ABSTRACT
Cellulomonas flavigena is a saprotrophic bacterium that encodes, within its genome, four predicted lytic polysaccharide monooxygenases (LPMOs) from Auxiliary Activity family 10 (AA10). We showed previously that three of these cleave the plant polysaccharide cellulose by oxidation at carbon-1 (J. Li, L. Solhi, E.D. Goddard-Borger, Y. Mattieu et al., Biotechnol Biofuels 14:29, 2021, https://doi.org/10.1186/s13068-020-01860-3). Here, we present the biochemical characterization of the fourth C. flavigena AA10 member (CflaLPMO10D) as a chitin-active LPMO. Both the full-length CflaLPMO10D-Carbohydrate-Binding Module family 2 (CBM2) and catalytic module-only proteins were produced in Escherichia coli using the native general secretory (Sec) signal peptide. To quantify chitinolytic activity, we developed a high-performance anion-exchange chromatography–pulsed amperometric detection (HPAEC-PAD) method as an alternative to the established hydrophilic interaction liquid ion chromatography coupled with UV detection (HILIC-UV) method for separation and detection of released oxidized chito-oligosaccharides. Using this method, we demonstrated that CflaLPMO10D is strictly active on the β-allomorph of chitin, with optimal activity at pH 5 to 6 and a preference for ascorbic acid as the reducing agent. We also demonstrated the importance of the CBM2 member for both mediating enzyme localization to substrates and prolonging LPMO activity. Together with previous work, the present study defines the distinct substrate specificities of the suite of C. flavigena AA10 members. Notably, a cross-genome survey of AA10 members indicated that chitinolytic LPMOs are, in fact, rare among Cellulomonas bacteria.
IMPORTANCE Species from the genus Cellulomonas have a long history of study due to their roles in biomass recycling in nature and corresponding potential as sources of enzymes for biotechnological applications. Although Cellulomonas species are more commonly associated with the cleavage and utilization of plant cell wall polysaccharides, here, we show that C. flavigena produces a unique lytic polysaccharide monooxygenase with activity on β-chitin, which is found, for example, in arthropods. The limited distribution of orthologous chitinolytic LPMOs suggests adaptation of individual cellulomonads to specific nutrient niches present in soil ecosystems. This research provides new insight into the biochemical specificity of LPMOs in Cellulomonas species and related bacteria, and it raises new questions about the physiological function of these enzymes.
KEYWORDS: lytic polysaccharide monooxygenase, LPMO, AA10, carbohydrate-binding module, CBM2, Cellulomonas flavigena, chitin, HPAEC-PAD
INTRODUCTION
Chitin is a water-insoluble linear homopolysaccharide of β-(1,4)-N-acetylglucosaminyl residues which occurs in two predominant allomorphs, α (anti-parallel chain packing) and β (parallel chain packing) (1). Chitin functions primarily as a structural polysaccharide in the cell walls of fungi, the exoskeletons of arthropods and mollusks, and the gladius (“pen”) of cephalopods (2, 3). Due to this broad distribution, chitin is the second most abundant organic polymer on Earth, after cellulose (4, 5). Like cellulose, the regular structure of chitin gives rise to strong interchain hydrophobic and hydrogen-bonding interactions (6, 7), leading to high resistance to degradation. Despite the recalcitrance of cellulose and chitin, saprotrophic organisms have evolved efficient multienzyme systems to deconstruct and saccharify these substrates for growth. Specifically, many saprotrophs secrete complex suites of enzymes to catalyze both hydrolysis by glycosidases and oxidative cleavage by the more recently discovered lytic polysaccharide monooxygenases (LPMOs) (8–10).
LPMOs are small copper metalloenzymes that utilize either molecular oxygen and an external reducing agent (9, 11, 12) or hydrogen peroxide (H2O2) (13, 14) as a cosubstrate to catalyze oxidative cleavage of glycosidic bonds. The active site of all LPMOs is comprised of two histidine residues which coordinate a bound copper ligand in a characteristic “histidine-brace” motif (15). Oxidation occurs either at C1 (EC 1.14.99.54 on cellulose, EC 1.14.99.53 on chitin) and/or C4 (EC 1.14.99.56 on cellulose) of the polysaccharide chain, depending on the homolog.
LPMOs are widespread in nature, produced by prokaryotes, eukaryotes, and viruses, and their oxidative diversity has thus far been demonstrated on a plethora of poly- and oligosaccharides, including chitin, cellulose, hemicelluloses, starch, and pectins (16–18). LPMO sequences are classified into eight auxiliary activity families in the Carbohydrate-Active Enzyme (CAZy) database (19), namely, AA9 (9), AA10 (10), AA11 (20), AA13 (21), AA14 (22), AA15 (23) AA16 (24), and AA17 (17). In particular, the bacterial LPMOs of family AA10 comprise both chitin- (18, 25–38) and cellulose-active LPMOs (29, 39–46), of which Serratia marcescens CBP21 serves as the archetype that first defined these enzymes as polysaccharide oxidases (10).
Species from the genus Cellulomonas are characterized by a rich ability to degrade plant polysaccharides, the characterization of which has contributed greatly to current understanding of glycoside hydrolases (GHs) and carbohydrate-binding modules (CBMs) (47–78). The complete genome of Cellulomonas flavigena strain DSM 20109 was sequenced in 2010 (79) and was shown to encode four multimodular AA10 LPMO sequences, each consisting of an AA10 catalytic domain appended to a C-terminal CBM2 domain (with homologs known to bind insoluble cellulose, chitin, and xylan substrates [78–80]). This diversity distinguishes C. flavigena from the widely studied Cellulomonas fimi, which contains only one, cellulose-active LPMO (43, 78). We previously reported the biochemical characterization of the C. flavigena LPMOs CflaLPMO10A, CflaLPMO10B, and CflaLPMO10C, which showed that all three are strictly cellulose-active with primarily C1 oxidative regiospecificity (CflaLPMO10B and CflaLPMO10C also had some capacity to oxidize at C4) (43).
To complete the biochemical characterization of C. flavigena LPMOs, we report here the analysis of CflaLPMO10D (encoded by gene locus tag Cfla_0490) in full-length (including the C-terminal CBM2) and truncated forms (catalytic module only) following recombinant production in Escherichia coli. Substrate screening and product analysis by high-performance liquid chromatography (HPLC) and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) indicated that CflaLPMO10D has strict substrate specificity for β-chitin, which is oxidized regiospecifically at C1. To facilitate activity quantitation, we developed a new high-performance anion-exchange chromatography–pulsed amperometric detection (HPAEC-PAD) method to analyze oxidized chito-oligosaccharides as an alternative to the commonly utilized hydrophilic interaction liquid ion chromatography coupled with UV detection (HILIC-UV) method for determining the product profile of chitinolytic LPMOs. Notably, the presence of the CBM2 module was shown to boost binding toward chitin substrates and decrease the rate of futile cycling and autocatalytic inactivation. A phylogenetic survey of all characterized and putative AA10 sequences from Cellulomonas bacteria currently deposited into GenBank indicated the rarity of chitinolytic LPMOs.
RESULTS
Primary sequence analysis of CflaLPMO10D.
Primary sequence and phylogenetic analysis indicated that the catalytic module of the fourth C. flavigena AA10 member, encoded by Cfla_0490, is grouped within a clade of known chitin-active LPMOs (43) and contains the characteristic chitin-binding motif (Y[W]EPQSVE) (16) within its L2 loop (Fig. 1A). Furthermore, among characterized AA10 members, the catalytic module of CflaLPMO10D exhibited the highest sequence identity (50%) with the chitin-active LPMO from Jonesia denitrificans (JdLPMO10A) (30). The C-terminal CBM2 module had the highest sequence identity (39%) with a cellulose-binding CBM2 member from the Cellulomonas fimi GH10 xylanase (73, 81).
FIG 1.
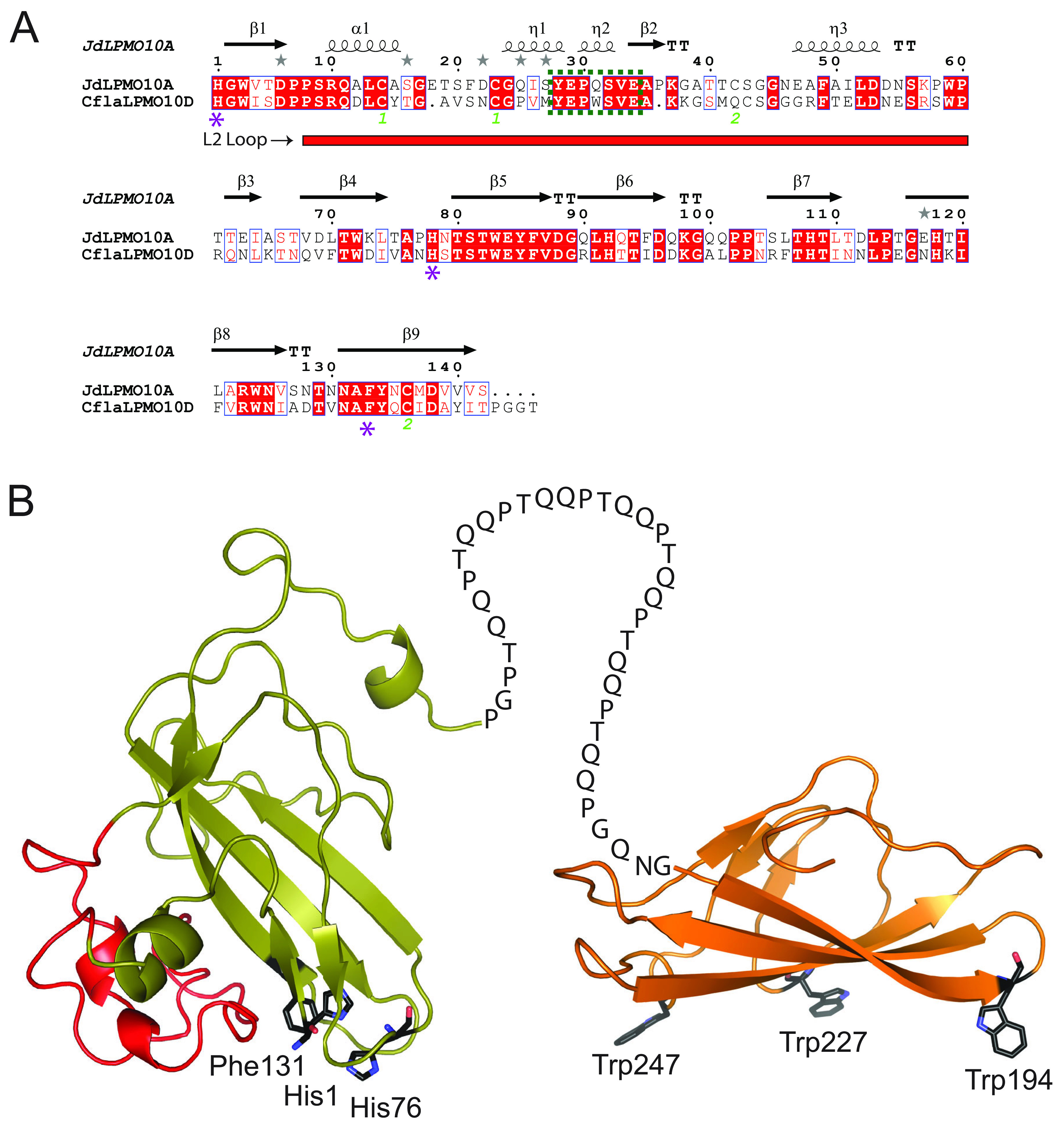
Primary sequence analysis and predicted tertiary structure of CflaLPMO10D. (A) Sequence alignment of CflaLPMO10D with chitin-active JdLPMO10A (30) in which it shares 50% sequence identity. All predicted secondary structure features are predicted and labeled above the alignment. The L2 loop region is indicated by a red rectangle under the alignment, the conserved chitin-binding motif (Y[W]EPQSVE) is indicated by a dotted green box, and the catalytic residues are indicated by a purple star. (B) Homology model of the full-length CflaLPMO10D showing the AA10 domain appended to a CBM2 domain. AA10 catalytic residues and CBM binding residues are labeled beside each respective residue. The L2 loop of the catalytic module, which contains the chitin-binding motif, is highlighted in red. The Phyre2 protein fold recognition server (104) was used to generate structural models. The catalytic and CBM2 domains utilized 5 AA10 templates with 100% confidence (PDB IDs 2BEM, 5L2V, 5AA7, 6T5Z, and 5WSZ [25, 30, 105–107]) and four CBM2 templates with 100% confidence (PDB IDs 3NDY, 1EXH, 5F7E, and 2RTT [73, 86, 108, 109]), respectively.
