Abstract
The development of styrene maleic acid (SMA) and diisobutylene maleic acid (DIBMA) copolymers provides an alternative to traditional detergent extraction of integral membrane proteins. By inserting into the membrane, these polymers can extract membrane proteins along with lipids in the form of native nanodiscs made by poly(styrene co-maleic anhydride) derivatives. Unlike detergent solubilization, where membrane proteins may lose annular lipids necessary for proper folding and stability, native nanodiscs allow for proteins to reside in the natural lipid environment. In addition, polymer-based nanodiscs can be purified using common chromatography methods similar to protocols established with detergent solubilization purification. Here we describe the solubilization screening and purification of an integral membrane protein using several commercial copolymers.
Keywords: ATP-binding cassette (ABC) transporter, Detergents, Purification, Polymers, Solubilization, Membrane proteins, Nanodiscs, Styrene maleic acid (SMA) copolymer, Diisobutylene-maleic acid (DIBMA) copolymer
1. Introduction
Integral membrane proteins are abundant in cells and play key sensory and regulatory roles, yet remain largely understudied due to purification challenges. In established membrane protein extraction protocols, the cell membrane is treated with detergents or other surfactants, often stripping away structurally and functionally essential annular lipids [1, 2]. An increasingly popular solubilization strategy employs amphipathic polymers of the styrene maleic acid (SMA) or diisobutylene-maleic acid (DIBMA) type to extract membrane proteins in their native lipid environment in the form of nanodiscs [3–5] (Fig. 1). Nanodisc-encapsulated protein targets can be purified with common chromatography techniques, enabling structure–function studies on the most physiologically relevant protein states [6–11]. The continued development of polymers with enhanced solubilization abilities is an active area of research in many laboratories [12, 13]. Several promising copolymers can now be purchased (see Table 1).
Fig. 1: Structure of SMA, SMI and DIBMA polymers.
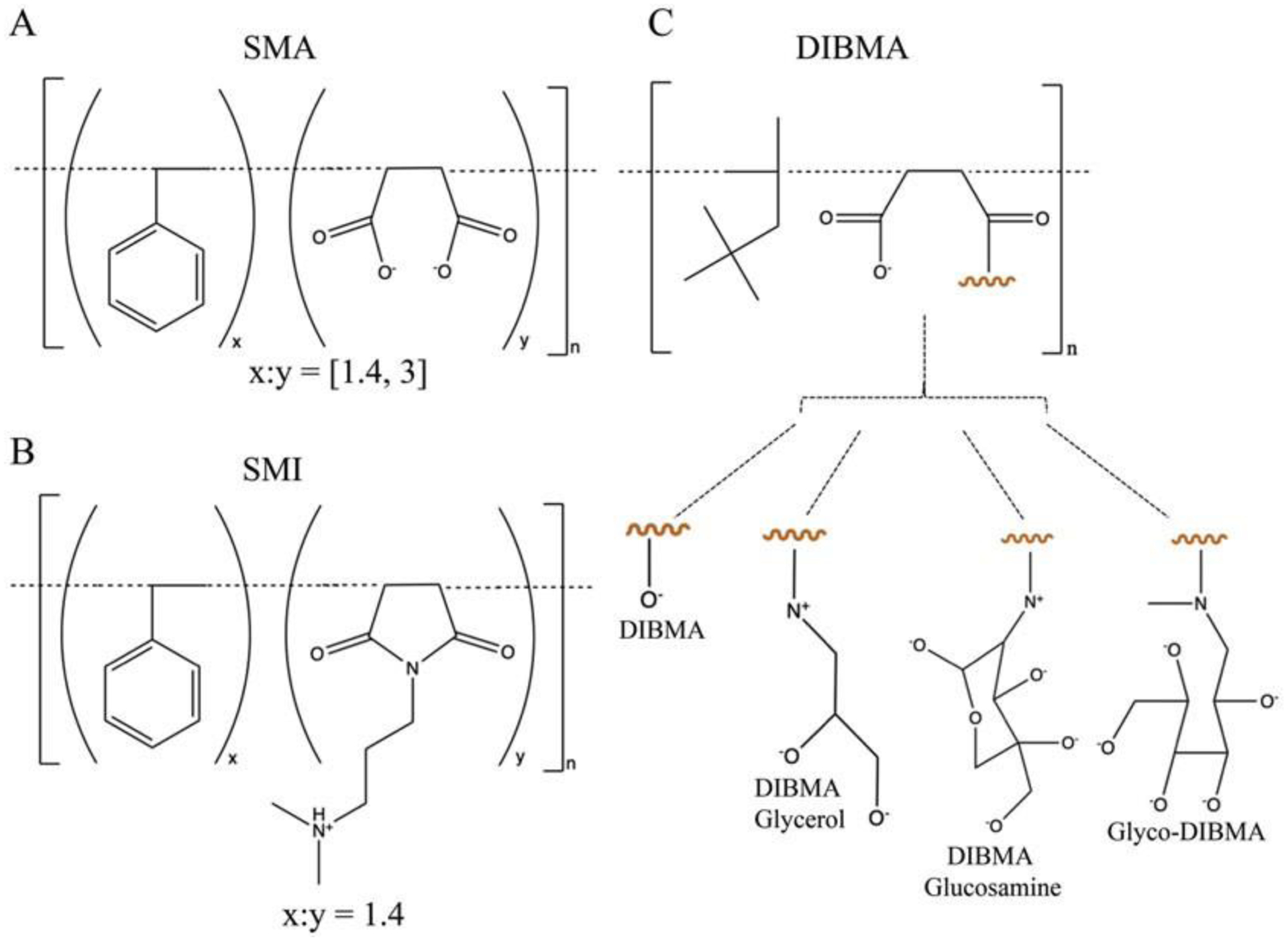
Base polymer structures and modifications of (A) styrene-co-maleic acid, (B) styrene-co-maleimide, and (C) di-isobutylene maleic acid co-polymers. All figures adapted from [28] and https://cube-biotech.com/us/products/nanodisc-products/synthetic-nanodisc-products/?p¼1.
