Abstract
A new gene, bkdR (formerly called yqiR), encoding a regulator with a central (catalytic) domain was found in Bacillus subtilis. This gene controls the utilization of isoleucine and valine as sole nitrogen sources. Seven genes, previously called yqiS, yqiT, yqiU, yqiV, bfmBAA, bfmBAB, and bfmBB and now referred to as ptb, bcd, buk, lpd, bkdA1, bkdA2, and bkdB, are located downstream from the bkdR gene in B. subtilis. The products of these genes are similar to phosphate butyryl coenzyme A transferase, leucine dehydrogenase, butyrate kinase, and four components of the branched-chain keto acid dehydrogenase complex: E3 (dihydrolipoamide dehydrogenase), E1α (dehydrogenase), E1β (decarboxylase), and E2 (dihydrolipoamide acyltransferase). Isoleucine and valine utilization was abolished in bcd and bkdR null mutants of B. subtilis. The seven genes appear to be organized as an operon, bkd, transcribed from a −12, −24 promoter. The expression of the bkd operon was induced by the presence of isoleucine or valine in the growth medium and depended upon the presence of the sigma factor SigL, a member of the sigma 54 family. Transcription of this operon was abolished in strains containing a null mutation in the regulatory gene bkdR. Deletion analysis showed that upstream activating sequences are involved in the expression of the bkd operon and are probably the target of bkdR. Transcription of the bkd operon is also negatively controlled by CodY, a global regulator of gene expression in response to nutritional conditions.
In Bacillus subtilis, the sigL gene encodes a sigma factor homologous to members of the RpoN family of sigma factors (5). Promoters recognized by an RNA polymerase associated with RpoN have common features: (i) they are devoid of typical −10 and −35 sequences but contain a strongly conserved TGGCAC N5 TTGCA sequence centered at positions −12 and −24, and (ii) they require a positive regulatory protein with a central domain (the catalytic domain) which includes a conserved nucleotide-binding pocket. This positive regulatory protein interacts with upstream activating sequences (UAS) to stimulate the isomerization of closed complexes between RNA polymerase and the promoter DNA sequences to open complexes (21). RpoN differs from other alternative sigma factors in that it is needed for the transcription of genes whose products have diverse physiological roles.
In B. subtilis, sigL mutants have a pleiotropic phenotype: transcription of the levanase operon is strongly reduced, and catabolism of several amino acids (arginine, ornithine, isoleucine, and valine) is abolished (5). Most studies of the catabolism of branched-chain amino acids have been done with Pseudomonas, Streptomyces, and Streptococcus species. In these bacteria, the catabolism of branched-chain amino acids requires the cooperation of two sequential series of reactions. The enzymes of the first series constitute a common pathway and catalyze the conversion by deamination or dehydrogenation of Leu, Val, and Ile to their respective 2-keto acids. Branched-chain 2-keto acid dehydrogenase, which catalyzes the second step in this initial process, is a multienzyme complex (BCDH complex) involved in the oxidation of the 2-keto acid derivatives of all three branched-chain amino acids. The acyl coenzyme A metabolites formed at the end of the common pathway are catabolized by a series of enzymes, one specific for each initial amino acid (19).
The BCDH complex from several sources has been characterized; these include Pseudomonas aeruginosa (20), Pseudomonas putida (32), B. subtilis (12), rabbit liver (25), and rat and bovine kidneys (24, 26). The purified complexes from B. subtilis, P. putida, P. aeruginosa, and several mammals are all composed of four polypeptides: a dehydrogenase (E1α), decarboxylase (E1β), a dihydrolipoamide acyltransferase (E2), and a dihydrolipoamide dehydrogenase (E3). In B. subtilis, the BCDH complex is involved in the biosynthesis of branched-chain fatty acids, which are the major acyl constituents of the cell membrane (37). A bfmB mutant of B. subtilis requiring short branched-chain carboxylic acids for growth has been described. It is defective in branched-chain 2-keto acid dehydrogenase. Three genes, bfmBAA, bfmBAB, and bfmBB, encode the E1α, E1β, and E2 components, respectively, of the BCDH complex involved in the biosynthesis of branched-chain fatty acids (35). Little is known concerning the regulation of the expression of the genes involved in isoleucine and valine catabolism in B. subtilis. A sigL mutant cannot use isoleucine or valine as a source of nitrogen, suggesting that the expression of one or several enzymes of the isoleucine and valine degradation pathway is controlled by a transcriptional regulator with a central domain (5).
In this paper, we characterize an operon containing seven genes involved in the isoleucine and valine degradation pathway. Transcription of this operon is induced by the presence of isoleucine or valine in the growth medium and strongly depends on SigL. We also describe a new transcriptional regulator, BkdR, which has a central domain and which is an activator of the transcription of this operon.
MATERIALS AND METHODS
Bacterial strains and culture media.