To support primary structure analysis, tertiary structure homology models were produced for both the AA10 and CBM2 modules using the Phyre2 server (82) (Fig. 1B). The model of CflaLPMO10D clearly illustrated the active site consisting of His1 and His76, which form the hallmark “histidine brace” motif, as well as a phenylalanine residue (Phe131). CflaLPMO10D has this aromatic residue in common with CflaLPMO10A, but not CflaLPMO10B and CflaLPMO10C, which both contain a tyrosine (all three are cellulose active) (43). Additionally, homology modeling of the C-terminal CBM2 module revealed three tryptophan residues (Trp194, Trp227, and Trp194), which are generally conserved and constitute the substrate-binding surface (Fig. 1B) (81, 83).
Heterologous production of CflaLPMO10D-CBM2 and CflaLPMO10D in E. coli.
Analogous to our previous work on CflaLPMO10A, CflaLPMO10B, and CflaLPMO10C (43), CflaLPMO10D (catalytic module only) and the full-length CflaLPMO10D-CBM2 gene products were successfully produced by recombinant expression in Rosetta(DE3) E. coli by using the native secretion signal peptide (general secretory [Sec] pathway [84]). High levels of production allowed isolation directly from media supernatants due to leakage from the periplasm. Following purification (see Fig. S1 in the supplemental material), we achieved soluble protein yields of 5 to 10 mg/L of media for both the full-length and catalytic module variants. The correct processing of the native signal peptide, which is critically important to liberate His-1 for copper coordination, was confirmed via intact-protein mass spectrometry (MS) (Fig. S1). Specifically, we observed masses of 18,066.5 Da for the catalytic module (calculated 18,071.0 Da, based on protein sequence) and 30,737.5 for the full-length protein (calculated 30,743.7, based on protein sequence), which correspond to the correctly processed proteins with fully formed disulfides (i.e., loss of 4 mass units by oxidation of 2 cysteine pairs in the catalytic domain and 2 mass units by oxidation of 1 cysteine pair in the CBM2 module).
Differential scanning fluorimetry experiments showed that both the CflaLPMO10D catalytic module and the full-length CflaLPMO10D-CBM2 proteins were maximally thermostable over the pH range 5 to 7 (melting temperature [Tm] values, 54 to 58°C), with the CflaLPMO10D catalytic module exhibiting marginally less thermostability at pH 5 (Fig. S2). Unlike CflaLPMO10A, CflaLPMO10B, and CflaLPMO10C (43), neither protein evidenced a secondary melting event after cooling, suggesting an inability to refold spontaneously. Likewise, no activity on β-chitin (see below) was observed for either protein following boiling and cooling (a second melting event was observed for the full-length CflaLPMO10D in 50 mM glycine buffer, pH 3.6; however, no LPMO activity was detected at this pH).
Substrate specificity.
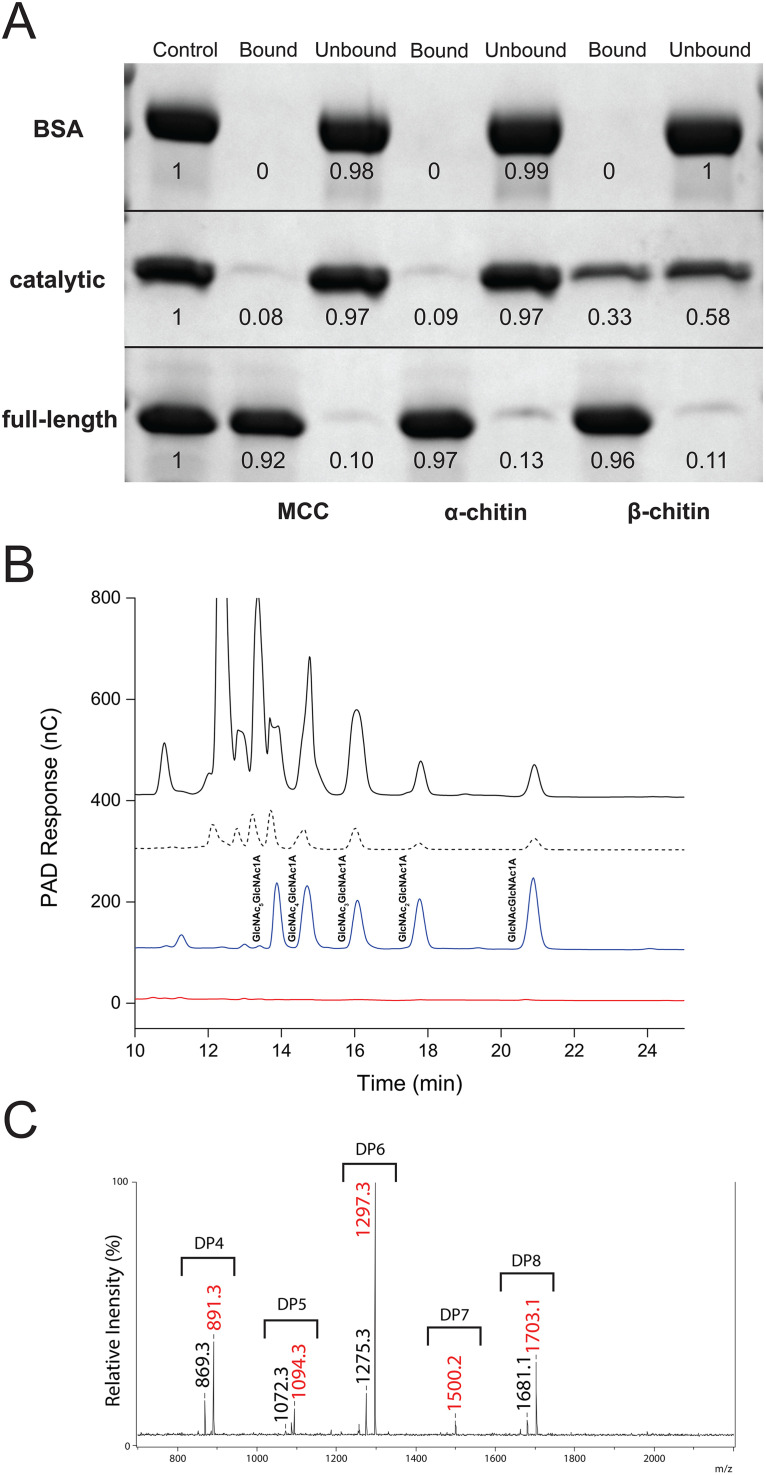
To qualitatively assess the capacity of CflaLPMO10D and the full-length CflaLPMO10D-CBM2 proteins to bind to insoluble substrates, we incubated purified proteins with excess (1:100 ratio of protein to substrate) microcrystalline cellulose (MCC), α-chitin, or β-chitin and analyzed the bound and unbound fractions by SDS-PAGE. The full-length CflaLPMO10D-CBM2 (Fig. 2A) bound strongly to both allomorphs of chitin, as well as MCC, with essentially identical affinities. Conversely, the isolated CflaLPMO10D catalytic domain showed a limited capacity to bind to MCC and α-chitin due to the lack of the carbohydrate-binding module. The ability of the CBM2 members to bind insoluble chitin and cellulose is well precedented (49, 57, 85; reviewed in reference 81), and the ability of a CBM2 module to potentiate the binding of an AA10 LPMO to cellulose has been specifically demonstrated (86). However, the catalytic module on its own bound to β-chitin to a significant extent, which is commensurate with its catalytic activity on this substrate.
FIG 2.
Substrate preference and product profile of CflaLPMO10D. (A) SDS-PAGE analysis of bound and unbound fractions following incubation of full-length and catalytic-only CflaLPMO10D with insoluble cellulose and chitin substrates (1:100 [wt/wt] protein-to-substrate loading; an equivalent amount of sample was loaded in each well). Bovine serum albumin (BSA) used as a negative control. The first lane of each experiment contains a reference control (100%) for pixel densitometry analysis (given below each protein band). (B) HPAEC-PAD chromatogram of soluble assay supernatant when 1 μM CflaLPMO10D-CBM2 (full-length, solid black trace) or CflaLPMO10D (catalytic module only, dotted black trace) was incubated with 0.1% β-chitin and 1 mM ascorbate reducing agent. C1-oxidized chito-oligosaccharide standards (blue trace) were produced using an AA7 member from Polyporus brumalis (PbChi7A) (117). The chromatogram for each protein variant shows the absolute PAD response (nC, unscaled) and is commensurate with the higher oxidizing activity of the full-length protein (Fig. 3). The red trace represents a no-enzyme control reaction. (C) MALDI-TOF MS spectrum of CflaLPMO10D assay supernatant indicating presence of oxidized chito-oligosaccharides. Masses in black are monosodiated aldonic acid forms ([M+Na]+), and masses in red are disodiated aldonic acid forms ([M−H+2Na]+).
In turn, activity screening of both proteins against a panel of insoluble and soluble substrates indicated that both were strictly active on β-chitin from squid pen, as evidenced by detection of oxidized chito-oligosaccharides by HPAEC-PAD and MALDI-TOF MS (Fig. 2B and C). No activity was observed on a range of other insoluble and soluble carbohydrates, including α-chitin, chito-oligosaccharides (DP2 to DP6), MCC, phosphoric acid swollen cellulose (PASC), unbleached pulp fibers, cellulose nanofibrils, cellulose nanocrystals, bacterial cellulose, hydroxyethyl cellulose, carboxymethyl cellulose, cello-oligosaccharides (DP2 to DP6), xylan, xyloglucan, mixed-linkage β-glucan, and soluble starch.
The release of soluble products by chitin-active LPMOs is typically quantified by hydrophilic interaction liquid ion chromatography coupled with UV detection (HILIC-UV) (26, 31, 33, 34, 36, 87). On the other hand, HPAEC-PAD is widely used for product analysis from cellulose-active LPMOs. Hence, to unify and streamline our analyses of C. flavigena LPMOs, we optimized a new HPAEC-PAD method to resolve C1-oxidized chito-oligosaccharides (DP2 to DP6) on a CarboPac PA1 column (Fig. 2B). A linear, concentration-dependent PAD response was observed for oxidized products between 10 and 90 μM. To facilitate quantitation by minimizing the number of integrated peaks, a GH18 chitinase from Clostridium thermocellum was used to simplify the soluble product profile to oxidized and native chitobiose (DP2) and chitotriose (DP3).
