Abstract
Neisseria meningitidis uses hemoglobin (Hb) as an iron source via two TonB-dependent outer membrane receptors, HmbR and HpuB. Analysis of 25 epidemiologically unrelated clinical isolates from serogroups A, B, C, and Y revealed that 64% strains possessed both Hb receptor genes. Examination of the hmbR expression pattern in strains in which the hpuB gene was genetically inactivated revealed two distinct Hb utilization phenotypes. Five strains retained the ability to grow as a confluent lawn, while seven grew only as single colonies around Hb discs. The single-colony phenotype observed for some hpuB mutants is suggestive of phase variation of hmbR. The length of the poly(G) tract starting at position +1164 of hmbR absolutely correlated with the two Hb utilization phenotypes. All five strains that grew as confluent lawns around Hb discs possessed either 9 or 12 consecutive G residues. All seven strains that grew as single colonies around Hb discs had poly(G) tracts of a length other than 9 or 12. These single-colony variants that arose around the Hb discs had poly(G) tracts with either 9 or 12 consecutive G residues restoring the hmbR reading frame. Inactivation of hmbR in these strains resulted in a loss of Hb utilization, demonstrating that the change in the hmbR gene was responsible for the phenotypic switch. The switching rates from hmbR phase off to phase on were ∼5 × 10−4 in four serogroup C strains, 2 × 10−2 in the serogroup A isolate, and 7 × 10−6 in the serogroup B isolate.
The gram-negative diplococcus Neisseria meningitidis is a major cause of bacterial meningitis and is a significant public health problem throughout the world (7, 38). The incidence of meningococcal disease in developing countries ranges between 10 and 25 per 100,000, with periodic epidemics having attack rates of up to 1,000 per 100,000 (6, 12, 45). Approximately 2,600 cases of meningococcal disease occur annually in the United States, predominantly in children less than 2 years old (7). Since the success of the current vaccine against Haemophilus influenzae type b, N. meningitidis is the leading cause of meningitis in children in the United States (7).
Despite its relatively low incidence, several facts about meningococcal disease make it a potential public health problem of major importance. Mortality from meningococcal meningitis remains, despite optimal antibiotic and supportive therapy, about 5% in children and 10 to 15% in adults (33). There have been recent reports from Europe, Canada, South Africa, and the United States of N. meningitidis isolates that show moderate resistance to penicillin (48, 51). The occurrence of a new, more virulent strain of N. meningitidis serogroup C associated with an increase in both incidence and mortality of meningococcal disease in Canada is a reminder that the evolution of N. meningitidis virulence is ongoing (49).
The best preventive measure against meningococcal disease is a polyvalent polysaccharide vaccine. The vaccine is not protective against serogroup B meningococcus, however, nor can it protect children under 2 years of age (7, 14). Among new vaccine targets against N. meningitidis being explored are iron-regulated outer membrane proteins (2, 24). These proteins are attractive vaccine candidates due to their important role in virulence and surface accessibility (2, 24, 39). N. meningitidis expresses several outer membrane receptors that enable the bacterium to use human transferrin, lactoferrin, hemoglobin (Hb), heme, and haptoglobin-hemoglobin complexes (Hpt-Hb) as sources of iron (11, 13, 22, 23, 36, 42). Heme and Hb are the most abundant sources of iron in the body (29). Recently, two neisserial Hb-binding outer membrane receptors have been identified. HmbR is an iron-regulated, 89.5-kDa protein that binds Hb, extracts the heme from it, and transports the heme into the periplasm (42–44). The hmbR mutant of N. meningitidis was attenuated in an infant rat infection model, confirming the importance of Hb acquisition in meningococcal virulence (42). A second Hb-binding protein is the HpuAB bipartite receptor involved in use of Hb and Hpt-Hb complexes in N. meningitidis and Neisseria gonorrhoeae (8, 22, 23). In addition to Hb, the HpuAB receptor binds Hpt-Hb complexes and apo-Hpt (22). The HpuAB bipartite receptor is most likely the main Hb receptor of gonococci, since the hmbR gene contains a premature stop codon in all gonococcal strains tested (8, 43). Recently, the expression of the hpuAB receptor genes in N. gonorrhoeae was shown to undergo phase variation (9).
A limited analysis of meningococcal clinical isolates revealed functional redundancy in their Hb utilization systems (43). In this communication, we show that this functional redundancy is a common phenomenon in clinical isolates of N. meningitidis. Moreover, we show that the expression of hmbR undergoes phase variation due to the slipped-strand mispairing (SSM) of a poly(G) tract beginning at position +1164 in the hmbR coding sequence. The rate of switching between the off and on phases of hmbR differs more than 1,000-fold between different clinical isolates. These results establish a rational explanation for the existence of functional redundancy in Hb utilization systems and support a novel hypothesis correlating the rates of phase variation and virulence.
MATERIALS AND METHODS
Plasmids, bacteria, and media.
