Abstract
Rapid, accurate and frequent detection of the RNA of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) and of serological host antibodies to the virus would facilitate the determination of the immune status of individuals who have Coronavirus disease 2019 (COVID-19), were previously infected by the virus, or were vaccinated against the disease. Here we describe the development and application of a 3D-printed lab-on-a-chip that concurrently detects, via multiplexed electrochemical outputs and within 2 h, SARS-CoV-2 RNA in saliva as well as anti-SARS-CoV-2 immunoglobulins in saliva spiked with blood plasma. The device automatedly extracts, concentrates and amplifies SARS-CoV-2 RNA from unprocessed saliva, and integrates the Cas12a-based enzymatic detection of SARS-CoV-2 RNA via isothermal nucleic acid amplification with a sandwich-based enzyme-linked immunosorbent assay on electrodes functionalized with the Spike S1, nucleocapsid and receptor-binding-domain antigens of SARS-CoV-2. Inexpensive microfluidic electrochemical sensors for performing multiplexed diagnostics at the point of care may facilitate the widespread monitoring of COVID-19 infection and immunity.
Subject terms: Lab-on-a-chip, Diagnostic markers, Viral infection
A 3D-printed lab-on-a-chip allows for the concurrent rapid electrochemical detection of SARS-CoV-2 RNA in saliva and of anti-SARS-CoV-2 antibodies in saliva spiked with blood plasma.
Main
The COVID-19 pandemic has highlighted the need for cost-effective diagnostics for SARS-CoV-2 RNA as well as for the detection of antibodies generated by the host in response to infection. This type of multifunctional detection method will be particularly useful for diagnosis of both acute and convalescent infections, as well as for assessing patient immunization status following vaccination. The clinical timeline of SARS-CoV-2 infection consists of an acute phase, when viral RNA is detectable in clinical samples, such as saliva or nasopharyngeal swabs, followed by a convalescent phase when serology biomarkers, such as IgG antibodies, are present in saliva and serum1. Therefore, simultaneous analysis of these different biomarkers in clinical samples as the disease progresses could provide more accurate results for disease monitoring and management.
Molecular (nucleic acid) diagnostics that detect the presence of viral RNA are key to detecting the virus during the first 5 d of infection, with a viral load peak around day 4 (Supplementary Fig. 1)2–4. Following the first few days of infection, the host produces IgM, IgA and IgG antibodies in a process known as seroconversion. These antibodies often become stable after the first 6 d of seroconversion and their titres can remain stable over months5,6. The presence of different antibody types varies during infection7 and correlates with disease severity8,9. In particular, extensive cohort studies in hospitalized patients show that IgG antibodies against different viral proteins (for example, nucleocapsid or spike proteins) correlate with disease severity and outcomes. For example, antibodies against the spike (S) protein are more specific10 and correlate with virus neutralization8, while antibodies against the nucleocapsid (N) protein have longer clearance rates11 and might appear earlier during infection12. Therefore, serological assays that detect the host’s antibodies developed after an infection can widen the testing window for SARS-CoV-2 beyond the molecular diagnostic timeframe and provide insights into the patient’s progression. Such assays may also be used to determine whether titres are maintained over time and thus the effectiveness of vaccination responses. For example, SARS-CoV-2 vaccine efficacy trials have highlighted a direct correlation between the titre of antibodies targeting the receptor-binding domain (RBD) found in the S1 subunit of the S protein, the neutralizing antibody titre and vaccine efficacy13. Thus, the development of serological assays targeted to individual SARS-CoV-2 viral antigens could have important implications for predicting the efficacy of vaccines and estimating the need for boosters.
Molecular diagnostics, on the other hand, commonly involve methods such as quantitative polymerase chain reaction (qPCR), which require rigorous sample preparation and temperature control, cold storage of reagents, expensive instrumentation (requiring routine maintenance) and trained personnel to run the tests. While there are home diagnostic tests approved for use by the US Food and Drug Administration (FDA), many of these tests either involve self-collection and mailing to a central laboratory or are based on rapid antigen tests, which have shown to be less accurate than nucleic acid tests, such as qPCR14. During the last few years, powerful diagnostic techniques that capitalize on CRISPR (clustered regularly interspaced short palindromic repeats) technology and associated programmable endonucleases have gained interest due in part to their high specificity, programmability and capacity to work at physiological conditions15–19. CRISPR-based diagnostics capitalize on endonucleases, such as Cas12a, which has a specific cleavage activity towards double-stranded DNA (dsDNA) fragments matching its guide RNA (gRNA) sequence. Once the Cas12a-gRNA complex binds to its dsDNA target, it activates and subsequently engages in indiscriminate collateral hydrolysis of nearby single-stranded DNA (ssDNA)20,21.
Electrochemical (EC) methods of CRISPR-based nucleic acid detection typically combine nucleic acid probes conjugated on an electrode with CRISPR-Cas effectors and have detection limits in the picomolar to femtomolar (10−12–10-15) range22–26. Unfortunately, these detection limits are inadequate for diagnosing SARS-CoV-2 in clinical samples, which require ultrasensitive RNA detection at the attomolar (10−18) scale. To overcome this limitation, amplification techniques such as loop-mediated isothermal amplification (LAMP) can be performed before Cas12a detection, which improves the sensitivity of fluorescent CRISPR-based assays by orders of magnitude15–17,19,27.
