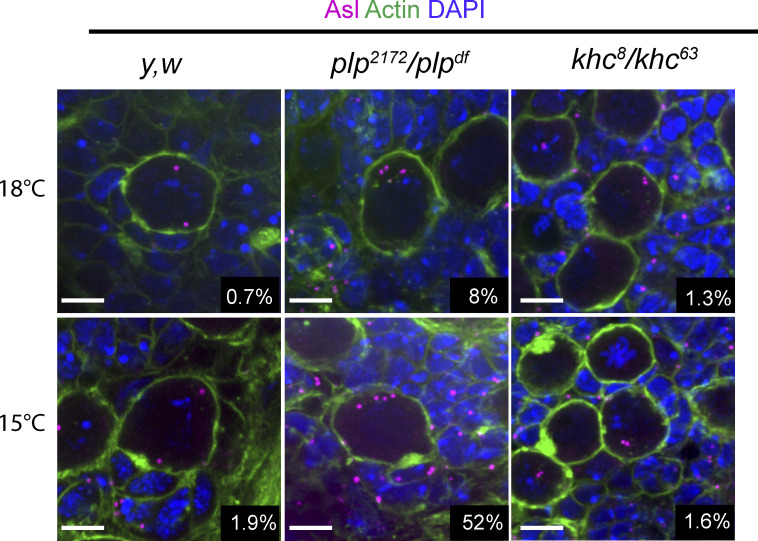
Secondly, we examined centriole number following PLP or KHC loss of function. We confirmed that plp mutants have supernumerary centrioles in 10% of NBs; however, khc mutants did not (Fig. 8 H). Importantly, even at low temperatures, which enhances the plp phenotype, khc NBs were comparable to controls in centriole number (Fig. S5). We concluded that the supernumerary centriole phenotype was due to a function of PLP beyond its role in interphase centriole motility. Indeed, live imaging suggests that the centriole number defect in plp mutants can arise from an inability to separate centrosomes in mitosis, resulting in both centrosomes being inherited by the NB after division (Fig. 8 I and Video 16). This is likely due to problems in the Kinesin-5/Dynein dependent centrosome separation pathway that is activated after nuclear envelope breakdown (Tanenbaum and Medema, 2010; Agircan et al., 2014). Loss of khc does not appear to affect the Kinesin-5/Dynein pathway in mitosis.
Figure S5.
Increased supernumerary centrioles at low temperature is only observed in plp mutant NBs. Fixed NBs stained for Asterless (Asl, magenta) to label centrioles, Phalloidin (green), and DAPI (blue). Centrioles were counted by counting the number of Asl positive puncta in each cell. Raising flies in lower temperatures only increased in the percentage of NBs carrying more than two Asl puncta in plp mutants (plp2172/plpdf). 18°C: Control = 0.7% ± 0.8 (n = 4 brains, 230 NBs), plp = 8.8% ± 3.6 (n = 4 brains, 330 NBs), khc = 1.3% ± 2.7 (n = 4 brains 235 NBs). 15°C: Control = 1.9% ± 1.5 (n = 4 brains, 205 NBs), plp = 52% ± 6.7 (n = 4 brains, 161 NBs), khc = 1.6% ± 2.3 (n = 4 brains, 197 NBs). Data = mean ± SD. Scale bars: 5 µm.
Video 16.
Supernumerary centrioles following PLP knockdown is a result of incomplete prophase centrosome separation. Spinning disk confocal timelapse showing maximum projection of an isolated neuroblast expressing UAS-PLPRNAi, SAS-4::Neon (Centrioles, cyan) and mCherry::Jupiter (MTs, red). As the cell enters mitosis, the two centrosomes collapse to the apical spindle pole resulting in the segregation of both into the NB following mitosis. This then leads to four in the following cell cycle. Time stamp: hh:mm. Scale bar: 5 µm.
Thirdly, we examined PCM asymmetry on NB centrosomes. If mother centriole motility away from the apical domain was important for centrosome asymmetry, then the knockdown of PLP or Kinesin-1 would perturb the asymmetric localization of PCM on centrioles. Indeed, the asymmetry index (ASI) in NBs expressing PLP or KHC RNAi was significantly reduced compared to controls (Fig. 8, J and K). In both cases, the reduction in asymmetry was due to increased levels of PCM on the mother centriole rather than loss of PCM from the daughter centriole, suggesting that PLP and KHC may be working together for centrosome asymmetry.
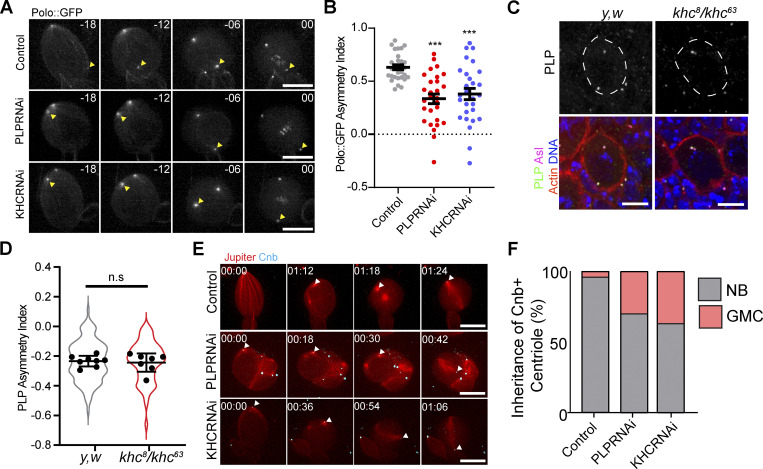
Previously, the PCM asymmetry defects observed in plp mutants were proposed to be due to PLP inhibiting the localization of the PCM regulator Polo kinase, which functions to promote PCM recruitment (Lerit and Rusan, 2013; Singh et al., 2014). Our finding that KHC knockdown results in defective PCM asymmetry suggest that Kinesin-1 is also involved in regulating Polo. Consistent with this, live cell imaging of endogenous Polo::GFP localization in isolated NBs revealed that, like PLP, KHC is necessary for Polo asymmetry in early prophase (Fig. 9, A and B; and Video 17). We hypothesized that KHC could regulate Polo localization by regulating PLP recruitment and asymmetric localization to the centrioles. However, in KHC mutants the asymmetric localization of PLP to the mother centriole was unaffected, demonstrating that Kinesin-1 does not regulate PLP levels, and therefore the normal distribution of PLP is not sufficient to prevent precocious PCM recruitment (Fig. 9, C and D). How might KHC regulate Polo? One possibility is that Kinesin-1 could directly promote the removal of Polo from the mother centriole. Alternatively, the Kinesin-1 dependent movement of the mother centriole away from the apical MT aster could prevent exposure to Polo activity and thereby prevent precocious PCM recruitment.
Figure 9.
Loss of KHC or PLP causes early Polo exposure and defective age dependent centriole segregation. (A) Polo::GFP is recruited early to the ganglion mother cell (GMC) inherited centrosome (yellow arrowhead) in isolated NBs expressing KHC or PoloRNAi. (B) Quantification of Polo asymmetry ∼18 min before metaphase (Control ASI: 0.63 ± 0.12, n = 29. PLPRNAi ASI: 0.33 ± 0.24 n = 28, KHCRNAi ASI: 0.37 ± 0.28 n = 28, ANOVA: P < 0.0001, Tukey pairwise comparison: Control vs PLPRNAi P < 0.0001, Control vs. KHCRNAi P = 0.0002). Error bars = SEM. (C) Example projections from whole mount brains showing that PLP localization is not perturbed in khc mutants (khc8/khc63). (D) Quantification of PLP asymmetry in khc mutant NBs (Control ASI: −0.23 ± 0.03 n = 112 cells, eight brains. khc ASI: −0.024 ± 0.06 n = 123 cells, seven brains. Unpaired, two tailed, t test P = 0.7). Violins show ASI from all cells measured; points show averaged ASI per brain imaged. Errors bars = SD. (E) Isolated NBs expressing Cnb::GFP (cyan, arrowhead) to label the daughter centriole. In NBs also expressing KHC or PLP RNAi, the daughter centriole is more frequently segregated into the GMC. (F) Quantification of Cnb::GFP + centriole inheritance (% Cnb + centriole inherited by NB: Ctrl: 96%, PLPRNAi: 74%, KHCRNAi: 63%, 40 NBs were imaged per condition). Scale bars: 10 µm.