Strains and plasmids used in this study are listed in Table 1. The meningococcal strains in this study were epidemiologically unrelated clinical isolates obtained from Michael Reeves at the Centers for Disease Control and Prevention, Atlanta, Ga. (3, 10). The meningococci were grown on GCB (Difco) agar containing Kellog’s supplements and incubated at 37°C with 5% CO2. Escherichia coli was grown in LB (Luria broth). When necessary, the following antibiotics were used in work with neisseriae: kanamycin at 100 mg/liter and erythromycin at 3 mg/liter (300 mg/liter in E. coli). Inactivation of hpuB and hmbR was performed by transforming meningococci with pARR1500 and pIRS525, respectively. pARR1500 was linearized by digestion with XbaI, and pIRS525 was linearized by digestion with BamHI. One microgram of linearized DNA was incubated with ∼107 bacteria in GCB broth supplemented with 5 mM MgCl2 for 30 min. Reaction mixtures were transferred to 500 μl of GCB plus Kellog’s supplements and allowed to grow for 6 h. Transformants were selected on GCB plates with the appropriate antibiotic overnight. Southern blot analysis was performed to verify the inactivation of hmbR and hpuB.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source(s) |
|---|---|---|
| Strains | ||
| IR2781 | N. meningitidis serogroup B clinical isolate | D. Stephens |
| IR2848 to IR2856 | N. meningitidis serogroup A clinical isolates | M. Reeves |
| IR2857 to IR2863 | N. meningitidis serogroup C clinical isolates | M. Reeves |
| IR2843 to IR2847 | N. meningitidis serogroup Y clinical isolates | M. Reeves |
| IR1072 to IR1075 | N. meningitidis strains | Lab collection |
| IR1113, IR1114, IR1311, IR2359, IR2864, and IR2865 | N. gonorrhoeae strains | Lab collection |
| IR2363 | N. lactamica O9238 | Lab collection |
| IR2364 | N. lactamica E6040 | Lab collection |
| IR2365 | N. lactamica O1748 | Lab collection |
| IR2366 | N. kochii 31291 | Lab collection |
| IR2367 | N. kochii 31292 | Lab collection |
| IR2368 | N. flava NS-6 | Lab collection |
| IR2369 | N. ovis 72-B | Lab collection |
| IR2370 | N. polysaccharea N462 | Lab collection |
| IR2371 | N. mucosa A7895 | Lab collection |
| IR2372 | N. cinerea 34382 | Lab collection |
| IR2373 | N. cinerea 33683 | Lab collection |
| IR2375 | N. subflava B886 | Lab collection |
| IR2376 | N. subflava NS-6 | Lab collection |
| IR3261 to IR3277 | hpuB::erm derivatives of the following strains in order: IR2781, IR2844 to IR2846, IR2848, IR2850 to IR2853, IR2855 to IR2860, IR2862, and IR2863 | This study |
| IR3287 to IR3292 | hmbR phase-on revertants of the following hmbR phase-off strains in order: IR3261, IR3270, IR3272, IR3273, IR3275, and IR3277 | This study |
| IR3297 to IR3318 | hmbR::kan derivatives of the following strains in order: IR1074, IR2781, IR2849, IR2851, IR2855 to IR2860, IR2862, IR2863, IR3267, IR3271, IR3274, IR3276, and IR3287 to IR3292 | This study |
| Plasmids | ||
| pDS85 | DraI-StuI fragment of hpuB cloned into pBluescript | D. Dyer and L. Lewis |
| pARR1500 | Ermr from Tn1545 cloned into ClaI fragment of pDS85 | This study |
| pIRS523 | EcoRI-SalI fragment of hmbR cloned into pUC18 | Lab collection |
| pIRS525 | Kanr cassette cloned into NotI site of pIRS523 | Lab collection |
| pIRS876 | PstI-SalI fragment of hmbR cloned into pBluescript | Lab collection |
Hb utilization assays.
To test the ability of neisseriae to use Hb and/or hemin as the sole iron sources, a suspension of bacteria was plated onto GCB agar containing 50 μM deferoxamine mesylate (Desferal; Ciba Geigy). Filter discs (1/4-in. diameter; Schleicher & Schuell, Inc., Keene, N.H.) supplemented with 10 μl of either 5.0-mg/ml human Hb (Sigma) or 6.5-mg/ml bovine hemin (Sigma) stock solutions and placed onto plates inoculated with N. meningitidis. Zones of growth around the discs were recorded after overnight incubation at 37°C and 5% CO2.
Chromosomal DNA preparations.
Approximately one-eighth plate full of N. meningitidis or one quarter-plate full of other Neisseria species was resuspended in 500 μl of saline-EDTA (150 mM NaCl, 100 mM EDTA [pH 8.0]) and frozen at −80°C. The samples were thawed in the presence of 500 μl of Tris-sodium dodecyl sulfate (SDS) (100 mM Tris [pH 9.0], 100 mM NaCl, 1% SDS) at 50°C. DNA was extracted with saline-EDTA-equilibrated phenol and centrifuged at 14,000 × g for 20 min. The supernatant was removed, and DNA was precipitated with isopropanol and washed with 70% ethanol. DNA was resuspended in double-distilled H2O (ddH2O) and stored at 4°C.
Outer membrane preparations.
Cells of N. meningitidis grown on one plate were resuspended in 20 ml of GCB medium containing Kellog’s supplements minus iron but with 50 μM deferoxamine mesylate and incubated for 4 h at 37°C with 5% CO2. Cells were pelleted by centrifugation at 500 × g and placed on ice. The pellet was resuspended in 1.6 ml of 600 mM sucrose–6 mM EDTA–200 mM Tris (pH 8.0). Spheroplasts were generated by treating the cells with 200 μg of lysozyme (2 mg/ml in 200 mM Tris [pH 8.0]) in 5 ml of ddH2O. Outer membranes were extracted from spheroplasts with 2% Triton X-100 in 50 mM Tris (pH 8.0) in the presence of 10 mM MgCl2 and 100 μg of DNase. Outer membranes were sedimented by centrifugation at ∼40,000 × g for 60 min. Outer membranes were washed and resuspended in ddH2O.
Southern blot hybridization, PCR, and DNA sequencing.
Standard methods for plasmid DNA preparation, restriction endonuclease analysis, and ligation were performed by the methods of Sambrook et al. (33a). Southern blot analysis was done with a digoxigenin (DIG) nonradioactive DNA labeling and detection kit (Genius system; Boehringer, Mannheim, Germany) under medium-stringency conditions (two washes with 2× SSC–0.1% SDS [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] at room temperature followed by two washes in 0.5× SSC–0.1% SDS at 60°C). The internal hpuB-specific probe was obtained via random priming of the DraI-StuI fragment of pDS85. The internal hmbR-specific probe was obtained as described above with the PstI-SalI fragment from pIRS876. PCRs were performed with Taq polymerase. The ∼600-bp fragment containing the poly(G) tract of hmbR was amplified by using primers hmbrpg1a (5′ GCTTATCAAATCAACGACAACCACC 3′) and hmbrpg1b (5′ GGTGTTTTGTCACAAGCATGAC 3′) and an annealing temperature of 60°C. This fragment was sequenced by dye terminator cycle sequencing on an ABI model 377 automated sequencer with primers hmbrpg1a and hmbrpg1b to obtain the nucleotide sequence of both DNA strands.
Determination of the rate of switching.
