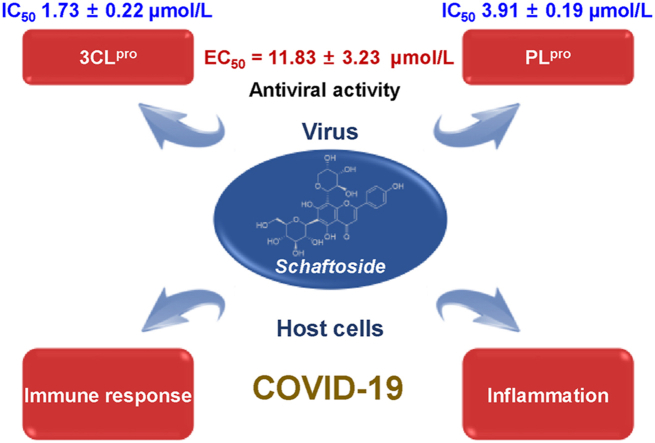
Abstract
It is an urgent demand worldwide to control the coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. The 3-chymotrypsin-like protease (3CLpro) and papain-like protease (PLpro) are key targets to discover SARS-CoV-2 inhibitors. After screening 12 Chinese herbal medicines and 125 compounds from licorice, we found that a popular natural product schaftoside inhibited 3CLpro and PLpro with IC50 values of 1.73 ± 0.22 and 3.91 ± 0.19 μmol/L, respectively, and inhibited SARS-CoV-2 virus in Vero E6 cells with EC50 of 11.83 ± 3.23 μmol/L. Hydrogen–deuterium exchange mass spectrometry analysis, quantum mechanics/molecular mechanics calculations, together with site-directed mutagenesis indicated the antiviral activities of schaftoside were related with non-covalent interactions with H41, G143 and R188 of 3CLpro, and K157, E167 and A246 of PLpro. Moreover, proteomics analysis and cytokine assay revealed that schaftoside also regulated immune response and inflammation of the host cells. The anti-inflammatory activities of schaftoside were confirmed on lipopolysaccharide-induced acute lung injury mice. Schaftoside showed good safety and pharmacokinetic property, and could be a promising drug candidate for the prevention and treatment of COVID-19.
Key words: COVID-19, SARS-CoV-2, 3-Chymotrypsin-like protease, Papain-like protease, Licorice, Schaftoside, Immune response, Inflammation
Graphical abstract
Schaftoside from the Chinese herbal medicine licorice can inhibit the SARS-CoV-2 virus in Vero E6 cells with EC50 of 11.83 ± 3.23 μmol/L, and regulate immune response and inflammation.
1. Introduction
Currently, it is a world-wide urgent requirement to control the pandemic of the coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since its first report in late 2019, COVID-19 has caused over 6.30 million deaths globally as of June 20221,2. While vaccines have been developed, SARS-CoV-2 variants exhibit higher infectivity and contain mutations that cause partial immune escape3.
The major therapeutic targets to discover SARS-CoV-2 inhibitors include 3-chymotrypsin-like protease (3CLpro or Mpro, nsp5) and papain-like protease (PLpro, nsp3). The 3CLpro is a highly conserved cysteine protease indispensable for coronavirus replication4. It processes two viral polyproteins pp1a and pp1ab into 16 non-structural proteins, which engage in the production of subgenomic RNAs to encode four main structural proteins5, 6, 7, 8, 9. PLpro plays an important role in viral maturation, dysregulation of host inflammation, and antiviral immune responses10, 11, 12. As the crystal structures of SARS-CoV-2 3CLpro and PLpro have been resolved6,10, a number of small molecule inhibitors have been reported. For instance, 11a (IC50, 0.053 μmol/L against 3CLpro; EC50, 0.53 μmol/L against SARS-CoV-2 virus) and 11b (IC50, 0.040 μmol/L; EC50, 0.72 μmol/L) formed covalent bonding with Cys145 of 3CLpro (Mpro) through an aldehyde group6. Other potent 3CLpro inhibitors include GC376 (IC50, 0.15 μmol/L; EC50, 0.70 μmol/L)7, MI30 (IC50, 17.2 nmol/L; EC50, 0.54 μmol/L in Vero E6 cells and 1.1 nmol/L in HPAEpiC cells), and PF07321332 (IC50, 3.11 nmol/L; EC50, 74.5 nmol/L)8,9. Only a few PLpro inhibitors have been reported including GRL0617 (IC50, 2.2 μmol/L; EC50, 21 μmol/L)10, 11, 12.
Recently, molnupiravir and paxlovid [nirmatrelvir (PF-07321332) tablets and ritonavir tablets] have been approved as the first small molecule oral drugs for the treatment of COVID-19. They reduced the risk of hospitalization or death by 50% and 89%, respectively13,14. However, their prices are high, around 700 and 500 US dollars for five days treatment, respectively15,16. Molnupiravir must be given within five days after symptoms appear, and it is ineffective if it is taken after the patient is hospitalized13, 14, 15, 16. Therefore, it is still highly demanded to develop effective and less expensive drugs against COVID-19.
Aside from synthetic chemicals, traditional Chinese medicines can ameliorate clinical symptoms of COVID-19 and lower the risk of in-hospital mortality, and have been widely used for the prevention and treatment of COVID-19 in China17, 18, 19. However, only a few bioactive phytochemicals have been discovered from Chinese herbal medicines, thus far, though natural products are an important source for drug discovery. Salvianolic acid C (EC50, 3.41 μmol/L) from Salvia miltiorrhiza20 and kobophenol A (EC50, 71.6 μmol/L) from Caragana sinica21 inhibited SARS-CoV-2 infection by blocking the formation of six-helix bundle core of spike protein and the interaction between the ACE2 receptor and S1-RBD, respectively. Recently, Su et al.4 reported that myricetin (EC50, 8.0 μmol/L) and dihydromyricetin (EC50, 13.5 μmol/L) inhibited the SARS-CoV-2 3CLpro with IC50 of 0.63 and 1.14- μmol/L, respectively.
In the present work, we screened 12 frequently used Chinese herbal medicines and 125 compounds from licorice for their inhibitory activities against SARS-CoV-2 3CLpro and PLpro by enzymatic and antiviral assays. We discovered that schaftoside can be a potent 3CLpro/PLpro dual-target SARS-CoV-2 inhibitor, and also remarkably regulate immune response and inflammation of host cells in a mice model.
2. Materials and methods
2.1. Chemicals and reagents
All licorice compounds were purified by our laboratory from the roots and rhizomes of Glycyrrhiza uralensis Fisch22. GC376 and GRL0617 were purchased from Shanghai Yuanye Bio-Techno-logy Co., Ltd. Dabcyl-KTSAVLQSGFRKME-Edans and (E-EDANS)RELNGGAPI(K-DABCYL)S were synthesized from GL Biochem (Shanghai) Ltd. SARS-CoV-2 3CLpro and PLpro were respectively purchased from Novoprotein Technology Co., Ltd., and Sino Biological Inc. IL-1, IL-1β, IL-6, IL-7, IL-8, TNF-α, IFN-β, and IFN-γ were measured by ELISA kits from MEIMIAN (www.mmbio.cn). Antibodies of IL-1β, IL-6, and TNF-α were purchased from ABCAM (https://www.abcam.cn). The animal facilities and protocols were approved by the Animal Care and Use Committee of Peking University Health Science Center (SYXK 2016-0041). All animal care and experimental procedures in this work were in accordance with Guide for the Care and Use of Laboratory Animals (National Institutes of Health).