The combination of serological and nucleic acid diagnostics can improve the overall accuracy of SARS-CoV-2 diagnosis28 and provide qualitative data on the patient’s disease severity and state of progression. However, workflows for SARS-CoV-2 diagnosis that integrate serological and nucleic acid tests are currently limited as these assays require laboratory equipment such as pipettes, centrifuges and heating blocks, as well as specialized technical skills. Despite the advances to make SARS-CoV-2 tests as widely available as possible, a lab-on-a-chip (LOC) platform for point-of-care (POC) use that can detect both SARS-CoV-2 RNA and serological markers is not yet available.
EC biosensors offer a particularly promising solution to achieve ultrasensitive, selective, multiplexed, quantitative and cost-effective LOC detection of both nucleic acids and proteins; they also offer the potential to interface with electronic medical records, integrated cloud systems and telemedicine. Despite their potential, EC diagnostics have only been used to detect either nucleic acids or proteins individually26,29 and have been limited to methods that require multiple liquid handling steps and specialized equipment. Here we describe a low-cost, three-dimensional (3D)-printed, self-contained LOC diagnostic platform that is capable of concurrent detection of SARS-CoV-2 nucleic acids and host antibodies directed against the virus from unprocessed saliva samples. This device integrates microfluidics that enable automated liquid handling for sample preparation with a simple and sensitive readout for both viral RNA as well as host antibodies. The simplicity of this device makes it user-friendly and should enable its use for POC testing within hospitals and in COVID-19 testing clinics.
Results
Design of the microfluidic electrochemical lab-on-a-chip platform
In considering the design of an LOC diagnostic device, we focused on accuracy, ease of use and a capacity to integrate with digital health platforms. Towards this goal, we fabricated a microfluidic chip capable of processing untreated saliva to detect both viral RNA and host antibodies on the same EC sensor chip (Fig. 1). We chose saliva over the more common nasopharyngeal swab specimens on our device due to saliva’s ease of self-collection and increased clinical sensitivity due to a high viral load30,31. Because unprocessed saliva is viscous and contains nucleases that could inhibit a downstream reaction, viral RNA is typically purified from saliva using costly and complicated nucleic acid purification kits. However, to eliminate the need for purification before reaction, we experimented with various sample preparation workflows that would allow for viral lysis and nuclease inactivation (Supplementary Fig. 2). Among different buffers tested, we found that incubating the saliva sample with a proteinase K32 solution at 55 °C followed by a 95 °C inactivation effectively lysed the sample and eliminated false positive signals without inhibiting the performance of the downstream LAMP and CRISPR-based detection steps. Subsequent to the sample preparation step, we needed a method to capture the nucleic acids from the saliva sample. We found that a polyethersulfone (PES) membrane was able to concentrate the RNA without inhibiting the downstream reactions33–36.
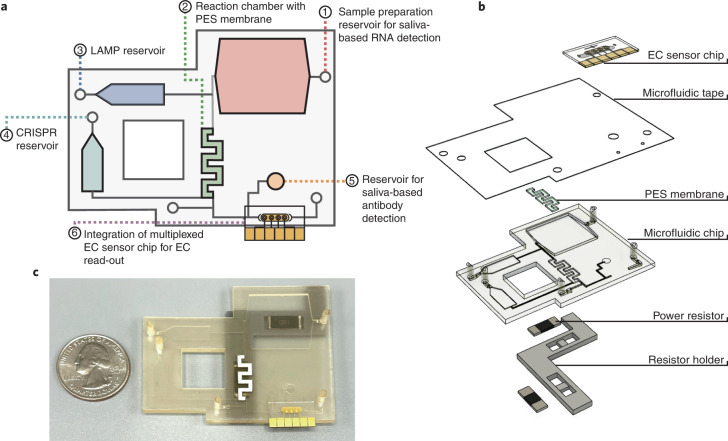
Fig. 1. Overview of multiplexed electrochemical sensor system.
a, Overview of the microfluidic chip designed for an LOC sample-to-answer saliva detection of SARS-CoV-2 RNA and antibodies. (1) The user inputs saliva onto the antibody detection reservoir and a saliva and proteinase K mixture into the sample preparation reservoir, where it incubates. (2) The saliva is pumped over the PES membrane inside the reaction chamber for RNA capture and heated to denature potential reaction inhibitors. (3) The LAMP solution is then pumped from the reservoir into the reaction chamber and incubated. (4) The CRISPR mixture is pumped into the reaction chamber, incubated and then pumped over the EC sensor chip. (5) The saliva for antibody detection is pumped over the EC sensor chip. (6) After the addition of polystreptavidin-HRP and TMB, results from the EC sensor chip are read with a potentiostat. b, An exploded view of the multiplexed system, which includes a heater system, a sealed microfluidic chip and a multiplexed EC sensor chip. c, Photograph of the microfluidic system with a quarter dollar for scale.
Overall, the device incorporates a 500 µl sample preparation chamber that is used for the saliva-based RNA detection, a 50 µl reaction chamber that contains the PES membrane, a 25 µl LAMP reservoir, a 22 µl CRISPR reservoir and a 20 µl reservoir for direct saliva-based antibody detection. The compact design allows for the use of two high-power resistors as heating elements (Fig. 1b), one beneath the sample preparation reservoir and the other beneath the reaction chamber. The LAMP and CRISPR reservoirs are isolated from the sample preparation reservoir and reaction chamber to ensure no carryover heating from the heated elements during sample preparation and concentration.