Single colonies from hpuB::erm strains that were hmbR phase off were isolated on nonselective GCB plates and resuspended in 200 μl of GCB broth. Samples (50 μl) of 10−1 to 10−3 serial dilutions were plated on GCB plates containing 50 μM deferoxamine mesylate and 100 mg of Hb per liter. Samples (50 μl) of 10−4 to 10−6 serial dilutions were plated on nonselective GCB plates. The hmbR-off to -on switching rates were determined by dividing the number of colonies on GCB-deferoxamine mesylate-Hb plates by the number of colonies on the nonselective GCB plates. Medians of eight independent measurements were used for comparisons between strains. Statistical significance of measurements was determined by nonparametric Wilcoxon signed-rank and rank sum tests. Validity of the assay was determined by plating hmbR phase-on strains and determining the ratio of colonies on restrictive versus nonrestrictive GCB plates.
Western blot analysis.
Samples (2 μl) of outer membrane preparations were boiled in SDS buffer and loaded in a 10% polyacrylamide gel. Samples were transferred to OPTITRAN BA-S 83 filter by using a Schleicher & Schuell Pronto Semi-Dry Electroblotter and discontinuous buffers. The membrane was probed with a 1:1,000 dilution of a polyclonal rabbit anti-glutathione S-transferase fused to HmbR′ N terminus (anti-2532#2) and washed with phosphate-buffered saline containing 0.05% Tween 20. Samples were probed with a 1:10,000 dilution of goat anti-rabbit antibody conjugated to horseradish peroxidase. Signals were detected by fluorography, using equal volumes of Enhanced Luminol Reagent and Oxidizing Reagent (NEN Life Science Products Renaissance Western Blot Chemiluminescence Reagent).
RESULTS
Distribution of hmbR and hpuB among Neisseria species.
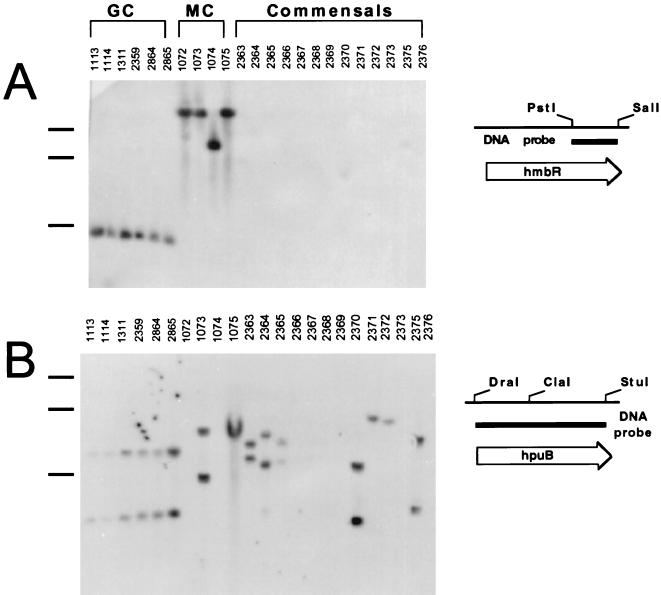
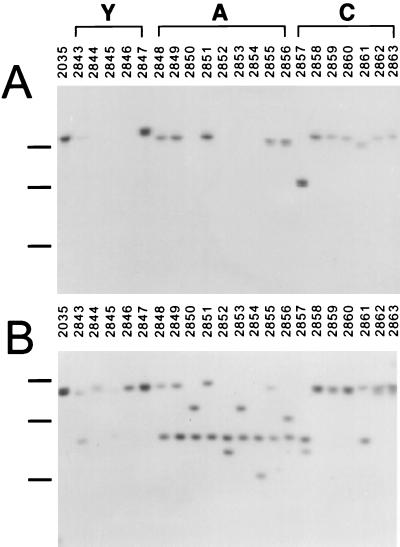
N. meningitidis can express two receptors, HmbR and HpuB, capable of utilizing free Hb as an iron source (23, 42). The distribution of these two receptors in different species and serotypes was investigated by DNA hybridization. Chromosomal DNA from six strains of N. gonorrhoeae hybridized with both hmbR and hpuB probes (Fig. 1A and B). Chromosomal DNA from only 2 of the 13 tested commensal Neisseria strains hybridized with the hmbR probe. These two commensal strains, 2370 (Neisseria polysaccharea N462) and 2375 (Neisseria subflava B886), yielded only very weak signals (Fig. 1A). In contrast, the hpuB probe hybridized strongly with chromosomal DNA from 7 of the 13 commensal strains: 2363 (Neisseria lactamica O9238), 2364 (N. lactamica E6040), 2365 (N. lactamica O1748), 2370 (N. polysaccharea N462), 2371 (Neisseria mucosa A7895), 2372 (Neisseria cinerea 34382), and 2375 (N. subflava B886) (Fig. 1B). Chromosomal DNA from twenty-five N. meningitidis clinical isolates, representing four serotypes, was tested with hmbR and hpuB probes. The hmbR-specific probe hybridized with 18 of the 25 chromosomal DNAs, and the hpuB probe hybridized with 23 (Fig. 1 and 2). All nine serogroup C meningococcal strains tested possessed both hmbR and hpuB with the exception of strain 1072, which did not yield a signal with the hpuB probe. The single serogroup B isolate, strain 1074, possessed only hmbR (Fig. 1). The largest divergence in the distribution of hmbR and hpuB was found in serogroups A and Y. All of the 10 serogroup A isolates contained hpuB, but only 6 possessed hmbR (Fig. 1 and 2). Similarly, all five serogroup Y strains possessed hpuB, while only two contained hmbR (Fig. 2). The absence of hmbR in the serogroup A and Y isolates was confirmed by PCR (data not shown).
FIG. 1.
Distribution of hmbR and hpuB genes in isolates of N. gonorrhoeae (GC), N. meningitidis (MC), and commensal Neisseria species. N. meningitidis 1072 and 1075 represent serotype C, strain 1074 represents serotype B, and strain 1073 represents serotype A. ClaI-digested chromosomal DNA from indicated strains was hybridized with hmbR-specific (A) or hpuB-specific (B) DIG-labeled DNA probes. The positions of 3-, 6-, and 12-kb marker bands are indicated by the black bars to the left of the gel (from the bottom to the top).
