Abstract
In the genome of the gram-positive bacterium Lactococcus lactis MG1363, we have identified three genes (clpC, clpE, and clpB) which encode Clp proteins containing two conserved ATP binding domains. The proteins encoded by two of the genes belong to the previously described ClpB and ClpC families. The clpE gene, however, encodes a member of a new Clp protein family that is characterized by a short N-terminal domain including a putative zinc binding domain (-CX2CX22CX2C-). Expression of the 83-kDa ClpE protein as well as of the two proteins encoded by clpB was strongly induced by heat shock and, while clpC mRNA synthesis was moderately induced by heat, we were unable to identify the ClpC protein. When we analyzed mutants with disruptions in clpB, clpC, or clpE, we found that although the genes are part of the L. lactis heat shock stimulon, the mutants responded like wild-type cells to heat and salt treatments. However, when exposed to puromycin, a tRNA analogue that results in the synthesis of truncated, randomly folded proteins, clpE mutant cells formed smaller colonies than wild-type cells and clpB and clpC mutant cells. Thus, our data suggest that ClpE, along with ClpP, which recently was shown to participate in the degradation of randomly folded proteins in L. lactis, could be necessary for degrading proteins generated by certain types of stress.
The ClpA, ClpB, ClpC, ClpD, ClpX, and ClpY proteins constitute a large family of closely related proteins that are found in both prokaryotic and eukaryotic cells (31). Several members of the Clp family are chaperones that also can target specific proteins for degradation by association with ClpP (13, 34, 41). By itself, ClpP has only peptidase activity, but when it is associated with other members of the Clp family, the resulting Clp complex has serine protease activity (23). The first substrate found to be degraded in vitro by the Clp protease was casein, thus, the designation Clp, for caseinolytic protease (19). Later, it was shown that several Escherichia coli proteins were degraded in vivo by either ClpAP or ClpXP complexes (10).
Members of the Clp family of proteins display a modular structure, with both invariant and variant modules (Fig. 1A). They are classified based on the presence of either one or two ATP binding domains as well as on the occurrence of specific signature sequences (12, 31). The class 2 Clp proteins, such as ClpX and ClpY, have one nucleotide binding domain (ATP-2 domain) and a C-terminal domain with two conserved regions (signature sequences IV and V; Fig. 1A). The larger, class 1 proteins (ClpA, ClpB, ClpC, and ClpD) have one additional nucleotide binding domain (ATP-1 domain) and are usually distinguished by the size of the middle region, which separates the two ATP binding domains.
FIG. 1.
Type-specific signature sequences in L. lactis ClpC and ClpE. (A) The class 1 Clp proteins contain N-terminal and C-terminal domains (white bars), two highly conserved ATP binding domains (ATP-1 and ATP-2, shaded bars), and a variably sized middle domain (white bars in center). The presence of signature sequences is indicated by black boxes and numbering as described previously (31). (B) Comparison of the amino acid sequences of the L. lactis ClpC and ClpE proteins with those of homologous proteins from various organisms (GenBank accession numbers): ClpC—A. aeolicus (AE000733), S. hyodysenteriae (X73140), and B. subtilis (D26185); ClpE—B. subtilis (BSUB0008) and L. sake (OrfX; partial sequence, U82366). The ClpC and ClpE sequences are aligned with signature sequences I, II, and III and with the consensus sequence of a domain present in both the UvrB and the UvrC proteins from a variety of bacteria (24). A putative coiled-coil heptad motif (abcdefg) in which the first and fourth amino acids are hydrophobic (bold) is indicated. Also shown is the ClpE-specific signature sequence (El) and a PDZ-like domain (believed to be involved in protein-protein interactions) deduced from the E. coli ClpA (M31045) and ClpB (M29364) proteins and from the S. cerevisiae Hsp104 (M67479) and Hsp78 (L16533) proteins (21). Residues identical to the signature sequences are indicated by gray shading, whereas amino acids matching the UvrB or UvrC sequences or the PDZ-like consensus sequence are in boldface. h, hydrophobic amino acid; e, D or E; t, S or T.
Bacteria contain a plentiful and variable complement of Clp proteins that have diverse functions often associated with stress adaptation. Of the ClpA family, the E. coli member is by far the best studied (28, 42). While the expression of ClpA is unaffected by stress, the expression of both ClpB and ClpX in E. coli is induced by heat shock. However, only mutants lacking clpB are phenotypically different from wild-type cells, as they show impaired growth at high temperatures (35). This effect is not likely to be mediated through proteolytic activity, as ClpB, in contrast to both ClpA and ClpX, does not associate with ClpP. While members of the ClpB family are found in many organisms, members of the ClpC family are generally found only in gram-positive bacteria and plants (31). In Bacillus subtilis, which does not carry a clpB allele, the expression of both clpC and clpX is induced by general stress conditions, and mutants lacking either of these genes are affected in sporulation, competence development, and growth at high temperatures (9, 26). Similar phenotypes were observed for a B. subtilis clpP null mutant, suggesting that the effects could be mediated through a proteolytic complex (25). In general, ClpC proteins appear to be able both to function as molecular chaperones (27) and to target proteins for degradation by the ClpP protease (32).