To optimize chip functionality, we tested a variety of reservoir and channel dimensions and placements to ensure uniform heating and precise fluid flow (Supplementary Fig. 3). The chip was designed with different channel dimensions to establish resistors to the fluid flow, while the fluids were controlled by a small, fingertip-sized DC peristaltic pump with a maximum flow rate of 200 µl min−1 controlled by an Arduino microcontroller. The channels leading to the reservoirs were designed with higher resistance to avoid any backflow or cross-contamination with the reservoirs (400 ×150 µm) (Supplementary Fig. 4), while the ends of the channels leading to the reservoirs were designed with a stepped stop valve to stop any flow from the reaction chamber to the reservoirs. We tested a variety of reaction chamber geometries to ensure that the PES membrane surface area was adequate for nucleic acid capture and that the chamber had optimal fluid retention over the course of the reaction, settling on a serpentine shape (Supplementary Fig. 3), and designed higher resistance on the flow path to the EC sensor chip to avoid unwanted flow before the desired time point.
The workflow of the sample-to-answer microfluidic detection device with fluid flow driven by an Arduino-controlled small peristaltic pump is as follows. First, the user manually inputs saliva onto the antibody detection reservoir (Fig. 1a(5)) and the RNA sample preparation reservoir (Figs. 1a(1)). In the RNA sample preparation chamber, the saliva combines with a proteinase K solution and is heated to 55 °C for 15 min followed by 95 °C for 5 min using integrated high-power resistors to allow for virus lysis and nuclease inactivation32. The saliva sample is then pumped over the PES membrane within the reaction chamber, where the RNA binds to the membrane. The reaction chamber is heated to 95 °C for an additional 3–5 min to ensure denaturation of potential reaction inhibitors. The LAMP solution is then pumped from the reservoir into the reaction chamber and incubated at 65 °C for 30 min, followed by the CRISPR mixture and incubation at 37 °C for an additional 30 min, after which it is pumped over the EC sensor chip to incubate. Saliva for antibody detection is then directly pumped over the EC sensor chip and incubated. The chip is washed with phosphate buffered saline with Tween 20 (PBST), followed by the addition of polystreptavidin-horseradish peroxidase (HRP) and a precipitating form of tetramethylbenzidine (TMB), after which the chip is read using a potentiostat. A more in-depth visual of the microfluidic chip and its workflow is provided in Supplementary Fig. 5.
Integrated CRISPR-based molecular diagnostic assay
The CRISPR-Cas RNA detection experiments reported here capitalize on Cas12a from Lachnospiraceae bacterium ND2006, which has specific cleavage activity towards dsDNA fragments matching its gRNA sequence. Upon target binding, activated Cas12a-gRNA engages in collateral cleavage of nearby ssDNA20,21, which can be read optically as an increase in fluorescence due to the hydrolysis of a fluorophore-quencher-labelled ssDNA reporter probe (RP). As a result, the signal obtained from the fluorescent assay will be high in the presence of its SARS-CoV-2 target and low when it is not detected. LAMP primers37–41 and Cas12a-gRNAs were evaluated from a range of conserved regions in the SARS-CoV-2 genome to determine the most sensitive combinations using commercially available synthetic full-length SARS-CoV-2 genomic RNA. The open reading frame 1a (ORF1a) assay, which targets a highly conserved region in the SARS-CoV-2 viral genome, had a limit of detection (LOD) of 2.3 viral RNA copies per µl, with a reaction time of 50 min (Supplementary Table 1, and Figs. 6 and 7). This LOD is comparable to high-performance SARS-CoV-2 reverse transcription quantitative polymerase chain reaction (RT–qPCR) assays42, with half the time to result. To confirm that the assay was able to detect SARS-CoV-2 active virus, the ORF1a assay primers and guide RNA were further validated using 11 SARS-CoV-2 RT–qPCR negative patient saliva samples and 19 SARS-CoV-2 RT–qPCR positive saliva samples with a range of cycle threshold (CT) values (Supplementary Figs. 8 and 9, and Table 2).
To integrate the CRISPR-based molecular assay onto the EC sensor platform, we designed a biotinylated ssDNA RP that partially hybridized to peptide nucleic acid (PNA) capture probes immobilized on the surface of the antifouling composite-coated gold electrodes43 (Fig. 2a). Functionalized EC biosensors were incubated with samples containing the LAMP/Cas12a mix which includes the biotinylated ssDNA RP. In the presence of SARS-CoV-2 target RNA, Cas12a collaterally cleaved the biotinylated ssDNA RP, leading to a reduction in binding of polystreptavidin-HRP and thus, a reduction in the precipitation of TMB deposited locally on the surface of the electrode43. Reduced precipitation of TMB was recorded as peak current, which was measured using cyclic voltammetry (CV) by sweeping the voltage between −0.5 and 0.5 V (Fig. 2b). As a result, the signal obtained from the EC platform demonstrated an inversely proportional relationship with target concentrations. It should be noted that the LAMP amplification before the CRISPR-based sample detection increases the sensitivity of the sensor, allowing for a consistently distinguishable signal from samples that are at or above the LOD of the sensor (Fig. 2c).