Video 17.
Polo::GFP asymmetry is disrupted following PLP or KHC knockdown. Spinning disk confocal timelapse showing maximum projection of Polo::GFP in isolated neuroblasts. Movie pauses to show precocious activation of centrosomes following PLP (center) and KHC (right) knockdown relative to controls (left). Arrowheads indicate centrosome that gets segregated into the GMC. Time stamp: hh:mm. Scale bar: 5 µm.
NBs consistently retain the daughter centriole during asymmetric cell division. Since PCM asymmetry is perturbed in plp− and khc− NBs, we hypothesized that the age-dependent segregation of centrioles would be disrupted. To examine this, we performed live cell imaging of NBs expressing GFP tagged Centrobin (Cnb), a marker for the daughter centriole (Januschke et al., 2011, 2013). Observing the segregation of the Cnb + centriole revealed that in the absence of PLP or KHC the daughter centriole was more likely to be segregated to the GMC compared to controls (Fig. 9, E and F; and Video 18). We conclude that KHC and PLP are similarly important to generate correct PCM asymmetry which facilitates daughter centriole retention in NBs.
Video 18.
Age-dependent centrosome segregation is disrupted following PLP or KHC knockdown. Spinning disk confocal time lapse showing maximum projection of isolated neuroblasts expressing ubi-Cbn::GFP (daughter centriole, cyan, Arrowhead) and UAS-mCherry::Jupiter (MTs, red). Note that following PLP (center) or KHC (right) knockdown the Cbn + centriole ends up segregating to the basal side of the cell in mitosis rather than being retained by the NB at the apical side. Time stamp: hh:mm. Scale bar: 5 µm.
Discussion
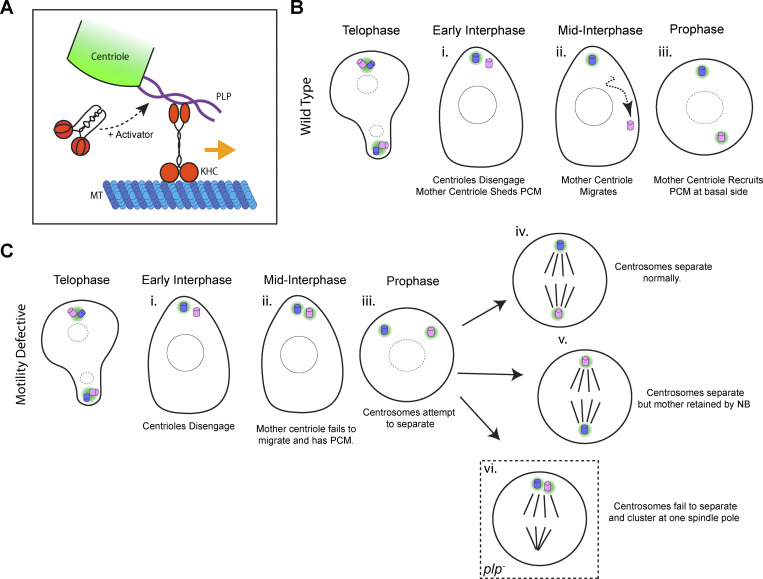
The movement of centrioles is critical for an array of cellular processes. Most of the work in this research area has focused on the separation of active centrosomes (not inactive centrioles), which move by indirect transport using motors anchored to the cortex or anchored to other MTs (sliding [Tanenbaum and Medema, 2010; Agircan et al., 2014]). In our work, we have leveraged Drosophila to understand how centrioles can move when they are not functioning as a MTOC. In multiple cell types, we show that centrioles move on interphase MTs as cargo, independently of MT dynamics (Fig. 3). Several of our results support a model whereby centrioles are cargo for the motor protein Kinesin-1 (Fig. 10 A). Firstly, the knockdown of Kinesin-1 components inhibits motility, independent of KHC’s role in MT sliding (Fig. 4). Secondly, KHC localizes to the outer centriole. Thirdly, KHC interacts directly with PLP, and their interaction is necessary for centriole motility (Figs. 5, 6, and 7). Interestingly, PLP is not necessary for the localization of KHC to the centriole, suggesting that another unknown centriolar protein is involved in recruiting KHC.
Figure 10.
Model. (A) Diagram showing proposed complex assembled on the outer centriole for transport. Kinesin-1 interacts with the central region of PLP (aa584-1811). A yet-to-be-identified activator relieves autoinhibition of the Kinesin heavy chain, thereby enhancing MT landing rate as well as PLP interaction. (B) Diagram illustrating the centriole cycle of asymmetrically dividing neuroblasts (Fig. 8 A). (C i–vi) Diagram showing the consequence of defective centriole motility on the centriole cycle. (i) In early interphase, the centrioles disengage but remain at the apical side of the cell (Fig. 8, C–E). (ii) In mid interphase, both centrioles remain at the apical side of the cell, and the mother centriole (normally inactive) recruits PCM (Fig. 8 J and Fig. 9 A). (iii) The consequence of this is that at prophase, both centrosomes are activated at the apical side of the cell. They attempt to separate via the prophase centrosome separation pathway (Fig. 8, C–E). (iv) In some cells, the prophase centrosome separation and spindle alignment machinery is able to rescue the defective motility, resulting in a normal division with the neuroblast retaining the daughter centrosome. (v) In ∼30% of cells, the prophase centrosome separation pathway is able to rescue the centrosome separation; however, the NB will retain the mother centrosome (Fig. 9, E and F). (vi) In plp− neuroblast, some cells fail to separate the two centrosomes by the prophase centrosome separation pathway. In this case, both centrosomes cluster at one spindle pole (Fig. 8 I); this will result in supernumerary centrioles in the next cell cycle (Fig. 8 H).
A critical step in Kinesin-1–dependent transport is the relief of Kinesin-1 autoinhibition (Verhey and Hammond, 2009). Although PLP584-1811 was able to co-migrate with KHC on MTs in vitro, it alone was not sufficient to enhance the basal activity of the Kinesin-1 motor in vitro (Fig. 6). Interestingly, our data show that Kinesin activation is important for PLP interaction. Therefore, it is likely that an unknown component is required to promote PLP–KHC interaction by activating the motor (Fig. 6, H and J; and Fig. 10 A). It is not uncommon that Kinesin-1 requires multiple interactors for full activation. One example of this is the scaffolding protein JNK-interacting protein 1 (Jip-1), which is insufficient alone for Kinesin activation but instead co-operates with secondary factors for activation (Blasius et al., 2007; Hammond et al., 2008; Sun et al., 2011).
The complexity of motor activation is likely a mechanism by which cells can control the directionality of intracellular transport in cases where a single cargo can bind multiple types of motors (Fu and Holzbaur, 2014). For example, many adaptor molecules can interact with both plus-end-directed Kinesin-1 and minus-end-directed dynein, which interestingly has been implicated in the movement of centrioles into the oocyte during Drosophila oogenesis (Grieder et al., 2000; Bolvar et al., 2001). Moreover, in mammalian cells, Dynein intermediate chain interacts with the PLP ortholog Pericentrin (Purohit et al., 1999; Young et al., 2000; Sepulveda et al., 2018). Given that we were searching for a plus-end directed motor for centriole transport in NBs, we have not investigated Dynein in this study. However, the potential for PLP–Dynein interaction is an interesting avenue for future study.
We propose that PLP and Kinesin-1 function together for centriole separation in NBs. During asymmetric cell division of NBs, the mother centriole sheds PCM following cytokinesis and migrates away from the daughter, which recruits PCM and becomes an MTOC (Fig. 10 B). We show that the motile mother centriole is associated with the MT network (Fig. 3 B) and appears to move along it directionally (Fig. 3 D). Kinesin-1 and PLP directly interact (Fig. 5) and their interaction is required for the timely separation of centrioles prior to prophase (Lerit and Rusan, 2013; Gallaud et al., 2014; Fig. 7, F and G).