The B. subtilis strains used in this work are listed in Table 1. Escherichia coli TGI [K-12 Δ(lac-pro) supE thi hsd5/F′ traD36 proA+B+ lacIq lacZΔM15] was used for cloning experiments. E. coli was grown in LB broth (27), and B. subtilis was grown in SP medium (8 g of nutrient broth/liter, 1 mM MgSO4, 10 mM KCl, 0.5 mM CaCl2, 10 μM MnCl2, 2 μM FeSO4) or MM minimal medium [60 mM K2HPO4, 44 mM KH2PO4, 15 mM (NH4)2SO4, 3 mM trisodium citrate, 2 mM MgSO4, 2.2 mg of ferric ammonium citrate per liter] supplemented with carbon sources (0.1%) and auxotrophic requirements (at 100 mg/liter). TPK minimal medium contains 50 mM glucose, 100 mM potassium phosphate (pH 7.0), 0.5 mM MgSO4, 0.01 mM MnSO4, 0.02 mM FeCl, 50 μg of tryptophan per ml, and 20 mM nitrogen source.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype or descriptiona | Source or referenceb |
|---|---|---|
| 168 | trpC2 | 1 |
| QB5505 | trpC2 sigL::aphA3 | 5 |
| QB7501 | trpC2 amyE::ptb′-′lacZ (ΔA) | This work |
| QB7502 | trpC2 amyE::ptb′-′lacZ sigL::aphA3 | QB5505 DNA→QB7501 |
| QB7505 | trpC2 amyE::ptb′-′lacZ tnrA::erm | SF706T DNA→QB7501 |
| QB7507 | trpC2 lpd::pMutin4 | This work |
| QB7511 | trpC2 amyE::ptb′-′lacZ bkdR::aphA3 | pBkdR2 lin→QB7501 |
| QB7512 | trpC2 bkdR::aphA3 | pBkdR2 lin→168 |
| QB7513 | trpC2 lpd::pMutin4 bkdR::aphA3 | pBkdR2 lin→QB7507 |
| QB7514 | trpC2 bcd::pMutin4 | This work |
| QB7515 | trpC2 bcd::pMutin4 bkdR::aphA3 | pBkdR2 lin→QB7514 |
| QB7517 | trpC2 amyE::ptb′-′lacZ (ΔB) | This work |
| QB7518 | trpC2 amyE::ptb′-′lacZ (ΔC) | This work |
| QB7519 | trpC2 amyE::ptb′-′lacZ (ΔD) | This work |
| QB7520 | trpC2 amyE::ptb′-′lacZ (ΔE) | This work |
| QB7521 | trpC2 bkdB::pMutin4 | This work |
| QB7522 | trpC2 amyE::ptb′-′lacZ codY::erm | PS258 DNA→QB7501 |
| QB7530 | trpC2 amyE::ptb′-′lacZ bkdR::aphA3 pBkdR1 | This work |
| PS258 | trpC2 codY::erm | A. L. Sonenshein |
| SF706T | trpC2 tnrA::Tn917 amyE::gabP-lacZ | 38 |
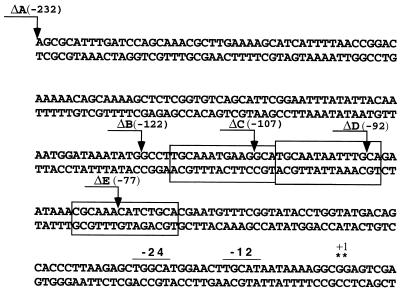
ΔA, ΔB, ΔC, ΔD, and ΔE are defined in Fig. 5.
lin, linearized. pBkdR1 and pBkdR2 were constructed as discussed in Materials and Methods.
Transformation and phenotype characterization.
Standard procedures were used to transform E. coli (27), and transformants were selected on LB broth plates containing ampicillin (100 μg/ml). B. subtilis was transformed with plasmid or chromosomal DNA as previously described (16), and transformants were selected on SP medium plates containing chloramphenicol (5 μg/ml), kanamycin (5 μg/ml), erythromycin (1 μg/ml), and lincomycin (10 μg/ml). Amylase activity in B. subtilis was detected after growth on tryptose blood agar base (Difco) containing 10 g of hydrolyzed starch per liter (Connaught). Starch degradation was detected by sublimating iodine onto the plates.
DNA manipulation.
Standard procedures were used to extract plasmid from E. coli (27). Restriction enzymes, phage T4 DNA polymerase, phage T4 DNA ligase, and T4 polynucleotide kinase were used as recommended by the manufacturers. DNA fragments were purified from the agarose gel with a Prep-A-Gene kit (Bio-Rad Laboratories, Richmond, Calif.). The PCR technique with Thermus aquaticus DNA polymerase was used for amplification. The oligonucleotide primers used included mismatches allowing the creation of EcoRI and BamHI restriction sites or HindIII and BamHI restriction sites.
Plasmid constructions.
pAC5 (17), a derivative of pAF1 (6), carries the pC194 chloramphenicol resistance gene cat and a lacZ gene between two fragments of the B. subtilis amyE gene. PCR was used to introduce EcoRI restriction sites at various positions upstream from ptb. PCR was performed with one oligonucleotide (5′-GGAGGATCCTCAGCATGAGCAAC-3′) corresponding to the coding sequence of the ptb gene (codons 20 to 25) and the other one corresponding to various positions in the ptb promoter region. The EcoRI-BamHI restriction fragments generated in this way were inserted between the EcoRI and BamHI restriction sites of pAC5, creating translational fusions between codon 25 of ptb and codon 8 of lacZ. The DNA sequences of the different PCR fragments were verified by direct sequencing of the various corresponding plasmids. The resulting plasmids were linearized at the single PstI restriction site and integrated into the chromosome of strain 168 by homologous recombination at the amyE locus by use of chloramphenicol selection. Integrants carrying the translational fusions were named QB7501, QB7517, QB7518, QB7519, and QB7520 (see Fig. 5 and Table 1).
FIG. 5.
Nucleotide sequence of the ptb upstream region. Deletion end points are indicated by arrows and numbered with respect to the transcriptional start site (indicated by asterisks). The sequences at positions −12 and −24 are indicated. Boxed regions indicate putative UAS. The effects of upstream deletions on the expression of the ptb′-′lacZ translational fusions are expressed as β-galactosidase specific activities, which were determined with extracts prepared from cells growing exponentially in MM minimal medium containing glucose and 20 mM isoleucine as the inducer. In the absence of isoleucine, the basal level was about 5 U/mg of protein for each strain. For deletion end points ΔA (−232), ΔB (−122), ΔC (−107), ΔD (−92), and ΔE (−77), β-galactosidase activities were 2,350, 2,135, 205, 5, and 10 U/mg of protein, respectively.