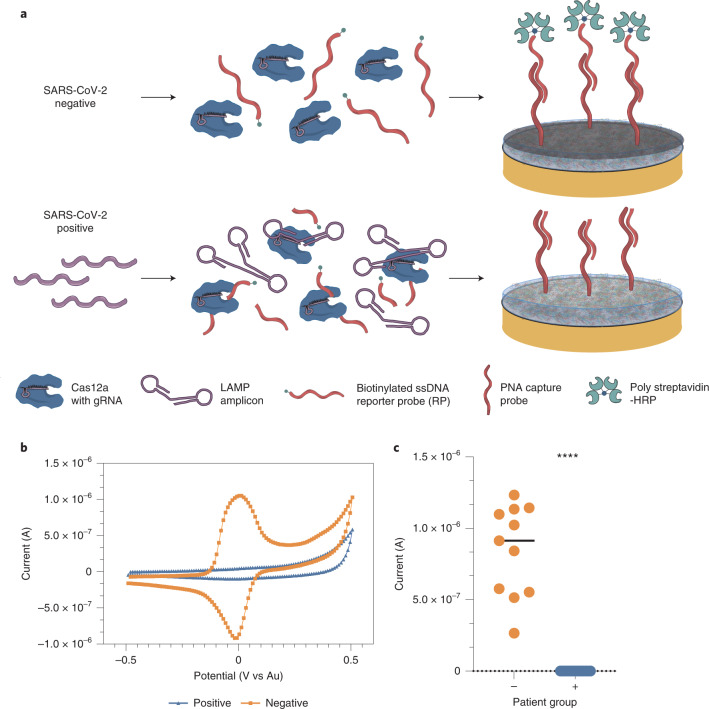
Fig. 2. Schematic of the CRISPR electrochemical assays and assay performance using clinical samples.
a, Schematic illustrating the surface chemistry of the EC assay. Without viral RNA (top row), the biotinylated ssDNA RP probe is not cleaved; therefore, the polystreptavidin-HRP binds to the PNA/biotin-DNA duplex when added to the EC sensor chip and consequently precipitates TMB, resulting in an increase in current. In contrast, the biotinylated RP ssDNA is hydrolysed in the presence of viral target RNA (bottom row), cleaving the biotin group. Consequently, polystreptavidin-HRP does not bind to the surface of the chips, resulting in no TMB precipitation and no increase in current. b, Cyclic voltammogram showing the typical current peak signal achieved after incubation of samples from both SARS-CoV-2 negative (orange) and positive (blue) clinical samples. c, Clinical samples that contained SARS-CoV-2 viral RNA (+, blue circles, 19 samples) had low signals in our device and were clearly distinguishable from the high signals obtained for samples that did not contain viral RNA (−, orange circles and black line representing the median value, 11 samples). Student’s t-test ****P < 0.001.
To optimize the binding efficiency of the PNA-based CRISPR-EC sensor platform, we varied the concentration and incubation time of the RP to obtain a rapid, high signal-to-noise ratio (Supplementary Figs. 10 and 11). Among all the concentrations tested, 1 nM RP and 5 min incubation produced a high signal with no background. Interestingly, the CRISPR-EC sensor platform gave a single molecule LOD of 0.8 copies per µl, which was nearly four times more sensitive than the initial fluorescence-based assays used to validate the primer and guide pairs (Supplementary Table 1 and Figs. 7 and 12). To determine the potential clinical value of the optimized CRISPR-EC sensor platform, we extracted RNA from 19 patient saliva samples that were positive for SARS-CoV-2 based on RT–qPCR with a range of CT values and 11 RT–qPCR negative clinical saliva samples (Supplementary Fig. 8). The current measured in the form of the output signal from the electrodes was clearly distinguishable (P < 0.0001) when comparing the SARS-CoV-2 positive and negative samples (Fig. 2c). In addition, receiver operating characteristic (ROC) curve analysis demonstrated an impressive correlation with RT–qPCR and CRISPR-based fluorescent detection, with 100% accuracy and areas under the curve (AUC) of 1 (Table 1).
Table 1.
ROC curve analysis for electrochemical assays
| Viral RNA | Serology (IgG) | ||||
|---|---|---|---|---|---|
| Electrode | ORF1a RNA | S1 | S1-RBD | N | Combined |
| Area under the curve | 1.00 | 0.98 | 1.00 | 0.98 | 1.00 |
| 95% CI | (1.00, 1.00) | (0.95, 1.00) | (1.00, 1.00) | (0.95, 1.00) | (1.00, 1.00) |
| P value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Cut-off | 8,859 | >1.9 × 10–7 | >1.7 × 10–7 | >3.1 × 10–8 | >2.4 × 10–7 |
| Sensitivity, 95% CI | 1.00, (0.83, 1.00) | 0.95, (0.83, 0.97) | 1.00, (0.94, 1.00) | 0.95, (0.86, 0.98) | 1.00, (0.94, 1.00) |
| Specificity, 95% CI | 1.00, (0.74, 1.00) | 1.00, (0.93, 1.00) | 1.00, (0.93, 1.00) | 1.00, (0.93, 1.00) | 1.00, (0.93, 1.00) |
| Number of positive samples | 19 | 54 | 54 | 54 | 54 |
| Number of negative samples | 11 | 58 | 58 | 58 | 58 |
Summary of the numerical values of the ROC curve analysis of the patient sample data collected for the SARS-CoV-2 biomarker assays for detection of (left to right) ORF1a RNA, S1 subunit, S1-RBD, N and a combined assay. Positive and negative SARS-CoV-2 samples were confirmed by RT–qPCR. CI, confidence interval.