Using this new quantitative assay, we demonstrated that the presence of the CBM in the full-length CflaLPMO10D-CBM2 facilitated the release of over 2-fold-more oxidized chito-oligosaccharides than the catalytic module alone under aerobic conditions after incubation for 24 h (Fig. 3A). The data also indicate that both proteins had succumbed to autoinactivation (11, 87, 88) by this time point; the addition of β-chitin at this time failed to cause the release of additional soluble products (Fig. 3B).
FIG 3.
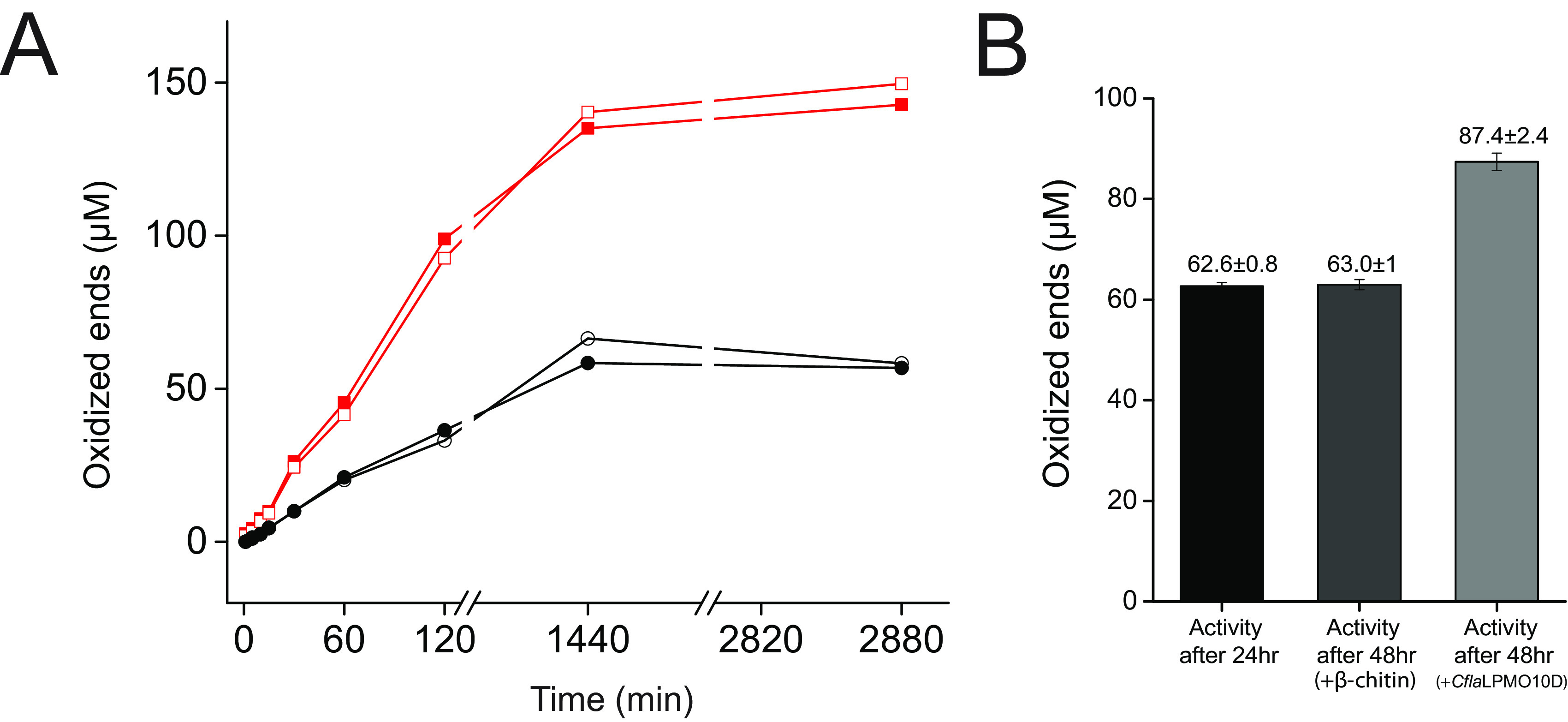
Quantitation of CflaLPMO10D activity on β-chitin. (A) Release of oxidized ends following incubation on β-chitin, as determined by HPAEC-PAD. Duplicate assays were performed in parallel, and the individual results at each time point are shown. Red trace, CflaLPMO10D-CBM2 activity; black trace, CflaLPMO10D catalytic domain activity. (B) Inactivation check of CflaLPMO10D catalytic domain comparing activity at 24 h following addition of either fresh β-chitin or fresh enzyme.
Further analysis of the isolated catalytic module, using β-chitin as a substrate, indicated that CflaLPMO10D was most active at pH 6, with no activity detected at pH 3.6 and pH 9 (Fig. 4A). Using representatives of four structurally unique, plant-derived reducing agent types, as defined by Fromhaggen et al. (89), ascorbic acid (nonsulfur, nonphenol) was observed to be superior to l-cysteine (sulfur based), hydroquinone (benzenediol based), and gallic acid (benzenetriol based) (Fig. 4B). For the latter three reducing agents, the time course experiments indicate that CflaLPMO10D exhibited slower initial rates and ultimately succumbed to autocatalytic inactivation at a similar point in comparison with ascorbic acid as an electron donor. Despite numerous reports indicating peroxygenase activity among LPMOs, including AA10 members (90), we were unable to observe chitinolytic activity under anaerobic conditions in the presence of hydrogen peroxide (excess β-chitin, 1 μM CflaLPMO10D, 1 mM ascorbic acid, 100 μM H2O2, anaerobic chamber; see Materials and Methods).
FIG 4.
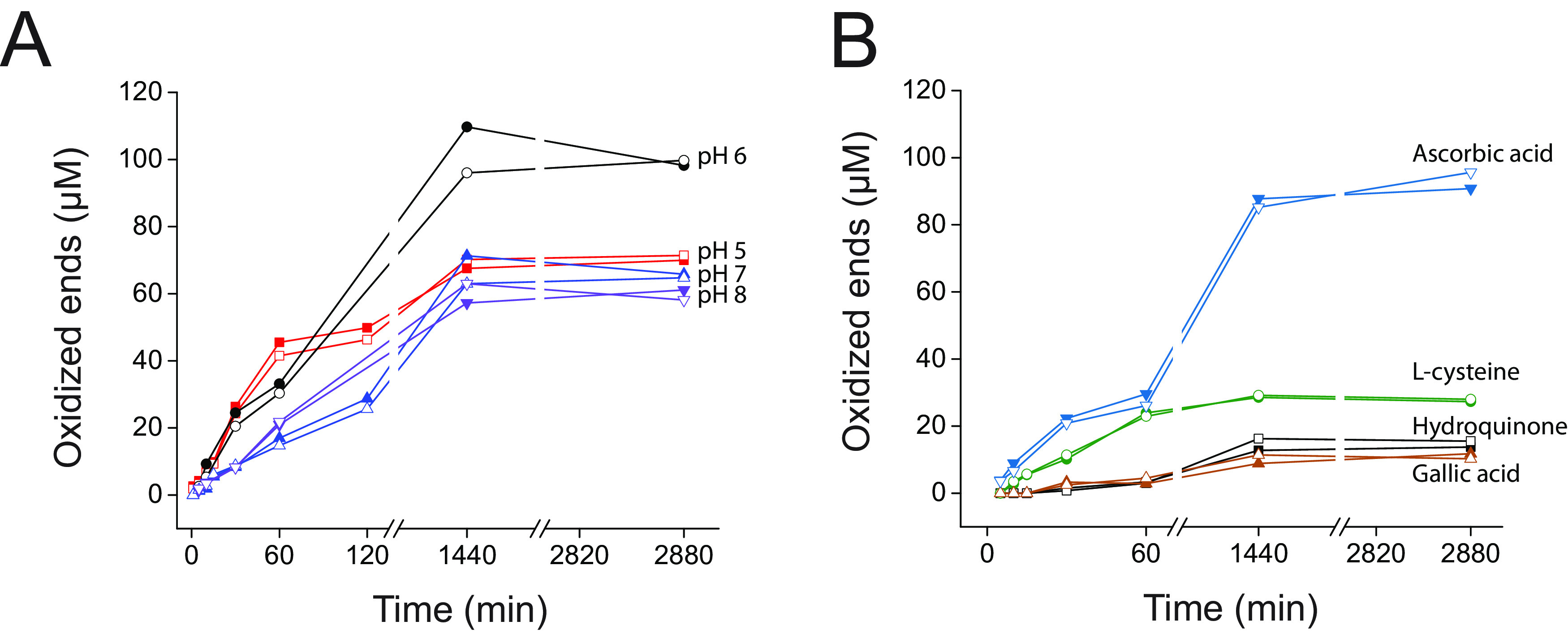
pH dependence and reducing agent specificity of CflaLPMO10D on β-chitin. (A) Quantitative pH dependence time course assays over 48 h. (B) Quantitative reducing agent specificity assays over 48 h. Duplicate assays were performed in parallel, and the individual results at each time point are shown.
DISCUSSION
C. flavigena encodes, in total, four AA10 sequences, three of which we previously reported to have strict cellulose-oxidizing activity (43). Our phylogenetic analysis indicated that the fourth AA10 sequence (CflaLPMO10D encoded by Cfla_0490) was putatively a chitin-active LPMO, despite prior secretomic data implicating a role in cellulose and xylan utilization (78). Here, biochemical characterization of CflaLPMO10D definitively demonstrated a strict preference for the β-allomorph of chitin.
All AA10 LPMOs contain a variable disordered region found between the first and third β-sheet, termed the L2 loop/region, which has been demonstrated to be critical for substrate recognition (16). A signature chitin-binding motif was indeed present in the L2 loop of CflaLPMO10D, commensurate with the observed substrate specificity. Additionally, the active site of all LPMOs comprises a T-shaped histidine-brace surrounding the copper ion, with an axial phenylalanine or tyrosine in AA10 LPMOs (in all other LPMO families, this aromatic residue is a tyrosine [91]). In CflaLPMO10D, this residue is a phenylalanine (Phe131) (Fig. 1). The catalytic implications of this are not fully known, whereas studies have shown that tyrosine in this position may be important in facilitating formation of oxidized intermediates or playing a protective role against oxidative damage (92).
There is ongoing discussion on whether the primary cosubstrate of LPMOs is molecular oxygen (O2) (11, 26, 91, 93) or hydrogen peroxide (H2O2) (13, 14, 90). In our previous study of the three cellulose-active AA10 members from C. flavigena, we observed that only one, CflaLPMO10A, had demonstrable activity with H2O2 as a cosubstrate. Our inability to observe chitinolytic activity with CflaLPMO10D upon incubation with β-chitin and H2O2 under anaerobic conditions suggests that hydrogen peroxide is not a universal cosubstrate for LPMOs from this bacterium.
We independently produced the full-length CflaLPMO10D-CBM2 and the catalytic module alone to explore the role of the C-terminal CBM. Together, substrate binding studies and time course activity assays demonstrated the importance of the CBM2 domain in localizing CflaLPMO10D to chitin substrates and attenuating rate of futile cycling/autocatalytic inactivation. The demonstrable binding of the catalytic domain to β-chitin, but not α-chitin, was commensurate with its catalytic specificity.