Gene disruptions and transcriptional fusions with the pMutin4 vector (34) were constructed by PCR amplification of an internal segment of the target gene, ligation of the amplified DNA fragment into pMutin4 in E. coli, and insertion of the plasmid into the B. subtilis chromosome. PCR amplifications were done with the following oligonucleotides: 5′-GCCGAAGCTTGGCCGCTATATTCAAGG-3′ and 5′-CGCGGATCCGGCTACGTTCCCAACAC-3′ for the bcd gene; 5′-GCCGAAGCTTCTGTTGAGCGGGGAAAT-3′ and 5′-CGCGGATCCGGCAGCACTTTTGCCCC-3′ for the lpd gene; and 5′-GCCGAAGCTTGACCTCGATCAAGTGAC-3′ and 5′-CGCGGATCCAATTTTGTCCCCCGCCC-3′ for the bkdB gene. Target genes were interrupted by Campbell-type crossover integrations. The integration of the recombinant plasmids places the downstream genes under the control of the spac promoter regulated by isopropyl-β-d-thiogalactopyranoside (IPTG) and fuses the target gene to the lacZ gene, leading to a transcriptional fusion.
pBkR1, which contains the wild-type bkdR regulatory gene, was constructed as follows. Two DNA fragments encoding the amino-terminal part and the carboxy-terminal part of BkdR were synthesized by PCR with the following respective pairs of oligonucleotides: (i) 5′-CCGGAATTCGTGGTAACATAGGGTTG-3′ and 5′-TCCCCCGGGTTCCCGGCATGACAATG-3′ and (ii) 5′-TCCCCCGGGTATCCGATCTCCATCC-3′ and 5′-CGCGGATCCGGGTGCTGTCATACCAGG-3′. The two DNA fragments were ligated into pHT315 (2) between the EcoRI and BamHI restriction sites and on each side of a DNA fragment containing the aphA3 gene (33), leading to pBkR2.
The interrupted gene was introduced into the chromosomes of strain 168 to give strain QB7512 and strain QB7501 to give strain QB7511. The wild-type bkdR gene was cloned by homologous recombination as follows. Strain 168 containing pBkR2 was grown in LB medium containing erythromycin. Plasmid DNA was extracted and used to transform strain QB7511 containing the ptb′-′lacZ fusion. Transformants were selected on SP medium plates containing chloramphenicol and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). One blue colony was reisolated, and plasmid DNA (pBkR1) was extracted. This plasmid contains the entire bkdR gene.
Reverse transcriptase mapping of the mRNA start point in the ptb gene.
Total RNA was isolated from B. subtilis 168 grown in MM medium supplemented with glucose and tryptophan and with or without 20 mM isoleucine as the inducer. Exponentially growing cells were harvested at an optical density at 600 nm of 1, and RNA was extracted as previously described (8). Two oligonucleotides, O1 (5′-CCTCAGCATGAGCAACCGCAATGGTC-3′) and O2 (5′-GTGTATCGACGCTTTGCCGATTAAATC-3′), complementary to the ptb coding sequence were labelled with 10 U of polynucleotide kinase and 0.37 MBq of [γ-32P]ATP (15 TBq/mmol; Amersham). DNA primers were elongated, and the products were analyzed as previously described (15) and shown to have the same transcriptional start sites.
β-Galactosidase assays.
B. subtilis cells containing lacZ fusions were grown to an optical density at 600 nm of 1. β-Galactosidase specific activities were determined as previously described and are expressed as Miller units per milligram of protein (5). The values reported represent averages from at least three independent assays.
RESULTS
Identification of an activator involved in isoleucine utilization in B. subtilis.
Members of the family of activators of ς54-dependent transcription are composed of three distinct functional domains, an NH2-terminal domain, a central domain, and a COOH-terminal domain, involved in signal reception, transcriptional activation, and DNA binding, respectively. The central domain and the C-terminal domain are the most highly conserved among these regulators (30). Degenerate oligonucleotide primers have been used to amplify DNA fragments encoding central domains from the genomes of diverse bacteria, including B. subtilis. This procedure led to the identification of both known genes (levR and rocR) and two novel gene fragments, called 70-Bsu and 81-Bsu (9). The 70-Bsu DNA fragment (a gift from I. Kaufman and T. Nixon) was cloned in pHT181, an integrative plasmid (11). The recombinant plasmid was used to transform B. subtilis 168. Since the 70-Bsu DNA fragment is an internal fragment of the gene, the integration led to a null mutation in the corresponding chromosomal gene. The B. subtilis mutant strain containing pHT181::70-Bsu inserted by homologous recombination in the corresponding gene was tested for growth in TPK minimal medium containing isoleucine (20 mM) as the sole nitrogen source. This strain grew much more slowly than the wild-type strain in the same medium containing isoleucine (data not shown). The mutant strain and the wild-type strain grew well with (NH4)2SO4 as the sole nitrogen source. These results strongly suggest that the DNA fragment encoding a new central domain is part of a regulatory gene controlling isoleucine utilization in B. subtilis.
A new gene, bkdR (formerly yqiR), was found during the B. subtilis genome sequencing project (10). This gene encodes a protein with a central domain identical to the peptide deduced from the 70-Bsu fragment (9). The polypeptide deduced from the DNA sequence of bkdR contains 692 residues with a calculated molecular weight of 77,747. We constructed a B. subtilis strain in which bkdR was disrupted by the aphA3 gene: this disruption was created by transformation and recombination into the chromosome of wild-type B. subtilis 168, leading to strain QB7512.