Multiplexed serology electrochemical assay
Multiplexed assays that diagnose disease by combining viral RNA with serology markers can lead to a more robust understanding of the progression of diseases, including SARS-CoV-244. The primary antigens that elicit antibodies during coronavirus infection are the N and S proteins10. The N protein is the most abundant viral protein and is highly conserved among the coronavirus family45, with several studies indicating that IgG antibodies targeting the N protein may be more sensitive due to their early appearance and longer clearance rates during and post infection7,11,12. While the S protein is less conserved than the N protein, it is highly immunogenic due to the S1 subunit that mediates attachment to target cells46. Studies indicate that IgG antibodies against S1 are more specific and strongly correlate with virus neutralization47,48. Antibodies targeting S1 and the ribosomal binding domain within this subunit (S1-RBD) may also be used to assess the effectiveness of vaccination responses and titre maintenance over time, with SARS-CoV-2 vaccine efficacy trials highlighting a direct correlation between the titre of antibodies targeting the RBD of the S protein, the neutralizing antibody titre and vaccine efficacy13. Therefore, to maximize our assay’s accuracy, we leveraged the multiplexing capabilities of our EC sensor and fabricated a multiplexed serology assay capable of measuring antibodies against the S1, S1-RBD and N proteins for detection of a patient’s immunity, whether through previous viral infection or vaccination.
We began by using an enzyme-linked immunosorbent assay (ELISA) to optimize the reagents before assembling the multiplexed serology EC sensor chip (Supplementary Figs. 13–17). Initially, a small set of high-titre and low-titre positive clinical plasma samples were used to optimize the assay over a broad range of IgG titres. The best-performing capture antigens were S1, S1-RBD and N, and the best-performing detection antibodies were biotinylated goat anti-human IgG, rabbit anti-human IgM and goat anti-human IgA. We validated the ELISA accuracy with 58 SARS-CoV-2 plasma samples from patients with a previous positive SARS-CoV-2 RT–qPCR result and 54 SARS-CoV-2 negative samples. Out of the 54 negative SARS-CoV-2 samples, 22 were collected before the onset of the SARS-CoV-2 pandemic. The ROC curve analysis of the ELISA results yielded AUC values between 0.68 and 0.89 for IgG, IgM and IgA (Supplementary Figs. 15–17).
Next, we used the optimized reagents to develop a multiplexed EC sensor to measure the humoral response against SARS-CoV-2 from patient plasma samples. We fabricated an EC sensor with an antifouling coating composed of bovine serum albumin and reduced graphene crosslinked with glutaraldehyde (BSA/rGOx/GA)43,49,50, where each electrode was individually functionalized with S1 (electrode 1), S1-RBD (electrode 2), N (electrode 3), or BSA as an on-chip negative control (electrode 4) to perform a multi-antigen sandwich EC ELISA (Fig. 3a). We used an affinity-based sandwich strategy for the EC sensor assay, meaning that when SARS-CoV-2 antibodies were present, they bound to both the surface antigen and secondary antibody, leading to an increase in the EC signal. Each immunoglobulin isotype (IgG, IgM and IgA) was detected individually on the multiplexed EC sensor chips. Figure 3b–e show typical CV results obtained with the multiplexed EC sensor chips for detection of anti-SARS-CoV-2 IgG using SARS-CoV-2 positive and negative clinical samples.
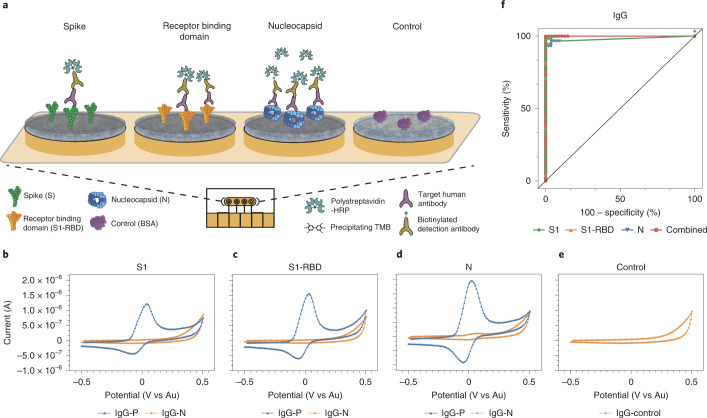
Fig. 3. Schematic and representative raw cyclic voltammetry data of the multiplexed serology assay.
a, Schematic illustrating the multiplexed EC serological assay to assess host antibody responses on electrodes functionalized with SARS-CoV-2 antigens. Host antibodies bind to the SARS-CoV-2 antigens immobilized on the chips. Subsequently, biotinylated anti-human IgG secondary antibodies bind, followed by polystreptavidin-HRP binding and TMB precipitation on the chips. b–e, Typical cyclic voltammograms for the four different electrodes that target host antibodies against S1 subunit (b), S1-RBD (c), N (d), and BSA negative control (e). f, ROC curves generated from the patient sample data obtained for the IgG EC serology assay.
We optimized the assay conditions of the serology EC assays to obtain the highest signal-to-noise ratios on both high- and low-titre clinical samples by varying factors such as plasma dilutions, sample incubation times and TMB precipitation times (Supplementary Figs. 18–20). We found that a 30 min sample incubation at a 1:9 plasma dilution with a 3 min TMB precipitation time resulted in the highest sensitivity and specificity for detection of SARS-CoV-2 antibodies in clinical plasma samples (Supplementary Fig. 19c).
We then evaluated the accuracy of the EC serology platform using plasma samples from patients with a previous SARS-CoV-2 RT–qPCR positive result. We performed an ROC curve analysis utilizing the 58 SARS-CoV-2 positive plasma samples and 54 SARS-CoV-2 negative samples used to optimize the ELISA assay (Fig. 3f and Supplementary Fig. 21a). The clinical samples had a wide range of IgG levels, which fall within the IgG detection sensitivity range of our platform (Supplementary Figs. 22 and 23), and there was excellent correlation between IgG levels detected and the clinical status of the patients. Overall, the AUC for anti-SARS-CoV-2 IgG (Table 1) was higher than that for IgM (Supplementary Fig. 21b), whereas IgA’s AUC was lowest (between 0.57 and 0.78) and did not add diagnostic value (Supplementary Fig. 21b). The specificity of each individual sensor modified with either S1, N or RBD was over 95% for IgG. Further analysis of the multiplexed assay’s ROC curves showed that S1-RBD was the most accurate capture probe (AUC = 1 for IgG), followed by N and S1 (Table 1 and Supplementary Fig. 21a).