Previously, it was reported that delayed centriole separation resulted in supernumerary centrioles (Lerit and Rusan, 2013). However, we found here that supernumerary centrioles are specific to PLP loss of function as centrioles did not accumulate in khc mutant NBs (Fig. 8 H). This suggests that the supernumerary centrioles in plp− mutants are not entirely caused by defective interphase centriole motility. Instead, our live imaging shows that in plp− mutants, centrosomes sometimes fail to push each other apart in mitosis and therefore cluster at one spindle pole (Fig. 8 I), likely due to the role that PLP has in organizing mitotic PCM and MTs (Galletta et al., 2014; Lerit et al., 2015; Varadarajan and Rusan, 2018).
The motility of the mother centriole away from the daughter is critical to maintain the PCM asymmetry. Normally through interphase, PCM is restricted to the daughter centriole by the asymmetric localization of the PCM regulator Polo (Rebollo et al., 2007; Rusan and Peifer, 2007; Conduit and Raff, 2010; Lerit and Rusan, 2013; Singh et al., 2014). Previously it was proposed that the asymmetric localization of PLP to the centriole drove centrosome asymmetry by inhibiting Polo on the mother centriole. However, KHC loss of function causes defective PCM asymmetry, without changing PLP localization (Fig. 9, C and D). Two potential models could explain why PCM asymmetry is defective. We support a model in which when the mother is unable to migrate (khc, plp), it maintains its position at the apical pole, and is exposed to higher levels of Polo. Normally, Polo localizes to both the daughter centriole and the MT network, where it moves toward the apical MTOC (Ramdas Nair et al., 2016). We show that in the absence of centriole motility, Polo::GFP is present on both mother and daughter centrioles earlier in the prophase than normal (Fig. 9, A and B).
A second possibility is that PLP and Kinesin-1 function separately to antagonize Polo. Given the minus end-directed transport of Polo along the MTs (Ramdas Nair et al., 2016), Kinesin-1 may oppose this independently of centriole transport. Indeed, work in C.elegans showed that Kinesin-1 was important to prevent the precocious activation of the sperm centriole during female Meiosis (McNally et al., 2012). Kinesin-1 is also important for centrosome clustering in the Drosophila oocyte, a phenotype that could be mimicked by the artificial activation of Polo at the centrosome (Loh et al., 2022 Preprint).
Finally, the interphase migration of the mother centriole away from the daughter is critical for the NB to consistently retain the daughter centriole. We have observed that failed centriole separation in interphase leads to failed centrosome age-dependent segregation (Fig. 9, E and F). Although age-dependent segregation is a conserved phenomenon (Chen and Yamashita, 2021), it is unclear why the age asymmetry of centrosomes is so tightly regulated. Reports have shown that fate determinants (Ramat et al., 2017; Tozer et al., 2017), damaged proteins (Fuentealba et al., 2008), and foreign DNA (Wang et al., 2016) are all associated with centrosomes of a determined age. However, without an experimental system to trigger inverse age segregation in every cell cycle it will be difficult to determine the true functionality of age-dependent centriole segregation.
In human cells, centrosomes have been observed to be motile at the end of mitosis, when the centrosomes reorient themselves towards the midbody (Piel et al., 2001). Interestingly, recent work has demonstrated that this centrosome motility involves the Rab11-dependent localization of PCNT to the centrosomes (Krishnan et al., 2022). It is not known whether Kinesin-1 is involved in midbody-directed movement, but Kinesin does localize to centrosomes (Neighbors et al., 1988) and interacts with PCNT (Fig. S3). It is therefore likely that Kinesin-1–mediated centriole motility is a conserved process.
In summary, we have dissected a novel mechanism for centriolar transport where PLP interacts with Kinesin-1 to facilitate direct MT transport of centrioles. Importantly, we have shown that this process is important for centriole separation in asymmetric cell division. Future studies will be required to uncover how Kinesin-1 is activated at the centriole to ensure proper centriole motility.
Materials and methods
Plasmids and molecular cloning
Sequences were amplified from cDNA clones by PCR and inserted into the pEntr gateway vector using the pEntr D-Topo kit (Life Technologies). Gateway reactions were used to recombine the cDNA into the destination vectors. ANW: Actin promoter with N-term mNeon tag. PAT20RW: Actin promoter with N-term TOM20 mitochondrial localization domain and TagRFP. PPWG: UAS promoter with C-term GFP. pDEST-pGADT7: Y2H bait plasmid, GAL4 binding domain. pDEST-pGBKT7: Y2H prey plasmid with GAL4 activation domain. PCNT and KIF5B fragments were generated using DNA synthesis (Twist Biosciences). The following published plasmids were used: pEntr-PLP1-583, pEntr-PLP584-1375, pEntr-PLP1376-1810, pEntr-PLP1811-2537, pEntr-PLP2538-2895, pEntr-PLP583-1810, and pEntr-Sas6 (Galletta et al., 2016); pEntr-PLP (Galletta et al., 2014); and F-Tractin::mCherry (Liu et al., 2021). KHC fragments were amplified with the following primers: KHC1-341 Fwd: 5′-CACCATGTCCGCGGAACGAGAGATTCC-3′, Rev: 3′-CTCGTTAACGCAGACCACGTTCTTCACTGTCTTG-5′; KHC342-600 Fwd: 5′-CACCATGGAGCTTACTGCCGAGGAATGGAAG-3′, Rev: 3′-AGCCAGAGCACTCATCTTAAGGTCGATGCTGG-5′; KHC601-849 Fwd: 5′-CACCATGGGCACGGATGCCAGCAG-3′, Rev: 3′-CGCGAGTGATCCACCGTCCTCCTC-5′; KHC850-975 Fwd: 5′-CACCATGCAGAAACAGAAGATTTCCTTCTTGGAGAACAACC-3′, Rev: 3′-CGAGTTGACAGGATTAACCTGGGCCAGC-5′.
Fly stocks
Flies were maintained on Bloomington Recipe Fly Food (LabExpress). Crosses were performed at 25°C unless otherwise stated in the text. Transgenic Flies were generated using standard P-element transformation methods (Bestgene, Inc.). The following stocks were used: ubi-gfp::sas6 (this study), sas4::neon (Galletta et al., 2020), UAS-lifeact::rfp (#58362; BDSC), UAS-jupiter::mcherry (gift from C. Cabernard, University of Washington, Seattle, WA), worniu-GAL4 (#56553; BDSC), AGIR-GAL4 (#6773; BDSC), tubulin-GAL4 (#5138; BDSC), ubi-tubulin::gfp (gift from T. Avidor-Reiss, University of Toledo, Toledo, OH), ubi-sas6::mcherry (Rogers et al., 2008), ubi-cbn::gfp (Lerit and Rusan, 2013), polo::gfp (Ramdas Nair et al., 2016; gift from C. Cabernard, University of Washington, Seattle, WA), UAS-plpRNAi296 (#101296; VDRC, [Singh et al., 2014]), UAS-plpRNAi645 (#101645; VDRC, [Dietzl et al., 2007]), UAS-khcRNAi (#44338; VDRC, [Gallaud et al., 2014]), UAS-KhcRNAi (#35770; BDSC, [Kelliher et al., 2018]), UAS-klcRNAi (#42597; BDSC, [Lu et al., 2018]), UAS-ensRNAi (#40825; BDSC, [Perkins et al., 2015]), UAS-mcherryRNAi (#35785; BDSC, [Ito et al., 2019]), UAS-luciferaseRNAi (#31603; BDSC, [Pareek and Pallanck, 2020]), UAS-eb1::gfp (Swider et al., 2019), UAS-plp::gfp, UAS-plpPD::gfp (this study), UAS-plpΔ741-970::gfp (this study), plp2172 (#12089; BDSC), Df(3L)Brd15 (#5354; BDSC), khc8 (#1607; BDSC), khc63 (Djagaeva et al., 2012; gift from B. Saxton, University of California Santa Cruz, Santa Cruz, CA), khcmut.A (79036; BDSC), and ubi-mNG::KHC (this study).