FIG. 2.
Distribution of hmbR and hpuB in N. meningitidis clinical isolates representing serogroups Y, A, and C. ClaI-digested chromosomal DNA from indicated strains was hybridized with hmbR-specific (A) or hpuB-specific (B) DIG-labeled DNA probes. The positions of 3-, 6-, and 12-kb marker bands are indicated by the black bars to the left of the gel (from the bottom to the top). Due to the small amount of DNA loaded in lane 2845, the hpuB-hybridizable band is relatively weak.
Characterization of different Hb utilization phenotypes in N. meningitidis.
All wild-type N. meningitidis strains tested were able to use human Hb as an iron source with the exception of strain 2844. Furthermore, nearly all strains that used Hb grew as a uniform lawn around Hb-supplemented discs, strain 2781 being the only exception (Table 2). In order to study the function of HmbR, the hpuB gene was inactivated by insertion of an erythromycin cassette. The hpuB-negative strains displayed two distinct Hb utilization phenotypes in the disc diffusion assay: (i) single-colony growth around a disc supplemented with Hb (3261, 3270, 3272, 3273, 3275, 3276, and 3277), and (ii) confluent growth around the Hb disc (1074, 3265, 3267, 3271, and 3274) (Table 2). The hpuB::erm strains that did not use Hb as a source of iron (3262, 3263, 3264, 3266, 3268, and 3269) were found not to possess hmbR (Fig. 2A and Table 2). Similar phenotypes with regard to growth around Hb discs were observed when the function of HpuB was tested in an hmbR-negative genetic background. Eleven strains were still able to grow as confluent lawns despite the loss of hmbR, whereas six grew only as single colonies (Table 2).
TABLE 2.
Genotypic and phenotypic characterization of N. meningitidis clinical isolates and isogenic hpuB::erm and hmbR::kan derivatives
| Serotype | Clinical isolates
|
hpuB::erm derivatives
|
hmbR::kan derivatives
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | hmbRa | hpuBa | PHb | Strain | No. of Gc | PH | Strain | PH | |
| C | 2857 | + | + | LW | 3272 | 8 | SC | 3303 | LW |
| 2858 | + | + | LW | 3273 | 13 | SC | 3304 | LW | |
| 2859 | + | + | LW | 3274 | 12 | LW | 3305 | SC | |
| 2860 | + | + | LW | 3275 | 10 | SC | 3306 | LW | |
| 2861 | + | + | LW | NTd | * | NA | NT | NA | |
| 2862 | + | + | LW | 3276 | 13 | SC | 3307 | LW | |
| 2863 | + | + | LW | 3277 | 13 | SC | 3308 | LW | |
| A | 2848 | + | + | LW | 3265 | 9 | LW | NT | NA |
| 2849 | + | + | LW | NT | 9 | NA | 3299 | SC | |
| 2850 | − | + | LW | 3266 | * | 0 | NA | LW | |
| 2851 | + | + | LW | 3267 | 9 | LW | 3300 | SC | |
| 2852 | − | + | LW | 3268 | * | 0 | NA | LW | |
| 2853 | − | + | LW | 3269 | * | 0 | NA | LW | |
| 2854 | − | + | LW | NT | * | NA | NA | LW | |
| 2855 | + | + | LW | 3270 | 8 | SC | 3301 | SC | |
| 2856 | + | + | LW | 3271 | 9 | LW | 3302 | SC | |
| Y | 2843 | + | + | LW | NT | 10 | NA | NT | NA |
| 2844 | − | + | 0 | 3262 | * | 0 | NA | 0 | |
| 2845 | − | + | LW | 3263 | * | 0 | NA | LW | |
| 2846 | − | + | LW | 3264 | * | 0 | NA | LW | |
| 2847 | + | + | LW | NT | 10 | NA | NT | NA | |
| B | 1074 | + | − | LW | NAe | 9 | LW | 3297 | 0 |
| 2781 | + | + | SC | 3261 | 8 | SC | 3298 | SC | |
Presence (+) or absence (−) of homologous sequence determined by Southern blot hybridization.
The Hb utilization phenotype (PH) of the strain is as follows: LW, confluent lawn; SC, single colony; NA, not applicable; 0, no growth.
Number of consecutive G residues in the poly(G) tract of hmbR. An asterisk indicates that no sequence was obtained.
NT, no transformants obtained.
NA, not available.
These data suggest that some meningococcal strains, although possessing both Hb receptor genes, were expressing only one receptor. The single-colony phenotype observed in some hmbR::kan and hpuB::erm genetic backgrounds may be due to the selection for rare switching-on events in hmbR and hpuB genes. Alternatively, Hb+ revertants may be the result of activation of some currently unknown Hb utilization mechanism in meningococci.
Correlation between hmbR genotypes and different Hb utilization phenotypes in N. meningitidis.
Recently, the HpuAB receptor of N. gonorrhoeae was shown to undergo phase variation due to the SSM at the poly(G) tract in the hpuA gene (9). Since the N. meningitidis hpuA and hpuB genes are highly homologous to N. gonorrhoeae hpuAB genes, it is very likely that the observed Hb+ revertants in the hmbR::kan background arose in hpuAB by a phase variation mechanism (9, 23).
The single-colony phenotype of the hpuB::erm strains could be also explained by the phase variation of hmbR. Indeed, the hmbR gene has a poly(G) tract located at position +1164 relative to the hmbR start codon that could be a substrate for SSM (42). Poly(G) tracts containing 9 or 12 consecutive G residues would allow hmbR to remain in frame with respect to the start codon. In order to determine the number of G residues in the poly(G) tract of hmbR, a 600-bp region surrounding the poly(G) tract of the wild-type isolates was PCR amplified and sequenced. The length of the poly(G) tract in the hmbR genes of 12 clinical isolates (Table 2) was compared to the Hb utilization phenotypes of their hpuB::erm derivatives (Table 2). All five strains (1074, 3265, 3267, 3271, and 3274) possessing either 9 or 12 G residues grew as a uniform lawn around Hb discs (putative hmbR phase on). All seven strains possessing poly(G) tracts of a length other than 9 or 12 (3261, 3270, 3272, 3273, 3275, 3276, and 3277) grew as single colonies around Hb discs (putative hmbR phase off). The length of poly(G) tracts in three clinical isolates, 2843, 2847, and 2849, were not correlated with the phenotypes of their hpuB::erm derivatives because these mutants could not be isolated (Table 2). These data, though merely a correlation, support the hypothesis that hmbR undergoes phase variation via SSM at a run of consecutive G residues within the coding region.