In addition, we found that combining the results of the three antigens as a multiplexed readout resulted in a slight increase in accuracy when assessing a patient’s immune status (AUC = 1 for IgG) (Fig. 3f and Table 1, combined). The combined EC IgG assay had an excellent correlation with previous SARS-CoV-2 infection and was shown to be 100% accurate (100% sensitivity and specificity); we therefore used this for subsequent experiments on the multiplexed EC sensor chips. We also found the EC sensor-based serology platform to be more accurate in detecting samples from patients with previous SARS-CoV-2 infection than the ELISA platform. The high specificity of the EC sensor platform may be attributed to the low non-specific binding on our nanocomposite-coated EC electrodes43,49,50.
A multiplexed viral RNA and antibody diagnostic in a 3D-printed LOC platform
We next worked to combine both EC sensors to create a multiplexed assay for simultaneous viral RNA and serological biomarker detection on-chip to facilitate increased sensitivity and specificity of SARS-CoV-2 detection44. Saliva is an excellent source of both viral RNA as well as host antibodies (IgG, IgM and IgA) in SARS-CoV-2 patients51, and hence is an ideal sample for a multiplexed assay for viral RNA and serology. Unfortunately, the patient saliva samples used for this study had to be heat-inactivated before use as saliva from SARS-CoV-2 infected patients are of a highly contagious nature. Because high heat denatures the antibodies1, we spiked a National Institute for Biological Standards and Control (NIBSC) SARS-CoV-2 IgG calibrant into heat-inactivated saliva samples at 1:20 dilution to simulate the ratio of IgG present in saliva. After confirming that the signal outputs from the spiked saliva samples were consistent with the signals generated from the plasma samples (Supplementary Fig. 24), we modified the EC sensor chip so that the four electrodes could be used individually to detect the three antigens (S1, N and S1-RBD) and PNA (Fig. 4a).
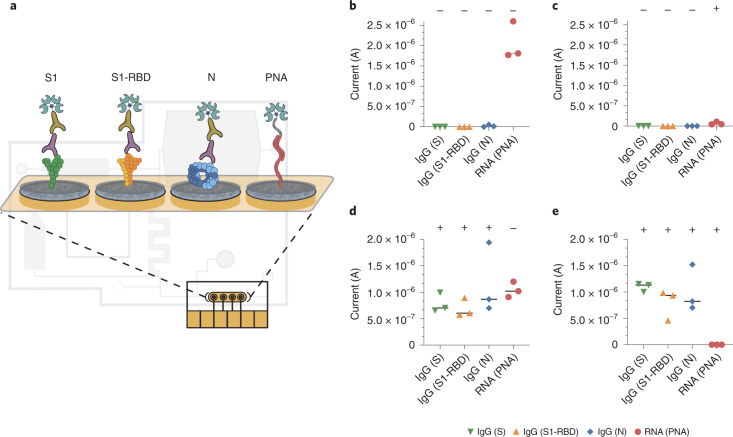
Fig. 4. Electrochemical platforms can be used for multiplexed sensing with simultaneous detection of SARS-CoV-2 viral RNA and host antibodies against the virus.
a, Schematic of the multiplexed chip surface conjugated with SARS-CoV-2 antigens: S1, S1-RBD and N, as well as PNA for the detection of SARS-CoV-2 viral RNA on the LOC microfluidic system. b–e, Current (A) EC readout from the LOC microfluidic chip for a triplicate of clinical samples containing different host antibody and viral RNA combinations: b, Clinical samples negative for both serology and viral RNA. c, Clinical samples with negative host antibody levels but are positive for viral RNA. d, Clinical samples that contain host antibodies against SARS-CoV-2 but are negative for viral RNA. e, Clinical samples positive for both host antibodies and viral RNA. Clinical RNA samples showed clear signal difference between positive and negative clinical samples (Student’s t-test, P = 0.0002) and the clinical IgG showed clear signal difference for all three antigen tests (Student’s t-test for S1 and S1-RBD, P < 0.0001 and for N, P = 0.0004, datapoints represent independent replicates, black lines represent the median value).
To study the performance of our multiplexed EC sensor, we first conducted a two-step assay where antibody-spiked saliva was split into two volumes: 15 µl was incubated on the chip for multiplexed serological detection of the host’s anti-SARS-CoV-2 antibodies and 280 µl was used for RNA extraction followed by the LAMP-CRISPR-based assay and incubation on the same chip. Following incubation of both samples on the electrodes, we simultaneously measured the SARS-CoV-2 viral RNA and host antibodies on-chip with the EC sensor readout of precipitating TMB following the addition of polystreptavidin-HRP. We validated the assay performance by testing the four possible combinations of serology and RNA-positive and RNA-negative clinical samples (Supplementary Fig 25). We achieved a very high probability (Student’s t-test P < 0.0001) of distinguishing positive from negative samples with an ultra-low background signal in all the combinations tested. Taken together, these results showed excellent multiplexing capacity for SARS-CoV-2 viral RNA and host antibodies on the chips, with 100% correlation in specificity and sensitivity.