Lactococci are gram-positive bacteria that are widely used in the dairy industry as acidifiers. Dairy strains of Lactococcus lactis are auxotrophic for a number of amino acids and have acquired the ability to utilize casein, the major protein found in milk, as the source of amino acids in dairy fermentations. When L. lactis grows in milk, the degradation of casein takes place outside the cells and is mediated by the PrtP protease (33). However, as the Clp protease was originally identified as a caseinolytic protease, we found it intriguing to identify Clp proteins and investigate their role in L. lactis. Recently, we reported the analysis of the clpP gene in L. lactis (7). The ClpP protease was found to be required for survival at high temperatures and growth in the presence of the tRNA analogue puromycin. Here we report the identification of three clp genes in L. lactis, namely, clpB, clpC, and clpE, the last encoding a member of a new Clp protein family.
MATERIALS AND METHODS
Strains and growth media.
L. lactis subsp. cremoris MG1363 (8) cells were grown in either M17 (38) supplemented with 1% glucose (GM17) or minimal morpholinepropanesulfonic acid (MOPS)-based SA medium (18) supplemented with 1% glucose (GSA medium). E. coli XL1-Blue (Stratagene) grown in Luria broth was used for cloning purposes. Puromycin was obtained from Sigma and used at various concentrations.
DNA manipulations and construction of clp disruption strains.
MG1363 chromosomal DNA was isolated as described previously (1), and clp-like sequences were amplified with the following degenerate oligonucleotide primers hybridizing to the conserved regions: primer 1, 5′-GGTGAAYCNGGTGT(A/C)GGTAAAACYGC-3′; primer 2, 5′-TCGTCGATYAAATCRATRGCTTTATCYGG-3′; and primer 3, 5′-TC(A/T)GTYTTYCC(T/G)AC(G/C)CCRGT(A/T)GGGCC-3′. In these sequences, Y represents C or T and R represents A or G. The conditions for PCR amplifications were 30 rounds of 30 s at 94°C, 30 s at 49°C, and 45 s at 72°C. For amplification of specific clp alleles, we used the following oligonucleotides: primer B, 5′-GTATTGGTCACTGAGCCTACCGTTG-3′ (nucleotide positions 1011 to 1035 from the clpB ATG start codon); primer C, 5′-GCGCAGTGACACTTAGTGTTCGG-3′ (nucleotide positions 1159 to 1181 from the clpC ATG start codon); and primer E, 5′-GATGAGGCTATTGAAGCAGCTGC-3′ (nucleotide positions 934 to 956 from the clpE ATG start codon).
PCR products were purified with a Qiagen gel extraction kit or PCR purification kit. Mutant strains HI1615 (MG1363 clpEΔ1), HI1632 (MG1363 clpCΔ1), and HI1635 (MG1363 clpBΔ1) were obtained by cloning the PCR products obtained with primer 1 and primer 2 (clpC and clpB) or primer 1 and primer 3 (clpE) (see Fig. 2) into the SmaI site of pBluescript II SK(+) (Stratagene), followed by insertion of the erythromycin resistance gene (4) into the BamHI site. The resulting plasmids (pHI1613 carrying clpE, pHI1629 carrying clpC, and pHI1630 carrying clpB fragments), which cannot be maintained in L. lactis, were transformed into MG1363 as described previously (16), and erythromycin-resistant colonies (2 μg/ml) were selected. Another clpE disruption mutant, HI1882 (MG1363 clpEΔ2), carrying only the N-terminal region and ATP-1 domain, was constructed by digesting pHI1613 with Bsu36I and partially with BamHI, religating, and transforming the resulting plasmid (pHI1874) into MG1363 as described above. DNA restriction enzyme digestions were carried out as described by the manufacturer (New England BioLabs). DNA sequence analysis of PCR products was carried out with an ALFexpress DNA sequencer (Pharmacia Biotech).
FIG. 2.
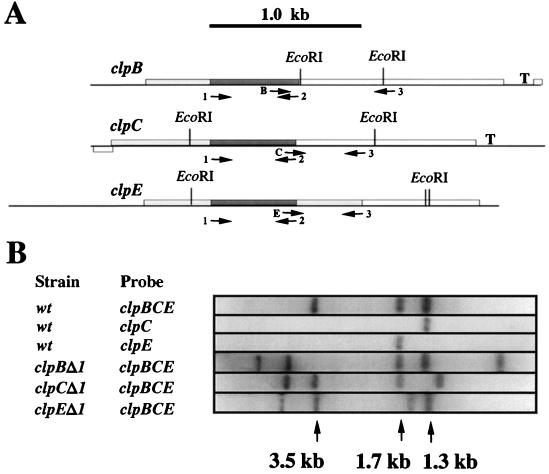
L. lactis clp genes. (A) The L. lactis clpB, clpC, and clpE genes are shown as open boxes, and the EcoRI restriction endonuclease sites as well as putative Rho-independent transcription terminator structures (T) are indicated. The dark grey boxes indicate the DNA amplified with degenerate primers 1 and 2. The allele-specific primers are indicated by B, C, and E; for generating probes specific for each of the clp genes, these primers were used in PCR with degenerate primer 3. The dark and light grey boxes together indicate the portions of the clp genes expressed in each of the mutants HI1635 (clpBΔ1), HI1632 (clpCΔ1), and HI1615 (clpEΔ1). (B) Chromosomal DNA isolated from wild-type (wt) (L. lactis MG1363) cells or HI1635 (clpBΔ1), HI1632 (clpCΔ1), and HI1615 (clpEΔ1) mutant cells was separated on a 1% agarose gel and hybridized with probes specific for either clpC (obtained with primers C and 3) or clpE (obtained with primers E and 3) or with a probe that recognizes all three alleles (obtained with primers 1 and 2).