Table 1:
SMA, SMI and DIBMA commercial polymers used in the screen
| Commercially available polymer | Subunit | Ratio | MW (g/mol) | Source company | Prior applications |
|---|---|---|---|---|---|
| SMALP 140-I | S, MI | 1.4:1 | 5000 | Cube Biotecha, Orbiscopeb | [16] |
| SMALP 300 | S, MA | 3:1 | 10,000 | Cube Biotech, Orbiscope | [8, 29, 30, 35, 36] |
| SMALP 200 | S, MA | 2:1 | 6500 | Cube Biotech, Orbiscope | [5, 6, 9–11, 34, 37–39] |
| SMALP 140 | S, MA | 1.4:1 | 5000 | Cube Biotech, Orbiscope | [37] |
| DIBMA 10 | DIB, MA | 1:1 | 10,000 | Cube Biotech | n/a |
| DIBMA 12 | DIB, MA | 1:1 | 12,000 | Cube Biotech | [4, 18, 31, 32] |
| DIBMA Glycerol | DIB, MAGlyc | 1:1 | 10,000 | Cube Biotech | n/a |
| DIBMA Glucosamine | DIB, MAGluc | 1:1 | >15,000 | Cube Biotech | n/a |
| Glyco-DIBMA | DIB, MAGlyco | 1:1 | 15,701c | Glycon Biochemicals | [33] |
Polymers supplied by Cube Biotech are in powder form. DIBMA polymers from this vendor are buffered with HEPES or Tris to pH 7.5.
Orbiscope offers SMA polymers as 20% w/v solutions in water.
Molecular weight as reported by the Glycon Biochemicals certificate analysis. Most of the listed vendors supply the ready-to-use acid form of SMA and DIBMA polymers. Sigma cells the pre-hydrolyzed styrene maleic anhydride (SMAnh) copolymers, which can be chemically converted in-house to the SMA final product (25, 27). Several different molecular weights of DIBMA are commercially available: Cube Biotech offers 10 kDa and 12 kDa versions, while Anatrace and BASF provide a 15.3 kDa version of DIBMA.
Polymer hydrophobicity and charge distribution are critical for efficient membrane solubilization, and research indicates that solubilization can be improved upon by changing the polymer composition [14, 15]. The most hydrophobic SMA has styrene-to-maleic acid (S:MA) ratio of 3:1 (Fig. 1), whereas less hydrophobic SMA polymers have a reduced S:MA ratio or a modified maleic acid group [16]. Hydrophobicity is further reduced in DIBMA polymers, which, unlike SMAs, have a more even charge distribution, an increased tolerance for divalent cations, and a lower absorbance at 260–280 nm [17, 18]. Polymer performance can be enhanced by adjusting solubilization parameters such as polymer concentration, polymer-to-membrane ratio, pH, ionic strength, incubation temperature, and duration [19–22]. Importantly, the specific lipid/protein composition of the membrane greatly influences the solubilization efficiency, therefore different proteins may require different polymers and/or buffers for optimal extraction.
The protocol below details a screening process aimed at identifying a commercially available polymer that solubilizes membrane protein targets. For this study, SapC—a transmembrane domain in the Sap ABC transporter was utilized as a target of interest [23, 24]. We describe the polymer screening and selection, large-scale purification of SapC with the SMALP 200 polymer and demonstrate that SMA-bound SapC nanodiscs can assemble with the separately purified nucleotide binding domain SapF to build the SapCF complex.
2. Materials
Prepare all solutions using ultrapure water (prepared by purifying deionized water, to attain a sensitivity of 18 MΩ-cm at 25°C) and analytical grade reagents and stored at room temperature (unless indicated otherwise).
2.1. Membrane isolation and solubilization
40 g E. coli cell pellet with the expressed membrane protein of interest.
Tris stock solution: 1 M Tris–HCl, pH 8.0. In a glass beaker, pour 800 mL of water and add 121.14 g Tris base while stirring on a magnetic stir plate. Once the powder has fully dissolved, adjust the pH to 8.0 by adding concentrated HCl (see Note 1). Transfer the beaker contents into a 1 L graduated cylinder and add water to 1 L. Filter the stock solution into a glass bottle, seal tightly, and store at room temperature.