Immunofluorescence and fixed cell microscopy
Larvae were selected at the third instar stage and dissected in PBS before being fixed in 4% formaldehyde at room temperature for 20 min. Fixed samples were washed and then blocked in PBS + 0.5% Triton X-100 (PBST) + 5% normal goat serum. Samples were then incubated in PBST + primary antibody overnight at 4°C. S2 cells were cultured in serum-free SF900 II media and plated onto coverslips precoated with Concanalavin-A. After being allowed to adhere for 30 min, SF900 media was removed and replaced with 4% formaldehyde in PBS for 20 min. Staining was then performed as with tissue, but with 0.1% Triton X-100 rather than 0.5%. The following antibodies were used: anti-Cnn (Galletta et al., 2016; rabbit, 1:10,000), anti-Asterless (Klebba et al., 2013; guinea pig, 1:30,000), anti-PLP (Rogers et al., 2008; rabbit, 1:12,000), anti-gamma-tubulin (mouse, 1:500, GTU-88; Sigma-Aldrich), and anti-tubulin (mouse, 1:200; Sigma-Aldrich). Samples were washed in PBST and then incubated in secondary antibody (1:500; Life Technologies) for 2 h at room temperature. Following secondary antibody incubation, samples were washed in PBS before being mounted in a small drop of vectashield within a reinforcement label used as a spacer. All slides were mounted underneath a #1.5 coverslip. Phalloidin-Alexa488 (5:200; Life Technologies) was added for 20 min after the secondary antibody step.
Standard resolution imaging was performed using a Yokogawa Csu-W-1 spinning disk mounted on a Nikon Ti-2 Eclipse equipped with a Prime BSI CMOS camera (Photometrics) and a 100× Silicone immersion objective (N.A 1.4; Nikon). The microscope was controlled using Elements (Nikon).
Structured illumination microscopy was performed using an OMX4 (GE Healthcare) using immersion oil RI 1.516. Images were reconstructed using SoftWoRx (GE Healthcare). Stimulated emission depletion (STED) images were acquired using a Leica SP8 3X STED microscope, a white-light laser for fluorescence excitation (470–670 nm), a Leica HyD SMD time-gated photomultiplier tube, and a Leica 100× (NA 1.4) STED White objective (Leica Microsystems, Inc.). ATTO 647 and Alexa 594 were excited at 647 and 575 nm with fluorescence emission collected over a bandwidth of 658–755 and 583–700 nm, respectively. A 25-slice z-stack for both colors was acquired with a pinhole size of 0.7 airy unit (A.U.), a scan speed of 600 Hz, a pixel format of 1,024 × 1,024 (pixel size, 20 nm), an interslice distance of 0.16 μm, four line averages, and time gating on the HyD SMD set to a range of 0.7–6.5 ns. HyD SMD gains were set to 100 and 150% for ATTO 647 and Alexa Fluor 594, respectively. STED depletion was accomplished for both labels at 775 nm (pulsed at 80 MHz) at powers of ∼192 mW at the back aperture for ATTO 647-labeled MTs (40% full laser power) and 99 mW for Alexa Fluor 594-labeled centrioles (20% full laser power). STED images were deconvolved using the software Huygens Professional (v.19.1; Scientific Volume Imaging) using an idealized point spread functions and the classic maximum-likelihood estimation deconvolution algorithm.
Sample preparation and live cell microscopy
Wing discs were dissected from third instar larvae in Schneider’s medium supplemented with glucose (1g/l). They were mounted in a drop of the same media on a 50-mm gas permeable dish (Lumox). A drop of halocarbon oil (Sigma-Adrich) was added around the drop of media, and a 22 × 22-mm coverslip (#1.5; Thermo Fisher Scientific) was gently lowered on top. Centriole movement was imaged by mounting the wing discs with the peripodial membrane facing the coverslip.
For live imaging of neuroblasts, brains were dissected in collagenase buffer and incubated in collagenase for 20 min. Brains were then washed in Schneider’s medium supplemented with FCS, fly extract (DGRC), and insulin (Sigma-Aldrich). They were then dissociated as previously described (Pampalona et al., 2015) and plated onto a 35-mm glass bottom dish (Fluorodish, World Precision Instruments) precoated with Poly-L-Lysine (Sigma-Aldrich). Cells were allowed to adhere for 45 min prior to imaging.
S2 cells were plated directly onto 35-mm Glass bottom dishes coated with Concanalavin-A and allowed to adhere for 30 min. Cells were then washed with Schneider’s medium to remove SF900 II.
Imaging was performed at room temperature using a Nikon 100× oil objective (n.a 1.49), on an inverted microscope (Eclipse Ti, Nikon) fitted with a Csu-22 spinning disk confocal head (Yokogawa), and a sCMOS camera (Orca Flash 4, Hamamatsu). The acquisition was set up through Metamorph software (Molecular Devices), unless otherwise stated in the figure legend. Wing disc movies were collected by taking images of a single z slice at 2-s time intervals for 10 min. The focus was adjusted manually during acquisition when necessary. Neuroblast movies were acquired by taking a 10–12 μm volume every 180 s using 800 nm z-intervals. S2 cell movies were performed by acquiring a 2.4 μm volume every 2 s using 800-nm intervals.
Particle tracking
Movies were processed by subtracting the background and then applying a 1 px Gaussian filter. There is very little movement of the peripodial cells during the time frame of our movies (10 min, Fig. 1 C), so we did not correct for cell motility. However, visual inspection of a time projection was used before tracking to remove any movies in which the entire tissues were drifting through the movie. This is generally due to technical issues from sample mounting, not true tissue movement. Tracking was performed using a modified program in IDL (Harris Geospatial) based on a previous particle-tracking pipeline (Crocker and Grier, 1996) which can be found here for IDL and other implementations (http://www.physics.emory.edu/faculty/weeks//idl/).
Drug treatments
To depolymerize MTs, wing discs were dissected and placed in Schneider’s medium in an ice/ethanol slurry for 1 h. Wing discs were then transferred to room temperature media and allowed to recover in the presence of DMSO or 50 µM Colcemid (Tocris Bioscience). To block MT polymerization, wing discs were incubated in 200 µM Colchicine at room temperature for 20 min before imaging. To block actin polymerization, wing discs were incubated in 10 µM Latrunculin-A (Sigma-Aldrich) for 20 min before imaging. For drug experiments imaging was performed in the presence of the inhibitor.
Cell extract preparation
Cell lysate-based motility assays were performed as described previously (Ayloo and Holzbaur, 2015). Briefly, Drosophila S2 cells were transfected with mNeonGreen-tagged Kinesin-1 full-length heavy chain (KHC) and Halo-tagged PLP584-1811 (Effectene, Qiagen). The lysates were harvested for TIRF motility assays at ∼48 h post-transfection. Before extraction, HaloTag was labeled with tetramethylrhodamine (TMR) by incubating the transfected cells with 2.5 mM HaloLigand-TMR (Promega) for 15 min. Any unbound ligand was washed out with PBS, and the cells were lysed in buffer containing 120 mM NaCl, 0.1% Triton, 1 mM ATP, 1 mM EGTA, and 40 mM HEPES (pH 7.4) supplemented with protease inhibitor cocktail (Roche). The cell extract was centrifuged at 100,000 g for 10 min at 4°C and the supernatant was kept on ice until use.
TIRF motility assays
Unlabeled (T240), HiLyte-647 (TL670M), and Biotin (T333P)-labeled porcine tubulins were purchased from Cytoskeleton. Tubulins were resuspended according to the manufacturer’s instructions and mixed at a ratio of 50:1 (Unlabeled: Biotin) or 50:1:1 (Unlabeled: Biotin:Hilyte-647), with a total tubulin concentration of 10 mg/ml, in ice-cold BRB80 (80 mM K-PIPES, 2 mM MgCl2, 1 mM EGTA [pH 6.8 with KOH]) with 1 mM GTP. Polymerization was performed by adding warmed (37°C) BRB80 + 1 mM GTP + 10% glycerol and incubating at 37°C for 30 min. For stabilization, Taxol was added stepwise (0.5, 5, and 50 µM after addition) with 10 min incubation at 37°C for each step.