2.2. Herbal materials and extracts
The Chinese herbal medicines were purchased from Beijing Tong-Ren-Tang drug store. A total of 12 herbal medicines were studied, including Isatidis Radix (Ban-Lan-Gen), Ephedrae Herba (Ma-Huang), Astragali Radix (Huang-Qi), Glycyrrhizae Radix et Rhizoma (Gan-Cao), Lonicerae Flos (Jin-Yin-Hua), Scutellariae Radix (Huang-Qin), Forsythiae Fructus (Lian-Qiao), Platycodonis Radix (Jie-Geng), Armeniacae Semen Amarum (Ku-Xing-Ren), Atractylodis Macrocephalae Rhizoma (Bai-Zhu), Pogostemonis Herba (Guang-Huo-Xiang), and Citri Reticulatae Pericarpium (Chen-Pi). To prepare the herbal extracts, the powder (5 g) of each herb was extracted with 50 mL of 95% ethanol in a water bath at 90 °C for 30 min for three times. The extract was concentrated and then freeze dried.
2.3. Protein expression, purification, and crystallization
The full-length cDNA of SARS-CoV-2 3CLpro (Genbank No. MN908947.3) was cloned into the pET28a vector. A 6 × His tag followed by TEV protease cleavage sequence was added before the N-terminus of the target protein to facilitate purification. The His-TEV-3CLpro protein was expressed in Escherichia coli BL21(DE3) strain and purified by Ni-NTA affinity chromatography. After purification, the recombinant protein was digested by TEV protease to cut the His tag. The sample was added onto Ni-NTA affinity beads for the second time to purify the protein. The flow-through was concentrated and then applied to size-exclusion chromatography on a Superdex 200 increase 10/300 GL prepacked column for further purification. The elution buffer was 10 mmol/L Tris–HCl pH 7.5 and 100 mmol/L NaCl. Fractions containing 3CLpro were collected and concentrated to 10 mg/mL (Supporting Information Fig. S1). Crystals of 3CLpro were obtained after 5 days at 16 °C in hanging drops containing 2 μL of protein solution and 2 μL of reservoir solution (0.05 sodium citrate tribasic dihydrate, 0.12 mol/L potassium chloride, 0.08 mol/L Bis–Tris, 14% PEG 4000, pH = 6.0). The crystals were frozen in a reservoir solution with 30% glycerol.
2.4. Enzymatic activity assay
The proteolytic activity of SARS-CoV-2 3CLpro and PLpro were measured using the fluorogenic substrate Dabcyl-KTSAVLQ-SGFRKME-Edans and (E-EDANS)RELNGGAPI(K-DABCYL)S, respectively (https://www.novoprotein.com.cn/). The reaction mixture contained 12.5 μg/mL purified enzyme, 50 μg/mL herbal extract or 8 μmol/L pure compound, 20 mmol/L Tris–HCl buffer (pH 7.0) for 3CLpro or 50 mmol/L HEPES (pH 7.5) buffer for PLpro, and 3 mmol/L substrate for 3CLpro or 5 mmol/L substrate for PLpro. The inhibition kinetics for both 3CLpro and PLpro were determined at a constant substrate concentration with different concentrations of samples (1, 2, 4, 8, and 16 μmol/L). The reactions were conducted at 25 °C with continuous monitoring of fluorescence for 10 min. The enzyme activity was demonstrated by monitoring the increase of emission fluorescence at 535 nm upon excitation at 340 nm on a FlexStation 3 Multi-Mode Microplate Reader. The activity was calculated according to the following equation23,24:
| A = [(ΔODtest)df]/[ΔODcontrol(2.204092 × C × Vs)] |
where A is enzyme activity, df is dilution factor, ΔOD is absorption change/min, Vs is sample volume, and C is concentration of 3CLpro.
2.5. Surface plasmon resonance (SPR) assay
The binding affinity of samples to SARS-CoV-2 3CLpro, PLpro, and their mutants were determined using the SPR biosensor technology (Biacore 8K). The protein was immobilized onto the sensor chip CM5 by the standard primary amine coupling reaction. SARS-CoV 3CLpro, PLpro, and their mutants were diluted with 10 mmol/L sodium acetate (pH 4.5) to 25 μg/mL. The final immobilized level was typically above 10,000 RU. The compounds were injected as analytes at various concentrations at a flow rate of 30 μL/min with a contact time of 60 s and a dissociation time of 60 s, using PBS containing 5% DMSO and 0.05% Surfactant P20 as running buffer. Data were analyzed by the Biacore evaluation software. The equilibrium dissociation constants (KD) evaluating the protein–ligand binding affinity were determined by kinetics analysis of the Biacore data.
2.6. Cell viability assay
Cell Counting Kit-8 (CCK-8, Dojindo, Japan) assay was conducted to determine cell viability. Briefly, Vero E6 cells were seeded in 96-well plates at a density of 1 × 104 cells per well (in DMEM + 10% FBS), and were incubated for 24 h at 37 °C, 5% CO2. Then, the compounds (maximum concentration at 200 μmol/L, <1% DMSO) were diluted in DMEM culture medium containing 10% FBS and were added to each well. The organic solvent in DMEM was less than 0.1%. After 24 h, contents of the wells were replaced with fresh medium containing 10% CCK-8 solution and were incubated at 37 °C for 3 h. The final optical density at OD450 was measured using a Synergy H1 Microplate Reader.
2.7. Antiviral assay
Vero E6 cells were co-cultured with various concentrations of drugs overnight in 24-well plates at a density of 1 × 105 cells/well, and then infected by SARS-CoV-2 (nCoV-2019BetaCoV/Wuhan/WIV04/2019) at a multiplicity of infection (MOI) of 0.01. After 24 h of infection, RNA was extracted from the cell supernatant according to the manufacturer's instructions using QIAamp viral RNA mini kit (Qiagen, 52906). qRT-PCR was used to quantify the viral RNA with primers ORF1ab-F (5′-CCCTGTGGGTTTTACACTTAA-3′) and ORF1ab-R (5′-ACGATTGTGCATCAGCTGA-3′) combined with the probe 5′-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′ by Luna Universal Probe One-Step RT-PCR Kit (Invitrogen, E3006).
2.8. Immunofluorescence assay
Vero E6 cells were seeded on Biocoat Coverslips for 24 h and then infected with SARS-CoV-2 in the presence of 10 or 30 μmol/L of schaftoside. After 24 h of infection, cells were fixed with 4% paraformaldehyde for 15 min. Cells were incubated with 1:1000 diluted primary antibody against the spike protein of SARS-CoV-2 for 1 h. After a thorough wash for 15 min to remove unbound antibodies, cells were then incubated with 1:500 diluted FITC-labeled goat anti-rabbit IgG antibodies (ThermoFisher Scientific, Invitrogen) for 1 h. The coverslips were washed for another 15 min, followed by nuclei staining with 4,6-diamidino-2-phenylindole (DAPI, Sigma), and then imaged by a confocal fluorescence microscope (PerkinElmer, UltraVIEW VoX, USA).