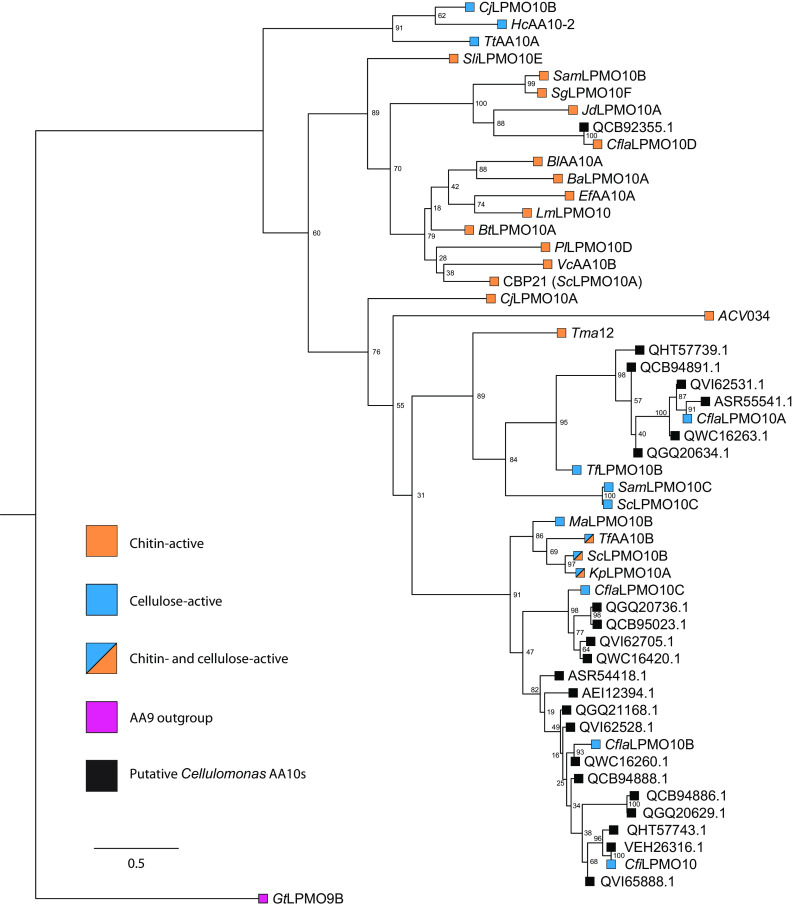
C. flavigena was originally isolated from soil (79) and, like other Cellulomonas species, is classically associated with the breakdown of plant biomass (78, 94). As such, the observation of a chitin-specific LPMO among the four C. flavigena AA10 members is biologically intriguing, especially because the well-studied C. fimi contains only a single, cellulose-specific AA10 LPMO (43, 95). Furthermore, a survey of 11 Cellulomonas species for which completed genomes have been deposited in GenBank (as compiled in CAZy [96]) revealed that among 29 AA10 members, only 2 are predicted to be chitinolytic (Fig. 5; Table 1). Of these, CflaLPMO10D and an AA10 member encoded by Cellulomonas shaoxiangyii strain Z28 (CsLPMO10A; GenPept accession no. QCB92355) share 91.5% and 95.1% sequence identity and similarity, respectively, which further highlights the rarity of chitinolytic LPMOs among Cellulomonas species (Fig. 5).
FIG 5.
Maximum-likelihood phylogenetic analysis of the catalytic domains of all characterized and putative Cellulomonas AA10 members from complete genomes deposited in GenBank as of November 2021 (see http://www.cazy.org/bC.html). G. trabeum LPMO9 (GtLPMO9B) (111) was used as the outgroup. GenBank accession numbers for each Cellulomonas AA10 member are given in Table 1. The following biochemically characterized AA10 members were used: SliLPMO10E (32), SgLPMO10F (34), SamLPMO10B (29), JdLPMO10A (30), BtLPMO10A (28), EfLPMO10A (123), LmLPMO10 (33), BaLPMO10A (37), BlLPMO10 (26), PtLPMO10D (25), VcLPMO10B (36), CBP21 (SmLPMO10A) (10), CjLPMO10B (45), HcAA10-12 (39), TtAA10A (42), CjLPMO10A (31), CflaLPMO10A-C and CfiLPMO10 (43), TfLPMO10B (46), SamLPMO10C (29), ScLPMO10C (46), Tma12 (27), ACV034 (35), MaLPMO10B (44), ScLPMO10B (46) KpLPMO10 (41), and TfLPMO10B (46).
TABLE 1.
Census of all characterized and predicted Cellulomonas AA10 LPMOs from complete genomes deposited in GenBank
| Strain | NCBI taxonomy ID | Genome accession no. (reference) | Cellulose-active LPMOs GenPept accession no.(s) (characterization reference)d | Chitin-active LPMOsGenPept accession no.(characterization reference)d |
|---|---|---|---|---|
| Cellulomonas flavigena DSM 20109 | 446466 | CP001964 (79) | ADG73094 (43), ADG73091 (43), ADG73234 (43) | ADG73405 (this study) |
| Cellulomonas shaoxiangyii Z28 | 2566013 | CP039291 (124) | QCB94886, QCB94888, QCB94891, QCB95023 | QCB92355 a |
| Cellulomonas fimi ATCC 484 | 590998 | CP002666 (94) | AEE44415 (43, 95) | |
| Cellulomonas fimi NCTC7547 | 1708 | LR134387 | VEH26316 b | |
| Cellulomonas gilvus ATCC 13127 | 593907 | CP002665 (94) | AEI12394 | |
| Cellulomonas sp. strain H30R-01 | 2566013 | CP048210 | QHT57739, QHT57743 | |
| Cellulomonas sp. strain JZ18 | 2654191 | CP045245 (125) | QGQ21153,c QGQ20629, QGQ21168, QGQ20634, QGQ20736 | |
| Cellulomonas sp. strain PSBB021 | 2003551 | CP021430 | ASR54418, ASR55541 | |
| Cellulomonas sp. strain zg-ZUI157 | 2819979 | CP076023 | QWC16260, QWC16263, QWC16420, QWC17684c | |
| Cellulomonas sp. strain zg-ZUI188 | 2819978 | CP074404 | QVI65888 | |
| Cellulomonas sp. strain zg-ZUI222 | 2816956 | CP074405 | QVI62528, QVI62531, QVI62705 |
Shares 91.5% sequence identity and 95.1% similarity with C. flavigena LPMO10D (ADG73405).
Shares 100% sequence identity with C. fimi AA10 (AEE44415).
Deposited coding DNA sequence incomplete.
For sequences lacking a literature reference, substrate specificity has been predicted based on sequence similarity (see Discussion).
Among other bacterial species with characterized AA10 members, Thermobifida fusca, Serratia marcescens, and Cellvibrio japonicus encode both cellulose and chitin-active LPMOs (31, 46). Of these, only C. japonicus encodes both strictly chitin-active (CjLPMO10A) and strictly cellulose active (CjLPMO10B) AA10s, with the two homologs demonstrating clear biological functions in either chitin or cellulose utilization (31). Yet, the physiological role of CflaLPMO10D in C. flavigena is enigmatic, but may be a vestigial or recently acquired activity related to insect biomass degradation (3) in soil. Fungal cell walls, which are also present in soil, are unlikely to be a natural substrate, as these are predominantly composed of α-chitin (1). Curiously, we were unable to identify related putative endo-chitinases (GH18 and GH19 members) or endo-chitosanases (GH46, GH75, or GH80 members) encoded by the C. flavigena genome (79), which could potentially work in concert with CflaLPMO10D. Commensurate with these observations, we were unable to detect the ability of C. flavigena ATCC 482 to utilize colloidal chitin to support growth (data not shown; growth on xylan was robust, as reported previously [78]).
Cellulomonadaceae are not generally known to grow on chitin (97, 98), and only very limited information is available on potential chitinolytic enzyme systems in these bacteria. C. shaoxiangyii, which contains a nearly identical orthologue to CflaLPMO10D, CsLPMO10A (Fig. 5), encodes a single predicted GH18 chitinase appended to an N-terminal CBM2 domain (GenBank accession no. QCB92763). However, there is presently no information on the ability of this isolate to use chitin as a carbon source. An exo-N,N’-diacetylchitobiohydrolase has been purified from cultures of the isolate C. flavigena NTOU 1. Although inclusion of chitin in the culture medium increased the native production of this chitobiohydrolase, C. flavigena NTOU 1 does not appear to be able to use chitin as a sole carbon source (99). This is analogous to our observations of C. flavigena ATCC 482. Unfortunately, no sequence information exists for the exo-N,N’-diacetylchitobiohydrolase, nor the C. flavigena NTOU 1 genome. Of note, the isolation of an eponymous chitin-utilizing cellulomonad, Cellulomonas chitinilytica, was reported in 2008 (100). Likewise unfortunate, no genome sequence information is available for this species, thus precluding analysis of its AA10 and related chitinolytic CAZyme content.
Interestingly, a recent study has reported that an S. lividans AA10 LPMO (SliLPMO10E) can potentiate lysozyme activity against peptidoglycan (comprising an alternating β-1,4-linked GlcNAc/MurNAc backbone), with potential physiological implications for bacterial cell wall remodeling (101). This LPMO was previously thought to be strictly specific for β-chitin (32). Alignment of the catalytic domains of SliLPMO10E and CflaLPMO10D reveals limited (ca. 40%) sequence identity (Fig. 5), perhaps suggesting divergent activities. Moreover, the extremely restricted distribution of putative chitinolytic AA10 LPMOs among Cellulomonas species argues against a general role in peptidoglycan remodeling in this genus. However, further studies are necessary to fully elucidate the biological role of CflaLPMO10D and other chitin-active LPMOs in cellulomonads.
MATERIALS AND METHODS
Sequence alignment and homology modeling.
Protein alignments were performed using MUSCLE (102), and alignment visualization was created using ESPript 3.0 (103). Structural homology modeling was performed via the Phyre2 protein fold recognition server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) (104). For both the catalytic and CBM2 domains of CflaLPMO10D, the intensive modeling mode was utilized in the structural search. The CflaLPMO10D catalytic domain was modeled with 100% confidence using 5 AA10 templates (PDB IDs 2BEM, 5L2V, 5AA7, 6T5Z, and 5WSZ [25, 30, 105–107]). The corresponding CBM2 domain was modeled with 100% confidence using 4 CBM2 templates (PDB IDs 3NDY, 1EXH, 5F7E, and 2RTT [73, 86, 108, 109]).
Phylogenetic analysis.
Full-length AA10 sequences, comprising all biochemically characterized members (http://www.cazy.org/AA10_characterized.html) and all genomic sequences from Cellulomonas spp. (http://www.cazy.org/bC.html) as of November 2021, were retrieved from GenBank via the CAZy database (96). The N-terminal signal peptide and any C-terminal CBM2 domains (including any disordered C-terminal extension [110]) were manually removed prior to alignment using MUSCLE (102). The catalytic domain of a Gloeophyllum trabeum AA9 (GtLPMO9B) (111) was included in the alignment as an outgroup for phylogenetic analysis. A maximum-likelihood phylogeny was generated using RAXML 8.2.10 within the CIPRES Science Gateway v3.1 1 (112) using a JTT matrix-based nucleotide substitution model (113) of 25 discrete rate categories and rapid bootstrapping with automatic halting enabled (600 bootstraps). FigTree was used to visualize the resulting phylogenetic tree (http://tree.bio.ed.ac.uk/software/figtree).
Carbohydrates.