Phylogenetic analysis.
A phylogenetic tree was constructed from the analysis of ATP-1 domains of 22 Clp or Hsp100 proteins by the neighbor-joining method (30). The ATP-1 domains were defined after alignment with the E. coli ClpA protein, and the regions corresponding to the ClpA sequence from amino acids 186 to 405 were included in the phylogenetic analysis. The tree was constructed from 1,000 bootstrap sample variants by use of the neighbor-joining algorithm in the CLUSTAL W program (39), followed by the DRAWGRAM program included in the PHYLIP package from Joseph Felsenstein. The tree was faithfully redrawn manually with the TOPDRAW program (version 3) for the addition of text.
The amino acid sequences used in the phylogenetic analysis can be found under the following GenBank or SWISS-PROT accession numbers: ClpA, Rhodobacter blastica (PO5444), E. coli (M31045), Borrelia burgdorferi (AE001142), Brassicus napus (actually ClpC, X75328), and Helicobacter pylori (AE000525); ClpB, Corynebacterium glutamicum (U43536), L. lactis (AF016634), Synechococcus sp. strain PCC 7942 (U97124), E. coli (M29364), and Aquifex aeolicus (AE000750); ClpC, Arabidopsis thaliana (Z29026), Synechococcus (U16134), B. subtilis (U02604), Listeria monocytogenes (U40604), Serpulina hyodysenteriae (X73140), A. aeolicus (AE000733), and L. lactis (AF023422); ClpD, A. thaliana (D17582); ClpE, B. subtilis (BSUB0008) and L. lactis (AF023421); and ClpL, L. lactis (Q06716) and Bos taurus (partial sequence, L34677).
Southern and Northern hybridizations.
Southern and Northern hybridizations were performed at 65°C as described previously (5). Northern analysis was performed as previously described (1) with RNA isolated from cells grown exponentially in defined GSA medium. Northern blots were quantitated with a Packard Instant Imager and a PDI Imagequant densitometer. For Southern analysis, chromosomal DNA was separated on a 1% agarose gel and transferred to a Hybond immobilizing membrane (Amersham). Probes specific for either clpC or clpE were generated from MG1363 chromosomal DNA by PCR amplification with degenerate primer 3 and specific primer B, C, or E (see above). The probes were labeled with 32P-dATP (Amersham) by random priming (Pharmacia).
Western blot analysis.
Proteins from cells lysed as described previously (20) were separated on sodium dodecyl sulfate-polyacrylamide (7.5 or 10%) gels. Transfer and Western blot analysis were performed (17) with a 1:4,000 dilution of primary antibody Hsp104 (kindly supplied by S. Lindquist) and a 1:10,000 dilution of anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Promega). Retained secondary antibodies were detected with a Renaissance detection kit (Dupont, NEN Research Products). 35S-methionine labeling of proteins, sample preparation, and two-dimensional protein gel electrophoresis were performed as described by Kilstrup et al. (20).
Stress tolerance.
Tolerance of puromycin was examined by plating appropriate dilutions of exponentially growing wild-type and mutant cells on GM17 plates containing various concentrations of puromycin. After incubation at 30°C for either 1 or 2 days, the colonies were photographed at a 15- to 60-fold magnification; the colony size (in millimeters) was measured for at least 15 colonies from plates on which the colony size appeared homogeneous and for 30 colonies when wild-type cells were plated in the presence of 20 μg of puromycin per ml. Tolerance of heat and salt was examined by plating exponentially growing cells in either the presence or the absence of 4.0% NaCl and incubating the plates at 30°C or plating in the absence of salt and incubating the plates at temperatures of 30 to 37°C. The number of colonies and the colony size were evaluated.
Nucleotide sequence accession numbers.
The partial clpB nucleotide sequence has been submitted to the GenBank database under accession no. AF023423. The clpC sequence has been submitted to GenBank under accession no. AF023422. The clpE nucleotide sequence has been submitted to GenBank under accession no. AF023421.
RESULTS
Identification of clpB and two additional clp-like genes in L. lactis MG1363.
We took advantage of the conservation among Clp proteins in designing degenerate oligonucleotide primers located in either of the two ATP binding regions, ATP-1 and ATP-2, present in class 1 Clp family members. Primer pair 1 (primers 1 and 2 in Fig. 2A) was designed to amplify a 580-bp fragment covering most of the ATP-1 region (Fig. 1A), whereas primer pair 2 (primers 1 and 3), in addition to covering the ATP-1 region, covered the middle region separating ATP-1 and ATP-2 (Fig. 1A). By PCR amplification of chromosomal DNA from L. lactis MG1363 with primer pair 1, we obtained a 580-bp PCR product. Subcloning and sequence analysis of this product revealed that it represented three clp-like sequences. With primer pair 2, we obtained a 1-kb PCR product which represented a single Clp homologue already identified with primer pair 1.
When we analyzed the PCR products obtained with primer 3 and each of the primers specific for the three clp alleles (B, C, and E in Fig. 2A), we found that the distance between the ATP-1 and ATP-2 domains in two of the alleles (designated clpC and clpE) was as expected for the ClpC family (data not shown). In the third clp allele, the distance indicated that it belongs to the ClpB family (data not shown), and the gene was named accordingly. Recently, the DNA sequence of the entire clpB gene was released (GenBank accession no. AF16639), showing that it encodes a 97-kDa protein and is followed by a Rho-independent terminator structure.