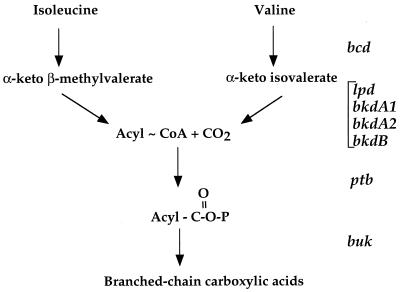
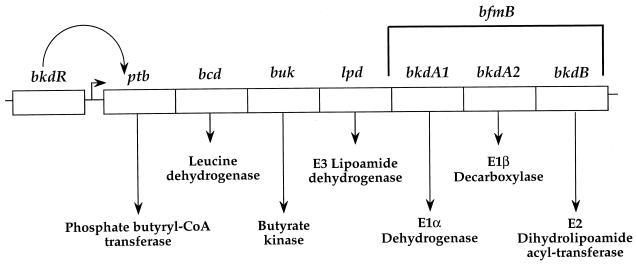
Strain QB7512 was tested for growth in TPK minimal medium containing the branched-chain amino acid isoleucine or valine as the sole nitrogen source (Table 2). It did not grow in the presence of isoleucine or valine as the sole nitrogen source, confirming that the product of the bkdR gene is involved in the control of the catabolism of these amino acids. bkdR and sigL null mutants can grow with (NH4)2SO4 as the sole nitrogen source. However, the bkdR mutant grows slightly more slowly than the sigL mutant (see Discussion). The products of several genes located downstream from bkdR are presumably involved in the metabolism of branched-chain amino acids, since they show strong similarities to phosphate butyryltransferase, leucine dehydrogenase, butyrate kinase, and the E3, E1α, E1β, and E2 components of the BCDH complex (Fig. 1 and 2) (22). As shown below, these genes probably form an operon containing the following genes: ptb, bcd, buk, lpd, bkdA1, bkdA2, and bkdB (formerly yqiS, yqiT, yqiU, yqiV, bfmBAA, bfmBAB, and bfmB, respectively) (Fig. 2).
TABLE 2.
Doubling time of bkd mutants in TPK minimal medium containing ammonium, isoleucine, or valine as a nitrogen source
| Strain | Relevant genotype | Doubling time (min) with indicated nitrogen source at 20 mM:
|
||
|---|---|---|---|---|
| (NH4)2SO4 | Isoleucine | Valine | ||
| 168 | 85 | 130 | 155 | |
| QB7512 | bkdR::aphA3 | 150 | >600 | >600 |
| QB5505 | sigL::aphA3 | 70 | >600 | >600 |
| QB7514 | ||||
| Without IPTG | bcd::pMutin4 | 80 | >600 | >600 |
| With IPTG | 76 | >600 | >600 | |
| QB7507 | ||||
| Without IPTG | lpd::pMutin4 | 75 | 170 | 200 |
| With IPTG | 115 | 160 | 190 | |
FIG. 1.
Proposed pathway for the degradation of branched-chain amino acids in B. subtilis.
FIG. 2.
Organization of the structural genes of the bkd operon of B. subtilis. The proposed functions of the gene products are based on similarities to the corresponding genes from Enterococcus faecalis (36). CoA, coenzyme A.
To test whether the transcription of these genes is induced by isoleucine or valine in the growth medium, a ptb′-′lacZ translational fusion was constructed and integrated at the amyE locus of B. subtilis 168, leading to strain QB7501. The level of β-galactosidase expression in this strain was assayed (Table 3), and the transcription of ptb was indeed induced by isoleucine or valine in the growth medium. Transcription was also induced, albeit to a lesser extent, when α-keto acids corresponding either to isoleucine (α-keto-β-methylvalerate) or to valine (α-ketoisovalerate) were present in the growth medium. A sigL::aphA3 null mutation was introduced by transformation of fusion strain QB7501 to give strain QB7502. The expression of the ptb′-′lacZ translational fusion strongly depended upon the sigL gene product (Table 3). The product of the bkdR gene shows similarities to transcriptional activators required to stimulate −12, −24 promoters. A bkdR::aphA3 null mutation was introduced by transformation into the chromosome of strain QB7501, leading to strain QB7511. The β-galactosidase activity in cultures of this strain in MM glucose minimal medium containing 20 mM isoleucine or valine as the inducer was assayed. The expression of the ptb′-′lacZ fusion was extremely low in the absence of BkdR (Table 3). BkdR is therefore required for full induction of ptb transcription. pBkdR1, containing the entire coding sequence of the positive regulator, was introduced by transformation into strain QB7511, leading to strain QB7530. Transformants were grown in MM glucose minimal medium containing isoleucine or valine. The intact bkdR gene restored ptb transcription (Table 3), confirming that BkdR stimulates transcription via the synthesis of an activator protein.
TABLE 3.
Effects of bkdR and sigL mutations on ptb′-′lacZ expression
| Strain | Relevant genotype | β-Galactosidase activity (Miller units/mg of protein) in the following minimal mediuma:
|
||||
|---|---|---|---|---|---|---|
| MM | MM + isoleucine (20 mM) | MM + α-keto-β-methylvalerate (20 mM) | MM + valine (20 mM) | MM + α-keto-isovalerate (20 mM) | ||
| QB7501 | amyE::ptb′-′lacZ | 55 | 2,265 | 950 | 1,145 | 400 |
| QB7502 | amyE::ptb′-′lacZ sigL::aphA3 | 5 | 5 | ND | 5 | ND |
| QB7511 | amyE::ptb′-′lacZ bkdR::aphA3 | 5 | 1 | ND | 5 | ND |
| QB7530 | amyE::ptb′-′lacZ bkdR::aphA3 pBkdR1 | 140 | 2,440 | ND | 2,120 | ND |
ND, not determined.