β-Chitin was kindly donated by Paul Walton (University of York, Heslington, York, UK). Avicel and α-chitin were purchased from Sigma-Aldrich (St. Louis, MO, USA). Starch and glucose were purchased from Fisher Scientific (Hampton, NH, USA). Chito-oligosaccharides, tamarind xyloglucan, barley mixed-linkage β-glucan, carboxymethyl cellulose, hydroxyethyl cellulose, beechwood xylan, and cellohexaose were purchased from Megazyme International (Bray, Ireland). Northern bleached softwood kraft pulp (NBSKP) was donated by Canfor Pulp Innovation (Burnaby, BC, Canada). Cellulose nanocrystals (CNC) and cellulose nanofibrils (CNF) were produced from NBSKP by acid hydrolysis and low-consistency homogenization, respectively. Bacterial cellulose was grown and harvested from Komagataeibacter xylinus (i.e., Acetobacter xylinum, Gluconacetobacter xylinus) according to a previously published protocol (114). Phosphoric acid-swollen cellulose (PASC) was prepared from Avicel as described previously (115). Colloidal chitin was prepared from α-chitin according to a previously published protocol (116).
Oxidized chito-oligosaccharide standards were individually produced by treating chito-oligosaccharides (DP2 to DP6; Megazyme, Bray, Ireland) with an AA7 chito-oligosaccharide dehydrogenase from Polyporus brumalis (PbAA7) (117) (kindly donated by Jean-Guy Berrin, INRAE, Aix-Marseille University). We added 0.5 μM PbAA7 to 5 mM chito-oligosaccharide in 100 mM Tris-HCl, pH 8, and it was incubated overnight in an Eppendorf ThermoMixer C (Hamburg, Germany) set at 37°C with constant orbital rotation of 500 rpm. A second addition of 0.5 μM PbAA7 was added and incubated overnight to ensure full oxidation of chito-oligosaccharides, as confirmed by HPAEC-PAD (see below).
Cloning and heterologous protein production.
The full-length Cfla_0490 gene from C. flavigena strain DSM 20109 (GenBank accession no. ADG73405) encoding CflaLPMO10D-CBM2 was codon optimized for expression in E. coli and obtained by synthesis through Bio Basic Inc. (Markham, ON, Canada). Both the full-length (CflaLPMO10D_FL) and catalytic module variants (CflaLPMO10D_catalytic) of CflaLPMO10D were amplified by PCR from the synthetic gene template and independently cloned into the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible pMCGS53 expression vector via restriction-free cloning (118). Briefly, primary PCR primers (see Table S1 in the supplemental material) were designed to amplify either the CflaLPM10D_catalytic or the full-length CflaLPMO10D with flanking overhanging regions complementary to pMCGS53 vectors. For both constructs, the reverse primary PCR primer was designed to introduce a C-terminal thrombin tag prior to the C-terminal hexahistidine tag. The primary PCR fragments were subsequently used as a primer pair in the secondary PCR step to directly insert the gene into pMCGS53 plasmid. The secondary PCR products were first treated with DpnI (NEB) to digest template pMCGS53 prior to transformation into DH5α E. coli cells and recircularization through homologous recombination. Both the primary and secondary PCR sequences were synthesized by IDT, and all nucleotides and PCR polymerases (Phusion) were purchased from New England Biolabs (NEB). Cloned vector sequences were verified through Sanger sequencing service provided by Genewiz. PFAM (119), and BLASTP analyses were used to guide module boundary identification and primer design.
Heterologous production of full-length and catalytic CflaLPMO10 proteins was performed in Rosetta(DE3) E. coli cells as previously described (43). Briefly, protein overexpression was achieved in LBE-5052 autoinduction media supplemented with 100 μM copper chloride grown at 25°C for 19 h. Soluble protein was purified out of supernatant (following leakage from the periplasm) via immobilized-metal affinity chromatography (IMAC) on an NGC chromatography system (Bio-Rad). Soluble LPMOs were incubated in 1 mM CuCl2 for 1 h at room temperature prior to buffer exchange into 20 mM Tris-HCl, pH 8, via size exclusion chromatography. Protein purity was assessed by SDS-PAGE, and translational fidelity was confirmed by intact protein MS (120). Protein aliquots were flash frozen and stored at −70°C for subsequent use.
Thermostability assays.
ThermoFluor protein denaturation assays were conducted as described previously (43). Briefly, 5 μM enzyme was incubated with 50 mM buffer (sodium acetate in the range of pH 3.6 to 5, Bis-Tris in the range of pH 6 to 8, or glycine at pH 9) and 10× SYPRO orange dye (diluted 5,000× of stock SYPRO orange supplied by Invitrogen) and analyzed in a 7500 Fast real-time PCR system (Applied Biosystems). We used 1 mM d-penicillamine (Alfa Aeser, Heysham, Lancashire, UK) in copper chelation experiments.
Insoluble substrate-binding assays.
LPMO binding to insoluble substrates was performed according to a previously published protocol (121). Briefly, 100 μg of protein was combined with 10 mg of substrate in 50 mM Bis-Tris, pH 7, in a total volume of 200 μL and allowed to mix for 4 h at 4°C and end-to-end rotation. Following binding, the soluble unbound fraction was removed through centrifugation, and the insoluble pellet was washed three times with 200 μL of 50 mM Bis-Tris, pH 7, via resuspension and centrifugation. The insoluble pellet was resuspended in 200 μL 1× SDS running buffer and heated to 95°C for 10 min prior to centrifugation and isolation of the soluble bound fraction. All fractions were analyzed on SDS-PAGE. Protein band intensities were quantified using pixel densitometry analysis via the ImageJ software (https://imagej.nih.gov/ij/) (122).
LPMO activity assays.
LPMO assays were performed in an Eppendorf ThermoMixer C (Hamburg, Germany) held at 37°C with constant orbital rotation at 1,000 rpm. Typical assays were performed in 500 μL total volume on 0.1% (wt/vol) substrate using either 1 mM ascorbic acid (Fisher Scientific), gallic acid (Fisher Scientific), l-cysteine (Sigma-Aldrich), or hydroquinone (Sigma-Aldrich) as the reducing agent. Substrate and enzyme were combined first and allowed to equilibrate in an Eppendorf ThermoMixer for 5 min prior to addition of reducing agent to start the reaction. The reactions were stopped via removal of the insoluble substrate through 0.22-μm cellulose acetate spin filters (VWR); the assay supernatant was stored at 4°C until analysis on high-performance anion-exchange chromatography coupled with pulsed amperometric detection (HPAEC-PAD) as described below.
Peroxide assays were performed in an anaerobic chamber (model 358; Coy Lab Products, Grass Lake, MI, USA) under an atmosphere of 80% N2, 10% CO2, and 10% H2. Assay components were degassed prior to entering the anaerobic chamber for further deoxygenation overnight. The LPMO enzyme was degassed and allowed to deoxygenate in the chamber for an hour before addition to Eppendorf assay tubes. Typical assays were performed essentially as described above (0.1% PASC and 1 mM ascorbate in 50 mM Bis-Tris, pH 7, with 1 μM CflaLPMO10D), and 100 μM H2O2 was added to start the reaction. Reactions were stopped through centrifugal filtration through 0.22-μm cellulose acetate filters anaerobically, and the soluble assay supernatant was analyzed via HPAEC-PAD.
HPAEC-PAD analysis.
Soluble LPMO reaction products were analyzed via HPAEC using an ICS-5000 HPLC system coupled to a gold electrochemical detector for PAD. Samples were injected on a CarboPac PA1 2- by 250-mm ion chromatography (IC) analytical column preceded by a CarboPac PA1 2- by 50-mm guard column (Dionex, Sunnyvale, CA, USA). Chito-oligosaccharide separation was achieved in 100 mM NaOH at a constant flow rate of 0.25 mL/min and an initial linear gradient toward 100 mM NaOAc over 35 min, followed by a linear gradient to 200 mM NaOAc over 10 min, and then a final a linear gradient to 500 mM NaOAc over 5 min before dropping to 0 mM NaOAc over 5 min, for a total running time of 55 min. The injection volume of 25-μL column compartment was held at 30°C.
For quantitative assays, a Clostridium thermocellum GH18 chitinase (Megazyme, Bray, Ireland) was used to convert LPMO assay supernatant containing oxidized chito-oligosaccharides to C1-oxidized diacetyl chitobiose (GlcNAcGlcNAc1A) and C1-oxidized triacetyl chitotriose [(GlcNAc)2GlcNAc1A]. In total, 0.01 U of CtGH18 was added (2.5 μL of a 3.8 U/mL stock) to 90 μL of assay supernatant and incubated for 16 h at 37°C. For assays performed outside the optimal range of CtGH18, the pH was adjusted to pH 6 to 7 prior to incubation. Following initial CtGH18 treatment, an extra 0.01 U of chitinase was added and allowed to incubate for an additional 16 h at 37°C to ensure full hydrolysis of the assay supernatant. NaOH was added to a final concentration of 50 mM (5 μL of 1 M NaOH) to stop the reaction.
MALDI-TOF MS.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) was performed essentially as previously reported (43). Briefly for MALDI-TOF MS, the LPMO assay supernatant was mixed with matrix solution (9 mg/mL 2,5-dihydroxybenzoic acid [DHB] in 50% acetonitrile) at a 1:1 ratio and analyzed in positive-ion mode on a Bruker Daltonics autoflex MALDI-TOF MS system (Billerica, MA, USA).
Growth of C. flavigena on chitin.
C. flavigena ATCC 482 was grown in low-salt Luria broth supplemented with either 0.5% glucose, 0.2% soluble birchwood xylan, or colloidal chitin (prepared as described above) at 30°C with shaking at 150 rpm (78). Growth was monitored spectrophotometrically at 600 nm.
Data availability.
All data generated or analyzed during this study are included in this published article and its supplementary information file.
ACKNOWLEDGMENTS
J.L. thanks Changqing Wang (Brumer group, MSL, UBC) for assistance with MALDI-TOF MS operation, Hila Behar (Brumer group, MSL, UBC) for assistance with bioinformatics, Paul Walton (University of York) for donating β-chitin, Jean-Guy Berrin (INRAE, Aix-Marseille University) for donating PbChi7A, and Paul Bicho for donating unbleached kraft pulp fibers. We thank Stephen Withers (UBC) for his early support of this study.
This work was supported by grants to H.B. from the NSERC Discovery Grants program (RGPIN-2018-03892) and the NSERC Strategic Partnership Grants for Projects program (STPGP 479088, F15-01751). This work was also supported by a grant to E.R.M. and H.B. from Genome Canada, Genome BC, and Ontario Genomics (project number 10405, SYNBIOMICS-Functional genomics and techno-economic models for advanced biopolymer synthesis; www.synbiomics.ca). The funding sources had no role in the experiment design, data collection and interpretation, or the decision to submit this work for publication.
J.L. cloned optimized LPMO gene constructs, performed recombinant protein production, performed all biochemical/biophysical studies, and performed all bioinformatics analyses. E.D.G.-B. performed initial cloning and method development for recombinant protein production. O.R. contributed to HPAEC-PAD method development under the guidance of E.R.M. H.S. performed C. flavigena growth studies. L.S. produced bacterial cellulose, CNC, and CNF. Y.M. produced PASC. H.B., E.D.G.-B., and W.W.W. conceptualized the study and supervised the research. J.L. drafted the manuscript, which was revised together with H.B. All authors read and approved the final version of the manuscript.
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Harry Brumer, Email: brumer@msl.ubc.ca.