Demonstration of hmbR switching in N. meningitidis.
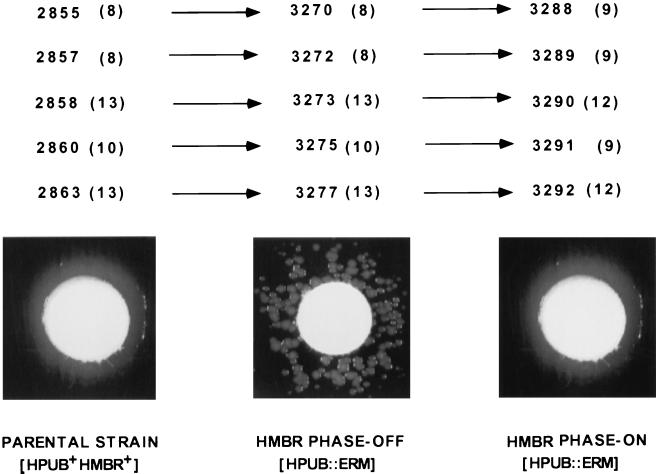
When the Hb+ revertant colonies (3288, 3289, 3290, 3291, and 3292) of hpuB::erm mutants (3270, 3272, 3273, 3275, and 3277) were assayed for Hb utilization, all grew as confluent lawns (Fig. 3, right). The inactivation of hmbR in these hpuB::erm strains eliminated Hb utilization, indicating that the HmbR receptor is responsible for the Hb+ phenotype. Southern blot hybridization confirmed that the hpuB gene in each revertant was still inactive (data not shown). These data demonstrated that the single-colony phenotype originates from changes within hmbR, most likely due to the SSM in the poly(G) tract of hmbR. Indeed, when the poly(G) tracts of the putative phase-on hmbR revertants were analyzed, all possessed either 9 or 12 consecutive G residues, while the parent strains had 8, 10, or 13 (Fig. 3). The only exception was strain 2855. This strain 2855 grew as a confluent lawn, while both hpuB::erm (3270) and hmbR::kan (3301) derivatives grew as single colonies around Hb discs (Table 2). In addition, the hmbR poly(G) tract contained eight G residues, indicating that the gene is switched off. However, the single-colony phenotype of hpuB and hmbR derivatives of strain 2855 was different from single-colony phenotypes of other studied strains. While an average single-colony phenotype is characterized by a few dozen colonies growing around Hb discs (Fig. 3, middle), several hundred colonies were growing around Hb discs when hpuB::erm and hmbR::kan derivatives of 2855 strain were analyzed. Due to the large number of single colonies growing around Hb discs, the single-colony phenotype of strain 2855 appeared on the plates as a confluent lawn.
FIG. 3.
Phase variation of hmbR. The number of G residues in the poly(G) tract of hmbR is shown in parentheses for each strain analyzed. The photographs shown are examples of the phenotypes seen for each group of strains. Parental strains (left) all demonstrate a phase-on phenotype. After inactivation of hpuB, the phase-off phenotype of hmbR can be seen as a single-colony phenotype (middle). These single colonies, representing rare phase-on cells, show a confluent lawn phenotype when tested again for Hb utilization (right). The changes in the Hb utilization phenotypes were accompanied by changes in the number of G residues in the hmbR poly(G) tracts.
Confirmation of hmbR expression patterns in N. meningitidis.
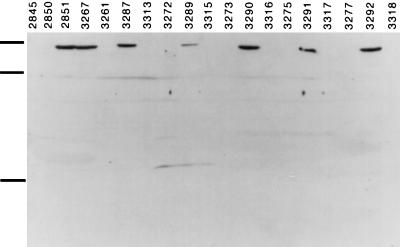
To confirm that the observed Hb utilization phenotypes corresponded with the actual expression of hmbR, a Western blot analysis was performed. HmbR was detected in outer membrane preparations from several N. meningitidis strains by using rabbit polyclonal anti-HmbR antibody (Fig. 4). DNA from strains 2845 and 2850 did not hybridize with the hmbR probe (Fig. 2), and these strains were used as negative controls. The strong signal from outer membranes of strain 2851 and its hpuB::erm derivative 3267 (Fig. 4) reinforced earlier observations that these strains contain a phase-on copy of hmbR (Fig. 2 and Table 2). Strains 3261, 3272, 3273, 3275, and 3277 all hybridized with the hmbR probe (Fig. 2) but were all in the off phase (Table 2). None of these strains gave a signal when probed with anti-HmbR antibody (Fig. 4). However, the phase-on revertants of these strains (3287, 3289, 3290, 3291, and 3292, respectively) all gave strong signals with the anti-HmbR antibody (Fig. 4). Conversely, the outer membrane preparations of the hpuB::erm hmbR::kan double mutants (3313, 3315, 3316, 3317, and 3318, respectively) gave no signals with the antibody. This result confirmed that the Hb+ phenotype of the phase-on hmbR revertants was dependent on HmbR expression.
FIG. 4.
Western blot analysis of N. meningitidis outer membrane preparations isolated from strains with different HmbR-dependent Hb utilization phenotypes. Strains 2845 and 2850 are both HmbR negative controls. Strain 2851 and its hpuB::erm derivative 3267 are hmbR phase on. Strains 3261, 3272, 3273, 3275, and 3277 are hmbR phase off. Strains 3287, 3289, 3290, 3291, and 3292 are hmbR phase-on revertants of strains 3261, 3272, 3273, 3275, and 3277, respectively. Strains 3313, 3315, 3316, 3317, and 3318 are hmbR::kan derivatives of strains 3287, 3289, 3290, 3291, and 3292, respectively. The positions of 97.4-, 68-, and 18.4-kDa marker bands are indicated by the black bars to the left of the gel (from the top to the bottom).
Determination of the rate of switching of the hmbR gene.