Promoter and control regions located upstream from the ptb gene.
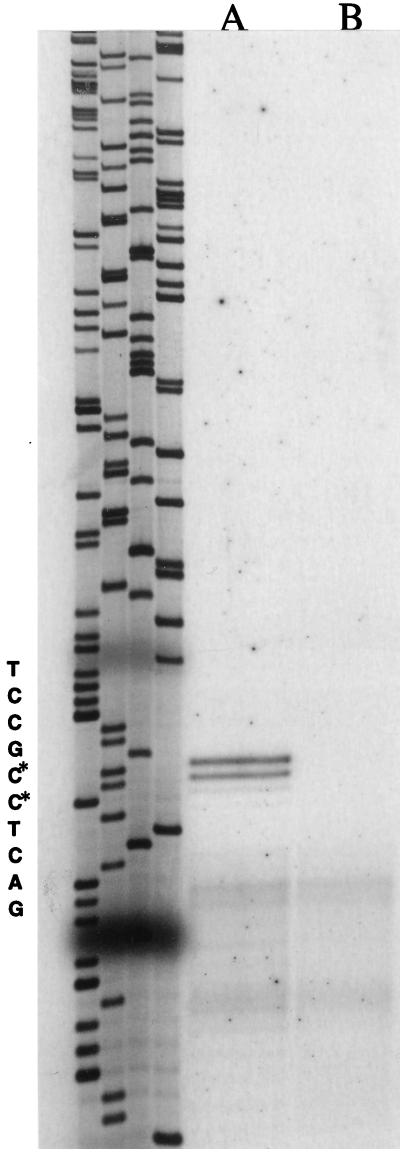
In gram-negative and gram-positive bacteria, all promoters recognized by holoenzymes containing ς54 possess at least a conserved GG doublet located at position −24 upstream from the transcriptional start site, followed by a GC doublet at position −12, with a spacing region of 10 bp. They do not have the typical −10 and −35 hexamers recognized by the major housekeeping sigma factor (21). The transcriptional start site of ptb was mapped by primer extension with reverse transcriptase and two oligonucleotides, O1 and O2. The same 5′ ends were identified with each oligonucleotide (Fig. 3). Transcription started at two adjacent nucleotides. The promoter region contains a region consistent with the consensus sequence (−12 and −24) of ς54-dependent promoters (Fig. 4).
FIG. 3.
Reverse transcriptase mapping of the transcriptional start site of the ptb gene in B. subtilis 168 grown in the presence (lane A) or absence (lane B) of 20 mM isoleucine. The positions of the cDNA-extended fragments identified with oligonucleotide O1 were compared with those obtained by sequencing of an M13 recombinant phage containing the promoter region with the same oligonucleotide used as a primer (lanes at left, TCGA, respectively, from left to right). Transcriptional start sites are indicated by the asterisks.
FIG. 4.
Nucleotide sequences of promoter regions of the levanase operon (plev), the rocABC operon (procABC), the rocDEF operon (procDEF), and the bkd operon (pbkd). The transcriptional start sites are indicated by arrows. Boxes indicate strictly conserved DNA sequences around positions −12 and −24 with respect to the transcriptional start sites.
ς54-dependent promoters contain regulatory sequences, called UAS, which bind to specific activators. Many of the binding sequences are inverted repeats that can be moved away from their original positions without losing their ability to allow transcriptional activation (4, 13, 17). To determine if such sequences were necessary for the expression of the ptb gene, a set of DNA fragments from which parts of the upstream region were deleted were constructed by PCR. These fragments, which contain the beginning of the ptb gene and the promoter region, were cloned upstream from the lacZ gene (see Materials and Methods). Deletion end points are indicated in Fig. 5. Fusions were reintroduced as single copies at the amyE locus of B. subtilis 168. lacZ expression in the resulting strains in MM glucose minimal medium containing isoleucine was assayed (Fig. 5).
lacZ expression was higher for ΔB than for ΔC deletions, indicating that DNA sequences located between the ΔB and ΔC end points are necessary for full promoter activation. There was no lacZ expression in ΔD (or ΔE) strains. Thus, DNA sequences near position −107 (the ΔC end point) with respect to the transcriptional start site are necessary for full induction of the ptb gene. Interestingly, three copies of an imperfect palindromic DNA sequence (Fig. 5) are present upstream from the ptb −12, −24 promoter. Since deletion analysis showed a sharp decrease in the rate of β-galactosidase synthesis when these palindromes were absent, the product of bkdR may interact with these repeated sequences to stimulate transcription.
Induction of the expression of the bcd, lpd, and bkdB genes.
pMutin4 (34) was used to construct transcriptional fusions with the lacZ gene. These constructions were integrated by homologous recombination into the bcd, lpd, and bkdB genes, leading to strains QB7514, QB7507, and QB7521, respectively. The recombination event was mediated by DNA fragments generated by PCR, inserted into the vector, and corresponding to the internal part of each gene. Integration of the plasmids (i) inactivates the corresponding gene; (ii) fuses the target gene to the lacZ gene; and (iii) allows the expression of downstream genes from an IPTG-inducible promoter, pspac. The β-galactosidase activities of strains QB7514, QB7507, and QB7521 were assayed after culturing in MM glucose minimal medium containing or not containing 20 mM isoleucine as the inducer (Table 4). The β-galactosidase expression in strains QB7514, QB7507, and QB7521 was induced by 20 mM isoleucine in the growth medium. The uninduced level of β-galactosidase expression was about 300 U/mg of protein. The addition of 1 mM IPTG to the growth medium led to a strong decrease in lacZ expression from the bcd′-lacZ fusion, suggesting that the product of one or several genes downstream from bcd represses the −12, −24 promoter (see Discussion). A bkdR::aphA3 null mutation was introduced into the bcd′-lacZ strain and into the lpd′-lacZ strain by transformation, leading to strains QB7515 and QB7513, respectively. β-Galactosidase was not expressed in cultures of these strains, whether or not they contained isoleucine as the inducer. Attempts to combine a bkdB::pMutin4 null mutation with either a sigL or a bkdR null mutation were unsuccessful, suggesting that three of the four subunits of the BCDH complex are essential for cell viability.