Then, 10-μl flow chambers with biotin-PEG functionalized coverslips were constructed as described previously (Tripathi et al., 2021). Chambers were washed with BRB80 (3 × 10 µl) followed by 5 mg/ml BSA + 5 mg/ml casein in BRB80 (3 × 10 µl). The final chamber volume was incubated in the chamber for 1 min. Then 2 mg/ml Neutravidin (1 × 10 µl) in BRB80 was incubated in the chamber for 1 min and then washed with BRB80 + 20 µM Taxol (3 × 10 µl); 0.5 µM MTs in BRB80 + 20 µM Taxol + 50 mM DTT were added and incubated for 1–3 min to allow for sufficient surface attachment. The chamber was washed with BRB80 + 20 µM Taxol + 50 mM DTT (3 × 10 µl). KHC, PLP584-1811, or KHC + PLP584-1811 cell lysates were diluted into assay buffer (BRB80 + 20 µM Taxol, 50 mM DTT, 1 mM ATP, 100 μg/ml glucose oxidase, 40 μg/ml catalase, 2.5 mg/ml glucose) and flowed into the chamber for imaging (3 × 10 µl). Optimal dilutions and mixing ratios were determined empirically based on the quantity of motility observed.
Movies were collected on an inverted Nikon Eclipse Ti-E microscope with H-TIRF module attachment, CFI60 Apochromat TIRF 100× oil Iimmersion objective lens (N.A. 1.49, W.D. 0.12 mm, F.O.V 22 mm), and an EM-CCD camera (Andor iXon Ultra 888 EMCCD, 1,024 × 1,024 array, 13 μm pixel). For Video 12, a maximum intensity projection of the KHC channel was performed to highlight the position of the MT track (depicted in blue). Particles were tracked using the FIJI plugin Trackmate (Tinevez et al., 2017), and plotting/analysis was performed in Graphpad Prism. Pauses between movements were included in this analysis. Data from three separate movies were combined to produce each of the KHC and PLP584-1811 datasets shown in Fig. 5. All quantifications relate to the mixture of both proteins.
Analysis of neuroblast phenotypes
ImageJ was used for the analysis of all neuroblast phenotypes. The asymmetry index between the mother and the daughter centriole was performed according to published methods (Lerit and Rusan, 2013). The index is defined by the equation (A − B)/(A + B). A is equal to the mean gray value of the more apical centrosome. B is equal to the mean gray value of the more basal centrosome. A positive ASI indicates a higher signal on the apical centrosome. Negative ASI indicates a higher signal on the basal centrosome. The time taken to cross the cell midline shown in Fig. 8 G was determined by following the centriole until it had migrated beyond the halfway point of the cell and determining the time from the cytokinesis of the previous cell cycle. The prophase angle between the two centrosomes measured in Fig. 7 G and Fig. 8 F was determined by measuring the angle between the MTOC relative to the nucleus. The more apical MTOC was taken as 0°.
Protein expression and purification
cDNAs encoding for KHC or PLP584-1811 were inserted into a modified pFastBac1 vector, which expresses a fusion FLAG-tag for purification. All KHC constructs contain an N-terminal GFP tag while PLP584-1811 has an N-terminal HaloTag. Transposition and the generation of recombinant baculovirus were performed following manufacturer’s protocols (Thermo Fisher Scientific). For producing Kinesin-1 HC-LCs complex, Sf9 insect cells were co-infected with recombinant baculovirus encoding for GFP::KHC and Kinesin Light Chain (KLC), and the complex was purified via the FLAG tag on KHC.
To purify Kinesin-1, infected Sf9 cells were harvested and homogenized in extraction buffer containing 300 mM NaCl, 4 mM MgCl2, 1 mM EGTA, 1 mM ATP, and 10 mM MOPS (pH 7.2) with protease inhibitor cocktail (Roche). The lysate was centrifuged at 48,000 g for 30 min and the clarified supernatant was allowed to incubate with FLAG-resins for 2 h at 4°C. Bound proteins were washed with buffer containing 10 mM MOPS, 300 mM NaCl, 1 mM ATP, and 0.1 mM EGTA (pH 7.2) for three times. The protein was eluted by adding 300 μg/ml FLAG peptide (GenScript). Eluted proteins were dialyzed overnight against the buffer containing 10 mM MOPS, 300 mM NaCl, 0.1 mM EGTA, 2 mM MgCl2, and 1 mM DTT (pH 7.2). Kinesins were further concentrated by low-speed centrifugation (4,000 g, 15 min) with Amicon filter units (Millipore Sigma) and flash-frozen with liquid nitrogen for future use. PLP584-1811 was purified with the same method except that the buffers did not contain ATP.
Single-molecule mass photometry
Mass photometry assays were performed as previously described (Liu et al., 2021) using the Refeyn OneMP mass photometer. Briefly, microscope coverslips (#1.5; Thermo Fisher Scientific) were cleaned and assembled into simple flow chambers. 20 nM of PLP584-1811 protein was flowed into the chamber in buffer containing 150 mM NaCl, 10 mM MOPS (pH 7.2). Images were processed using manufacturer supplied software (Refeyn DiscoverMP). The conversion between molecular mass and interferometric contrast was calibrated with protein standards of known molecular weight.
Biolayer interferometry
Binding analyses between Kinesin-1 and PLP584-1811 were performed with Octet RED96 (ForteBio, Pall Corporation). Streptavidin (SA) dip-and-read biosensors (ForteBio) were used for all interaction studies. A 20 µg/ml biotinylated GFP antibody (600-406-215; Rockland) was first coated to the biosensor surfaces via biotin-streptavidin interactions. Then, 10 µg/ml GFP-tagged Kinesin proteins were adhered in a “tail-up” orientation by binding to the surface-bound antibodies with its N-terminal GFP moiety. Finally, 0–1,000 μM PLP584-1811 were incubated with immobilized Kinesin for 5 min followed by 10 min dissociations. All interactions were carried out in buffers containing 150 mM NaCl, 2 mM MgCl2, 0.1 mM EGTA, 10 mM MOPS, 1% BSA (pH 7.2). The experimental temperature is 30°C.
Steady-state ATPase assay
Steady-state ATPase activities were measured in SpectraMax iD3 microplate reader (Molecular Devices) at 37°C in buffers containing 80 mM PIPES, 10 µM Taxol, 2 mM MgCl2, and 2 mM ATP. The buffer also contained an NADH-coupled, ATP regenerating system including 40 U/ml lactate dehydrogenase, 200 U/ml pyruvate kinase, 200 µM NADH, and 1 mM phosphoenolpyruvate. Then 20 nM Kinesin-1 and 2 μM PLP584-1811 were used as indicated. The rate of ATP hydrolysis was measured from the decrease in absorbance at 340 nm caused by the oxidation of NADH.
Landing rate quantification
A static image of surface adhered biotinylated MTs was first captured to allow MT length measurement. A movie of 25 nM Kinesin landing on the MTs was acquired in the same region at five frames per second. To quantify the landing rates, a FIJI macro based on the Ridge Detector plugin was used to measure the total length of MTs in the field. The FIJI Trackmate plugin was used to determine the total number of landing events during the movie (track filters = start > 0.2 s, minimum spots in tracks = 3, track displacement >3 pixels). Frame 1 was excluded from the calculation of total movie length (since tracks present in frame 1 are excluded) as were the two final frames (landing here creates a track with too few time-points to be included). The number of events was divided by total MT length and adjusted movie length to give the landing rate in events μm-1 min-1. For each condition, a minimum of three movies was quantified to produce the mean and SD shown in Fig. 6.