EDTA stock solution: 0.5 M EDTA, pH 8.0. Weigh 146.12 g of EDTA powder and stir into 800 mL of water (see Note 2). Adjust to pH 8.0 by adding a few NaOH pellets at a time, waiting for them to fully dissolve; add more pellets as needed. Once pH 8.0 is reached, pour the beaker contents into a 1 L graduated cylinder and fill with water to 1 L. Filter and store in a 1 L glass bottle at room temperature.
NaCl stock solution: 5 M NaCl. Dissolve 292.2 g of NaCl into 800 mL of water by stirring in a 1 L beaker (see Note 3). Pour the beaker contents into a 1 L graduated cylinder and use water to adjust the final volume to 1 L. Filter and store the stock solution in a glass bottle at room temperature.
PMSF stock solution: 100 mM PMSF. Dissolve 871 mg of PMSF powder into 50 mL 100% ethanol. Store at −20 °C tightly sealed tube.
Glycerol stock solution: 50% glycerol. Pour 900 mL of water into a 2 L beaker, containing a magnetic stir bar. Measure 1 L glycerol with a graduated cylinder and add to the beaker while stirring (see Note 4). Transfer the mixed beaker contents into a 2 L graduated cylinder and adjust the volume to 2 L with water. Store this stock solution in a capped bottle at room temperature.
Buffer A: 20 mM Tris–HCl, pH 8.0, 0.5 mM EDTA. In a 500 mL graduated cylinder, add 10 mL of 1 M Tris–HCl solution and 0.5 mL of EDTA stock solution, then swirl to mix. Add water to the 500 mL mark, then transfer into a capped glass bottle and store at 4 °C until needed.
Microfluidizer (Model M-110 L Microfluidizer Processor, Microfluidics®) for cell disruption.
Centrifuge bottles: 50 mL round-bottom tubes with leakproof caps, 70 mL polycarbonate ultracentrifuge bottle assemblies.
Centrifuge rotors: Sorvall® SS-34, Beckman Ti-45.
Buffer B: 50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 10% glycerol. In 500 mL graduated cylinder add 25 mL of 1 M Tris–HCl stock solution, 50 mL of 5 M NaCl stock solution and 100 mL of 50% glycerol stock solution. Adjust the volume to 500 mL, then store in a tightly capped bottle at 4 °C (see Note 5).
40 mL glass tissue grinder with a loose piston.
Synthetic nanodisc polymers (see Table 1).
Orbital laboratory mixer.
2.2. Protein purification and quality control
Magnetic stir plate and stir bars.
Imidazole stock solution: 4 M imidazole, pH 8.0. In a 250 mL beaker add 200 mL water and stir in 68.08 g imidazole. Once the powder has fully dissolved, adjust to pH 8.0 with HCl. Transfer the solution to a 250 mL graduated cylinder and adjust the final volume to 250 mL with water. Filter and store the stock in a capped bottle at 4 °C.
HisPur™ Ni-NTA resin (Thermo fisher scienctific, USA) (see Note 6).
Gravity-flow purification column.
Buffer C: 50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 250 mM imidazole, 10% glycerol. In a 50 mL graduated cylinder, combine 2.5 mL of 1 M Tris–HCl, pH 8.0, 5 mL of 5 M NaCl, 3.125 mL of 4 M imidazole, and 10 mL of 50% glycerol stock solution. Add water to the 50 mL mark, mix well, and store in a 50 mL conical tube at 4 °C.
SDS-PAGE gels.
PAGE apparatus.
SDS-PAGE running buffer.
SDS-PAGE loading dye.
Coomassie brilliant blue R-250 powder.
Gel developing solution: 10% acetic acid, 40% methanol. In a 1 L graduated cylinder, combine 100 mL glacial acetic acid, 400 mL methanol, and water to the 1 L mark. Pour into a 1 L bottle, cap well, and store at room temperature.
Buffer D: 50 mM Tris–HCl, pH 8.0, 150 mM NaCl. In a 500 mL graduated cylinder, add 25 mL of 1 M Tris–HCl pH 8.0, 15 mL of 5 M NaCl, and water up to the 500 mL mark. Cap the cylinder and invert several times to mix. Place a magnetic stir bar in a clean bottle, then transfer the buffer into the bottle through a vacuum filter. Allow thorough buffer degassing by leaving the vacuum on while stirring the bottle contents for at least 30 min (see Note 7). Cap the bottle and store at 4 °C until needed
Superdex® 200 10/300 column (GE healthcare, USA) (see Note 8), size exclusion chromatography system.
Means to test the functionality of the extracted protein (see Note 9).
3. Methods
In this protocol, 40 g of E. coli cell pellet is processed. All buffer volumes can be scaled up or down according to the amount of cell mass being used. Carry out all procedure steps on ice or at 4 °C unless otherwise specified. Sample foaming should be avoided as much as possible.
3.1. Membrane isolation
Place a 250 mL metal beaker on ice and add 150 mL of Buffer A and 1.5 mL of the 100 mM PMSF stock for a final concentration of 1 mM. Fully resuspend the 40 g bacterial cell pellet into the buffer (see Note 10).