TABLE 4.
Induction of the expression of bcd, lpd, and bkdB genes
| Strain | Relevant genotype | β-Galactosidase activity (Miller units/mg of protein) in the following minimal mediuma:
|
|||
|---|---|---|---|---|---|
| MM + glucose
|
MM + glucose + isoleucine (20 mM)
|
||||
| Without IPTG | With IPTG | Without IPTG | With IPTG | ||
| QB7514 | bcd::pMutin4 | 355 | 10 | 1,170 | 1,100 |
| QB7515 | bcd::pMutin4 bkdR::aphA3 | 2 | 2 | 3 | 2 |
| QB7507 | lpd::pMutin4 | 320 | 280 | 2,800 | 2,630 |
| QB7513 | lpd::pMutin4 bkdR::aphA3 | 5 | 5 | 3 | 4 |
| QB7521 | bkdB::pMutin4 | 290 | ND | 1,150 | ND |
ND, not determined.
The results presented above indicated that the expression of the ptb, bcd, lpd, and bkdB genes is induced by isoleucine in the growth medium. Since transcriptional activation of ptb, bcd, and lpd also requires the product of bkdR, it is likely that these genes are cotranscribed as an operon.
Disruption of bkdR and phenotypes of bcd, lpd, and bkdB mutants.
B. subtilis QB7512 (containing the aphA3 gene inserted into bkdR) cannot grow in the presence of the branched-chain amino acids isoleucine and valine as the sole nitrogen sources. Thus, the product of the bkdR gene is involved in the control of the catabolism of these amino acids (Table 2). A similar phenotype was observed for strain QB7514, in which the pMutin4 vector was integrated into the bcd gene (Table 2). This strain was unable to use isoleucine or valine as a sole nitrogen source. Moreover, the doubling time with ammonia as a nitrogen source was unaffected. This result indicates that the bcd gene product, which shows similarities to leucine dehydrogenases, is involved in isoleucine and valine catabolism.
pMutin4 was also integrated into the lpd gene (strain QB7507). The integration of the plasmid prevents the production of a polypeptide showing similarities to the E3 component of the BCDH complex. As expected, the growth of this strain was unaffected when isoleucine or valine was used as a nitrogen source in the growth medium. Indeed, in this case, the functional bcd gene product led to the production of ammonia and α-keto acids. This result shows that the lpd gene is not essential in B. subtilis. However, two other lipoamide dehydrogenase homologs exist in B. subtilis, and we cannot exclude complementation of the lpd null mutant. Moreover, the pspac promoter is not tightly controlled by the product of lacI, leading to a low level of transcription of the downstream genes (34). Strain QB7521 (bkdB::pMutin4) grew slowly, and the cells lysed in both rich and minimal media, indicating that the product of bkdB is necessary for normal growth.
Regulation of the operon in response to nitrogen or amino acid availability.
Two B. subtilis proteins, TnrA and CodY, are known to be involved in regulation by the nitrogen source and by an excess of amino acids, respectively. TnrA is a transcriptional factor required for global nitrogen regulation in B. subtilis (38). During nitrogen limitation, the TnrA protein is required for activation of the transcription of gabP, nasB, and nrgAB and negatively regulates glnRA expression. We introduced a trnA::erm mutation into strain QB7501 to give strain QB7505. lacZ expression in cultures of this strain during growth with the poor nitrogen source glutamate was assayed. In the absence of an inducer, lacZ expression in MN (7) glucose minimal medium containing glutamate was 4.5-fold higher in strain QB7505 than in strain QB7501. These results suggest that the tnrA gene product may act as a repressor of the operon. The presence of 20 mM isoleucine did not affect the induced level of expression in tnrA mutants (data not shown; see Discussion).
The expression of several genes and operons, including those for histidine degradation (hut), proline degradation, aconitase, and dipeptide permease (dpp), is repressed when the medium contains a mixture of amino acids. To assess the role of nutritional repression by amino acids, strain QB7501 was grown in MM glucose minimal medium with and without Casamino Acids (0.2% final concentration). The presence of Casamino Acids in MM glucose minimal medium containing 20 mM isoleucine as the inducer led to a 3.5-fold decrease in lacZ expression (Table 5). CodY is a global regulator of gene expression that mediates amino acid repression of the hut and dpp operons and the genetic competence pathway (28, 29, 31). We thus introduced a codY::erm mutation into strain QB7501 to give strain QB7522. Strain QB7522 was grown in MM minimal glucose medium containing isoleucine or Casamino Acids. The codY::erm mutation led to derepression of the ptb′-′lacZ fusion (Table 5). The mixture of amino acids did not repress expression in the codY::erm strain. The codY mutation appears, therefore, to relieve amino acid repression of the bkd operon, as previously observed for the dpp and hut operons. The codY mutation did not alter the expression of the fusion in MM glucose minimal medium containing isoleucine. However, partial constitutive expression of the ptb′-′lacZ fusion was observed in the absence of isoleucine.
TABLE 5.