Yeast-2-hybrid screening
Yeast-2-hybrid experiments were performed using a previously described modified version of the Matchmaker Gold system (Galletta and Rusan, 2015; Galletta et al., 2016). Constructs listed were recombined into bait and prey plasmids through gateway recombination. Bait plasmids were transformed into the Y2H Gold strain and Prey plasmids into Y187. Strains were individually mated in 2× Yeast extract Peptone Adenine dextrose. Plates were incubated at 30°C before being plated on DDO (SD-Leu-Trp) plates to select diploids carrying both prey and bait plasmids. Colonies were then replica plated onto DDO, QDO (Sd-Ade-His-Trp), and DDOXA (DDO + Auereobasidin A + X − α − Gal) for selection. Plates were scored based on the presence or absence of robust blue yeast growth on DDOXA plates.
Identification of PLP–KHC interaction mutations
Three fragments of PLP (PLP584-1376, PLP1377-1811, and PLP2539-2895) were found to interact with KHC. To identify mutants that disrupted these interactions, we first used a method previously described (Galletta et al., 2016). To induce random mutagenesis, PLP fragments were amplified by low fidelity PCR, a result of limiting dATP concentration in the PCR reaction (0.06 mM dATP, 0.25 mM dCTP, dGTP, and dTTP). This will induce a mutation approximately every 250bp. Primers used: Fwd: 5′-CGGAATTAGCTTGGCTGC-3′, Rev: 5′-TAATACGACTCACTATAGGGCG-3′. Amplification products were co-transformed into Y2H Gold with linearized pGBKT7. Approx. 2,000 clones were isolated of each mutagenesis reaction and arrayed in 96-well plates before being mated with the corresponding KHC fragment (in pGADT7, Y187 strain). The Array was then screened on plates for loss interaction (no growth on QDOXA and DDOXA plates). Clones that did not interact were isolated from the parent array and retested against previously identified interactors of PLP (Galletta et al., 2016). Using this approach, we were successful in identifying a PLP1377-1811 clone that retained interaction with most other identified interactors, but not KHC or Asl (Fig. S4 A). Sequencing revealed this clone carried mutations resulting in two substitutions (L1663P and G1699D). We referred to this mutant as PLPPD. The yeast mutagenesis approach did not yield useable mutations in either PLP584-1376 or PLP2539-2895.
The next approach was to screen a series of deletions within these two fragments for loss of KHC interaction. Previously a series of deletions had been generated within PLP584-1376 (Lerit et al., 2015). We found that a deletion corresponding to amino acids 741–970 inhibited interaction with KHC and the C-terminus of PLP without affecting other known interactors (Fig. S4 B). We were unable to identify any deletions within PLP2539-2895 that inhibited Kinesin interaction.
Insertion of the PlpΔ741-970 into the full-length PLP transgene was performed by amplifying the region downstream and upstream of the deletion and then assembling them using Gibson assembly into the pEntr plasmid. Primers used were as follows: F1 Fwd 5′-GCGGGAAAAACACATTCCTCCCTACCTCCA-3′, F1 Rev 3′-CAGAGTTTTAGACTCATCCAAGGAGAGGGA-5′, F2 Fwd 5′-TCCCTCTCCTTGGATGAGTCTAAAACTCTG-3′, F2 Rev 3′-TGGAGGTAGGGAGGAATGTGTTTTTCCCGC-5′.
Insertion of PLPPD into the full-length PLP transgene was performed by Gibson assembly of two fragments using the following primers: F1, Fwd 5′-GGTTGCGTGGAGCTTCAACATGAGC-3′, Rev, 3′-GATGCATTTCCCGCATGCTCTTGAAGATC-5′. F2, Fwd 5′-GCGGGAAAAACACATTCCTCCCTACCTCCAGATCTTCAAGAGCATGCGGGAAATGCATCA-3′, Rev, 3′-AGTCTGTTCCTCCATACGACCCTGCAGCGTATCCCGCTCATGTTGAAGCTCCACGCAACC-5′.
The cloning of PLPΔ2539-2895 was previously described (Lerit et al., 2015). PLPΔ2539-2895 was amplified using the following primers: 5′-CACCATGGCCATTAATATTGCTTTATTTACG-3′ and 5′-TTCATTGAAGTGTTCCAACTCTGTTTCGGC-3′ then inserted into pENTR by directional cloning
Statistical analysis
Statistical analysis and plots were generated in Graph pad Prism 9. To determine statistical significance, data were analyzed with either two-tailed t tests or one-way ANOVA followed by Tukey’s test for multiple comparisons unless otherwise specified in the figure legend. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows centriole motility is independent of the Actin network and centrioles associated with the interphase MT cytoskeleton. Fig. S2 shows that PLP knockdown does not cause precocious centriole activation in PCs. Fig. S3 shows PLP–Kinesin-1 interaction is conserved. Fig. S4 shows disrupting the PLP–Kinesin-1 interaction. Fig. S5 shows that increased supernumerary centrioles at low temperature are only observed in plp mutant NBs. Video 1 shows centriole motility in Drosophila neuroblasts. Video 2 shows that centrioles are motile in interphase S2 cells. Video 3 shows that centrioles are motile in interphase peripodial cells. Video 4 shows that centriole movement is independent of the actin cytoskeleton. Video 5 shows thatcentriole movement is dependent upon the MT network but not MT dynamics. Video 6 shows that centrioles move on the MT network in peripodial cells. Video 7 shows that centrioles move on the MT network in neuroblasts. Video 8 shows that centrioles switch between MTs at junctions. Video 9 shows that centriole movement requires Kinesin-1. Video 10 shows that centriole movement is dependent upon PLP. Video 11 shows that KHC and PLP584-1811 comigrate on MTs in vitro. Video 12 shows PLP motility. Video 13 shows that PLP584-1811 does not enhance KHC-landing rate. Video 14 shows that PLP–KHC interaction mutants fail to rescue centriole motility. Video 15 shows centriole separation in neuroblasts. Video 16 shows that supernumerary centrioles following PLP knockdown are a result of incomplete prophase centrosome separation. Video 17 shows that Polo::GFP asymmetry is disrupted following PLP or KHC knockdown. Video 18 shows that age-dependent centrosome segregation is disrupted following PLP or KHC knockdown.
Acknowledgments
We would like to thank Xufeng Wu and the National Heart, Lung and Blood Institute light microscopy core facility for technical assistance. We would like to thank all members of the Rusan lab for discussion. We thank Ryan O’Neill, Chaitali Khan, Alexander Kelly, and Greg Rogers for critically reading the manuscript. We would like to thank Sam Smith for discussions about the single molecule analysis of PLP. Finally, we thank Clemens Cabernard, Bill Saxton, Jill WiIdonger, and Tomer Avidor-Reiss for kindly sharing reagents.
This work is supported by the Division of Intramural Research at the National Institutes of Health/National Heart, Lung, and Blood Institute (1ZIAHL006126 to N.M. Rusan and ZIAACTHL006049 to J.R. Sellers)
The authors declare no competing financial interests.
Author contributions: M.R. Hannaford, R. Liu, N. Billington, Z.T. Swider., B.J. Galletta and C.J. Fagerstrom performed the experiments and analyzed the data. Christian Combs provided critical assistance in particle tracking and super resolution microscopy. M.R. Hannaford and Carey Fagerstrom designed and cloned all constructs. M.R. Hannaford and N.M. Rusan wrote the manuscript. J.R. Sellers and N.M. Rusan supervised and funded the project.