Lyse the bacterial suspension by passing the solution through a microfluidizer three times at pressure 16 kPa. After the final passage, flush the microfluidizer with 100 mL of Buffer A, combining with the processed cell lysate, then add 1 mL 100 mM PMSF stock for a final concentration of 1 mM. (see Note 11).
Transfer the cell lysate to round-bottom centrifuge tubes with leakproof caps and centrifuge in the SS-34 rotor at 23,430 × g for 30 min at 4 °C.
Transfer the supernatant into polycarbonate ultracentrifuge bottles and centrifuge at 205,075 × g for 1 h 15 min. Gently decant and transfer all supernatant without disturbing the pellet (see Note 12).
Place a prechilled glass tissue grinder on an analytical scale and zero the scale. Use a metallic spatula to scoop the ultracentrifugation pellets out of the centrifuge bottles and into the grinder, then determine the membrane weight in milligrams. Calculate the amount of Buffer B needed for a 40 mg/mL membrane suspension. Pour a portion of the calculated Buffer B amount into the tissue grinder and resuspend the membranes by gently running the piston up and down the grinding tube (see N Note 13). Transfer the fully mixed membrane suspension into a clean bottle and add the remaining Buffer B from the calculation above. Proceed immediately to do a membrane solubilization screen or large-scale protein purification.
3.2. Membrane solubilization screen
Make multiple 1 mL aliquots of the membrane sample produced in Subheading 3.1 or thaw out pre-frozen membrane aliquots (see Note 14).
Retrieve all synthetic polymers to be screened (Cube Biotech, Glycon Biochemicals Germany) (see Note 15).
Add 50 mg of each polymer per membrane aliquot (see Note 16). Leave one membrane aliquot untreated as a control.
Rock the membrane/polymer mix for 1 h on an orbital mixer set to 15 rpm (see Note 17).
Visually compare the opacity of each screened sample to that of the untreated membrane sample. Select all polymer-treated samples that appear significantly more transparent than the untreated control (see Note 18) (Fig. 2).
Fig. 2: Solubilization of SapC using several commercial polymers.
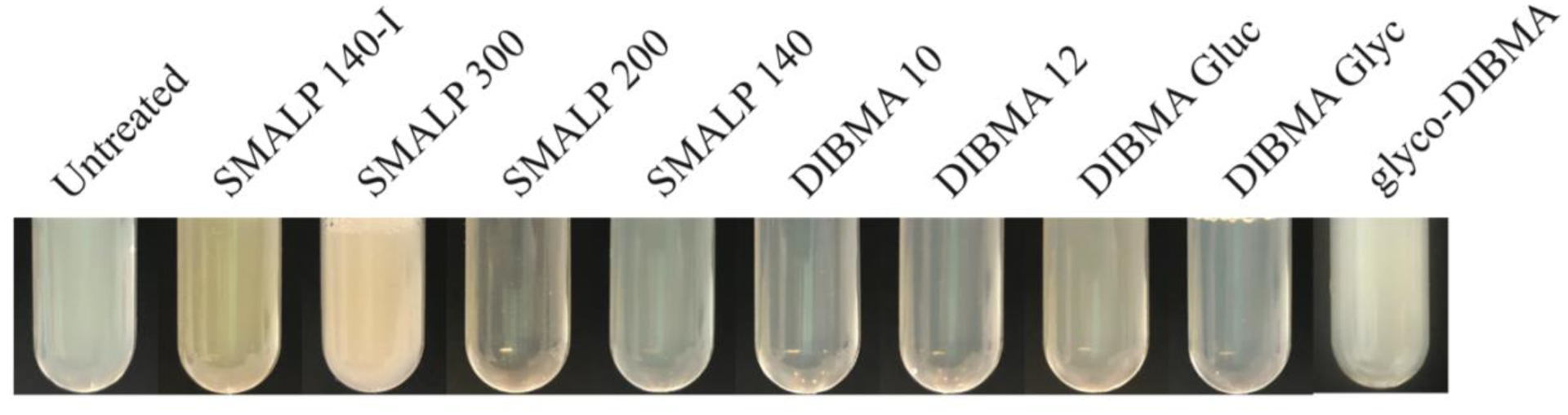
Cell membranes containing the transmembrane portion of the ABC Transporter (SapC) were individualy incubated with several different commercial polymers (SMALP 140-I (SMI)) SMALP300, SMALP 200, SMALP 140, DIBMA 10, DIBMA 12, DIBMA Glucosamine, DIBMA Glycerol, Glyco-DIBMA) for 1 hour at room temperature. In comparison to the untreated sample, SMALP 200, DIBMA 10 and DIBMA 12 increased sample transparency, indicating increased solubilization. Best solubilization was achieved with the SMALP 200 polymer.
3.3. Membrane protein purification with the SMALP 30010P polymer
Add SMALP 200 to the membrane extract generated in Subheading 3.1 (see Note 19) and gently stir on a magnetic plate at room temperature for 1 h (see Note 20).
Centrifuge the solubilized membrane sample at 205,075 × g for 1 h 15 minutes. Gently transfer the supernatant in a clean container.