Following these validation experiments, we integrated the multiplexed EC sensor chip into the compact LOC microfluidic platform (Fig. 1) to test the capabilities for automated viral RNA preparation before the EC sensor readout. Using the same functionalized chip assay as described above, each clinical saliva sample was split between the RNA and antibody reservoirs and the assay was performed as described in Fig. 1a and Supplementary Fig. 5. Similar to the initial validation of the multiplexed assay, the assay performance of the LOC microfluidic system with integrated EC sensor chip was validated by testing the four possible combinations of serology and RNA-positive and RNA-negative clinical samples (Fig. 4b–e). Clinical IgG negative samples showed no EC signal for the N, S1 and S1-RBD antigen-conjugated electrodes (Fig. 4b,c), whereas clinical samples from patients exposed to SARS-CoV-2 showed high IgG loads in all three antigen test areas (Student’s t-test for S1 and S1-RBD, P < 0.0001 and for N, P = 0.0004) (Fig. 4d,e). Moreover, we measured high signals on the PNA-conjugated electrode for all clinical samples that were negative for SARS-CoV-2 viral RNA (Fig. 4b,d) and low or no currents on the PNA electrode for SARS-CoV-2 RT–qPCR RNA-positive samples (Student’s t-test, P = 0.0002) (Fig. 4c,e). Taken together, these results show that the LOC system can effectively prepare and deliver unprocessed saliva samples containing both SARS-CoV-2 viral RNA and host antibodies for on-chip simultaneous multiplexed detection with 100% correlation in specificity and sensitivity. A cost analysis of the LOC multiplexed diagnostic is provided in Supplementary Table 4.
Discussion
Molecular diagnostics for detection of pathogen RNA and serological assays for assessment of host antibody responses are complementary tools that provide critical information to respond to disease outbreaks, assess vaccination status, and manage patient care and risks. While detection of SARS-CoV-2 RNA can be an important indicator of viral shedding during the infectious phase of the disease, factors such as sample types, virus variants and infection severity52–57 can impact diagnostic efficacy during the course of an infection and complicate patient management. Including serological assays can improve understanding of a patient’s clinical status following viral infection and can also be used to assess a patient’s response to vaccination13. For example, currently available SARS-CoV-2 vaccines induce antibody production against SARS-CoV-2 S and S1-RBD proteins, and vaccinated individuals who have not been infected with SARS-CoV-2 are expected to develop measurable antibodies against the S1, but not the N protein58. Therefore, multiplexed serology assays that target antibodies against several viral antigens can become key for seroprevalence studies to estimate the proportion of people in a population that have been infected, including those with asymptomatic infection, and/or those immunized with vaccines. This information is key for estimating herd immunity and vaccine efficacy13, which are critical in deciding to reopen economies59,60.
In the present study, we described a sample-in-answer-out diagnostic platform that integrates ultrasensitive and highly specific multiplexed EC sensors within an LOC microfluidic chip to rapidly and simultaneously detect both clinically relevant quantities of SARS-CoV-2 viral RNA and antibodies within patient saliva samples. This platform incorporates multiple innovations including: (1) engineering of a high-specificity and high-sensitivity multiplexed EC sensor chip that enables detection of both proteins and nucleic acids with clinically relevant samples of biological fluids perfused through a single microfluidic channel; (2) development of a CRISPR-based detection assay optimized to function in a multiplexed EC assay in parallel with antibody-based detection assays, the optimized assay being amenable to POC applications; (3) development of methods for automated microfluidic extraction and amplification of RNA from raw patient saliva samples; and (4) fluidic integration of the sample preparation process with the EC sensor chip.
While other CRISPR-based diagnostics (for example, for SARS-CoV-2) have been described61–63, they are typically limited to fluorescence and lateral flow readouts. Similarly, while there have been CRISPR-based EC sensors published22–26, none have been sensitive enough to detect clinically relevant quantities of target nucleic acid. A key difference between our work and that of other recently published POC CRISPR-based electrochemical detection platforms is our use of polystreptavidin-HRP/TMB-based reaction chemistry for readout, which enables further amplification of the EC signal for both the serological and CRISPR-based RNA sensors. Some EC-based sensors, such as the graphene field effect transistor-based EC biosensors for SARS-CoV-2, have displayed similar levels of sensitivity to the 0.8 copies μl−1 LOD shown here64; however, those assays were only validated for viral transport media-based nasopharyngeal swab samples and more importantly, do not have multiplexing capabilities.
In this study, we also designed a customized molecular assay for nucleic acid detection that combines isothermal nucleic acid amplification with CRISPR-based enzymatic detection to target SARS-CoV-2 viral RNA in saliva. This was possible through the development and optimization of RNA-dependent Cas12a cleavage of a biotinylated ssDNA RP, which interacts with the chip-based polystreptavidin-HRP/TMB-based reaction chemistry that allows for target detection more sensitively than fluorescence-based CRISPR-Cas12a assays, but with a similar time to result. The customizability of the EC chip’s BSA/rGOx/GA-based antifouling surface chemistry allowed for a novel multiplexed sensing system that generates high conductivity and low non-specific binding signals across a variety of biomolecular targets, aided by the sensitivity of the polystreptavidin-HRP/TMB reaction chemistry. Through optimization of reaction conditions, our EC assay has ultra-low background as well as improved sensitivity necessary for clinically relevant diagnostics that detect both nucleic acid and antibody targets37,49.