Effect of a codY mutation on the expression of the bkd operon
| Strain | Relevant genotype | β-Galactosidase activity (Miller units/mg of protein) in the following minimal medium:
|
|||
|---|---|---|---|---|---|
| MM + glucose | MM + glucose + isoleucine (20 mM) | MM + glucose + 0.2% Casamino Acids | MM + glucose + 0.2% Casamino Acids + isoleucine (20 mM) | ||
| QB7501 | ptb′-′lacZ | 20 | 1,975 | 45 | 575 |
| QB7522 | ptb′-′lacZ codY::erm | 140 | 2,255 | 765 | 2,325 |
DISCUSSION
The product of sigL is involved in the catabolism of isoleucine or valine present as the sole nitrogen source (5). In this work, we have identified an operon, the bkd operon, containing seven genes and involved in the deamination of isoleucine and valine and in the oxidative decarboxylation of branched-chain α-keto acids. Recently, a cluster of seven genes with a similar organization and involved in the breakdown of branched-chain amino acids was found in Enterococcus faecalis. Four of these genes, ldp, bkdA1, bkdA2, and bkdB, encode the E3, E1α, E1β, and E2 components of a BCDH complex. The genes preceding lpd in E. faecalis, ptb and buk, are similar to phosphotransbutyrylase and butyrate kinase genes, respectively, and are probably involved in ATP generation (36).
Here, we have shown that transcription of the bkd operon in B. subtilis is induced by the presence of isoleucine or valine in the growth medium. The α-keto acids α-keto-β-methylvalerate and α-ketoisovalerate are also inducers, a situation previously observed with P. putida (14). In a bcd null mutant (QB7514), the β-galactosidase expression observed in the absence of isoleucine in the growth medium was decreased by the presence of IPTG, which induces the expression of the downstream genes. As branched-chain amino acids or α-keto acids are probably internal inducers, it is likely that the intracellular levels of these inducers depend upon the expression of the downstream genes lpd, bkdA1, bkdA2, and bkdB. The bfmB mutant of B. subtilis requires branched short-chain carboxylic acids for growth. However, the lpd and bkdB genes could be interrupted without a loss of cell viability. Transcription of the operon is SigL dependent, and it was previously shown that mutants affected in the BCDH complex require branched-chain α-keto acids for growth. sigL and bkdR null mutants are viable, and it is therefore likely that the four genes encoding the different polypeptides of the BCDH complex are constitutively transcribed at a low level to ensure the synthesis of membrane branched-chain carboxylic acids. This basal level of transcription could be due to read-through transcription from the bkdR promoter upstream from the operon. The bkdR null mutant was constructed by inserting an antibiotic resistance cassette in bkdR. This cassette might affect the basal level of transcription of the operon, which could explain the slower rate of growth of the bkdR mutant than of the sigL mutant with ammonia as the sole nitrogen source. The expression of the operon is also subject to global regulation. During exponential growth in MM glucose minimal medium containing isoleucine, the expression of the bkd operon is slightly repressed by the addition of Casamino Acids, and this repression is codY dependent. The uninduced level of the bkd operon increases 3-fold in tnrA mutants, 7-fold in the codY mutant in MM minimal medium containing glucose, and 17-fold in the presence of Casamino Acids.
CodY is a repressor which binds to promoters in AT-rich regions, known to exhibit bending and curvature. Interestingly, looped DNA structures are involved in the formation of contacts between Eς54 holoenzymes and enhancer binding proteins. However, it would be interesting to test whether TnrA and CodY bind directly to the bkd promoter region. Another possibility is that the intracellular level of the inducer is higher in tnrA and codY mutants than in the wild-type strain, leading to a higher level of expression of the bkd operon in the mutants. Transcription of the operon at a high level requires the product of bkdR, a regulator with a central domain.
The different domains of members of this family of transcriptional activators have different functions and exhibit various degrees of evolutionary conservation (23). Typically, the amino-terminal domain is the signal reception domain of approximately 160 residues. There is only one known exception, the LevR protein of B. subtilis, in which the carboxy-terminal domain of the protein contains two signal reception domains (18). The amino-terminal part of BkdR is a large domain composed of 360 residues, a characteristic which is shared by two other regulators containing a central domain: AcoR from Alcaligenes eutrophus (9a) and AcoR from B. subtilis (10). This amino-terminal part contains two PAS-like domains (Fig. 6) (39). These domains, previously reported for proteins from mammals, insects, plants, fungi, and cyanobacteria, are involved in protein-protein interactions. They have been identified for a variety of sensor proteins that sense light, oxygen, and redox potential. In bacteria, the PAS domain is usually associated with the input domain of a histidine kinase or a sensor protein that regulates a histidine kinase. A PAS-like domain has also been found in the RocR regulator controlling arginine utilization in B. subtilis (39). Constitutive mutations located in the amino-terminal domain of RocR affect conserved residues of the PAS domain (3, 7), indicating that it is probably involved in the control of the activity of RocR. These PAS repeats may also be involved in the control of BkdR activity in response to the presence of the internal inducer. Site-directed mutagenesis of conserved residues in PAS domains of BkdR is under way as part of work to confirm their involvement in BkdR activity. Moreover, as nothing is known about the transcription of bkdR, further work is needed to define its regulation.
FIG. 6.
Alignment of PAS-like domains of RocR and BkdR from B. subtilis. Conserved amino acid residues are boxed. Asterisks indicated the locations of constitutive missense mutations in RocR.
ACKNOWLEDGMENTS
We thank Tracy Nixon and Ilene Kaufman for kindly giving us the DNA fragments encoding the central domain of BkdR; Susan Fisher, Linc Sonenshein, and Al Claiborne for helpful discussions; Joëlle Bignon for excellent technical assistance; Alex Edelman for correcting the manuscript; and Christine Dugast for expert secretarial assistance.