Analysis of clpC.
In order to classify the two L. lactis clpC-like genes, we determined the DNA sequences of both alleles from sets of overlapping PCR products generated by a new gene walking technique (15). One of the genes, clpC, which encodes a 90-kDa product, is followed by a putative Rho-independent terminator located 65 bp downstream of the gene. When examining the deduced amino acid sequence of ClpC, we found that the ATP-1 and ATP-2 domains are separated by approximately 100 amino acids, typical of ClpC and ClpD protein family members (31) (Fig. 1B). L. lactis ClpC carries two copies of amino-terminal signature sequence I, characteristic of the ClpC family, but no signature sequence II in the middle domain (Fig. 1B).
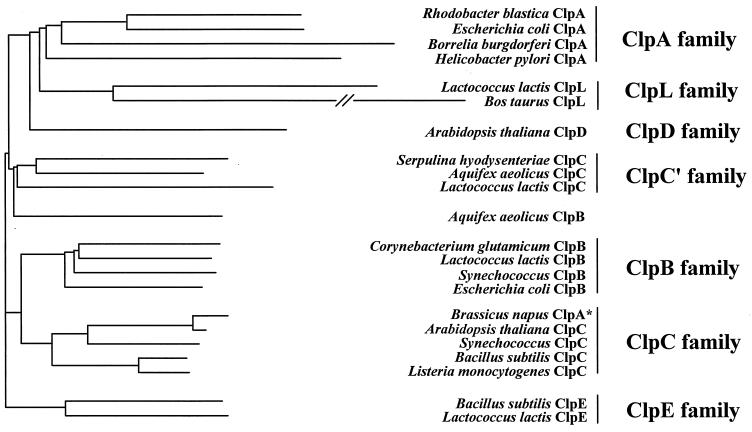
To clarify the relationship of L. lactis ClpC to other Clp proteins, we constructed a phylogenetic tree based upon a comparison of the amino acid sequences of ATP-1 domains from a number of Clp proteins by using the neighbor-joining method (30). The unrooted phylogenetic tree (Fig. 3) suggests that the L. lactis ClpC protein belongs to a subgroup of the ClpC family which includes members from the spirochete Serpulina and the evolutionary ancient thermophile Aquifex. When the phylogenetic relationship was analyzed by a parsimonious method, we found the same general organization (data not shown), which agrees well with a previously published parsimonious analysis of the ATP-1 domains for the class 1 Clp proteins (31). Thus, the L. lactis ClpC protein is a true member of the ClpC family despite its lack of a conserved ClpC signature sequence II.
FIG. 3.
ClpE proteins from L. lactis and B. subtilis form a separate branch in the Clp phylogeny. A phylogenetic tree was constructed by the neighbor-joining method (30) as described in Materials and Methods on the basis of an analysis of the ATP-1 domains of 22 Clp or Hsp100 proteins. The horizontal branch lengths are proportional to the genetic distance. The asterisk indicates that the B. napus ClpA protein belongs to the ClpC family when the taxonomy described by Schirmer et al. (31) is applied.
clpE encodes a member of a new Clp family.
The nucleotide sequence of the second clpC-like gene, clpE, was found to overlap the partial sequence of a previously identified clp gene of L. lactis. The gene is situated upstream of the gap gene, encoding glyceraldehyde-3-phosphate dehydrogenase (3). The intact clpE gene encodes an 83.3-kDa product followed by a putative transcription terminator (3). When analyzing ClpE, we found that the middle region resembles those of the ClpC and ClpD families and that both ClpC signature sequences II and III are present (Fig. 1B). However, in contrast to the ClpB, ClpC, and ClpD proteins, the ClpE protein has a very short N-terminal domain and, while lacking signal sequence I, contains a motif with the consensus sequence -CX2CX22CX2C- that may constitute a zinc binding domain (2). Furthermore, we noted a striking homology between signature sequence II of the ClpC and ClpE families and a domain present in both the UvrB and the UvrC proteins from a number of bacteria (Fig. 1B).
While the entire ClpE protein shows homology to a recently identified B. subtilis ClpE protein (GenBank accession no. Z99111), the N-terminal region resembles OrfX, encoded by a partially sequenced clp gene from Lactobacillus sake (36). In the phylogenetic analysis based on the ATP-1 domain, we found that the ClpE proteins of L. lactis and B. subtilis form a separate ClpE family distinct from all other families (Fig. 3). A similar organization was also found in a parsimonious analysis (data not shown). OrfX from L. sake was not included in the analysis, as the available sequence does not cover the ATP-1 domain. The combined results clearly show that the L. lactis ClpE protein is part of a new Clp family (ClpE).
clpE disruption reduces puromycin tolerance.
To gain further insight into the role of the Clp proteins in L. lactis, we insertionally inactivated either clpB, clpC, or clpE by transforming MG1363 cells with plasmids that, in addition to the erythromycin resistance gene, also carried PCR products internal to the clp genes. Since the E. coli plasmid vector carrying the PCR products cannot be maintained in L. lactis, erythromycin-resistant colonies should arise only after a single crossover recombination event with the corresponding chromosomal allele. The mutants were designated clpBΔ1, clpCΔ1, and clpEΝ1.