Transfer 4 mL of Ni-NTA slurry into a gravity column and let the column bed settle before allowing the storage solution to drip through by gravity. Wash the resin in the column with 10 column volumes (CV) of water and 10 CV of Buffer B.
Run the ultracentrifugation supernatant from Subheading 3.3, step 2 through the pre-washed nickel column. Wash the column with 50 CV Buffer B, then elute the protein with 10 CV Buffer C, collecting five fractions of equal volume. Collect small samples from all purification steps, and run on 12 % SDS-PAGE alongside a protein ladder. Stain the gel with 0.1% Coomassie brilliant blue in gel developing solution for 5 minutes, then destain for 15 minutes in dye-free gel developing solution. Inspect the gel to identify the elution fractions with highest protein purity. Combine the best fractions and concentrate with a centrifugal filter unit of the appropriate size cut-off.
Equilibrate a Superdex® 200 10/300 column with 2 CV of Buffer D. Subject the concentrated affinity-purified protein to size exclusion chromatography (SEC) (see Note 21). Take small samples from each of the fractions that build up the A280 peak and run on 12 % SDS-PAGE. Pool and concentrate the purest peak fractions.
Assess the functionality of the nanodisc-encapsulated protein (see Note 9).
4. Notes
Be aware that titrating the Tris stock with concentrated HCl will elevate the temperature of the solution and therefore affect the pH reading. We recommend allowing the beaker contents to equilibrate to room temperature prior to re-adjusting the pH and making up the final volume of the stock solution.
EDTA will not fully dissolve until pH has been properly adjusted.
To fully dissolve the NaCl powder, add as much water as possible without exceeding a final volume of 1 L.
Glycerol is very viscous and should be given enough time to fully drip out of the graduated cylinder. To facilitate complete transfer and avoid tedious waiting, pour as much of the measured glycerol into the beaker as possible, then use a large clamped stand to suspend the cylinder in an upside-down fashion above the stirring mixture. Return to retrieve the fully mixed sample in 20 minutes.
The composition of Buffer B is critical for efficient membrane solubilization and will likely need to be optimized by the user. When searching for the optimal buffer recipe, one should consider not only the stability of the protein of interest but also the effect of ionic strength, pH and divalent cations on the stability and solubilization efficiency of the screened polymers. Most SMA polymers are effective in the pH 7–9 range at salt concentrations 300 mM or above and precipitate in presence of millimolar concentrations of divalent cations or at pH < 6 [25]. The solubilization performance of DIBMA polymers is often enhanced at neutral to slightly acidic pH, high ionic strength and/or the presence of divalent cations [4,26].
This resin type has been chosen for affinity purification of His-tagged proteins. An alternative resin and elution strategy would be needed when utilizing a different affinity tag.
Degassing of the size exclusion buffer is necessary in order to prevent air entry into the gel filtration system and damaging the purification column. The degassing procedure must be repeated for buffers older than 1 week.
This column type is suitable for purification of proteins and protein complexes in the range of ~15–600 kDa. Significantly larger or smaller proteins may require a different column type.
Different strategies can be utilized to test whether the polymer-extracted protein is functional. Our approach involved testing whether the nanodisc-encapsulated transmembrane domain SapC interacts with the purified nucleotide binding domain SapF to form the physiologically relevant SapCF complex.
When working with protease-sensitive membrane proteins, additional protease inhibitors may be included besides PMSF. Cell pellets that have been stored at −80 °C should be thawed in Buffer A in presence of protease inhibitors to avoid proteolytic degradation. The cell resuspension process can be eased by the use of a cell disperser, set to a gentle mixing speed to avoid sample foaming. For the T25 digital ULTRA-TURRAX® model sold by IKA®, setting #6 allows full resuspension in less than 5 minutes with minimal sample foaming.
Additional PMSF/protease inhibitor tablets are needed at the end of this step in order to maintain the desired final concentration.
At the end of the ultracentrifugation step the membranes will be seen as a beige-brown gelatinous substance, firmly attached to the bottom of the bottle. It is important to remove as much of the supernatant as possible without disrupting the membranes.
For thorough membrane resuspension, run the loosely fitting piston up and down the grinding tube until no visible chunks remain, then perform 20 additional strokes.
The membrane aliquots can be used fresh or stored for future solubilization screens by snap-freezing in liquid nitrogen and placing in an −80 °C freezer. When ready to use, thaw the desired number of aliquots on ice for 15 min. Repeated freeze-thawing is strongly discouraged.
Comprehensive SMA and/or DIBMA screening kits are offered by Orbiscope and Cube Biotech.
Dissolving 50 mg of polymer in 1 mL membrane sample results in polymer concentration of 5 %, which is the highest value in the recommended 0.5–5 % polymer concentration range. In most cases 5 % polymer is an excessive amount and effective membrane protein solubilization can be achieved at lower concentrations. We recommend subjecting all hits from the initial solubilization screen to an additional polymer percentage screen designed to determine the lowest amount of polymer necessary to extract the membrane protein of interest. Such follow-up screens can also benefit from adjustments (pH, type of buffer, etc.) in the Buffer B recipe, factoring in the polymer identity (see Note 5).