This work was supported by research funds from the Institut Pasteur, the Centre National de la Recherche Scientifique, and the Ministère de l’Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 3.Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Débarbouillé M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol. 1994;176:1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Débarbouillé M, Martin-Verstraete I, Kunst F, Rapoport G. The Bacillus subtilis sigL gene encodes an equivalent of ς54 from Gram-negative bacteria. Proc Natl Acad Sci USA. 1991;88:9092–9096. doi: 10.1073/pnas.88.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouet A, Sonenshein L. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J Bacteriol. 1990;172:835–844. doi: 10.1128/jb.172.2.835-844.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardan R, Rapoport G, Débarbouillé M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol Microbiol. 1997;24:825–837. doi: 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- 8.Glatron M-F, Rapoport G. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie. 1972;54:1291–1301. doi: 10.1016/s0300-9084(72)80070-1. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman R I, Nixon B T. Use of PCR to isolate genes encoding ς54-dependent activators from diverse bacteria. J Bacteriol. 1996;178:3967–3970. doi: 10.1128/jb.178.13.3967-3970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Krüger N, Steinbüchel A. Identification of acoR, a regulatory gene for the expression of genes essential for acetoin catabolism in Alcaligenes eutrophus H16. J Bacteriol. 1992;174:4391–4400. doi: 10.1128/jb.174.13.4391-4400.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 11.Lereclus D, Arantes O. spbA locus ensures the segregational stability of pHT1030, a novel type of Gram-positive replicon. Mol Microbiol. 1992;6:35–46. doi: 10.1111/j.1365-2958.1992.tb00835.x. [DOI] [PubMed] [Google Scholar]
- 12.Lowe P N, Hodgson J A, Perham R N. Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched-chain 2-oxo acids in Bacillus subtilis. Biochem J. 1983;215:133–140. doi: 10.1042/bj2150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magasanik B. The regulation of nitrogen utilization in enteric bacteria. J Cell Biochem. 1993;51:34–40. doi: 10.1002/jcb.240510108. [DOI] [PubMed] [Google Scholar]
- 14.Marshall V P, Sokatch J R. Regulation of valine catabolism in Pseudomonas putida. J Bacteriol. 1972;110:1073–1081. doi: 10.1128/jb.110.3.1073-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Induction and metabolite regulation of levanase synthesis in Bacillus subtilis. J Bacteriol. 1989;171:1885–1892. doi: 10.1128/jb.171.4.1885-1892.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol. 1990;214:657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Verstraete I, Charrier V, Stülke J, Galinier A, Erni B, Rapoport G, Deutscher J. Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol Microbiol. 1998;28:293–303. doi: 10.1046/j.1365-2958.1998.00781.x. [DOI] [PubMed] [Google Scholar]
- 19.Massey L M, Sokatch J R, Conrad R S. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev. 1976;40:42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCully V, Burns G, Sokatch J R. Resolution of branched-chain oxo acid dehydrogenase complex of Pseudomonas aeruginosa. Biochem J. 1986;233:737–742. doi: 10.1042/bj2330737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 22.Mizuno M, Masuda S, Takemaru K, Hosono S, Sato T, Takeuchi M, Kobayashi Y. Systematic sequencing of the 283 kb 210°–232° region of the Bacillus subtilis genome containing the skin element and many sporulation genes. Microbiology. 1996;142:3103–3111. doi: 10.1099/13500872-142-11-3103. [DOI] [PubMed] [Google Scholar]
- 23.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odessey R. Purification of rat kidney branched-chain oxo acid dehydrogenase complex with endogenous kinase activity. Biochem J. 1982;204:353–356. doi: 10.1042/bj2040353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paxton R, Harris R A. Isolation of rabbit liver branched-chain α-ketoacid dehydrogenase and regulation by phosphorylation. J Biol Chem. 1982;257:14433–14439. [PubMed] [Google Scholar]
- 26.Pettit F H, Yeaman S J, Reed L J. Purification and characterization of branched chain-ketoacid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci USA. 1978;75:4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Serror P, Sonenshein A L. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996;178:5910–5915. doi: 10.1128/jb.178.20.5910-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serror P, Sonenshein A L. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol Microbiol. 1996;20:843–852. doi: 10.1111/j.1365-2958.1996.tb02522.x. [DOI] [PubMed] [Google Scholar]
- 30.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 31.Slack F J, Serror P, Joyce E, Sonenshein A L. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol. 1995;15:689–702. doi: 10.1111/j.1365-2958.1995.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 32.Sokatch J R, McCully V, Gebrosky J, Sokatch D J. Isolation of a specific lipoamide dehydrogenase for a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981;148:639–646. doi: 10.1128/jb.148.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 34.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 35.Wang G F, Kuriki T, Roy K L, Kaneda T. The primary structure of branched-chain α-oxo acid dehydrogenase from Bacillus subtilis and its similarity to other α-oxo acid dehydrogenases. Eur J Biochem. 1993;213:1091–1099. doi: 10.1111/j.1432-1033.1993.tb17858.x. [DOI] [PubMed] [Google Scholar]
- 36.Ward D, Claiborne A, de Kok A, Westphal A. Functional and regulatory studies on two distinct lipoamide dehydrogenases from Enterococcus faecalis. In: Stevenson K J, Massey V, Williams C H Jr, editors. Flavins and flavoproteins 1996. Calgary, Alberta, Canada: University of Calgary Press; 1997. pp. 673–676. [Google Scholar]
- 37.Willecke K, Pardee A B. Fatty acid-requiring mutant of Bacillus subtilis defective in branched-chain α-ketoacid dehydrogenase. J Biol Chem. 1971;246:5264–5272. [PubMed] [Google Scholar]
- 38.Wray L V, Jr, Ferson A E, Rohrer K, Fisher S H. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhulin I B, Taylor B L, Dixon R. PAS domain S-boxes in Archaea, bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]