By Southern blotting, we analyzed chromosomal DNA isolated from both wild-type and erythromycin-resistant cells with probes that were specific for either clpB, clpC, or clpE and that had been generated by PCR with primer 3 and either primer B, C, or E (Fig. 2A). In accordance with the DNA sequence, we found that for DNA isolated from wild-type cells, the clpC-specific probe hybridized specifically with a 1.3-kb DNA fragment and the clpE-specific probe reacted with a 1.7-kb DNA fragment (Fig. 2B), whereas the clpB-specific probe reacted with 500-bp, 3-kb, and 3.5-kb EcoRI DNA fragments (data not shown). When using a probe consisting of the 580-bp PCR product obtained with degenerate primers 1 and 2 and representing clpB, clpC, and clpE DNA (Fig. 2A), we observed hybridization to DNA fragments of 1.3, 1.7, and 3.5 kb for which mobility was altered in clpC, clpE, and clpB mutant cells, respectively (Fig. 2B). Since the clpE open reading frame in the clpE disruption mutant initially constructed (clpEΔ1) was larger than either the clpC or the clpB open reading frame in the respective mutants, we also constructed a mutant that carried a clpE gene truncated immediately after the ATP-1 domain (clpEΔ2; HI1882).
Next, we compared the phenotypes of the clpBΔ1 (HI1365), clpCΔ1 (HI1632), and clpEΔ1 (HI1615) mutant strains to that of the wild-type strain MG1363. While we did not detect altered heat or salt sensitivity, we found that clpE mutant cells were more sensitive to the tRNA analogue puromycin (Table 1). This antibiotic prematurely terminates translation, resulting in the synthesis of truncated, randomly folded proteins that in E. coli and L. lactis induce the heat shock response (7, 40). At 10 μg of puromycin per ml, the colony size was reduced equally in both wild-type and mutant cells, compared to the size obtained when cells were plated in the absence of puromycin (Table 1). However, at 14 μg of puromycin per ml, the colony size of clpEΔ1 mutant cells was reduced fourfold compared to those of wild-type, clpBΔ1 mutant, and clpCΔ1 mutant cells. At 20 μg/ml, clpEΔ1 mutant colonies were barely detectable, while wild-type cells and clpBΔ1 and clpCΔ1 mutant cells formed colonies. When we investigated the clpE disruption mutant carrying a larger deletion (clpEΔ2), we found the same phenotype as for the clpEΔ1 mutant (data not shown).
TABLE 1.
Colony size in the presence of puromycina
| Strain | Colony diam, in mm, after incubation with the following puromycin concn:
|
|||||
|---|---|---|---|---|---|---|
| 0 | 10 | 14 | 20 | 14 (2 days) | 20 (2 days) | |
| MG1363 (wild type) | 1.2 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 | 0.12 ± 0.02 | 1.1 ± 0.2 (2.5 × 108) | 0.6 ± 0.3 (2.0 × 107) |
| HI1635 (clpBΔ1) | 1.3 ± 0.1 | 0.9 ± 0.1 | 0.7 ± 0.05 | 0.30 ± 0.09 | 1.1 ± 0.1 (2.1 × 108) | 1.0 ± 0.1 (1.4 × 108) |
| HI1632 (clpCΔ1) | 1.2 ± 0.1 | 0.9 ± 0.1 | 0.6 ± 0.05 | 0.14 ± 0.03 | 1.2 ± 0.1 (2.4 × 108) | 1.1 ± 0.1 (1.2 × 108) |
| HI1615 (clpEΔ1) | 1.2 ± 0.1 | 0.9 ± 0.1 | 0.15 ± 0.02 | 0.04 ± 0.01 | 1.1 ± 0.1 (1.2 × 108) | 0.07 ± 0.04 (ND) |
Wild-type and mutant cells were plated in the presence of the indicated puromycin concentrations. After growth at 30°C (for 1 day, unless otherwise indicated), the colony diameter was determined. The numbers are the average ± standard deviation for at least 15 determinations. After 2 days of incubation, the numbers of colonies were also determined (numbers in parentheses; ND indicates that the colonies were too small to count).
Interestingly, after 2 days of incubation in the presence of 20 μg of puromycin per ml, colonies of clpEΔ1 mutant cells remained almost undetectable, whereas wild-type cells formed colonies that were larger but highly variable in size. In contrast, both the clpBΔ1 and the clpCΔ1 mutant cells formed colonies with five times greater efficiency than wild-type cells, and the colonies were homogeneous in size and almost twice as large as wild-type colonies (Table 1). Our data show that the disruption of clpE reduces the tolerance of L. lactis for puromycin, whereas the disruption of either clpB or clpC results in cells with a slightly higher tolerance for puromycin than wild-type cells.
clpB, clpC, and clpE expression is induced by heat shock.