References
- Agircan, F.G., Schiebel E., and Mardin B.R.. 2014. Separate to operate: Control of centrosome positioning and separation. Philos. Trans. R. Soc B Biol. Sci. 369:20130461. 10.1098/rstb.2013.0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayloo, S., and Holzbaur E.L.F.. 2015. Reconstitution of microtubule-based motility using cell extracts. Methods Cell Biol. 128:57–68. 10.1016/bs.mcb.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Azimzadeh, J., and Bornens M.. 2007. Structure and duplication of the centrosome. J. Cell Sci. 120:2139–2142. 10.1242/jcs.005231 [DOI] [PubMed] [Google Scholar]
- Bakhoum, S.F., and Cantley L.C.. 2018. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 174:1347–1360. 10.1016/j.cell.2018.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius, T.L., Cai D., Jih G.T., Toret C.P., and Verhey K.J.. 2007. Two binding partners cooperate to activate the molecular motor Kinesin-1. J. Cell Biol. 176:11–17. 10.1083/jcb.200605099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolvar, J., Huynh J.R., Lpez-Schier H., Gonzlez C., Johnston D.S., and Gonzlez-Reyes A.. 2001. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development. 128:1889–1897. 10.1242/dev.128.10.1889 [DOI] [PubMed] [Google Scholar]
- Burakov, A., Nadezhdina E., Slepchenko B., and Rodionov V.. 2003. Centrosome positioning in interphase cells. J. Cell Biol. 162:963–969. 10.1083/jcb.200305082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., and Yamashita Y.M.. 2021. Centrosome-centric view of asymmetric stem cell division. Open Biol. 11:200314. 10.1098/rsob.200314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching, K., Wang J.T., and Stearns T.. 2022. Long-range migration of centrioles to the apical surface of the olfactory epithelium. Elife. 11:e74399. 10.7554/elife.74399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., and Raff J.W.. 2010. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20:2187–2192. 10.1016/j.cub.2010.11.055 [DOI] [PubMed] [Google Scholar]
- Crocker, J.C., and Grier D.G.. 1996. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179:298–310. 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Dawe, H.R., Farr H., and Gull K.. 2007. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 120:7–15. 10.1242/jcs.03305 [DOI] [PubMed] [Google Scholar]
- Dietzl, G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448:151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Djagaeva, I., Rose D.J., Lim A., Venter C.E., Brendza K.M., Moua P., and Saxton W.M.. 2012. Three routes to suppression of the neurodegenerative phenotypes caused by kinesin heavy chain mutations. Genetics. 192:173–183. 10.1534/genetics.112.140798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin, D.L., and Vallee R.B.. 2002. Dynein at the cortex. Curr. Opin. Cell Biol. 14:44–49. 10.1016/s0955-0674(01)00292-7 [DOI] [PubMed] [Google Scholar]
- Friedman, D.S., and Vale R.D.. 1999. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat. Cell Biol. 1:293–297. 10.1038/13008 [DOI] [PubMed] [Google Scholar]
- Fu, M.m., and Holzbaur E.L.F.. 2014. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 24:564–574. 10.1016/j.tcb.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba, L.C., Eivers E., Geissert D., Taelman V., and Robertis E.M.D.. 2008. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc. Natl. Acad. Sci. USA. 105:7732–7737. 10.1073/pnas.0803027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaud, E., Caous R., Pascal A., Bazile F., Gagné J.-P., Huet S., Poirier G.G., Chrétien D., Richard-Parpaillon L., and Giet R.. 2014. Ensconsin/Map7 promotes microtubule growth and centrosome separation in Drosophila neural stem cells. J. Cell Biol. 204:1111–1121. 10.1083/jcb.201311094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaud, E., Nair A.R., Horsley N., Monnard A., Singh P., Pham T.T., Garcia D.S., Ferrand A., and Cabernard C.. 2020. Dynamic centriolar localization of Polo and Centrobin in early mitosis primes centrosome asymmetry. PLoS Biol. 18:e3000762. 10.1371/journal.pbio.3000762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta, B.J., Fagerstrom C.J., Schoborg T.A., McLamarrah T.A., Ryniawec J.M., Buster D.W., Slep K.C., Rogers G.C., and Rusan N.M.. 2016. A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat. Commun. 7:12476. 10.1038/ncomms12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta, B.J., Guillen R.X., Fagerstrom C.J., Brownlee C.W., Lerit D.A., Megraw T.L., Rogers G.C., and Rusan N.M.. 2014. Drosophila pericentrin requires interaction with calmodulin for its function at centrosomes and neuronal basal bodies but not at sperm basal bodies. Mol. Biol. Cell. 25:2682–2694. 10.1091/mbc.e13-10-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta, B.J., Ortega J.M., Smith S.L., Fagerstrom C.J., Fear J.M., Mahadevaraju S., Oliver B., and Rusan N.M.. 2020. Sperm head-tail linkage requires restriction of pericentriolar material to the proximal centriole end. Dev. Cell. 53:86–101.e7. 10.1016/j.devcel.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta, B.J., and Rusan N.M.. 2015. Methods in cell biology. Methods Cell Biol. 129:251–277. 10.1016/bs.mcb.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, M.C., Lehman D.A., and Schubiger G.. 2002. Lumenal transmission of decapentaplegic in Drosophila imaginal discs. Dev. Cell. 3:451–460. 10.1016/s1534-5807(02)00264-2 [DOI] [PubMed] [Google Scholar]
- Grieder, N.C., de Cuevas M., and Spradling A.C.. 2000. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 127:4253–4264. 10.1242/dev.127.19.4253 [DOI] [PubMed] [Google Scholar]
- Hammond, J.W., Griffin K., Jih G.T., Stuckey J., and Verhey K.J.. 2008. Co‐operative versus independent transport of different cargoes by Kinesin‐1. Traffic. 9:725–741. 10.1111/j.1600-0854.2008.00722.x [DOI] [PubMed] [Google Scholar]
- Ito, D., Zitouni S., Jana S.C., Duarte P., Surkont J., Carvalho-Santos Z., Pereira-Leal J.B., Ferreira M.G., and Bettencourt-Dias M.. 2019. Pericentrin-mediated SAS-6 recruitment promotes centriole assembly. Elife. 8:e41418. 10.7554/elife.41418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke, J., and Gonzalez C.. 2010. The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J. Cell Biol. 188:693–706. 10.1083/jcb.200905024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke, J., Llamazares S., Reina J., and Gonzalez C.. 2011. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2:243. 10.1038/ncomms1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke, J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J., and Gonzalez C.. 2013. Centrobin controls mother–daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15:241–248. 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- Jord, A.A., Spassky N., and Meunier A.. 2019. Motile ciliogenesis and the mitotic prism. Biol. Cell. 111:199–212. 10.1111/boc.201800072 [DOI] [PubMed] [Google Scholar]
- Kapitein, L.C., Peterman E.J.G., Kwok B.H., Kim J.H., Kapoor T.M., and Schmidt C.F.. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kelliher, M.T., Yue Y., Ng A., Kamiyama D., Huang B., Verhey K.J., and Wildonger J.. 2018. Autoinhibition of kinesin-1 is essential to the dendrite-specific localization of Golgi outposts. J. Cell Biol. 217:2531–2547. 10.1083/jcb.201708096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba, J.E., Buster D.W., Nguyen A.L., Swatkoski S., Gucek M., Rusan N.M., and Rogers G.C.. 2013. Polo-like kinase 4 autodestructs by generating its slimb-binding phosphodegron. Curr. Biol. 23:2255–2261. 10.1016/j.cub.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan, N., Swoger M., Rathbun L.I., Fioramonti P.J., Freshour J., Bates M., Patteson A.E., and Hehnly H.. 2022. Rab11 endosomes and Pericentrin coordinate centrosome movement during pre-abscission in vivo. Life Sci. Alliance. 5:e202201362. 10.26508/lsa.202201362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit, D.A., Jordan H.A., Poulton J.S., Fagerstrom C.J., Galletta B.J., Peifer M., and Rusan N.M.. 2015. Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J. Cell Biol. 210:79–97. 10.1083/jcb.201503117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit, D.A., and Rusan N.M.. 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202:1013–1022. 10.1083/jcb.201303141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R., Billington N., Yang Y., Bond C., Hong A., Siththanandan V., Takagi Y., and Sellers J.R.. 2021. A binding protein regulates myosin-7a dimerization and actin bundle assembly. Nat. Commun. 12:563. 10.1038/s41467-020-20864-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, M., Dauvet D., Sanchez-Garrido F., Sadaouli K., Bernard F., and Guichet A.. 20222022. Kinesin-1 promotes centrosome clustering and nuclear migration in the Drosophila oocyte. biorxiv. (Preprint posted February 16, 2022). 10.1101/2022.02.16.480671 [DOI] [PubMed] [Google Scholar]