2.9. Hydrogen–deuterium exchange mass spectrometry (HDX-MS) analysis
Deuterium labeling was initiated with a 20-fold dilution into D2O buffer (100 mmol/L phosphate, pH 7.0) of 3CLpro protein (1 mg/mL), 3CLpro (1 mg/mL) with schaftoside (1 mmol/L), PLpro protein (1 mg/mL), or PLpro (1 mg/mL) with schaftoside (1 mmol/L). After 0.25, 0.5, and 10 min of labeling, the labeling reaction was quenched with the addition of quenching buffer (100 mmol/L phosphate, 4 mol/L GdHCl, 0.5 mol/L TCEP, pH 2.0). Samples were then injected and online digested using a Waters ENZYMATE BEH pepsin column (2.1 mm × 30 mm, 5 μm). The peptides were trapped and desalted on a VanGuard Pre-Column trap (ACQUITY UPLC BEH C18, 1.7 μm) for 3 min, eluted from the trap using 15% acetonitrile at a flow rate of 100 μL/min, and then separated using an ACQUITY UPLC BEH C18 column (1.0 mm × 100 mm, 1.7 μm). All mass spectra were acquired on a Waters Xevo G2 mass spectrometer. Peptides from an unlabeled protein were identified using ProteinLynx Global Server (PLGS) searches of a protein database including 3CLpro and PLpro sequences only. Relative deuterium levels for each peptide were calculated by subtracting the mass of the undeuterated control sample from that of the deuterium-labeled sample. All mass spectra were processed using DynamX 3.0 (Waters Corporation). Deuterium levels were not corrected for back exchange and thus reported as relative.
2.10. Docking and quantum mechanics/molecular mechanics (QM/MM) simulations
Molecular structures of the natural product ligands were optimized by B3LYP/6-311G(d) method25, 26, 27. The crystal structure of 3CLpro (PDB ID: 6LZE)6 and PLpro (7CJM)10 were used as a reference for protein–ligand system. Docking calculations were performed by AutoDock Vina and AutoDocktools v1.5.6 software28. The extracted complex of the ligand and amino acid residues from molecular docking was optimized by QM/MM simulations. The QM atoms from schaftoside, H41, G143, C145, R188 and Q192 of 3CLpro, and K157, E167, A246 and Y268 of PLpro were described by DFT method B3LYP at 6-311G basis set, while the MM atoms from T25, T26, M49, M162, H163, F140, N142, H164, M165, E166, P168, D187 and Q189 of 3CLpro, and D164, P247, P248, Y264, Q269, Y273 and T301 of PLpro were simulated by the UFF force field29. The calculations were performed using the Gaussian16 suite of codes30.
2.11. Site-directed mutagenesis
Site-directed mutagenesis of 3CLpro and PLpro, including H41A, G143A and R188A mutants of 3CLpro, and K157A, E167A and Y268A mutants of PLpro, were constructed using Fast Mutagensis System kit (Transgen, China) according to the manufacturer's instructions. The primer pairs designed to construct the mutants are listed in Supporting Information Table S1. Enzyme assays of the mutated recombinant proteins were conducted under the same conditions as described above for native proteins (Fig. S1).
2.12. Proteomics analysis
Vero E6 cells were infected with SARS-CoV-2 virus, or with schaftoside (30 μmol/L) and SARS-CoV-2 for 24 h (MOI = 1), respectively. The samples were sonicated using a high intensity ultrasonic processor (Scientz) in lysis buffer (8 mmol/L urea, 1% protease inhibitor cocktail). The debris was removed by centrifugation. For the first digestion overnight, trypsin was added at 1:50 trypsin-to-protein mass ratio. Each channel of peptide was labeled by their respective TMT reagent (based on manufacturer's protocol, ThermoFisher Scientific), and incubated for 2 h at room temperature. The pooled samples were desalted with a Strata X C18 SPE column (Phenomenex) and dried by vacuum centrifugation. The tryptic peptides were dissolved in solvent A (0.1% formic acid and 2% acetonitrile in water), directly loaded onto a home-made reversed-phase analytical column (25 cm length, 75 μm i.d.). Peptides were separated with a gradient from 5% to 25% solvent B (0.1% formic acid in 90% acetonitrile) over 60 min, 25%–35% in 22 min, up to 80% in 4 min, and then held at 80% for 4 min, all at a constant flow rate of 450 nL/min on an EASY-nLC 1200 UPLC system (ThermoFisher Scientific). The separated peptides were characterized by a Q Exactive HF-X mass spectrometer (ThermoFisher Scientific) with a nano-electrospray ion source.
GO annotation proteome was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). Proteins were classified by Gene Ontology annotation based on three categories: biological process, cellular component, and molecular function. For functional enrichment, proteins were classified by GO biological process annotation. A two-tailed Fisher's exact test was employed to test the enrichment of the differentially expressed protein against all identified proteins. The GO with P value < 0.05 was considered significant.
2.13. Lipopolysaccharide (LPS)-induced acute lung injury (ALI) mice model
Male BALB/c mice (20 g) were provided by the Experimental Animal Center of Peking University Health Science Center (Beijing, China). Mice in the blank, control, and schaftoside groups were respectively administered with saline solution, LPS (2 mg/kg, i.n., intranasal administration), and LPS (2 mg/kg, i.n.) coupled with schaftoside (10, 20 mg/kg, i.g.). After 8 h of LPS administration, blood samples and lung tissues were collected.
2.14. Pathological analysis
Small pieces were obtained from lung tissues of the ALI model and fixed in 4% paraformaldehyde. Proper fixing was followed by dehydration of the specimens in graded ethanol, clearing in xylene, embedding in paraplast, and sectioning at 5-μm thickness. The sections were stained using the HE staining method to demonstrate the histological structure of testes in control, model, and schaftoside-treated mice. Images were taken using WISLEAP (WS-10).
2.15. Immunohistochemistry (IHC)
The lung tissue sections of ALI model were analyzed by IHC. After incubation with 3% H2O2 for 10 min, antigen retrieval was performed by incubating the samples in citrate buffer (pH 8.0) for 5 min high heat, and 15 min low heat. After blocking with goat serum, sections were incubated for 2 h with primary antibodies (Abcam, TNF-α, 1:20; IL-6, 1:50; IL-1β, 1:250). Sections were washed three times with phosphate-buffered saline and incubated with HRP goat anti-mouse IgG. After three times washing with phosphate-buffered saline, DAB was incubated for 2 min. Then, we stopped the reaction and stained using hematoxylin. Images were taken using WISLEAP (WS-10).
2.16. Acute toxicity test in mice
Male ICR mice (20 g) were randomly and evenly distributed into four groups (n = 5), including control (distilled water, i.g.) and different dosing groups (20, 40, and 300 mg/kg). Animals were treated for 7 days.
2.17. Statistical analysis
All the data are expressed as mean ± standard deviation (SD) and were analyzed using SPSS 20.0. Significant differences between groups were assessed by Student's t-test and one-way ANOVA. Differences were considered statistically significant at P < 0.05.