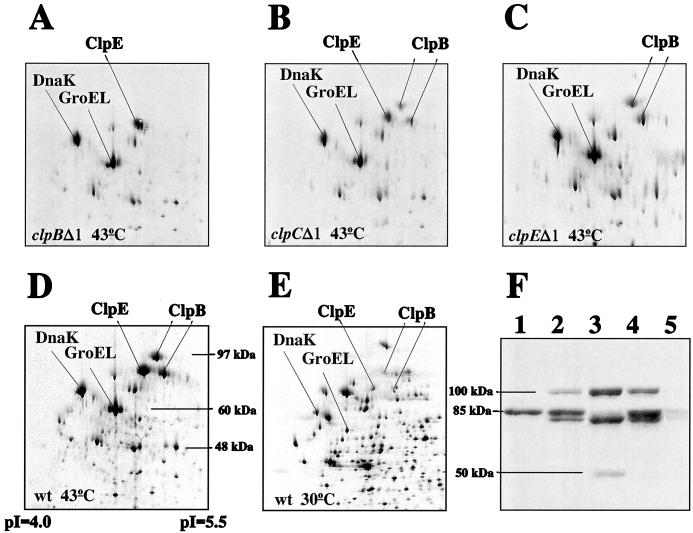
The expression of clp-like gene products is often induced by heat shock, suggesting a role for these proteins in stress adaptation. In order to evaluate if the L. lactis clp gene products also are induced by stress, we identified the Clp proteins (Fig. 4). By using two-dimensional protein gel electrophoresis, we found that in heat-treated clpBΔ1 mutant cells, proteins of 84 and 100 kDa were missing (Fig. 4A), while in clpEΔ1 cells, an 85-kDa protein spot was absent (Fig. 4C), in comparison with the protein pattern of heat-treated wild-type cells (Fig. 4D). These results were confirmed by Western blot analysis with an antibody raised against the Saccharomyces cerevisiae ClpB homologue Hsp104. Two proteins reacting with the antibody were absent in clpBΔ1 mutant cells (Fig. 4F, lane 1), while the intensity of an 85-kDa band was greatly reduced in clpEΔ1 cells (lane 3). Curiously, in clpEΔ1 mutant cells, a new protein of approximately 50 kDa reacted with the antibody. The size of this protein corresponds to the size of the expected product (54 kDa) of clpE when truncated at the position of primer 3 (corresponding to amino acid 486). The clpEΔ2 mutant (HI1882) also synthesized a ClpE protein (40 kDa) that was induced by heat shock (data not shown). Interestingly, the protein pattern of clpCΔ1 mutant cells resembled that of wild-type cells when investigated either by two-dimensional gel electrophoresis (Fig. 4B) or by Western blot analysis (Fig. 4F, lane 2). In cells which did not receive heat treatment, the clp gene products were barely visible (Fig. 4E and F, lane 5). Thus, both the clpB and the clpE gene products are strongly induced by heat shock.
FIG. 4.
Western blot and two-dimensional protein analyses of Clp expression in wild-type (wt) and clp mutant cells. (A to E) 35S-labeled proteins extracted from equal amounts of cells of L. lactis MG1363 (D and E), HI1635 (clpBΔ1; A), HI1632 (clpCΔ1; panel B), and HI1615 (clpEΔ1; C) grown at 30°C (E) or shifted to 43°C for 20 min (A to D) were separated in two dimensions. (F) Proteins extracted from equal amounts of L. lactis MG1363 (lane 4 and 5), HI1635 (clpBΔ1; lane 1), HI1632 (clpCΔ1; lane 2), and HI1615 (clpEΔ1; lane 3) grown at 30°C (lane 5) or shifted to 43°C for 20 min (lanes 1 to 4) were separated on a sodium dodecyl sulfate–10% polyacrylamide gel and reacted with anti-Hsp104 antibody diluted 1:4,000. The molecular masses deduced from known proteins or protein molecular mass markers (Gibco BRL) are indicated, together with the pIs.
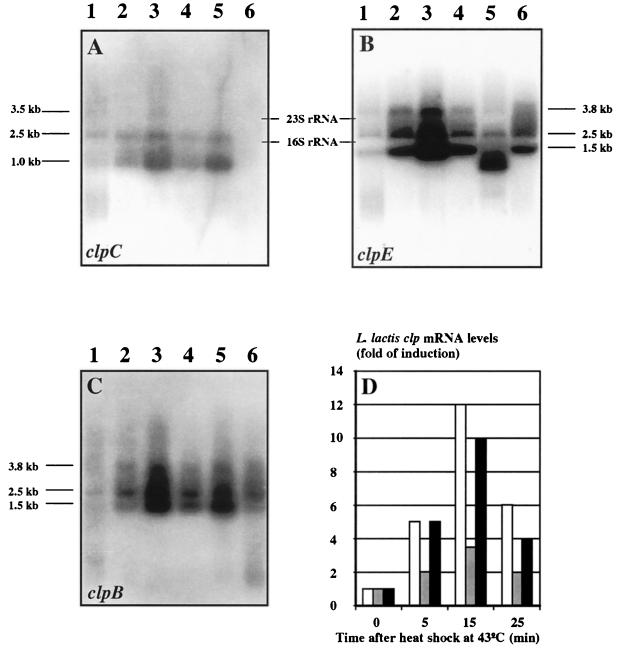
Next, we examined if stress regulation of the L. lactis clp genes occurs at the transcriptional level by reacting probes specific for each of the clp alleles (see above) with RNA isolated from cells grown at 30°C or shifted to 43°C for different periods of time (Fig. 5). By densitometric tracing of the Northern blots, we found that both clpB and clpE mRNA levels were induced approximately 10-fold (Fig. 5B and C), while the clpC-specific probe detected a transcript that was induced 3-fold by heat shock (Fig. 5A). The specificity of the probes was confirmed when mRNA isolated from clpC or clpE mutant cells was analyzed. In clpE mutant cells, the clpE transcript was reduced in size (Fig. 5B, lane 5); in clpC mutant cells, the clpC transcript was absent (Fig. 5A, lane 6); and the clpB-specific probe detected full-length transcripts in both clpC and clpE mutant cells (Fig. 5C, lanes 5 and 6). The temporal induction profiles were similar for all three genes, and the amount of clp mRNA was greatest 15 min after heat application (Fig. 5D). Curiously, more than one transcript size was detected for each of the three genes. Currently, we do not know if these transcripts have arisen from separate transcriptional initiation sites or if the primary transcripts are specifically processed.