The rate at which hmbR switches from phase off to phase on was determined for each of the six hmbR phase-off strains (hpuB::erm derivatives of wild-type clinical isolates). The switching rates for strains 3272, 3273, and 3275 ranged from 4.2 × 10−4 to 5.5 × 10−4. Strain 3277 switched at a lower rate of 3.4 × 10−5. Strain 3261 switched at a rate that was 40-fold lower than those of 3272, 3273, and 3275, namely, 7.4 × 10−6. Strain 3270 switched at a rate 70-fold higher than those of strains 3272, 3273, and 3275, namely, 2.0 × 10−2. All rates are median values for eight independent measurements.
DISCUSSION
It has been previously shown that 95% of tested meningococci and 60% of gonococci were able to use Hb, while all tested strains used free heme as a source of iron (27). The ability of Hb+ pathogenic neisseriae to bind Hb in vitro indicated the existence of cell surface Hb receptor(s) (20). This hypothesis has been confirmed by the independent discovery of two outer membrane Hb-binding proteins, HmbR and HpuB (22, 23, 42). Data presented in this study confirmed the universal ability of meningococci to use Hb and heme: only one serogroup Y isolate (2844) was unable to use Hb as a source of iron, although it contained hpuB-like nucleotide sequences. More interestingly, hybridization data showed that 64% (16 of 25) of strains possessed both receptor genes, indicating that the redundancy of Hb receptors is a common phenomenon in meningococci. The analysis established that 23 of 25 strains of meningococci possessed hpuB, while 7 of 25 isolates, clustered among serogroups A and Y, lacked the hmbR gene. This distribution pattern may indicate the relative importance of Hb receptors, since the more prevalent HpuAB receptor uses both Hpt-Hb and Hb, while HmbR is restricted to Hb use (22, 23, 42). The lack of hmbR and the high prevalence of hpuB in commensals suggest more recent acquisition of hmbR by meningococci.
The ability to use Hb has been shown to promote virulence of meningococci, thus categorizing Hb receptors as potential virulence factors (37, 42). The overlap in the functions of HmbR and HpuB is not unusual, and the functional redundancy of virulence factors has been observed in several bacterial pathogens (18, 31, 32, 40). The possession of redundant virulence factors by bacteria compromises the ability of the host immune system to control infection. If this were the main purpose of redundancy, bacteria should be able to regulate the expression of redundant virulence factors in order to minimize their exposure to immune response. One way of controlling the expression of redundant virulence functions in bacteria is by phase variation. Phase variation is the alteration of the gene expression between on and off phases as a result of reversible changes at the DNA level. It is a common mechanism of controlling the expression of surface-exposed virulence factors in many mucosal pathogens including Neisseria spp., H. influenzae, and possibly Haemophilus pylori (1, 9, 15, 16, 30, 31, 35, 40, 50). A variety of different mechanisms, including DNA inversion, recombination, and SSM have been proposed to account for phase variation in bacteria (21, 25, 26, 41, 53). SSM occurs as a result of illegitimate annealing of DNA containing multiple short tandem repeats. Subsequent replication or repair of this mispaired sequence may result in insertion or deletion of integral repeat units (21). Short repetitive sequences within coding regions can undergo SSM resulting in translational frameshifts, whereas SSM of repeats in promoter elements can result in various levels of transcription. Short polypyrimidine or polypurine tracts are capable of mediating SSM (21). The repetitive element (5′ CTCTT 3′)n has been implicated in the phase variation of several genes both transcriptionally and translationally (i.e., opacity proteins in N. gonorrhoeae and class V proteins and Opc in N. meningitidis) (4, 19, 28). Homopolymeric tracts are conducive to SSM as well. Poly(G) tracts mediate phase variation of several genes encoding surface-exposed molecules in neisseriae (15, 17, 18, 34, 47, 52).
In the course of studying hmbR and its role in Hb utilization, it was recognized that some strains that hybridized with an hmbR-specific probe did not express HmbR in the outer membranes. These observations led to the hypothesis that the expression of hmbR undergoes phase variation. Examination of Hb utilization phenotypes, HmbR expression, and nucleotide sequence in 15 strains representing four serogroups confirmed the role of the poly(G) tract in the phase variation of hmbR. While hmbR transcript levels were not examined, the position of the poly(G) tract in the middle of the hmbR coding region indicates translational regulation. In strains expressing functionally redundant Hb receptors, it would be advantageous for bacteria to phase vary both receptors. Indeed, the second Hb receptor, HpuAB, has been shown to phase vary in N. gonorrhoeae (9). The data presented in this study suggest that the expression of meningococcal HpuAB also undergoes phase variation. Another mucosal pathogen, H. influenzae, possesses at least two independent surface-exposed Hb or Hb-Hpt receptors, the expression of which are under the control of SSM-mediated phase variation (31, 32). Therefore, phase variation may be a common mechanism by which mucosal pathogens regulate the expression of surface-exposed molecules with redundant functions. Interestingly, 2 of 10 strains (2855 and 2781) examined had both receptors in off phase, while none of the strains had both receptors switched on. Despite having both receptors switched off, strain 2855 gave confluent growth phenotype around Hb discs due to the very high rate of switching of both genes.
The rates at which many of the phase-varying genes switch between different phases of expression has been determined to be ∼10−4 (4, 15, 28, 46). The rate of hmbR switching in all four serogroup C isolates tested was on the order of 10−4 to 10−5. However, serogroup A and B isolates gave significantly different rates, 10−2 and 10−6, respectively. Though the rate of phase-on to -off transition could not be measured, it is reasonable to assume that it occurs at a similar rate. One explanation for the different rates of switching in serogroup A and B isolates could be small differences in nucleotide sequence surrounding the poly(G) tract of hmbR. Some sequences may be more conductive to SSM than others. However, inspection of approximately 600 bp surrounding the poly(G) tract of hmbR revealed 94% identity between the strains of different serotypes. Another plausible explanation for differences in rates of switching between strains could be a variation in levels of hmbR transcription. Indeed, a recent study has indicated that promoter strength may influence the rate at which genes phase vary (5). The serogroup A strain found to have a higher rate of switching may have higher levels of hmbR transcription. Likewise, the low rate of switching of hmbR in the serogroup B isolate may be due to decreased transcription. However, Western blot analysis of outer membrane preparations from the serogroup B isolate did not show any decrease in HmbR expression.