3. Results
3.1. Enzymatic activity assay and molecular docking
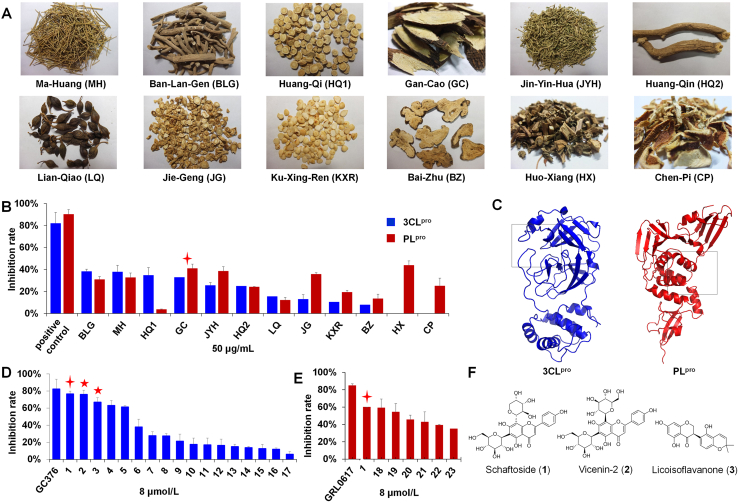
We tested the 12 most frequently used herbal medicines for the treatment of COVID-19 in China for their inhibitory activities against 3CLpro and PLpro (Fig. 1A)31. Of the herbal extracts (50 μg/mL), Isatidis Radix (BLG), Ephedrae Herba (MH), Astragali Radix (HQ1), and Glycyrrhizae Radix et Rhizoma (licorice, GC) showed the most potent activities against 3CLpro, with inhibition rates of 38.17%, 37.76%, 34.70%, and 32.85%, respectively (Fig. 1B and Supporting Information Table S2). Among them, licorice also showed high inhibitory activity against PLpro with an inhibition rate of 40.93% (Supporting Information Table S3). Thus, we focused on licorice for follow-up studies.
Figure 1.
The inhibitory activities against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) 3-chymotrypsin-like protease (3CLpro) and papain-like protease (PLpro) of traditional Chinese medicine herbs and compounds (n = 3). (A) Pictures of the 12 herbs. (B) The inhibitory activities against 3CLpro and PLpro of herbal extracts (50 μg/mL). (C) Active pockets of 3CLpro and PLpro for virtual screening. (D) The inhibitory activities against 3CLpro of licorice compounds (8 μmol/L). (E) The inhibitory activities against PLpro of licorice compounds (8 μmol/L). (F) Chemical structures of hit compounds 1–3. For structures of the other compounds, see Supporting Information Fig. S3.
Using AutoDock VINA, we conducted a virtual screening of 125 compounds we previously isolated from licorice (the roots and rhizomes of G. uralensis)22. The binding energies with 3CLpro and PLpro were determined using SARS-CoV-2 3CLpro (PDB ID: 6LZE)6 and PLpro (7CJM)10 crystal structures (Fig. 1C and Supporting Information Fig. S2), respectively. Finally, 17 compounds (1–17) for 3CLpro and 7 compounds (8–23) for PLpro with binding affinity energy below −8.0 kcal/mol were discovered (Supporting Information Tables S4 and S5).
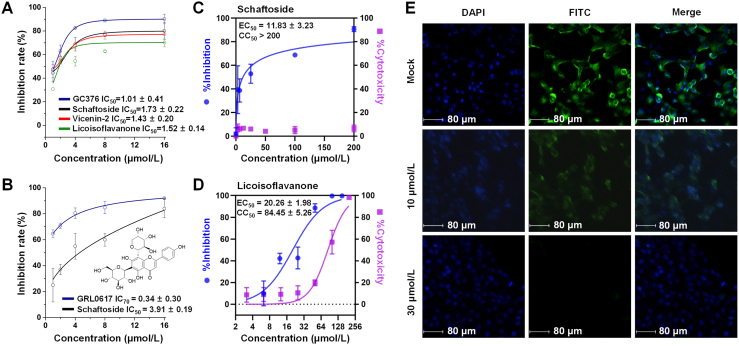
The above compounds were then evaluated by enzymatic assays. At 8 μmol/L, schaftoside (1), vicenin-2 (2), and licoisoflavanone (3) could inhibit 3CLpro by 75.9%, 76.53%, and 66.86%, respectively, with 82.29% for the positive control GC376 (Fig. 1D). Interestingly, schaftoside also demonstrated potent activity against PLpro with an inhibition rate of 60% at 8 μmol/L (positive control GRL0617, 84.75%) (Fig. 1E). The chemical structures are given in Fig. 1F and Supporting Information Fig. S3. The IC50 values of schaftoside (1.73 ± 0.22 μmol/L), vicenin-2 (1.43 ± 0.20 μmol/L), and licoisoflavanone (1.52 ± 0.14 μmol/L) against 3CLpro, and that of schaftoside (3.91 ± 0.19 μmol/L) against PLpro were all below 5 μmol/L (Fig. 2A and B). Moreover, schaftoside at 8 μmol/L also inhibited the 3CLpro and PLpro proteins of SARS-CoV, with inhibition rates of 68.4% and 53.1%, respectively (Supporting Information Fig. S4).
Figure 2.
Inhibitory activity profiles of licorice compounds against SARS-CoV-2 3CLpro and PLpro, and antiviral activities of schaftoside against SARS-CoV-2 in Vero E6 cells. (A, B) Half maximal inhibitory concentrations (IC50) against 3CLpro and PLpro. Data are shown as mean ± standard deviation, n = 3. GC376 and GRL0617 are the positive drugs for 3CLpro and PLpro, respectively. (C, D) Dose-dependent inhibition of schaftoside and licoisoflavanone against SARS-CoV-2 infection, n = 3. Cytotoxicities were determined by CCK-8 assay. Data are shown as mean ± standard deviation. (E) Immunofluorescence assay analysis of the inhibition of schaftoside against SARS-CoV-2 viral replication.
3.2. Antiviral assay
Next, the in vitro antiviral activities of schaftoside, vicenin-2, and licoisoflavanone against SARS-CoV-2 were measured in Vero E6 cells. The cells were inoculated with SARS-CoV-2 virus at a multiplicity of infection (MOI) of 0.01. At 24 h post-infection (dpi), viral RNA was extracted and determined by qRT-PCR. Schaftoside and licoisoflavanone showed remarkable antiviral activities with EC50 of 11.83 ± 3.23 and 20.26 ± 1.98 μmol/L, respectively (Fig. 2C and D). Vicenin-2 only showed weak activities, with an inhibition rate of around 40% at 30 μmol/L (Supporting Information Fig. S5). Given that licoisoflavanone showed obvious cytotoxicity with CC50 of 84.45 μmol/L, schaftoside was considered as the most promising inhibitor among the 125 licorice compounds. An indirect immunofluorescence assay was conducted to verify the antiviral effects. As shown in Fig. 2E, schaftoside inhibited the replication of SARS-CoV-2 in a dose-dependent manner, when compared to the drug-free mock cells.