FIG. 5.
Heat shock-induced expression of clpB, clpC, and clpE from L. lactis. (A to C) Total RNA was extracted from L. lactis MG1363 (wild type) growing exponentially at 30°C (lane 1) or at 5 min (lane 2), 15 min (lane 3), or 25 min (lane 4) after transfer to 43°C. RNA was also extracted from clpE (HI1615; lane 5) or clpC (HI1632; lane 6) mutant cells 15 min after a shift from 30 to 43°C. Ten micrograms each RNA preparation was separated on an agarose gel and transferred to a nylon membrane that was reacted with clpC-specific (A), clpE-specific (B), and clpB-specific (C) probes. (D) The resulting X-ray autoradiograms were quantitated by densitometric scanning. The mRNA synthesized at 43°C was normalized to the amount present at 30°C. White columns; clpB; grey columns, clpC; black columns, clpE.
DISCUSSION
L. lactis is a gram-positive bacterium frequently used in dairy fermentations. We have identified for L. lactis three genes, clpB, clpC, and clpE, that all encode Clp proteins with two conserved ATP binding domains (class 1). Based on a phylogenetic analysis of the first ATP binding domain of 22 Clp family members, we found that ClpE is part of a new Clp protein family. This family could be common to gram-positive bacteria, as clpE genes have recently been detected in B. subtilis and L. sake (GenBank accession no. Z99111) (36). A short N-terminal region including a motif (-CX2CX22CX2C-) that potentially can form a zinc finger (2) characterizes the ClpE family. A similar motif (-CX2CX18CX2C-) has also been found in ClpX (11), and recently it was reported that a zinc finger present in the molecular chaperone DnaJ is involved in binding to denatured protein substrates (37).
The middle domain of ClpE contains a signature sequence II resembling that characteristic of the ClpC and ClpD families. Curiously, we found that this signature sequence shows homology to a domain also present in the UvrB and UvrC proteins from several bacteria (Fig. 1). During DNA repair, this domain mediates the interaction between UvrC and UvrB (14, 22, 24); thus, it might constitute a site for interaction either between ClpE monomers or between ClpE and the DNA repair system.
In addition to clpE, we identified two other clp genes in L. lactis, the DNA sequence for one of which (encoding a ClpB family member) was recently released (GenBank accession no. AF016634). The third clp gene was designated clpC. Like other genes in the ClpC family, it encodes a product with two copies of signature sequence I in the N-terminal region; however, unlike that in other ClpC family members, signature sequence II in the middle domain is absent.
When investigating the expression of the clp genes by Northern blot analysis, we found that both clpB and clpE mRNA syntheses were strongly induced by heat shock (10-fold), while that of clpC mRNA was only moderately induced (3-fold). At the protein level, we also found that the clpE gene product and the two products encoded by the clpB gene were strongly induced by heat. Two protein products are encoded by the clpB genes of E. coli (29) and the cyanobacterium Synechococcus sp. strain PCC 7942 (6); translation of the smaller products in both cases is initiated at an internal GTG start codon. When we inspected the DNA sequence of the L. lactis clpB gene for alternative translational start sites, we found a GTG codon at position 151, preceded by a putative ribosome binding site (GGAGGTGA). Initiation at this position would result in an 80-kDa product in addition to the full-length product of 97 kDa, in agreement with the observed products of 84 and 100 kDa. We furthermore confirmed that one of the two proteins is not a processed form of the other by determining the ratio between the two clpB gene products at different times. After incubation of cells with 35S-methionine for 10 min at 43°C followed by a chase with a 100-fold excess of unlabeled methionine, we monitored the ratio between the two forms of ClpB at both 43 and 30°C by two-dimensional protein gel analysis (data not shown). Since at both temperatures we found the same ratio between the two ClpB proteins throughout the experiment, we conclude that the two forms are not interconverted or that processing is concomitant with translation.
When we examined the phenotypes of the clpB, clpC, or clpE disruption mutants, we found that the mutants were as tolerant of heat and salt as the wild type. However, while the clpE mutant cells were sensitive to puromycin, both clpB and clpC mutant cells were slightly more resistant to puromycin than wild-type cells. Since puromycin is an antibiotic that interferes with translation, resulting in the production of truncated, randomly folded proteins, ClpE could recognize such proteins, leading to subsequent degradation by the ClpP protease. In support of this proposal is the finding that disruption of the L. lactis clpP gene also results in sensitivity to puromycin (7).
ACKNOWLEDGMENTS
We thank T. Msadek for pointing out the putative zinc binding domain in ClpE; S. Lindquist, A. K. Clarke, and K. Keegstra for kindly supplying antibodies; K. Sørensen and D. Frees for stimulating discussions; and C. Nyholm and D. Overgaard Jensen for expert technical assistance.
FØTEK and MFF (Danish Dairy Research Board) supported this work.