Robert M. Kelly, North Carolina State University
REFERENCES
- 1.Fernando LD, Dickwella Widanage MC, Penfield J, Lipton AS, Washton N, Latgé J-P, Wang P, Zhang L, Wang T. 2021. Structural polymorphism of chitin and chitosan in fungal cell walls from solid-state NMR and principal component analysis. Front Mol Biosci 8:727053. 10.3389/fmolb.2021.727053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peniche C, Argüelles-Monal W, Goycoolea FM. 2008. Chitin and chitosan: major sources, properties and applications, p 517–542. In Belgacem MN, Gandini A (ed), Monomers, polymers and composites from renewable resources. Elsevier, Oxford, UK. [Google Scholar]
- 3.Merzendorfer H, Zimoch L. 2003. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol 206:4393–4412. 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 4.Elieh-Ali-Komi D, Hamblin MR. 2016. Chitin and chitosan: production and application of versatile biomedical nanomaterials. Int J Advanced Res 4:411–427. [PMC free article] [PubMed] [Google Scholar]
- 5.Klemm D, Heublein B, Fink H-P, Bohn A. 2005. Cellulose: fascinating biopolymer and sustainable raw material. Angew Chem Int Ed Engl 44:3358–3393. 10.1002/anie.200460587. [DOI] [PubMed] [Google Scholar]
- 6.Parthasarathi R, Bellesia G, Chundawat SPS, Dale BE, Langan P, Gnanakaran S. 2011. Insights into hydrogen bonding and stacking interactions in cellulose. J Phys Chem A 115:14191–14202. 10.1021/jp203620x. [DOI] [PubMed] [Google Scholar]
- 7.Jang M-K, Kong B-G, Jeong Y-I, Lee CH, Nah J-W. 2004. Physicochemical characterization of α-chitin, β-chitin, and γ-chitin separated from natural resources. J Polym Sci A Polym Chem 42:3423–3432. 10.1002/pola.20176. [DOI] [Google Scholar]
- 8.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. 2012. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5:45–45. 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen JCN, Johansen KS, Krogh KBRM, Jorgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA 108:15079–15084. 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sorlie M, Eijsink VGH. 2010. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330:219–222. 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 11.Hangasky JA, Iavarone AT, Marletta MA. 2018. Reactivity of O versus H2O2 with polysaccharide monooxygenases. Proc Natl Acad Sci USA 115:4915–4920. 10.1073/pnas.1801153115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Beeson WT, Phillips CM, Marletta MA, Cate JHD. 2012. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 20:1051–1061. 10.1016/j.str.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bissaro B, Varnai A, Røhr ÅK, Eijsink VGH. 2018. Oxidoreductases and reactive oxygen species in lignocellulose biomass conversion. Microbiol Mol Biol Rev 82:e00029-18. 10.1128/MMBR.00029-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bissaro B, Røhr ÅK, Müller G, Chylenski P, Skaugen M, Forsberg Z, Horn SJ, Vaaje-Kolstad G, Eijsink VGH. 2017. Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat Chem Biol 13:1123–1128. 10.1038/nchembio.2470. [DOI] [PubMed] [Google Scholar]
- 15.Walton PH, Davies GJ. 2016. On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr Opin Chem Biol 31:195–207. 10.1016/j.cbpa.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Zhu H. 2020. Current understanding of substrate specificity and regioselectivity of LPMOs. Bioresour Bioprocess 7:11. 10.1186/s40643-020-0300-6. [DOI] [Google Scholar]
- 17.Sabbadin F, Urresti S, Henrissat B, Avrova AO, Welsh LRJ, Lindley PJ, Csukai M, Squires JN, Walton PH, Davies GJ, Bruce NC, Whisson SC, McQueen-Mason SJ. 2021. Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes. Science 373:774–779. 10.1126/science.abj1342. [DOI] [PubMed] [Google Scholar]
- 18.Courtade G, Aachmann FL. 2019. Chitin-active lytic polysaccharide monooxygenases, p 115–129. In Yang Q, Fukamizo T (eds), Targeting chitin-containing organisms. Advances in experimental medicine and biology, vol 1142. Springer, Singapore. [DOI] [PubMed] [Google Scholar]
- 19.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. 2014. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol 10:122–126. 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo Leggio L, Simmons TJ, Poulsen J-CN, Frandsen KEH, Hemsworth GR, Stringer MA, Von Freiesleben P, Tovborg M, Johansen KS, De Maria L, Harris PV, Soong C-L, Dupree P, Tryfona T, Lenfant N, Henrissat B, Davies GJ, Walton PH. 2015. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun 6:5961. 10.1038/ncomms6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couturier M, Ladevèze S, Sulzenbacher G, Ciano L, Fanuel M, Moreau C, Villares A, Cathala B, Chaspoul F, Frandsen KE, Labourel A, Herpoël-Gimbert I, Grisel S, Haon M, Lenfant N, Rogniaux H, Ropartz D, Davies GJ, Rosso M-N, Walton PH, Henrissat B, Berrin J-G. 2018. Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat Chem Biol 14:306–310. 10.1038/nchembio.2558. [DOI] [PubMed] [Google Scholar]
- 23.Sabbadin F, Hemsworth GR, Ciano L, Henrissat B, Dupree P, Tryfona T, Marques RDS, Sweeney ST, Besser K, Elias L, Pesante G, Li Y, Dowle AA, Bates R, Gomez LD, Simister R, Davies GJ, Walton PH, Bruce NC, McQueen-Mason SJ. 2018. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat Commun 9:756. 10.1038/s41467-018-03142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filiatrault-Chastel C, Navarro D, Haon M, Grisel S, Herpoël-Gimbert I, Chevret D, Fanuel M, Henrissat B, Heiss-Blanquet S, Margeot A, Berrin J-G. 2019. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol Biofuels 12:55. 10.1186/s13068-019-1394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munzone A, El Kerdi B, Fanuel M, Rogniaux H, Ropartz D, Réglier M, Royant A, Simaan AJ, Decroos C. 2020. Characterization of a bacterial copper‐dependent lytic polysaccharide monooxygenase with an unusual second coordination sphere. FEBS J 287:3298–3314. 10.1111/febs.15203. [DOI] [PubMed] [Google Scholar]
- 26.Courtade G, Ciano L, Paradisi A, Lindley PJ, Forsberg Z, Sørlie M, Wimmer R, Davies GJ, Eijsink VGH, Walton PH, Aachmann FL. 2020. Mechanistic basis of substrate–O2 coupling within a chitin-active lytic polysaccharide monooxygenase: an integrated NMR/EPR study. Proc Natl Acad Sci USA 117:19178–19189. 10.1073/pnas.2004277117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yadav SK, Singh R, Singh PK, Vasudev PG. 2019. Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase. Planta 249:1987–1996. 10.1007/s00425-019-03135-0. [DOI] [PubMed] [Google Scholar]
- 28.Manjeet K, Madhuprakash J, Mormann M, Moerschbacher BM, Podile AR. 2019. A carbohydrate binding module-5 is essential for oxidative cleavage of chitin by a multi-modular lytic polysaccharide monooxygenase from Bacillus thuringiensis serovar kurstaki. Int J Biol Macromol 127:649–656. 10.1016/j.ijbiomac.2019.01.183. [DOI] [PubMed] [Google Scholar]
- 29.Valenzuela SV, Ferreres G, Margalef G, Pastor FIJ. 2017. Fast purification method of functional LPMOs from Streptomyces ambofaciens by affinity adsorption. Carbohydr Res 448:205–211. 10.1016/j.carres.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Mekasha S, Forsberg Z, Dalhus B, Bacik J-P, Choudhary S, Schmidt-Dannert C, Vaaje-Kolstad G, Eijsink VGH. 2016. Structural and functional characterization of a small chitin-active lytic polysaccharide monooxygenase domain of a multi-modular chitinase from Jonesia denitrificans. FEBS Lett 590:34–42. 10.1002/1873-3468.12025. [DOI] [PubMed] [Google Scholar]
- 31.Forsberg Z, Nelson CE, Dalhus B, Mekasha S, Loose JSM, Crouch LI, Røhr ÅK, Gardner JG, Eijsink VGH, Vaaje-Kolstad G. 2016. Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus. J Biol Chem 291:7300–7312. 10.1074/jbc.M115.700161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaplin AK, Wilson MT, Hough MA, Svistunenko DA, Hemsworth GR, Walton PH, Vijgenboom E, Worrall JAR. 2016. Heterogeneity in the histidine-brace copper coordination sphere in AA10 lytic polysaccharide monooxygenases. J Biol Chem 291:12838–12850. 10.1074/jbc.M116.722447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paspaliari DK, Loose JSM, Larsen MH, Vaaje-Kolstad G. 2015. Listeria monocytogenes has a functional chitinolytic system and an active lytic polysaccharide monooxygenase. FEBS J 282:921–936. 10.1111/febs.13191. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa YS, Kudo M, Loose JSM, Ishikawa T, Totani K, Eijsink VGH, Vaaje-Kolstad G. 2015. A small lytic polysaccharide monooxygenase from Streptomyces griseus targeting α- and β-chitin. FEBS J 282:1065–1079. 10.1111/febs.13203. [DOI] [PubMed] [Google Scholar]
- 35.Chiu E, Hijnen M, Bunker RD, Boudes M, Rajendran C, Aizel K, Oliéric V, Schulze-Briese C, Mitsuhashi W, Young V, Ward VK, Bergoin M, Metcalf P, Coulibaly F. 2015. Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization. Proc Natl Acad Sci USA 112:3973–3978. 10.1073/pnas.1418798112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loose JSM, Forsberg Z, Fraaije MW, Eijsink VGH, Vaaje-Kolstad G. 2014. A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett 588:3435–3440. 10.1016/j.febslet.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 37.Hemsworth GR, Taylor EJ, Kim RQ, Gregory RC, Lewis SJ, Turkenburg JP, Parkin A, Davies GJ, Walton PH. 2013. The copper active site of CBM33 polysaccharide oxygenases. J Am Chem Soc 135:6069–6077. 10.1021/ja402106e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaaje-Kolstad G, Bøhle LA, Gåseidnes S, Dalhus B, Bjørås M, Mathiesen G, Eijsink VGH. 2012. Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J Mol Biol 416:239–254. 10.1016/j.jmb.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 39.Ghatge SS, Telke AA, Waghmode TR, Lee Y, Lee K-W, Oh D-B, Shin H-D, Kim S-W. 2015. Multifunctional cellulolytic auxiliary activity protein HcAA10-2 from Hahella chejuensis enhances enzymatic hydrolysis of crystalline cellulose. Appl Microbiol Biotechnol 99:3041–3055. 10.1007/s00253-014-6116-6. [DOI] [PubMed] [Google Scholar]
- 40.Moser F, Irwin D, Chen S, Wilson DB. 2008. Regulation and characterization of Thermobifida fusca carbohydrate-binding module proteins E7 and E8. Biotechnol Bioeng 100:1066–1077. 10.1002/bit.21856. [DOI] [PubMed] [Google Scholar]
- 41.Corrêa TLR, Júnior AT, Wolf LD, Buckeridge MS, Dos Santos LV, Murakami MT. 2019. An actinobacteria lytic polysaccharide monooxygenase acts on both cellulose and xylan to boost biomass saccharification. Biotechnol Biofuels 12:117. 10.1186/s13068-019-1449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fowler CA, Sabbadin F, Ciano L, Hemsworth GR, Elias L, Bruce N, McQueen-Mason S, Davies GJ, Walton PH. 2019. Discovery, activity and characterisation of an AA10 lytic polysaccharide oxygenase from the shipworm symbiont Teredinibacter turnerae. Biotechnol Biofuels 12:232. 10.1186/s13068-019-1573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Solhi L, Goddard-Borger ED, Mathieu Y, Wakarchuk WW, Withers SG, Brumer H. 2021. Four cellulose-active lytic polysaccharide monooxygenases from Cellulomonas species. Biotechnol Biofuels 14:29. 10.1186/s13068-020-01860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsberg Z, Bissaro B, Gullesen J, Dalhus B, Vaaje-Kolstad G, Eijsink VGH. 2018. Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J Biol Chem 293:1397–1412. 10.1074/jbc.M117.817130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardner JG, Crouch L, Labourel A, Forsberg Z, Bukhman YV, Vaaje-Kolstad G, Gilbert HJ, Keating DH. 2014. Systems biology defines the biological significance of redox-active proteins during cellulose degradation in an aerobic bacterium. Mol Microbiol 94:1121–1133. 10.1111/mmi.12821. [DOI] [PubMed] [Google Scholar]