In some cases, longer incubations may be needed for efficient solubilization.
Membrane sample transparency can be assessed more quantitatively by measuring absorbance at 600 nm and comparing to the negative control. Sample clarity is often a good indicator that membrane solubilization was successful, however, there is always a chance that the protein of interest became solubilized, even though the bulk of the membrane did not. To verify extraction success, polymer-treated samples can be centrifuged at 535,000 × g for 40 min at 4 °C and a Western blot can be performed on the supernatant portion. The presence of high concentrations of synthetic polymers, especially DIBMA, may cause band blurring on SDS-PAGE gels. A methanol/chloroform extraction protocol can be performed on the solubilized protein fraction to precipitate the excess polymers and improve gel mobility [4,27].
For large-scale purification we recommend using the lowest amount of polymer that can successfully solubilize the protein of interest. This strategy would minimize the amount of free polymer after membrane solubilization is completed, thus improving downstream purification outcomes.
Depending on the efficiency of the chosen polymer, the solubilization step may require between 1 hour and > 24 hours. Ideally, the polymer selection and Buffer B composition should be optimized to complete the solubilization process within 1 hour. When a shorter incubation with the polymer is not an option or for temperature-sensitive protein targets, we recommend carrying the solubilization step at 4 °C to preserve protein integrity. Be mindful that nanodisc formation would slow down at decreased temperatures.
The shape and broadness of the SEC peak is a good indicator of sample homogeneity. SMA and DIBMA polymers often extract a range of nanodisc sizes, resulting in broad and/or multimodal SEC peaks (Fig. 3). Sample homogeneity can be further improved by pooling and concentrating only fractions that contain the protein of interest in the expected size range. The strong UV absorbance of SMA polymers, will significantly enhance the amplitude of the A280 peak.
Fig. 3: Purification of SapCF in polymer SMALP 200P.
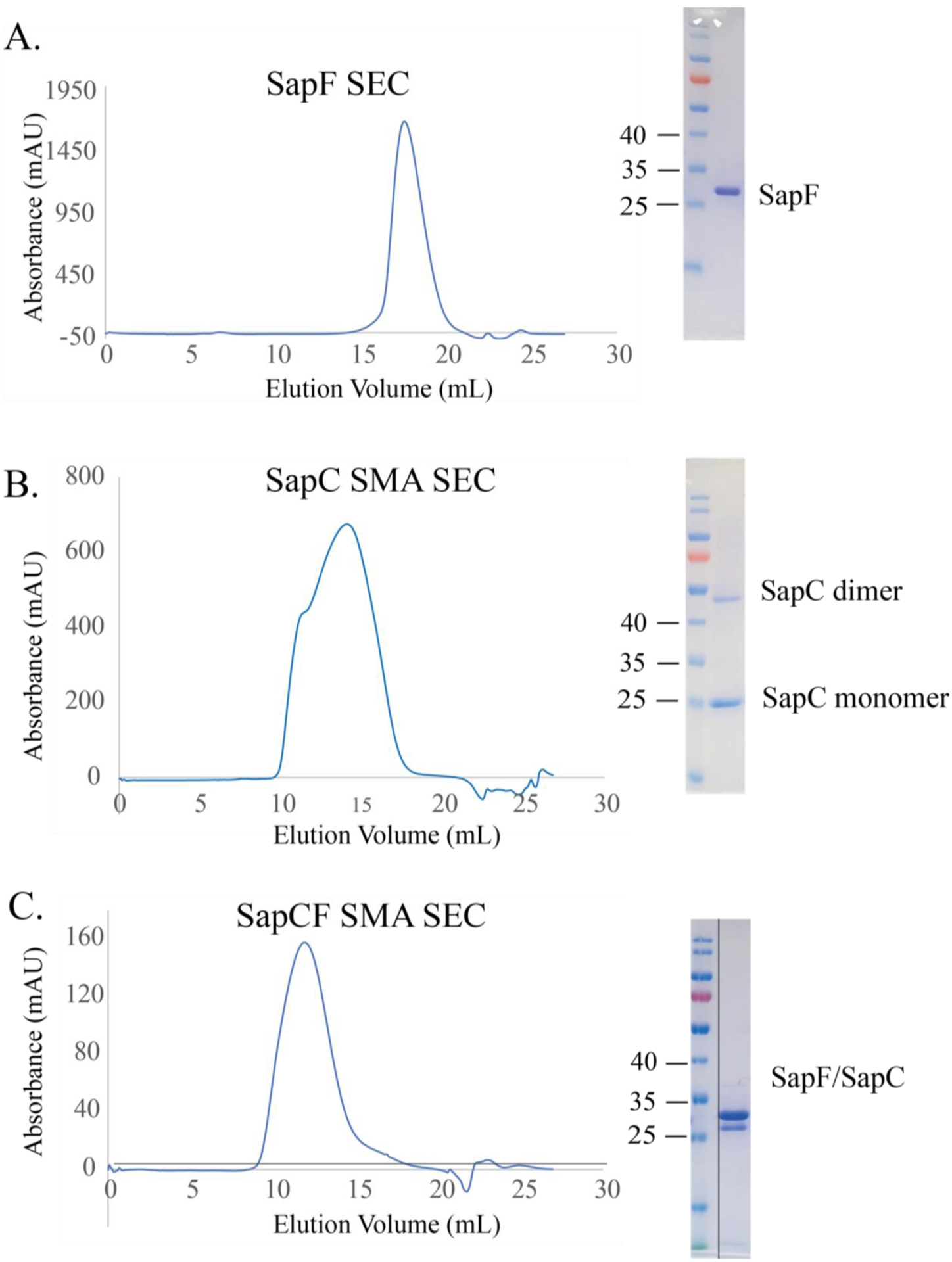
After affinity purification (A) SapF and (B) SapC were run on a size exclusion column, each resulting in a single peak at 95 % purity as indicated by SDS-PAGE. Both proteins were mixed to create a (C) SapCF complex before running on the size exclusion column to ensure complex formation of the transporter.