REFERENCES
- 1.Arnau J, Sørensen K I, Appel K F, Vogensen F K, Hammer K. Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology. 1996;142:1685–1691. doi: 10.1099/13500872-142-7-1685. [DOI] [PubMed] [Google Scholar]
- 2.Berg J M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990;265:6513–6516. [PubMed] [Google Scholar]
- 3.Cancilla M R, Hillier A J, Davidson B E. Lactococcus lactis glyceraldehyde-3-phosphate dehydrogenase gene, gap: further evidence for strongly biased codon usage in glycolytic pathway genes. Microbiology. 1995;141:1027–1036. doi: 10.1099/13500872-141-4-1027. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen B, Johnsen M G, Stenby E, Vogensen F K, Hammer K. Characterization of the lactococcal temperate phage TP901-1 and its site-specific integration. J Bacteriol. 1994;176:1069–1076. doi: 10.1128/jb.176.4.1069-1076.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eriksson M-J, Clarke A K. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1996;178:4839–4846. doi: 10.1128/jb.178.16.4839-4846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frees D, Ingmer H. ClpP participates in degradation of misfolded protein in Lactococcus lactis. Mol Microbiol. 1999;31:79–87. doi: 10.1046/j.1365-2958.1999.01149.x. [DOI] [PubMed] [Google Scholar]
- 8.Gasson M. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerth U, Krüger E, Derre I, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 12.Gottesman S, Clark W P, Maurizi M R. The ATP-dependent Clp protease of Escherichia coli: sequence of clpA and identification of a Clp-specific substrate. J Biol Chem. 1990;265:7886–7893. [PubMed] [Google Scholar]
- 13.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 14.Grossman L, Thiagalingam S. Nucleotide excision repair, a tracking mechanism in search of damage. J Biol Chem. 1993;268:16871–16874. [PubMed] [Google Scholar]
- 15.Harrison R W, Miller J C, D’Souza M J, Kampo G. Easy gene walking. BioTechniques. 1997;22:650–653. doi: 10.2144/97224bm17. [DOI] [PubMed] [Google Scholar]
- 16.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingmer H, Cohen S N. Excess intracellular concentration of the pSC101 RepA protein interferes with both plasmid DNA replication and partitioning. J Bacteriol. 1993;175:7834–7841. doi: 10.1128/jb.175.24.7834-7841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katayama Y, Gottesman S, Pumphrey J, Rudikoff S, Clark W P, Maurizi M R. The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning and mutational analysis of the ATP binding component. J Biol Chem. 1988;263:15226–15236. [PubMed] [Google Scholar]
- 20.Kilstrup M, Jacobsen S, Hammer K, Vogensen F K. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl Environ Microbiol. 1997;63:1826–1837. doi: 10.1128/aem.63.5.1826-1837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 22.Lin J-J, Sancar A. (A)BC exinuclease: the Escherichia coli nucleotide excision repair enzyme. Mol Microbiol. 1992;6:2219–2224. doi: 10.1111/j.1365-2958.1992.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 23.Maurizi M R, Clark W, Kim S H, Gottesman S. ClpP represents a unique family of serine proteases. J Biol Chem. 1990;265:12546–12552. [PubMed] [Google Scholar]
- 24.Moolenaar G F, Franken K L M C, Dijkstra D M, Thomas-Oates J E, Visse R, van de Putte P, Goosen N. The C-terminal region of the UvrB protein of Escherichia coli contains an important determinant for UvrC binding to the preincision complex but not the catalytic site for 3′-incision. J Biol Chem. 1995;270:30508–30515. doi: 10.1074/jbc.270.51.30508. [DOI] [PubMed] [Google Scholar]
- 25.Msadek T, Dartois V, Kunst F, Herbaud M-L, Denizot F, Rapoport G. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol Microbiol. 1998;27:899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- 26.Msadek T, Kunst F, Rapoport G. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and growth at high temperature. Proc Natl Acad Sci USA. 1994;91:5788–5792. doi: 10.1073/pnas.91.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp 100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pak M, Wickner S. Mechanism of protein remodeling by ClpA chaperone. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S K, Kim K I, Woo K M, Seol J H, Tanaka K, Ichihara A, Ha D B, Chung C H. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Schirmer E C, Glover J R, Singer M A, Lindquist S. Hsp100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 32.Shanklin J, Witt N D, Flanagan J M. The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell. 1995;7:1713–1722. doi: 10.1105/tpc.7.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smid E J, Poolman B, Konings W N. Casein utilization by lactococci. Appl Environ Microbiol. 1991;57:2447–2452. doi: 10.1128/aem.57.9.2447-2452.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Squires C, Squires C L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Squires C L, Pedersen S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stentz R, Lauret R, Ehrlich S D, Morel-Deville F, Zagorec M. Molecular cloning and analysis of the ptsHI operon in Lactobacillus sake. Appl Environ Microbiol. 1997;63:2111–2116. doi: 10.1128/aem.63.6.2111-2116.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szabo A, Korszun R, Hartl F U, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 38.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VanBogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wawrzynow A, Banecki B, Zylicz M. The Clp ATPase define a novel class of molecular chaperones. Mol Microbiol. 1996;21:895–899. doi: 10.1046/j.1365-2958.1996.421404.x. [DOI] [PubMed] [Google Scholar]
- 42.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenny K, Maurizi M R. A molecular chaperon, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]