We hypothesize that the differences in the rate of switching of hmbR among meningococcal isolates is attributed to different genetic backgrounds of these strains. N. meningitidis may possess a mechanism that can modulate the rate of switching of phase-varying genes. Strains with higher rates of switching would be able to adapt to the hostile environment of the host more successfully, leading to higher colonization, disease, and transmission rates. Additional work must be done to prove the connection between the rate of switching and virulence of meningococci, however.
ACKNOWLEDGMENTS
We thank Michael Reeves and Sally Berish for providing the strains used in this study. We also thank David Dyer and Lisa Lewis for the plasmid pDS85. We thank G. Churchward and M. Baer for suggestions and reading the manuscript.
This work was supported in part by Public Health Service grant AI472870-01A1.
REFERENCES
- 1.Aho E L, Murphy G L, Cannon J G. Distribution of specific DNA sequences among pathogenic and commensal Neisseria species. Infect Immun. 1987;55:1009–1013. doi: 10.1128/iai.55.4.1009-1013.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ala’Aldeen D A A, Stevenson P, Griffiths E, Gorringe A R, Irons L I, Robinson A, Hyde S, Borriello S P. Immune responses in humans and animals to meningococcal transferrin-binding proteins: implications for vaccine design. Infect Immun. 1994;62:2984–2990. doi: 10.1128/iai.62.7.2984-2990.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett S J, Sneath P H A. A numerical phenotypic taxonomic study of the genus Neisseria. Microbiology. 1994;140:2867–2891. doi: 10.1099/00221287-140-10-2867. [DOI] [PubMed] [Google Scholar]
- 4.Belland R J, Morrison S G, van der Lay P, Swanson J. Expression and phase variation of gonococcal P.II genes in Escherichia coli involves ribosomal frameshifting and slipped-strand mispairing. Mol Microbiol. 1989;3:777–786. doi: 10.1111/j.1365-2958.1989.tb00226.x. [DOI] [PubMed] [Google Scholar]
- 5.Belland R J, Morrison S G, Carlson J H, Hogan D M. Promoter strength influences phase variation of neisserial opa genes. Mol Microbiol. 1997;23:123–135. doi: 10.1046/j.1365-2958.1997.1971556.x. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Surveillance for diabetes mellitus-United States, 1980–1989, and laboratory-based surveillance for meningococcal disease in selected areas-United States, 1989–1991. Morbid Mortal Weekly Rep. 1993;42:21–30. [Google Scholar]
- 7.Centers for Disease Control and Prevention. Control and prevention of meningococcal disease and control and prevention of serogroup C meningococcal disease: evaluation and management of suspected outbreaks. Morbid Mortal Weekly Rep. 1997;46:1–21. [Google Scholar]
- 8.Chen C-J, Sparling P F, Lewis L A, Dyer D W, Elkins C. Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect Immun. 1996;64:5008–5014. doi: 10.1128/iai.64.12.5008-5014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C-J, Elkins C, Sparling P F. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect Immun. 1998;66:987–993. doi: 10.1128/iai.66.3.987-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun P K, Sensabaugh G F, Vedros N A. Genetic relationships among Neisseria species assessed by comparative enzyme electrophoresis. J Gen Microbiol. 1985;131:3105–3115. doi: 10.1099/00221287-131-11-3105. [DOI] [PubMed] [Google Scholar]
- 11.Cornellissen C N, Sparling P F. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 12.Cvjetanovic B. Cerebrospinal meningitidis, a disease with seasonal variations. Bull W H O. 1978;56:81–102. [Google Scholar]
- 13.Dyer D W, West E P, Sparling P F. Effects of serum carrier proteins on the growth of pathogenic neisseriae with heme-bound iron. Infect Immun. 1987;55:2171–2175. doi: 10.1128/iai.55.9.2171-2175.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold R, Lepow M L, Goldschneider T F, Draper T F, Gotschlich E C. Kinetics of antibody production to group A and group C meningococcal polysaccharide vaccines administered during the first six years of life: prospects for routine immunization of infants and children. J Infect Dis. 1979;140:690–697. doi: 10.1093/infdis/140.5.690. [DOI] [PubMed] [Google Scholar]
- 15.Hammerschmidt S, Muller A, Sillmann H, Muhlenhoff M, Borrow R, Fox A, van Putten J, Zollinger W D, Gerardy-Schahn R, Frosch M. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol. 1996;20:1211–1220. doi: 10.1111/j.1365-2958.1996.tb02641.x. [DOI] [PubMed] [Google Scholar]
- 16.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleischmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jennings M P, Hood D W, Peak I R A, Mumtaz V, Moxon E R. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 18.Jonsson A-B, Nyberg G, Normark S. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 1991;10:477–488. doi: 10.1002/j.1460-2075.1991.tb07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawula T H, Aho E L, Barritt D S, Klapper D G, Cannon J G. Reversible phase variation of expression of Neisseria meningitidis class 5 outer membrane proteins and their relationship to gonococcal proteins II. Infect Immun. 1988;56:380–386. doi: 10.1128/iai.56.2.380-386.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C B, Hill P. Identification of an outer-membrane haemoglobin-binding protein in Neisseria meningitidis. J Gen Microbiol. 1992;138:2647–2656. doi: 10.1099/00221287-138-12-2647. [DOI] [PubMed] [Google Scholar]
- 21.Levinson G, Gutman G A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 22.Lewis L A, Dyer D W. Identification of an iron-regulated outer membrane protein of Neisseria meningitidis involved in the utilization of hemoglobin complexed to haptoglobin. J Bacteriol. 1995;177:1299–1306. doi: 10.1128/jb.177.5.1299-1306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis L A, Gray E, Wang Y, Roe B A, Dyer D W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 24.Lissolo L, Maitre-Wilmotte G, Dumas P, Mignon M, Danve B, Quentin-Millet M-J. Evaluation of transferrin binding protein 2 within the transferrin-binding protein complex as a potential antigen for future meningococcal vaccines. Infect Immun. 1995;63:884–890. doi: 10.1128/iai.63.3.884-890.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer T F, Mlawer N, So M. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell. 1982;30:45–52. doi: 10.1016/0092-8674(82)90010-1. [DOI] [PubMed] [Google Scholar]