Acknowledgements:
This work was supported by NIH grants R01 AI139519 and R01 GM140584-01
References
- 1.Zoghbi ME, Cooper RS, Altenberg GA (2016) The lipid bilayer modulates the structure and function of an ATP-binding cassette exporter. J Biol Chem 291(9):4453–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gulati S, Jamshad M, Knowles TJ, Morrison KA, Downing R, Cant N, Collins R, Koenderink JB, Ford RC, Overduin M, Kerr ID, Dafforn TR, Rothnie AJ (2014) Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem J 461(2):269–278 [DOI] [PubMed] [Google Scholar]
- 3.Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M (2009) Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc 131(22): 7484–7485 [DOI] [PubMed] [Google Scholar]
- 4.Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S (2017) Solubilization of membrane proteins into functional lipid-bilayer Nanodiscs using a Diisobutylene/maleic acid copolymer. Angew Chem Int Ed Engl 56(7):1919–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamshad M, Grimard V, Idini I, Knowles TJ, Dowle MR, Schofield N, Sridhar P, Lin YP, Finka R, Wheatley M, Thomas OR, Palmer RE, Overduin M, Govaerts C, Ruysschaert JM, Edler KJ, Dafforn TR (2015) Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res 8(3):774–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EA, Dafforn TR, Baldus M, Killian JA (2014) Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc Natl Acad Sci U S A 111(52):18607–18612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broecker J, Eger BT, Ernst OP (2017) Crystal-logenesis of membrane proteins mediated by polymer-bounded lipid Nanodiscs. Structure 25(2):384–392 [DOI] [PubMed] [Google Scholar]
- 8.Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB (2018) Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 557(7703):123–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu W, Fu Z, Xu GG, Grassucci RA, Zhang Y, Frank J, Hendrickson WA, Guo Y (2018) Structure and activity of lipid bilayer within a membrane-protein transporter. Proc Natl Acad Sci U S A 115(51):12985–12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoder N, Gouaux E (2020) The his-Gly motif of acid-sensing ion channels resides in a reentrant “loop” implicated in gating and ion selectivity. Elife 9:e56527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Zhu H, Lape R, Greiner T, Du J, Lu W, Sivilotti L, Gouaux E (2021) Mechanism of gating and partial agonist action in the glycine receptor. Cell 184(4):957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bada Juarez JF, Harper AJ, Judge PJ, Tonge SR, Watts A (2019) From polymer chemistry to structural biology: the development of SMA and related amphipathic polymers for membrane protein extraction and solubilisation. Chem Phys Lipids 221:167–175 [DOI] [PubMed] [Google Scholar]
- 13.Brown CJ, Trieber C, Overduin M (2021) Structural biology of endogenous membrane protein assemblies in native nanodiscs. Curr Opin Struct Biol 69:70–77 [DOI] [PubMed] [Google Scholar]
- 14.Cunningham RD, Kopf AH, Elenbaas BOW, Staal BBP, Pfukwa R, Killian JA, Klumperman B (2020) Iterative RAFT-mediated copolymerization of styrene and maleic anhydride toward sequence- and length-controlled copolymers and their applications for solubilizing lipid membranes. Biomacromolecules 21(8): 3287–3300 [DOI] [PubMed] [Google Scholar]
- 15.Ball LE, Riley LJ, Hadasha W, Pfukwa R, Smith CJI, Dafforn TR, Klumperman B (2021) Influence of DIBMA polymer length on lipid Nanodisc formation and membrane protein extraction. Biomacromolecules 22(2): 763–772 [DOI] [PubMed] [Google Scholar]
- 16.Hall SCL, Tognoloni C, Charlton J, Bragginton É, Rothnie AJ, Sridhar P, Wheatley M, Knowles TJ, Arnold T, Edler KJ, Dafforn TR (2018) An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles. Nano-scale 10(22):10609–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulamhussein AA, Uddin R, Tighe BJ, Poyner DR, Rothnie AJ (2020) A comparison of SMA (styrene maleic acid) and DIBMA (di-isobutylene maleic acid) for membrane protein purification. Biochim Biophys Acta Biomembr 1862(7):183281. [DOI] [PubMed] [Google Scholar]
- 18.Oluwole AO, Klingler J, Danielczak B, Babalola JO, Vargas C, Pabst G, Keller S (2017) Formation of lipid-bilayer Nanodiscs by Diisobutylene/maleic acid (DIBMA) copolymer. Langmuir 33(50):14378–14388 [DOI] [PubMed] [Google Scholar]