- Lu, W., Lakonishok M., Serpinskaya A.S., Kirchenbüechler D., Ling S.-C., and Gelfand V.I.. 2018. Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination. J. Cell Biol. 217:3497–3511. 10.1083/jcb.201709174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W., Winding M., Lakonishok M., Wildonger J., and Gelfand V.I.. 2016. Microtubule-microtubule sliding by kinesin-1 is essential for normal cytoplasmic streaming in Drosophila oocytes. Proc. Natl. Acad. Sci. USA. 113:E4995–E5004. 10.1073/pnas.1522424113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius, G., Reymond L., D’Este E., Masharina A., Göttfert F., Ta H., Güther A., Fournier M., Rizzo S., Waldmann H., et al. 2014. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods. 11:731–733. 10.1038/nmeth.2972 [DOI] [PubMed] [Google Scholar]
- Martinez-Campos, M., Basto R., Baker J., Kernan M., and Raff J.W.. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165:673–683. 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, K.D., and Schubiger G.. 2005. Developmental analysis and squamous morphogenesis of the peripodial epithelium in Drosophila imaginal discs. Development. 132:5033–5042. 10.1242/dev.02092 [DOI] [PubMed] [Google Scholar]
- McNally, K.L.P., Fabritius A.S., Ellefson M.L., Flynn J.R., Milan J.A., and McNally F.J.. 2012. Kinesin-1 prevents capture of the oocyte meiotic spindle by the sperm aster. Dev. Cell. 22:788–798. 10.1016/j.devcel.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier, M., Monroy B.Y., Gallaud E., Caous R., Pascal A., Richard-Parpaillon L., Guichet A., Ori-McKenney K.M., and Giet R.. 2019. Dual control of Kinesin-1 recruitment to microtubules by Ensconsin in Drosophila neuroblasts and oocytes. Development. 146:dev171579. 10.1242/dev.171579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neighbors, B.W., Williams R.C., and McIntosh J.R.. 1988. Localization of kinesin in cultured cells. J. Cell Biol. 106:1193–1204. 10.1083/jcb.106.4.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalona, J., Januschke J., Sampaio P., and Gonzalez C.. 2015. Time-lapse recording of centrosomes and other organelles in Drosophila neuroblasts. Methods Cell Biol. 129:301–315. 10.1016/bs.mcb.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Pareek, G., and Pallanck L.J.. 2020. Inactivation of the mitochondrial protease Afg3l2 results in severely diminished respiratory chain activity and widespread defects in mitochondrial gene expression. PLoS Genet. 16:e1009118. 10.1371/journal.pgen.1009118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.-S., et al. 2015. The transgenic RNAi project at harvard medical school: Resources and validation. Genetics. 201:843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, M., Meyer P., Khodjakov A., Rieder C.L., and Bornens M.. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330. 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, M., Nordberg J., Euteneuer U., and Bornens M.. 2001. Centrosome-dependent exit of cytokinesis in animal cells. Science. 291:1550–1553. 10.1126/science.1057330 [DOI] [PubMed] [Google Scholar]
- Purohit, A., Tynan S.H., Vallee R., and Doxsey S.J.. 1999. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147:481–492. 10.1083/jcb.147.3.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramat, A., Hannaford M., and Januschke J.. 2017. Maintenance of miranda localization in Drosophila neuroblasts involves interaction with the cognate mRNA. Curr. Biol. 27:2101–2111.e5. 10.1016/j.cub.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas Nair, A., Singh P., Salvador Garcia D., Rodriguez-Crespo D., Egger B., and Cabernard C.. 2016. The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep. 14:1100–1113. 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Rebollo, E., Sampaio P., Januschke J., Llamazares S., Varmark H., and González C.. 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 12:467–474. 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Reiter, J.F., and Leroux M.R.. 2017. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18:533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G.C., Rusan N.M., Peifer M., and Rogers S.L.. 2008. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell. 19:3163–3178. 10.1091/mbc.e07-10-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque, H., Saurya S., Pratt M.B., Johnson E., and Raff J.W.. 2018. Drosophila PLP assembles pericentriolar clouds that promote centriole stability, cohesion and MT nucleation. PLoS Genet. 14:e1007198. 10.1371/journal.pgen.1007198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan, N.M., and Peifer M.. 2007. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177:13–20. 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg, T., Zajac A.L., Fagerstrom C.J., Guillen R.X., and Rusan N.M.. 2015. An Asp-CaM complex is required for centrosome-pole cohesion and centrosome inheritance in neural stem cells. J. Cell Biol. 211:987–998. 10.1083/jcb.201509054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda, G., Antkowiak M., Brust-Mascher I., Mahe K., Ou T., Castro N.M., Christensen L.N., Cheung L., Jiang X., Yoon D., et al. 2018. Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates. Elife. 7:e34959. 10.7554/elife.34959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth, W.T., Nardi I.K., Paul R., Mogilner A., and Cimini D.. 2012. Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell. 23:401–411. 10.1091/mbc.e11-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P., Ramdas Nair A., and Cabernard C.. 2014. The centriolar protein bld10/cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 24:1548–1555. 10.1016/j.cub.2014.05.050 [DOI] [PubMed] [Google Scholar]
- Spassky, N., and Meunier A.. 2017. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 18:423–436. 10.1038/nrm.2017.21 [DOI] [PubMed] [Google Scholar]
- Sun, F., Zhu C., Dixit R., and Cavalli V.. 2011. Sunday Driver/JIP3 binds kinesin heavy chain directly and enhances its motility. EMBO J. 30:3416–3429. 10.1038/emboj.2011.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swider, Z.T., Ng R.K., Varadarajan R., Fagerstrom C.J., and Rusan N.M.. 2019. Fascetto interacting protein ensures proper cytokinesis and ploidy. Mol. Biol. Cell. 30:992–1007. 10.1091/mbc.e18-09-0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum, M.E., and Medema R.H.. 2010. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell. 19:797–806. 10.1016/j.devcel.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Tang, N., and Marshall W.F.. 2012. Centrosome positioning in vertebrate development. J. Cell Sci. 125:4951–4961. 10.1242/jcs.038083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez, J.-Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., and Eliceiri K.W.. 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115:80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Tozer, S., Baek C., Fischer E., Goiame R., and Morin X.. 2017. Differential routing of Mindbomb1 via centriolar satellites regulates asymmetric divisions of neural progenitors. Neuron. 93:542–551.e4. 10.1016/j.neuron.2016.12.042 [DOI] [PubMed] [Google Scholar]
- Tripathi, A., Bond C., Sellers J.R., Billington N., and Takagi Y.. 2021. Myosin-specific adaptations of in vitro fluorescence microscopy-based motility assays. J. Vis. Exp. 10.3791/62180 [DOI] [PubMed] [Google Scholar]
- Varadarajan, R., and Rusan N.M.. 2018. Bridging centrioles and PCM in proper space and time. Essays Biochem. 62:793–801. 10.1042/ebc20180036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey, K.J., and Hammond J.W.. 2009. Traffic control: Regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Wang, X., Le N., Denoth-Lippuner A., Barral Y., and Kroschewski R.. 2016. Asymmetric partitioning of transfected DNA during mammalian cell division. Proc. Natl. Acad. Sci. USA. 113:7177–7182. 10.1073/pnas.1606091113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winding, M., Kelliher M.T., Lu W., Wildonger J., and Gelfand V.I.. 2016. Role of kinesin-1–based microtubule sliding in Drosophila nervous system development. Proc. Natl. Acad. Sci. USA. 113:E4985–E4994. 10.1073/pnas.1522416113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, A., Dictenberg J.B., Purohit A., Tuft R., and Doxsey S.J.. 2000. Cytoplasmic dynein-mediated assembly of pericentrin and γ tubulin onto centrosomes. Mol. Biol. Cell. 11:2047–2056. 10.1091/mbc.11.6.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]