- 26.Meyer T F, van Putten J P M. Genetic mechanisms and biological implications of phase variation in pathogenic neisseriae. Clin Microbiol Rev. 1989;2(Suppl.):S139–S145. doi: 10.1128/cmr.2.suppl.s139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mickelsen P A, Sparling P F. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from transferrin and iron compounds. Infect Immun. 1981;33:555–564. doi: 10.1128/iai.33.2.555-564.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy G L, Connell T D, Barritt D S, Koomey M, Cannon J G. Phase variation of gonococcal protein II: regulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell. 1989;56:539–547. doi: 10.1016/0092-8674(89)90577-1. [DOI] [PubMed] [Google Scholar]
- 29.Otto B R, Verweij-van Vught A M J J, MacLaren D M. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 30.Peak I R, Jennings M P, Hood D W, Bisercic M, Moxon E R. Tetrameric repeat units associated with virulence factor phase variation in Haemophilus also occur in Neisseria spp. and Moraxella catarrhalis. FEMS Microbiol Lett. 1996;137:109–114. doi: 10.1111/j.1574-6968.1996.tb08091.x. [DOI] [PubMed] [Google Scholar]
- 31.Ren Z, Jin H, Morton D J, Stull T L. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Evidence for slipped-strand mispairing as a mechanism of regulation of hemoglobin-binding proteins in Haemophilus influenzae, abstr. B-362; p. 116. [Google Scholar]
- 32.Ren Z, Jin H, Morton D J, Stull T L. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect Immun. 1998;66:4733–4741. doi: 10.1128/iai.66.10.4733-4741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riedo F X, Plikaytis B D, Broome C L. Epidemiology and prevention of meningococcal disease. J Infect Dis. 1995;14:643–657. doi: 10.1097/00006454-199508000-00001. [DOI] [PubMed] [Google Scholar]
- 33a.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Sarkari J, Pandit M, Moxon E R, Achtman M. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by the size variation of a promoter containing poly-cytidine. Mol Microbiol. 1994;13:207–217. doi: 10.1111/j.1365-2958.1994.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 35.Saunders N J, Peden J F, Hood D W, Moxon E R. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 36.Schryvers A B, Morris L J. Identification and characterization of the transferrin receptor from Neisseria meningitidis. Mol Microbiol. 1988;2:281–288. doi: 10.1111/j.1365-2958.1988.tb00029.x. [DOI] [PubMed] [Google Scholar]
- 37.Schryvers A B, Gonzalez G C. Comparison of the abilities of different protein sources of iron to enhance Neisseria meningitidis infection in mice. Infect Immun. 1989;57:2425–2429. doi: 10.1128/iai.57.8.2425-2429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz B, Moore P S, Broome C V. Global epidemiology of meningococcal disease. Clin Microbiol Rev. 1989;2(Suppl.):S118–S124. doi: 10.1128/cmr.2.suppl.s118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sparling P F, Elkins C, Wyrick P B, Cohen M S. Vaccines for bacterial sexually transmitted infections: a realistic goal? Proc Natl Acad Sci USA. 1994;91:2456–2463. doi: 10.1073/pnas.91.7.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stern A, Brown M, Nickel P, Meyer T F. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986;47:61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 41.Stern A, Meyer T. Common mechanism controlling phase and antigenic variation in pathogenic neisseriae. Mol Microbiol. 1987;1:5–12. doi: 10.1111/j.1365-2958.1987.tb00520.x. [DOI] [PubMed] [Google Scholar]
- 42.Stojiljkovic I, Hwa B, de Saint Martin L, O’Gaora P, Nassif X, Hefferon F, So M. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 43.Stojiljkovic I, Larson J, Hwa V, Anic S, So M. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J Bacteriol. 1996;178:4670–4678. doi: 10.1128/jb.178.15.4670-4678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stojiljkovic I, Srinivasan N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in Neisseriae. J Bacteriol. 1997;179:805–812. doi: 10.1128/jb.179.3.805-812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tikhomirov E. Meningococcal meningitis. Global situations and control measures. World Health Stat Q. 1987;40:98–108. [PubMed] [Google Scholar]
- 46.van Belkum A, Sherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Ende A, Hopman C T, Zaat S, Essink B B, Berkhout B, Dankert J. Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the −10 and −35 regions of the promoter. J Bacteriol. 1995;177:2475–2480. doi: 10.1128/jb.177.9.2475-2480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Esso D, Fontanals D, Uriz S. Neisseria meningitidis strains with decreased susceptibility to penicillin. Pediatr Infect Dis J. 1987;6:438–439. doi: 10.1097/00006454-198705000-00003. [DOI] [PubMed] [Google Scholar]
- 49.Whalen C M, Hockin J C, Ryan A, Ashton F. The changing epidemiology of invasive meningococcal disease in Canada, 1985 through 1992: emergence of a virulent clone of Neisseria meningitidis. JAMA. 1995;273:390–394. [PubMed] [Google Scholar]
- 50.Woods J P, Cannon J G. Variation in expression of class 1 and class 5 outer membrane proteins during nasopharyngeal carriage of Neisseria meningitidis. Infect Immun. 1990;58:569–572. doi: 10.1128/iai.58.2.569-572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woods C R, Smith A L, Wasilauskas B L, Campos J, Givner L B. Invasive disease caused by Neisseria meningitidis relatively resistant to penicillin in North Carolina. J Infect Dis. 1994;170:453–456. doi: 10.1093/infdis/170.2.453. [DOI] [PubMed] [Google Scholar]
- 52.Yang Q-L, Gotschlich E C. Variation of gonococcal lipooligosaccharide structure is due to alterations in poly-G tracts in lgt genes encoding glycosyl transferases. J Exp Med. 1996;183:323–327. doi: 10.1084/jem.183.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zieg J, Hilmen M, Simon M. Regulation of gene expression by site-specific inversion. Cell. 1978;15:237–244. doi: 10.1016/0092-8674(78)90098-3. [DOI] [PubMed] [Google Scholar]