- 19.Scheidelaar S, Koorengevel MC, Pardo JD, Meeldijk JD, Breukink E, Killian JA (2015) Molecular model for the solubilization of membranes into nanodisks by styrene maleic acid copolymers. Biophys J 108(2):279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheidelaar S, Koorengevel MC, van Walree CA, Dominguez JJ, Dorr JM, Killian JA (2016) Effect of polymer composition and pH on membrane Solubilization by styrene-maleic acid copolymers. Biophys J 111(9):1974–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grethen A, Oluwole AO, Danielczak B, Vargas C, Keller S (2017) Thermodynamics of nanodisc formation mediated by styrene/maleic acid (2:1) copolymer. Sci Rep 7(1): 11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopf AH, Dörr JM, Koorengevel MC, Antoniciello F, Jahn H, Killian JA (2020) Factors influencing the solubilization of membrane proteins from Escherichia coli membranes by styrene-maleic acid copolymers. Biochim Biophys Acta Biomembr 1862(2): 183125. [DOI] [PubMed] [Google Scholar]
- 23.Rice AJ, Park A, Pinkett HW (2014) Diversity in ABC transporters: Type I, II and III importers. Crit Rev Biochem Mol Biol 49(5): 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka KJ, Song S, Mason K, Pinkett HW (2017) Selective substrate uptake: the role of ATP-binding cassette (ABC) importers in pathogenesis. Biochim Biophys Acta 1860(4): 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopf AH, Koorengevel MC, van Walree CA, Dafforn TR, Killian JA (2019) A simple and convenient method for the hydrolysis of styrene-maleic anhydride copolymers to styrene-maleic acid copolymers. Chem Phys Lipids 218:85–90 [DOI] [PubMed] [Google Scholar]
- 26.Danielczak B, Meister A, Keller S (2019) Influence of mg. Chem Phys Lipids 221:30–38 [DOI] [PubMed] [Google Scholar]
- 27.Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR (2016) A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat Protoc 11(7):1149–1162 [DOI] [PubMed] [Google Scholar]
- 28.Overduin M, Esmaili M (2019) Structures and interactions of transmembrane targets in native Nanodiscs. SLAS Discov 24(10):943–952 [DOI] [PubMed] [Google Scholar]
- 29.Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A (2012) Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer Lipodisq particles for functional and biophysical studies. Nano Lett 12(9): 4687–4692 [DOI] [PubMed] [Google Scholar]
- 30.Dörr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schafer M, van Walree CA, Killian JA (2016) The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur Biophys J 45(1):3–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barniol-Xicota M, Verhelst SHL (2018) Stable and functional rhomboid proteases in lipid Nanodiscs by using Diisobutylene/maleic acid copolymers. J Am Chem Soc 140(44): 14557–14561 [DOI] [PubMed] [Google Scholar]
- 32.Dilworth MV, Findlay HE, Booth PJ (2021) Detergent-free purification and reconstitution of functional human serotonin transporter (SERT) using diisobutylene maleic acid (DIBMA) copolymer. Biochim Biophys Acta Biomembr 1863(7):183602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danielczak B, Rasche M, Lenz J, Patallo EP, Weyrauch S, Mahler F, Agbadaola MT, Meister A, Babalola JO, Vargas C, Kolar C, Keller S (2022) A bioinspired glycopolymer for capturing membrane proteins in native-like lipid-bilayer nanodiscs. Nanoscale 14(5): 1855–1867 [DOI] [PubMed] [Google Scholar]
- 34.Caveney NA, Workman SD, Yan R, Atkinson CE, Yu Z, Strynadka NCJ (2021) CryoEM structure of the antibacterial target PBP1b at 3.3A resolution. Nat Commun 12(1):2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P, Cymes GD, Grosman C (2021) Structure and function at the lipidprotein interface of a pentameric ligand-gated ion channel. PNAS USA 118(23):e2100164118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Han L, Vallese F, Ding Z, Choi SK, Hong S, Luo Y, Liu B, Chan CK, Tajkhorshid E, Zhu J, Clarke O, Zhang K, Tennis R (2021) Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site. PNAS USA 118(34): e2106750118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruggisser J, Iacovache I, Musson SC, Degiacomi MT, Posthaus H, Zuber B (2021) Cryo-EM structure of the octameric pore of Clostridium perfringens b-toxin. bioRxiv preprint 10.1101/2021.11.23.469684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollock NL, Lloyd J, Montinaro C, Rai M, Dafforn TR (2022) Conformational trapping of an ABC transporter in polymer lipid nanoparticles. Biochem J 479(2):145–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catalano C, Ben-Hail D, Qiu W, Blount P, des Georges A, Guo Y (2021) Cryo-EM structure of mechxnosensitive channel YnaI using SMA2000: challenges and opportunities. Membranes 11(11):849. [DOI] [PMC free article] [PubMed] [Google Scholar]