- 46.Forsberg Z, Mackenzie AK, Sørlie M, Røhr ÅK, Helland R, Arvai AS, Vaaje-Kolstad G, Eijsink VGH. 2014. Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc Natl Acad Sci USA 111:8446–8451. 10.1073/pnas.1402771111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J, Wakarchuk W. 2014. Characterization of five β-glycoside hydrolases from Cellulomonas fimi ATCC 484. J Bacteriol 196:4103–4110. 10.1128/JB.02194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hekmat O, Kim Y-W, Williams SJ, He S, Withers SG. 2005. Active-site peptide “fingerprinting” of glycosidases in complex mixtures by mass spectrometry. Discovery of a novel retaining beta-1,4-glycanase in Cellulomonas fimi. J Biol Chem 280:35126–35135. 10.1074/jbc.M508434200. [DOI] [PubMed] [Google Scholar]
- 49.McLean BW, Boraston AB, Brouwer D, Sanaie N, Fyfe CA, Warren RAJ, Kilburn DG, Haynes CA. 2002. Carbohydrate-binding modules recognize fine substructures of cellulose. J Biol Chem 277:50245–50254. 10.1074/jbc.M204433200. [DOI] [PubMed] [Google Scholar]
- 50.Stoll D, Boraston A, Stålbrand H, McLean BW, Kilburn DG, Warren RAJ. 2000. Mannanase Man26A from Cellulomonas fimi has a mannan-binding module. FEMS Microbiol Lett 183:265–269. 10.1111/j.1574-6968.2000.tb08969.x. [DOI] [PubMed] [Google Scholar]
- 51.Notenboom V, Birsan C, Warren RAJ, Withers SG, Rose DR. 1998. Exploring the cellulose/xylan specificity of the β-1,4-glycanase Cex from Cellulomonas fimi through crystallography and mutation. Biochemistry 37:4751–4758. 10.1021/bi9729211. [DOI] [PubMed] [Google Scholar]
- 52.Henrissat B, Teeri TT, Warren RAJ. 1998. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett 425:352–354. 10.1016/S0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- 53.Damude HG, Ferro V, Withers SG, Warren RAJ. 1996. Substrate specificity of endoglucanase A from Cellulomonas fimi: fundamental differences between endoglucanases and exoglucanases from family 6. Biochemical J 315:467–472. 10.1042/bj3150467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarke JH, Davidson K, Gilbert HJ, Fontes CMGA, Hazlewood GP. 1996. A modular xylanase from mesophilic Cellulomonas fimi contains the same cellulose-binding and thermostabilizing domains as xylanases from thermophilic bacteria. FEMS Microbiol Lett 139:27–35. 10.1111/j.1574-6968.1996.tb08175.x. [DOI] [PubMed] [Google Scholar]
- 55.Shen H, Gilkes NR, Kilburn DG, Miller RC, Warren RAJ. 1995. Cellobiohydrolase B, a second exo-cellobiohydrolase from the cellulolytic bacterium Cellulomonas fimi. Biochemical J 311:67–74. 10.1042/bj3110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen H, Tomme P, Meinke A, Gilkes NR, Kilburn DG, Warren RAJ, Miller RC. 1994. Stereochemical course of hydrolysis catalysed by Cellulomonas fimi CenE, a member of a new family of β-1,4-glucanases. Biochem Biophys Res Commun 199:1223–1228. 10.1006/bbrc.1994.1361. [DOI] [PubMed] [Google Scholar]
- 57.Ong E, Gilkes NR, Miller RC, Jr, Warren RAJ, Kilburn DG. 1993. The cellulose‐binding domain (CBDCex) of an exoglucanase from Cellulomonas fimi: production in Escherichia coli and characterization of the polypeptide. Biotechnol Bioeng 42:401–409. 10.1002/bit.260420402. [DOI] [PubMed] [Google Scholar]
- 58.Khanna S. 1993. Regulation, purification, and properties of xylanase from Cellulomonas fimi. Enzyme and Microbial Technology 15:990–995. 10.1016/0141-0229(93)90177-4. [DOI] [Google Scholar]
- 59.Gilkes NR, Kilburn DG, Miller RC, Jr, Warren RAJ, Sugiyama J, Chanzy H, Henrissat B. 1993. Visualization of the adsorption of a bacterial endo β-l,4-glucanase and its isolated cellulose-binding domain to crystalline cellulose. Int J Biological Macromolecules 15:347–351. 10.1016/0141-8130(93)90052-N. [DOI] [PubMed] [Google Scholar]
- 60.MacLeod AM, Gilkes NR, Escote-Carlson L, Warren RA, Kilburn DG, Miller RC. 1992. Streptomyces lividans glycosylates an exoglucanase (Cex) from Cellulomonas fimi. Gene 121:143–147. 10.1016/0378-1119(92)90173-M. [DOI] [PubMed] [Google Scholar]
- 61.Gebler J, Gilkes NR, Claeyssens M, Wilson DB, Béguin P, Wakarchuk WW, Kilburn DG, Miller RC, Jr, Warren RAJ, Withers SG. 1992. Stereoselective hydrolysis catalyzed by related β-1,4-glucanases and, β-1,4-xylanases. J Biol Chem 267:12559–12561. 10.1016/S0021-9258(18)42313-7. [DOI] [PubMed] [Google Scholar]
- 62.Meinke A, Braun C, Gilkes NR, Kilburn DG, Miller RC, Jr, Warren RAJ. 1991. Unusual sequence organization in cenb, an inverting endoglucanase from Cellulomonas fimi. J Bacteriol 173:308–314. 10.1128/jb.173.1.308-314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilkes NR, Claeyssens M, Aebersold R, Henrissat B, Meinke A, Morrison HD, Kilburn DG, Warren RA, Miller RC. 1991. Structural and functional relationships in two families of β-1,4-glycanases. Eur J Biochem 202:367–377. 10.1111/j.1432-1033.1991.tb16384.x. [DOI] [PubMed] [Google Scholar]
- 64.Moser B, Gilkes NR, Kilburn DG, Warren RA, Miller RC. 1989. Purification and characterization of endoglucanase C of Cellulomonas fimi, cloning of the gene, and analysis of in vivo transcripts of the gene. Appl Environ Microbiol 55:2480–2487. 10.1128/aem.55.10.2480-2487.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilkes NR, Kilburn DG, Miller RC, Jr, Warren RAJ. 1989. Structural and functional analysis of a bacterial cellulase by proteolysis. J Biol Chem 264:17802–17808. 10.1016/S0021-9258(19)84644-6. [DOI] [PubMed] [Google Scholar]
- 66.Owolabi JB, Béguin P, Kilburn DG, Miller RC, Jr, Warren RAJ. 1988. Expression in Escherichia coli of the Cellulomonas fimi structural gene for endoglucanase b. Appl Environ Microbiol 54:518–523. 10.1128/aem.54.2.518-523.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gilkes NR, Warren RAJ, Miller RC, Jr, Kilburn DG. 1988. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem 263:10401–10407. 10.1016/S0021-9258(19)81530-2. [DOI] [PubMed] [Google Scholar]
- 68.Langsford ML, Gilkes NR, Singh B, Moser B, Miller RC, Jr, Warren RAJ, Kilburn DG. 1987. Glycosylation of bacterial cellulases prevents proteolytic cleavage between functional domains. FEBS Lett 225:163–167. 10.1016/0014-5793(87)81150-X. [DOI] [PubMed] [Google Scholar]
- 69.Withers SG, Dombroski D, Berven LA, Kilburn DG, Miller RC, Warren RAJ, Gilkes NR. 1986. Direct 1H N.M.R. determination of the stereochemical course of hydrolyses catalysed by glucanase components of the cellulase complex. Biochem Biophys Res Commun 139:487–494. 10.1016/S0006-291X(86)80017-1. [DOI] [PubMed] [Google Scholar]
- 70.Rapp P, Wagner F. 1986. Production and properties of xylan-degrading enzymes from Cellulomonas uda. Appl Environ Microbiol 51:746–752. 10.1128/aem.51.4.746-752.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gilkes NR, Langsford ML, Kilburn DG, Miller RC, Jr, Warren RAJ. 1984. Mode of action and substrate specificities of cellulases from cloned bacterial genes. J Biol Chem 259:10455–10459. 10.1016/S0021-9258(18)90985-3. [DOI] [PubMed] [Google Scholar]
- 72.Whittle DJ, Kilburn DG, Warren RAJ, Miller RC, Jr.. 1982. Molecular cloning of a Cellulomonas fimi cellulase gene in Escherichia coli. Recombinant DNA; plasmid pBR322; immunoassay. Gene 17:139–145. 10.1016/0378-1119(82)90066-X. [DOI] [PubMed] [Google Scholar]
- 73.Xu G-Y, Ong E, Gilkes NR, Kilburn DG, Muhandiram DR, Harris-Brandts M, Carver JP, Kay LE, Harvey TS. 1995. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry 34:6993–7009. 10.1021/bi00021a011. [DOI] [PubMed] [Google Scholar]
- 74.Kenyon WJ, Esch SW, Buller CS. 2005. The curdlan-type exopolysaccharide produced by Cellulomonas flavigena KU forms part of an extracellular glycocalyx involved in cellulose degradation. Antonie Van Leeuwenhoek 87:143–148. 10.1007/s10482-004-2346-4. [DOI] [PubMed] [Google Scholar]
- 75.Sánchez-Herrera LM, Ramos-Valdivia AC, De La Torre M, Salgado LM, Ponce-Noyola T. 2007. Differential expression of cellulases and xylanases by Cellulomonas flavigena grown on different carbon sources. Appl Microbiol Biotechnol 77:589–595. 10.1007/s00253-007-1190-7. [DOI] [PubMed] [Google Scholar]
- 76.Pérez-Avalos O, Sánchez-Herrera LM, Salgado LM, Ponce-Noyola T. 2008. A bifunctional endoglucanase/endoxylanase from Cellulomonas flavigena with potential use in industrial processes at different pH. Curr Microbiol 57:39–44. 10.1007/s00284-008-9149-1. [DOI] [PubMed] [Google Scholar]
- 77.Amaya-Delgado L, Mejía-Castillo T, Santiago-Hernández A, Vega-Estrada J, Amelia F-GS, Xoconostle-Cázares B, Ruiz-Medrano R, Montes-Horcasitas MC, Hidalgo-Lara ME. 2010. Cloning and expression of a novel, moderately thermostable xylanase-encoding gene (Cflxyn11A) from Cellulomonas flavigena. Bioresour Technol 101:5539–5545. 10.1016/j.biortech.2010.02.057. [DOI] [PubMed] [Google Scholar]
- 78.Wakarchuk WW, Brochu D, Foote S, Robotham A, Saxena H, Erak T, Kelly J. 2016. Proteomic analysis of the secretome of Cellulomonas fimi ATCC 484 and Cellulomonas flavigena ATCC 482. PLoS One 11:e0151186. 10.1371/journal.pone.0151186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abt B, Foster B, Lapidus A, Clum A, Sun H, Pukall R, Lucas S, Glavina Del Rio T, Nolan M, Tice H, Cheng J-F, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Ovchinnikova G, Pati A, Goodwin L, Chen A, Palaniappan K, Land M, Hauser L, Chang Y-J, Jeffries CD, Rohde M, Göker M, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk H-P. 2010. Complete genome sequence of Cellulomonas flavigena type strain (134T). Stand Genomic Sci 3:15–25. 10.4056/sigs.1012662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sidar A, Albuquerque ED, Voshol GP, Ram AFJ, Vijgenboom E, Punt PJ.2020. Carbohydrate binding modules: diversity of domain architecture in amylases and cellulases from filamentous microorganisms. Front Bioeng Biotechnol 8:871. 10.3389/fbioe.2020.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.CAZypedia Consortium. 2018. Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology 28:3–8. https://pubmed.ncbi.nlm.nih.gov/29040563/. [DOI] [PubMed] [Google Scholar]
- 82.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gilbert Harry J, Ficko-Blean E. 2021. Carbohydrate binding module family 2. Cazypedia. http://www.cazypedia.org/index.php?title=Carbohydrate_Binding_Module_Family_2&oldid=16503. Accessed June 10, 2022.