3.3. Binding mechanisms of schaftoside with 3CLpro and PLpro
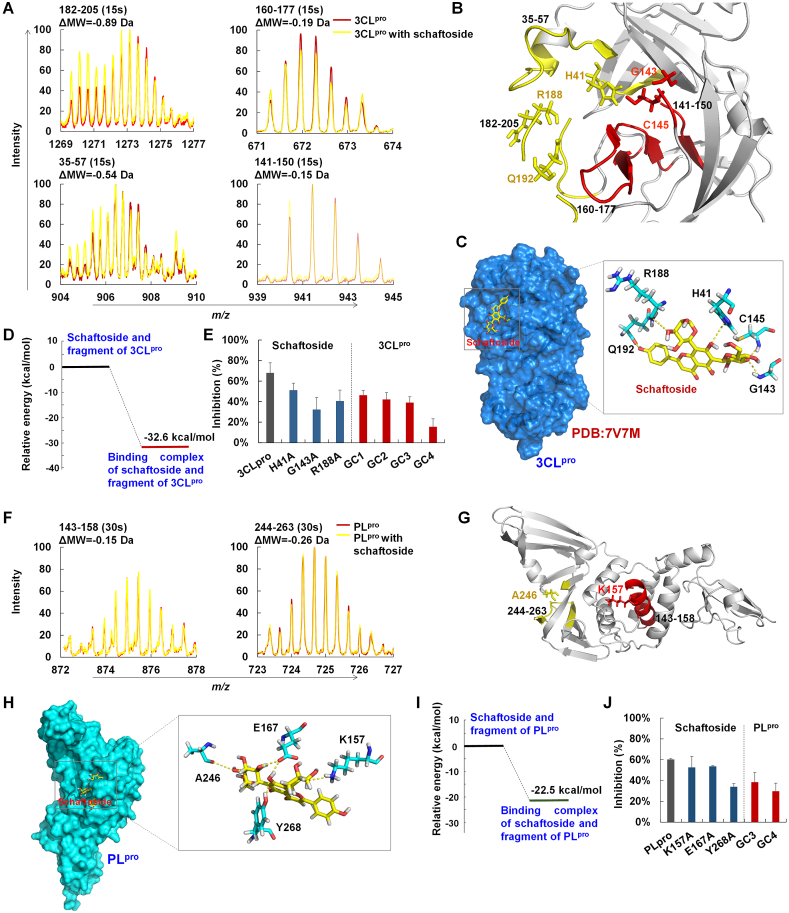
To elucidate the binding mechanisms of schaftoside with 3CLpro and PLpro, we conducted SPR analysis. The results indicate that schaftoside binded to 3CLpro and PLpro with KD values of 12.4 and 13.2 μmol/L, respectively (Supporting Information Fig. S6). Then, HDX-MS analysis was conducted to determine protein conformation changes due to interactions between 3CLpro and schaftoside. After co-incubation with schaftoside, peptides 182–205, 35–57, 160–177, and 141–150 of 3CLpro demonstrated 0.89, 0.54, 0.19 and 0.15 Da m/z loss, respectively (Fig. 3A). To further determine the specific binding sites (Fig. 3B), we solved the crystal structure of 3CLpro (PDB ID: 7V7M, 2.08 Å) (Supporting Information Tables S6), and simulated the schaftoside/3CLpro complex structure by molecular docking (Fig. 3C). The complex of schaftoside and the active sites (Fig. S2)6 was then computed by the QM/MM method. The calculated interaction energy of schaftoside with SARS-CoV-2 3CLpro residues was −32.6 kcal/mol (Fig. 3D). The strong binding may be due to the hydrogen bonding between 7-OH of schaftoside and the nitrogen atom of H41. The C8-arabinosyl moiety could interact with R188 through hydrogen bonding, and the C6-glucosyl moiety could bind to G143 and C145 through two hydrogen bonds. Moreover, 4′-OH of schaftoside could form a hydrogen bond with C O of Q192.
Figure 3.
Binding mechanisms of schaftoside with 3CLpro and PLpro. (A) Mass spectra of peptides 182–205 and 35–57 (15 s) of 3CLpro and 3CLpro with schaftoside determined by hydrogen–deuterium exchange mass spectrometry (HDX-MS). (B) Location of peptides 35–57, 182–205, and 141–150 in 3CLpro. (C) The crystal structure of 3CLpro (PDB ID: 7V7M), and the binding mode of schaftoside with active residues of 3CLpro by molecular docking. Hydrogen bonds (yellow dashes) are formed between schaftoside and residues H41, R188, Q192, G143 and C145 of 3CLpro. (D) Relative energy of schaftoside with 3CLpro computed by quantum mechanics/molecular mechanics (QM/MM). (E) Inhibitory activities of schaftoside (8 μmol/L) against SARS-CoV-2 3CLpro and 3CLpro mutants, and of flavonoid C-glycosides GC1, GC2, GC3 and GC4 (8 μmol/L) against SARS-CoV-2 3CLpro. SARS-CoV-2 3CLpro and 3CLpro mutants were expressed and purified by our laboratory. (F) Mass spectra of peptides 143–158 and 244–263 (15 s) of PLpro and PLpro treated with schaftoside determined by HDX-MS. (G) Location of peptides 143–158 and 244–263 in PLpro. (H) Binding mode of schaftoside with active residues of PLpro by molecular docking. Hydrogen bonds (yellow dashes) are formed between schaftoside and residues K157, E167, A246 and Y268 of 3CLpro. (I) Relative energy of schaftoside with PLpro computed by QM/MM. (J) Inhibitory activities of schaftoside (8 μmol/L) against SARS-CoV-2 PLpro and PLpro mutants, and of flavonoid C-glycosides GC3 and GC4 (8 μmol/L) against SARS-CoV-2 PLpro.
To verify the above key amino acid residues, we conducted site-directed mutagenesis of SARS-CoV-2 3CLpro. The Michaelis constants (Km) of 3CLpro mutants hydrolyzing the commercial substrate were measured (Supporting Information Fig. S7). For the H41A mutant, schaftoside showed an inhibition rate of 50.9%, which was remarkably lower than with the wild type (67.5%) (Fig. 3E). For the G143A and R188A mutants, the inhibition rates decreased substantially to 32% and 40.3%, respectively. These data were consistent with SPR measurements (Fig. S6). We further tested the inhibitory activities of schaftoside analogs GC1-GC4 (Supporting Information Fig. S8). When the 4′-OH group was absent, or only one C-glucosyl residue was substituted at the flavone aglycone, the inhibitory activities decreased to 15.2%–46.1% (Fig. 3E). These results support our proposed mechanisms.
Likewise, HDX-MS analysis indicated the peptides 143–158 and 244–263 of PLpro showed 0.15 and 0.26 Da m/z loss, respectively, upon the treatment of schaftoside (Fig. 3F and G). In molecular docking, A246 and E167 could interact with the C6-glucosyl moiety through hydrogen bonding, and K157 could bind to the C8-arabinosyl moiety through one hydrogen bond. In addition, 7-OH of schaftoside could form two hydrogen bonds with E167 and Y268 (Fig. 3H)10. These bindings were verified by QM/MM calculations (Supporting Information Fig. S9). The interaction energy of schaftoside with key residues K157, E167, A246 and Y268 was −22.5 kcal/mol (Fig. 3I).
Site-directed mutagenesis of SARS-CoV-2 PLpro K157A, E167A and Y268A was conducted, and Km and KD values of PLpro mutants were calculated (Fig. S6 and Supporting Information Fig. S10). Schaftoside could bind to all the three mutants, though around four times weaker with Y268A than with the wild type (Fig. S6). Consistently, schaftoside showed a low inhibitory activity against Y268A, with an inhibition rate of 33.7%. Meanwhile, the inhibitory activities of schaftoside with K157A and E167A decreased to 52.3% and 53.3%, respectively. These results indicate that Y268, K157 and E167 are critical residues for binding with schaftoside. For schaftoside analogs GC3 and GC4, where only one C-glucosyl residue is substituted at the flavone aglycone, the inhibitory activities against PLpro decreased to 38.2% and 29.5%, respectively (Fig. 3J). The above data are consistent with our proposed mechanisms.
3.4. Proteomic variations of SARS-CoV-2-infected host cells treated with schaftoside
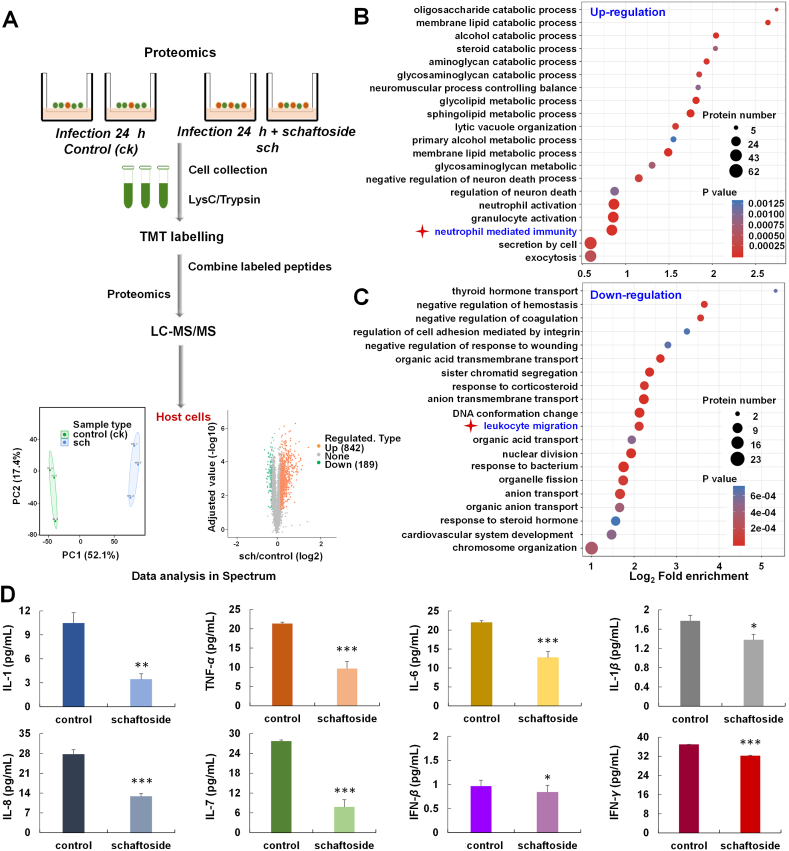
In order to understand the impact of schaftoside on the host cells, we conducted quantitative proteome analysis of the Vero E6 cells treated with SARS-CoV-2 coupled with 30 μmol/L schaftoside for 24 h (Fig. 4A). A total of 842 up-regulated and 189 down-regulated proteins were identified (fold change > 1.3). Then we performed enrichment analyses of biological processes based on Gene Ontology (GO) annotations (P < 0.05), and 174 up-regulated and 40 down-regulated proteins were related to immune system process (Supporting Information Figs. S11 and S12)32. In functional enrichment analysis, schaftoside treatment led to regulations of immune response (neutrophil mediated immunity, 25 up-regulated proteins) and inflammation (leukocyte migration, 11 down-regulated proteins) (Fig. 4B and C)33,34. Moreover, 60 other regulated proteins were related with leukocyte activation and T cell differentiation (Supporting information Table S7).
Figure 4.
Proteomic profiling and cytokine levels of SARS-CoV-2 virus-infected Vero E6 cells upon schaftoside treatment. (A) Experimental scheme. Vero E6 cells were infected with SARS-CoV-2, or with schaftoside and SARS-CoV-2 for 24 h (MOI = 1), respectively. (B, C) The GO-based enrichment analysis of biologic processes that are differentially regulated by schaftoside (B, up-regulated; C, down-regulated). A two-tailed Fisher's exact test was employed to test the enrichment of the differentially expressed protein against all identified proteins (P < 0.05). (D) IL-1, TNF-α, IL-6, IL-1β, IL-8, IL-7, IFN-β and IFN-γ levels in Vero E6 cells measured by monkey ELISA kit. For the control group, Vero E6 cells were treated with SARS-CoV-2 for 24 h and were sonicated three times on ice using a high intensity ultrasonic processor (Scientz) in lysis buffer (8 mmol/L urea, 1% protease inhibitor cocktail). For the schaftoside group, Vero E6 cells were treated with schaftoside and SARS-CoV-2 for 24 h. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 compared with the control group, n = 3.
It has been reported that T cell responses are induced after SARS-CoV-2 infection35,36. Moreover, the spike protein carries immunodominant epitopes against which humoral B cell responses are generated upon natural infection37. In these immune processes, cytokine storm can be caused by the overproduction of early response proinflammatory cytokines38,39. Our ELISA results show the levels of cytokines IL-1, TNF-α, IL-6, IL-1β, IL-8, IL-7, IFN-β, and IFN-γ in the SARS-CoV-2 infected host cells decreased remarkably upon the treatment of schaftoside (Fig. 4D). These results indicate that schaftoside regulated immune response and inflammation of the host cells. A proposed mechanism is shown in Supporting Information Fig. S13.
3.5. Anti-inflammatory activities of schaftoside in ALI mice model
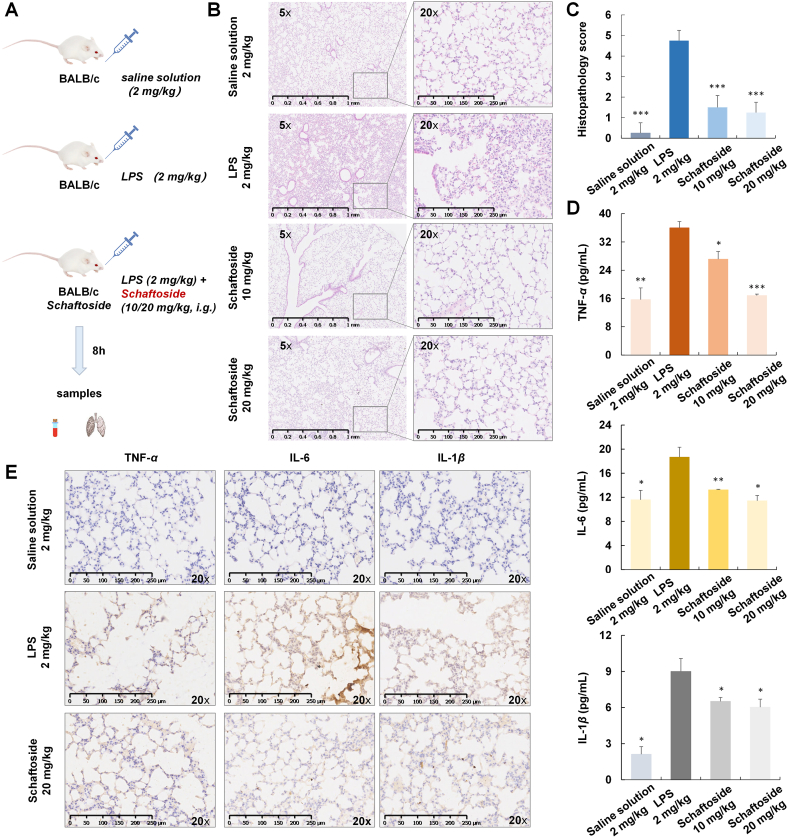
To further evaluate the anti-inflammatory activities of schaftoside, an ALI mice model was established by intranasal (i.n.) administration of 2 mg/kg LPS (Fig. 5A)40. After 8 h LPS stimulation, histopathology of the lung tissue changed remarkably. The locally widened alveolar septum and perivascular areas were infiltrated by lymphocytes and neutrophils. Moreover, microvascular hyperpermeability could lead to alveolar wall thickening, alveolar space congestion, and alveolar edema (Fig. 5B and Supporting Information Fig. S14). These injuries were alleviated upon the treatment of schaftoside (10 and 20 mg/kg) (Fig. 5C). The levels of proinflammatory cytokines TNF-α, IL-6, and IL-1β in mice blood samples decreased as well (Fig. 5D)41, which were consistent with lung tissue IHC staining (Fig. 5E). These results confirm that schaftoside possessed remarkable anti-inflammatory activities, and could down-regulate inflammatory cytokines.