- 84.Tsirigotaki A, De Geyter J, Šoštaric′ N, Economou A, Karamanou S. 2017. Protein export through the bacterial Sec pathway. Nat Rev Microbiol 15:21–36. 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 85.Nakamura T, Mine S, Hagihara Y, Ishikawa K, Ikegami T, Uegaki K. 2008. Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus. J Mol Biol 381:670–680. 10.1016/j.jmb.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 86.Courtade G, Forsberg Z, Heggset EB, Eijsink VGH, Aachmann FL. 2018. The carbohydrate-binding module and linker of a modular lytic polysaccharide monooxygenase promote localized cellulose oxidation. J Biol Chem 293:13006–13015. 10.1074/jbc.RA118.004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eijsink VGH, Petrovic D, Forsberg Z, Mekasha S, Røhr ÅK, Várnai A, Bissaro B, Vaaje-Kolstad G. 2019. On the functional characterization of lytic polysaccharide monooxygenases (LPMOs). Biotechnol Biofuels 12:58. 10.1186/s13068-019-1392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kadić A, Várnai A, Eijsink VGH, Horn SJ, Lidén G. 2021. In situ measurements of oxidation–reduction potential and hydrogen peroxide concentration as tools for revealing LPMO inactivation during enzymatic saccharification of cellulose. Biotechnol Biofuels 14:46. 10.1186/s13068-021-01894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frommhagen M, Koetsier MJ, Westphal AH, Visser J, Hinz SWA, Vincken JP, Van Berkel WJH, Kabel MA, Gruppen H. 2016. Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity. Biotechnol Biofuels 9:186. 10.1186/s13068-016-0594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kont R, Bissaro B, Eijsink VGH, Väljamäe P. 2020. Kinetic insights into the peroxygenase activity of cellulose-active lytic polysaccharide monooxygenases (LPMOs). Nat Commun 11:5786. 10.1038/s41467-020-19561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vu VV, Ngo ST. 2018. Copper active site in polysaccharide monooxygenases. Coord Chem Rev 368:134–157. 10.1016/j.ccr.2018.04.005. [DOI] [Google Scholar]
- 92.Hemsworth GR, Davies GJ, Walton PH. 2013. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol 23:660–668. 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 93.Wang B, Walton PH, Rovira C. 2019. Molecular mechanisms of oxygen activation and hydrogen peroxide formation in lytic polysaccharide monooxygenases. ACS Catal 9:4958–4969. 10.1021/acscatal.9b00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Christopherson MR, Suen G, Bramhacharya S, Jewell KA, Aylward FO, Mead D, Brumm PJ. 2013. The genome sequences of Cellulomonas fimi and “Cellvibrio gilvus” reveal the cellulolytic strategies of two facultative anaerobes, transfer of “Cellvibrio gilvus” to the genus Cellulomonas, and proposal of Cellulomonas gilvus sp. nov. PLoS One 8:e53954. 10.1371/journal.pone.0053954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crouch L, Labourel A, Walton PH, Davies GJ, Gilbert Harry J. 2016. The contribution of non-catalytic carbohydrate binding modules to the activity of lytic polysaccharide monooxygenases. J Biol Chem 291:7439–7449. 10.1074/jbc.M115.702365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, Kyrpides NC, Pukall R, Klenk H-P, Goodfellow M, Göker M. 2018. Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol 9:2007. 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bergey D, Harrison F, Breed R, Hammer B, Huntoon F. 1923. Bergey's manual of determinative bacteriology, 1st ed. Williams & Wilkins Co., Baltimore, MD. [Google Scholar]
- 99.Chen H-C, Hsu M-F, Jiang S-T. 1997. Purification and characterization of an exo-N,N′-diacetylchitobiohydrolase-like enzyme from Cellulomonas flavigena NTOU 1. Enzyme and Microbial Technology 20:191–197. 10.1016/S0141-0229(96)00111-1. [DOI] [Google Scholar]
- 100.Yoon MH, Ten LN, Im WT, Lee ST. 2008. Cellulomonas chitinilytica sp. nov., a chitinolytic bacterium isolated from cattle-farm compost. Int J Syst Evol Microbiol 58:1878–1884. 10.1099/ijs.0.64768-0. [DOI] [PubMed] [Google Scholar]
- 101.Zhong X, Zhang L, von Wezel GP, Vijgenboom E, Claessen D. 2022. Role for a lytic polysaccharide monooxygenase in cell wall remodelling. mBio 13:e0045622. 10.1128/mbio.00456-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4:363–373. 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 105.Light SH, Agostoni M, Marletta MA, Anderson WF, Center for Structural Genomics of Infectious Diseases (CSGID). 2017. Catalytic domain of LPMO Lmo2467 from Listeria monocytogenes. 10.2210/pdb5l2v/pdb. [DOI]
- 106.Vaaje-Kolstad G, Houston DR, Riemen AHK, Eijsink VGH, van Aalten DMF. 2005. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J Biol Chem 280:11313–11319. 10.1074/jbc.M407175200. [DOI] [PubMed] [Google Scholar]
- 107.Zhao Y, Zhang H, Yin H. 2017. Crystal structure of a lytic polysaccharide monooxygenase, BtLPMO10A, from Bacillus thuringiensis. 10.2210/pdb5wsz/pdb. [DOI]
- 108.Bianchetti CM, Smith RW, Bingman CA, Phillips GN, Jr.. 2010. The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans. 10.2210/pdb3ndy/pdb. [DOI]
- 109.Okumura A, Uemura M, Yamada N, Chikaishi E, Takai T, Yoshio S, Akagi K, Morita J, Lee Y, Yokogawa D, Suzuki K, Watanabe T, Ikegami T. 2013. Solution structure of the chitin-binding domain of Chi18aC from Streptomyces coelicolor. 10.2210/pdb2rtt/pdb. [DOI]
- 110.Tamburrini KC, Terrapon N, Lombard V, Bissaro B, Longhi S, Berrin J-G. 2021. Bioinformatic analysis of lytic polysaccharide monooxygenases reveals the pan-families occurrence of intrinsically disordered C-terminal extensions. Biomolecules 11:1632. 10.3390/biom11111632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hegnar OA, Petrovic DM, Bissaro B, Alfredsen G, Várnai A, Eijsink VGH. 2019. pH-Dependent relationship between catalytic activity and hydrogen peroxide production shown via characterization of a lytic polysaccharide monooxygenase from Gloeophyllum trabeum. Appl Environ Microbiol 85:e02612-18. 10.1128/AEM.02612-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, p 1–8. In Proceedings of the gateway computing environments workshop (GCE). IEEE, New Orleans, LA. [Google Scholar]
- 113.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 114.Wu S-C, Lia Y-K. 2008. Application of bacterial cellulose pellets in enzyme immobilization. J Molecular Catalysis B: Enzymatic 54:103–108. 10.1016/j.molcatb.2007.12.021. [DOI] [Google Scholar]
- 115.Zhang YHP, Cui J, Lynd LR, Kuang LR. 2006. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7:644–648. 10.1021/bm050799c. [DOI] [PubMed] [Google Scholar]
- 116.Souza CP, Burbano-Rosero EM, Almeida BC, Martins GG, Albertini LS, Rivera ING. 2009. Culture medium for isolating chitinolytic bacteria from seawater and plankton. World J Microbiol Biotechnol 25:2079–2082. 10.1007/s11274-009-0098-z. [DOI] [Google Scholar]
- 117.Haddad Momeni M, Fredslund F, Bissaro B, Raji O, Vuong TV, Meier S, Nielsen TS, Lombard V, Guigliarelli B, Biaso F, Haon M, Grisel S, Henrissat B, Welner DH, Master ER, Berrin J-G, Abou Hachem M. 2021. Discovery of fungal oligosaccharide-oxidising flavo-enzymes with previously unknown substrates, redox-activity profiles and interplay with LPMOs. Nat Commun 12:2132. 10.1038/s41467-021-22372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van den Ent F, Löwe J. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67:67–74. 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 119.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sundqvist G, Stenvall M, Berglund H, Ottosson J, Brumer H. 2007. A general, robust method for the quality control of intact proteins using LC–ESI-MS. J Chromatogr B Analyt Technol Biomed Life Sci 852:188–194. 10.1016/j.jchromb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 121.Tamura K, Foley MH, Gardill BR, Dejean G, Schnizlein M, Bahr CME, Louise Creagh A, Van Petegem F, Koropatkin NM, Brumer H. 2019. Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus. Cell Mol Life Sci 76:4319–4340. 10.1007/s00018-019-03115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Forsberg Z, Vaaje-Kolstad G, Westereng B, Bunæs AC, Stenstrøm Y, MacKenzie A, Sørlie M, Horn SJ, Eijsink VGH. 2011. Cleavage of cellulose by a CBM33 protein. Protein Sci 20:1479–1483. 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tian Z, Lu S, Jin D, Yang J, Pu J, Lai X-H, Ren Z-H, Wu X-M, Li J, Wang S, Xu J. 2020. Cellulomonas shaoxiangyii sp. nov., isolated from faeces of Tibetan antelope (Pantholops hodgsonii) on the Qinghai–Tibet Plateau. Int J Syst Evol Microbiol 70:2204–2210. 10.1099/ijsem.0.003939. [DOI] [PubMed] [Google Scholar]
- 125.Eida AA, Bougouffa S, Alam I, Hirt H, Saad MM. 2021. Complete genome sequence of Cellulomonas sp. JZ18, a root endophytic bacterium isolated from the perennial desert tussock-grass Panicum turgidum. Curr Microbiol 78:1135–1141. 10.1007/s00284-021-02429-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Fig. S1 and S2. Download aem.00968-22-s0001.pdf, PDF file, 0.8 MB (869.7KB, pdf)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information file.