Figure 5.
The anti-inflammatory activities of schaftoside on lipopolysaccharide (LPS)-induced acute lung injury (ALI) mice. (A) Experimental scheme40. (B) Effects of schaftoside (10 and 20 mg/kg, i.g.) on histological changes of the mice (n = 4). Representative images of mice lung tissue stained with HE after 8 h of LPS treatment. (C) Pathology evaluation for the therapeutic effect of schaftoside; n = 4 and ∗∗∗P < 0.00141. (D) TNF-α, IL-6, and IL-1β levels of blood samples measured by mice ELISA kit. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 compared with the control group, n = 4. (E) Immunohistochemistry of lung tissues of LPS-induced ALI mice for TNF-α, IL-6, and IL-1β analysis.
3.6. Druggability evaluation of schaftoside
The acute toxicity of schaftoside in mice was evaluated42. Male ICR mice (20 g) were treated with schaftoside (20, 40, and 300 mg/kg, i.g.) for 7 days, the mice weights remained at around 22 g without death (Supporting Information Fig. S15). No obvious histological changes were observed for the lung, heart, spleen, liver, and kidney tissues (Supporting Information Fig. S16). We also conducted a pharmacokinetic study of schaftoside in rats (i.p., 150 mg/kg). The maximum plasma concentration was 23.75 μmol/L, which was two-fold higher than the EC50 value (Supporting Information Fig. S17). The t1/2 value of schaftoside was 7.5 h. The above results indicated that schaftoside showed little toxicity and good pharmacokinetic property.
4. Discussion
A number of small molecule inhibitors of SARS-CoV-2 virus 3CLpro and PLpro have been reported in the past two years. Most of these compounds were obtained by protein structure-based rational design and chemical synthesis. In this work, we focus on the discovery of natural products from Chinese herbal medicines that have been widely used in clinical practice for the treatment of COVID-19. By screening 12 popular herbal medicines and 125 compounds from licorice, we find that schaftoside is a potent 3CLpro/PLpro dual-target inhibitor, and could remarkably inhibit SARS-CoV-2 virus with EC50 of 11.83 ± 3.23 μmol/L. Molecular docking, HDX-MS analysis, and site-directed mutagenesis indicated the mechanisms may be related with non-covalent interactions. This mechanism is different from most previously reported 3CLpro/PLpro inhibitors, which could form covalent bonding with protein residues like cysteine. More importantly, we find that schaftoside not only inhibits the virus, but also regulates the immune response and inflammation of host cells infected with SARS-CoV-2, according to proteomics analysis and cytokine assay. The anti-inflammatory activity of schaftoside was further confirmed on an LPS-induced acute-lung injury mice model.
Schaftoside is a popular natural product (Supporting Information Figs. S18 and S19). It is present in at least 184 species of higher plants from 39 families, and the contents could be as high as 2.78%43. These plants include medicinal herbs like licorice and Artemisia annua L. (Qinghao), which are components of many traditional Chinese medicine formulas for the treatment of COVID-19. Recently, we have discovered key C-glycosyltransferases involved in the biosynthesis of schaftoside, and established efficient in vitro biosynthetic routes. Given its easy accessibility and good safety and pharmacokinetic property, schaftoside may be a promising drug candidate for the treatment of COVID-19.
5. Conclusions
Our previous study has found that glycyrrhetinic acid and licorice-saponin A3 from licorice could potently inhibit SARS-CoV-2 spike protein, with EC50 of 3.17 and 0.075 μmol/L against SARS-CoV-2 virus in Vero E6 cells23, respectively. These compounds together with schaftoside are key effective components of licorice for the prevention and treatment of COVID-19. This work also demonstrates the multi-component, multi-target mode of action of herbal medicines.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Nos. 81891010/81891011, 81725023, 82003614, 82173950, 31770192, 32070187 and 82003681), China Postdoctoral Science Foundation (2022T150029, China), the National Key Research and Development Program of China (No. 2017-YFC1700405), and the Science & Technology Department of Xinjiang Uygur Autonomous Region (2018AB012, China). The authors thank Tao Du, Lun Wang, Jin Xiong, and the entire running team from Zhengdian Biosafety Level 3 Laboratory of Wuhan Institute of Virology for technical support, and Prof. Chao Zhong at the School of Basic Medical Sciences of Peking University for helpful discussions. We also thank the Wroclaw Center for Networking and Super Computing for providing generous computer time, and Jingjie PTM BioLab Co., Ltd. (Hangzhou, China) for proteomics mass spectrometry analysis.
Author contributions
Min Ye conceived this study. Yang Yi, Joachim Wlodarz and Szczepan Roszak conducted theoretical calculations. Yang Yi, Yi Kuang, Rong Yu and Yue Chai performed enzyme assay and pharmacokinetic (PK) studies. Heng Xue, Wei Hong, Junhua Li, Elishiba Muturi and Hongping Wei conducted antiviral and immunofluorescence assays. Meng Zhang, Yangoujie Bao and Yang Yi purified protein. Wen Ma conducted mass spectrometry analysis for PK experiments. Jing Wang completed SPR experiments. Xiaomeng Shi conducted hydrogen–deuterium exchange mass spectrometry analysis. Rong Yu and Yang Yi conducted anti-inflammation and toxicity tests in mice. Yang Yi and Wenzhe Li conducted immunohistochemistry analysis. Yang Yi, Xue Qiao, Hang Yang and Min Ye analyzed the data and wrote the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.07.017.
Contributor Information
Xue Qiao, Email: qiaoxue@bjmu.edu.cn.
Hang Yang, Email: yangh@wh.iov.cn.
Min Ye, Email: yemin@bjmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang C.L., Wang Y.M., Li X.W., Ren L.L., Zhao J.P., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . World Health Organization; 2022-06-15. WHO coronavirus (COVID-19) dashboard.https://covid19.who.int Available from: [Google Scholar]
- 3.Kimura I., Kosugi Y., Wu J.Q., Yamasoba D., Butlertanaka E.P., Tanaka Y.L., et al. SARS-CoV-2 lambda variant exhibits higher infectivity and immune resistance. BioRxiv. 2021 doi: 10.1016/j.celrep.2021.110218. Available from: https://doi.org/10.1101/2021.07.28.454085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su H.X., Yao S., Zhao W.F., Zhang Y.M., Liu J., Shao Q., et al. Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat Commun. 2021;12:3623. doi: 10.1038/s41467-021-23751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramajayam R., Tan K.P., Liang P.H. Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem Soc Trans. 2011;39:1371–1375. doi: 10.1042/BST0391371. [DOI] [PubMed] [Google Scholar]
- 6.Dai W.H., Zhang B., Jiang X.M., Su H.X., Jian Li, Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu L.F., Ye F., Feng Y., Yu F., Wang Q.S., Wu Y., et al. Both boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiao J.X., Li Y.S., Zeng R., Liu F.L., Luo R.H., Huang C., et al. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., et al. An oral SARS-CoV Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;24:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 10.Fu Z.Y., Huang B., Tang J.L., Liu S.Y., Liu M., Ye Y.X., et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat Commun. 2021;12:488. doi: 10.1038/s41467-020-20718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao X.P., Qin B., Chen P., Zhu K.X., Hou P.J., Wojdyla J.A., et al. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm Sin B. 2021;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osipiuk J., Azizi S.A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medicines and Healthcare products Regulatory Agency . GOV.UK; 2021-11-04. First oral antiviral for COVID-19, Lagevrio (molnupiravir), approved by MHRA.https://www.gov.uk/government/news/first-oral-antiviral-for-covid-19-lagevrio-molnupiravir-approved-by-mhra Available from: [Google Scholar]
- 14.Pfizer . Pfizer; 2021-12-22. Pfizer receives U.S. FDA emergency use authorization for novel COVID-19 oral antiviral treatment.https://www.pfizer.com/news/press-release/press-release-detail/pfizer-receives-us-fda-emergency-use-authorization-novel Available from: [Google Scholar]
- 15.Merck . MERCK; 2021-11-04. Merck and ridgeback’s molnupiravir, an oral COVID-19 antiviral medicine, receives first authorization in the world.https://www.merck.com/news/merck-and-ridgebacks-molnupiravir-an-oral-covid-19-antiviral-medicine-receives-first-authorization-in-the-world Available from: [Google Scholar]
- 16.Pfizer . worldpharmanews; 2021-11-18. Pfizer to provide U.S. government with 10 million treatment courses of investigational oral antiviral candidate to help combat COVID-19.https://www.worldpharmanews.com/pfizer/5863-pfizer-to-provide-u-s-government-with-10-million-treatment-courses-of-investigational-oral-antiviral-candidate-to-help-combat-covid-19 Available from: [Google Scholar]
- 17.Hu K., Guan W.J., Bi Y., Zhang W., Li L.J., Zhang B.L., et al. Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L.H., Zheng X., Bai X.K., Wang Q., Chen B.W., Qang H.B., et al. Association between use of Qingfei Paidu Tang and mortality in hospitalized patients with COVID-19: a national retrospective registry study. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2021.153531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X.F., Wu Y.L., Chen C., Gu Y.Q., Zhu C.Y., Wang S.P., et al. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen Capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm Sin B. 2021;11:222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C., Pan X.Y., Xu X.F., Cheng C., Huang Y., Li L., et al. Salvianolic acid C potently inhibits SARS-CoV-2 infection by blocking the formation of six-helix bundle core of spike protein. Signal Transduct Target Ther. 2020;5:220. doi: 10.1038/s41392-020-00325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suresh G., Badavath V.N., Thakur A., Yin N., De Jonghe S., Acevedo O., et al. Kobophenol A inhibits binding of host ACE2 receptor with spike RBD domain of SARS-CoV-2, a lead compound for blocking COVID-19. J Phys Chem Lett. 2021;12:1793–1802. doi: 10.1021/acs.jpclett.0c03119. [DOI] [PubMed] [Google Scholar]
- 22.Ji S., Li Z.W., Song W., Wang Y.R., Liang W.F., Li K., et al. Bioactive constituents of Glycyrrhiza uralensis (licorice): discovery of the effective components of a traditional herbal medicine. J Nat Prod. 2016;79:281–292. doi: 10.1021/acs.jnatprod.5b00877. [DOI] [PubMed] [Google Scholar]
- 23.Yi Y., Li J.H., Lai X.Y., Zhang M., Kuang Y., Bao Y.O.J., et al. Natural triterpenoids from licorice potently inhibit SARS-CoV-2 infection. J Adv Res. 2022;36:201–210. doi: 10.1016/j.jare.2021.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L.L., Chen K., Wang Z.L., Yi Y., Zhang M., Hasan A., et al. AmAT19, an acetyltransferase from Astragalus membranaceus, catalyses specific 6α-OH acetylation for tetracyclic triterpenes and steroids. Org Biomol Chem. 2021;19:7186–7189. doi: 10.1039/d1ob01106e. [DOI] [PubMed] [Google Scholar]
- 25.McLean A.D., Chandler G.S. Contracted Gaussian-basis sets for molecular calculations. J Chem Phys. 1980;72:5639–5648. [Google Scholar]
- 26.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 27.Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 28.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rappe A.K., Casewit C.J., Colwell K.S., III Goddard W.A., Skiff W.M.J. UFF. A full periodic table force field for molecular mechanics and molecular dynamics simulations. J Am Chem Soc. 1992;114:10024–10035. [Google Scholar]
- 30.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 09, revision E.01, Inc., Wallingford. 2013. Available from: https://gaussian.com.
- 31.Luo L., Jiang J.W., Wang C., Fitzgerald M., Hu W.F., Zhou Y.M., et al. Analysis on herbal medicines utilized for treatment of COVID-19. Acta Pharm Sin B. 2020;10:1192–1204. doi: 10.1016/j.apsb.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Geneset enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nomani M., Varahram M., Tabarsi P., Hashemian S.M., Jamaati H., Malekmohammad M., et al. Decreased neutrophil-mediated bacterial killing in COVID-19 patients. Scand J Immunol. 2021;94 doi: 10.1111/sji.13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu G., Qi F.R., Li H.J., Yang Q.T., Wang H.Y., Wang X., et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Disc. 2020;6:73. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breton G., Mendoza P., Hagglof T., Oliveira T.Y., Schaefer-Babajew D., Gaebler C., et al. Persistent cellular immunity to SARS-CoV-2 infection. bioRxiv. 2021 doi: 10.1101/2020.12.08.416636. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni L., Ye F., Cheng M., Feng Y., Deng Y.Q., Zhao H., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou F., Yu T., Du R.H., Fan G.H., Liu Y., Liu Z.B., et al. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet. 2020;395:1054–1062. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Veerdonk F.L., Giamarellos-Bourboulis E., Pickkers P., Derde L., Leavis H., van Crevel R., et al. A guide to immunotherapy for COVID-19. Nat Med. 2022;28:39–50. doi: 10.1038/s41591-021-01643-9. [DOI] [PubMed] [Google Scholar]
- 40.Bai Z.F., Li P.Y., Wen J.C., Han Y.Z., Cui Y.Y., Zhou Y.F., et al. Inhibitory effects and mechanisms of the anti-COVID-19 traditional Chinese prescription, Keguan-1, on acute lung injury. J Ethnopharmcol. 2022;285 doi: 10.1016/j.jep.2021.114838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szarka R.J., Wang N.D., Gordon L., Nation P.N., Smith R.H. A murine model of pulmonary damage induced by lipopolysaccharide via intranasal instillation. J Immunol Methods. 1997;202:49–57. doi: 10.1016/s0022-1759(96)00236-0. [DOI] [PubMed] [Google Scholar]
- 42.Chang X., Dong M., Mi X., Hu M., Lu J., Chen X. The protective effect of Trichilia catigua A. Juss. on DEHP-induced reproductive system damage in male mice. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.832789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z.L., Gao H.M., Wang S., Zhang M., Chen K., Zhang Y.Q., et al. Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc Natl Acad Sci U S A. 2020;117:30816–30823. doi: 10.1073/pnas.2012745117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.