We validated the EC sensor for serology and obtained an accuracy of 100% (100% sensitivity and 100% specificity) for IgG as compared with traditional ELISA, with S1-RBD showing the highest accuracy in detecting IgG in clinical samples. This increased specificity for S1-RBD may be explained by the RBD domain being a highly immunogenic epitope for development of neutralizing antibodies to SARS-CoV-2 during the humoral response47. In addition, we characterized a multiplexed EC sensor that detects antibodies against relevant viral structural proteins (S1-RBD, S1 and N) for a variety of antibody isotypes (IgG, IgM and IgA), and enables a more robust understanding of the humoral response in patients. Importantly, we also found that simultaneous multiplexed detection of different viral antigens led to increased diagnostic sensitivity.
Finally, we designed and tested a microfluidic LOC platform that integrates with the multiplexed EC sensor for simultaneous detection of both RNA and IgG in clinical saliva samples. Saliva is an excellent alternative to nasopharyngeal swabs and nasal swabs for SARS-CoV-2 diagnosis, as it is simple to collect, does not require extensive collection equipment, and has been shown to provide both nucleic acid and serological data during and post infection or vaccination1,30,65. The microfluidic chip eliminates the need for RNA extraction kits by automating raw saliva sample preparation, RNA amplification and CRISPR-based RNA detection steps. With its compact and sealed design, our LOC platform limits user steps to avoid possible sources of contamination or human-introduced error to allow for device use by untrained end-users, further increasing its potential as a POC testing system. With our integrated device, we are able to perform both serological and RNA detection from a saliva sample without requiring specialized collection reagents or equipment, and with results reported concurrently. To our knowledge, there has so far been no report of an EC diagnostic device that is multiplexed, highly sensitive and capable of processing raw biological samples such as saliva.
We tested our microfluidic LOC platform on four categories of SARS-CoV-2 clinical saliva samples: RNA negative and antibody negative; RNA positive and antibody positive; RNA negative and antibody positive; and RNA positive and antibody negative. Clear differentiation was seen between IgG negative and positive samples for N, S1 and S1-RBD antigen-conjugated electrodes (Student’s t-test for S1 and S1-RBD, P < 0.0001 and for N, P = 0.0004) as well as between samples positive and negative for SARS-CoV-2 viral RNA (Student’s t-test, P = 0.0002) within 2 h after inputting the unprocessed saliva sample, highlighting the utility of our platform.
Limitations to our study include the small set of clinical COVID-19 saliva samples available due to the difficulty in acquiring saliva through biorepositories that do not routinely collect this sample type. However, the fact that we were able to obtain 100% accuracy with clinical samples that contained a wide range of viral loads strongly suggests that our CRISPR-based EC sensor platform could become a faster, simpler and cheaper non-invasive strategy compared with RT–qPCR and traditional fluorescent diagnostics. Additionally, to ensure that our multiplexed system was sensitive for clinically relevant levels of the biomarkers in question, we devised a system that would provide simultaneous qualitative results to generate a more robust and accurate estimation of a patient’s infectious and immune status. Future work can be directed towards designing a multiplexed system capable of providing semi-quantitative or quantitative data on biomarkers of interest.
While our LOC platform is promising, there are bottlenecks that need to be addressed before this technology can be readily adopted for clinical POC settings. Currently, our system uses peristaltic pumping for fluid movement and a potentiostat for the readout of the EC sensor chips. Further integration of the electronics, peristaltic pumping and potentiostat-based readout would allow for a robust reusable system that could make this multiplexed diagnostic tool useful for healthcare and clinical POC settings. Similarly, the 3D-printed microfluidic chips can be made at scale by transitioning to injection moulding techniques, with the LAMP and the CRISPR detection reagents pre-measured, lyophilised and sealed within the cartridge for streamlined assay use.
As the COVID-19 pandemic has shown, there is a critical need to rapidly adapt current testing strategies to more quickly and easily monitor both the infection and immune status of patients. Knowledge of infection stages can help curb disease spread, while insights on antibody titre levels can help with understanding how novel variants may affect individuals with immune protection through infection, vaccination or a combination of both. The streamlined workflow and multiplexing capabilities of our EC sensor represent important steps towards building the infrastructure necessary to provide this information to clinicians and members of the public alike.
Methods
Chip preparation
Gold chips were custom manufactured by Telic Company using a standard photolithography process with deposition of 15 nm of chromium and 100 nm of gold on a glass wafer. The area of electrodes was controlled by depositing a layer of 2 µm of insulating layer (SU-8). Before use, gold chips were cleaned by 5 min sonication in acetone (Sigma Aldrich, 650501) followed by isopropanol (Sigma Aldrich, W292907). To ensure a clean surface, the chips were then treated with oxygen plasma using a Zepto Diener plasma cleaner (Diener Electronics) at 0.5 mbar and 50% power for 2 min.
Nanocomposite preparation and activation
Nanocomposite coating was prepared using the previously described method43. Briefly, amine-functional reduced graphene oxide (Sigma Aldrich, 805432) was dissolved in 5 mg ml−1 BSA (Sigma Aldrich, 05470) in 10 mM PBS solution at pH 7.4 (Sigma Aldrich, D8537), and ultrasonicated for 1 h using 1 s on/off cycles at 50% power. The solution was then denatured by heating at 105 °C for 5 min and centrifuged to remove the excess aggregates. The nanomaterial solution was then crosslinked by mixing with 70% glutaraldehyde (Sigma Aldrich, G7776) at a ratio of 69:1, deposited on the glass chip with the gold electrodes and incubated in a humidity chamber for 20–24 h to form a conductive nanocomposite50. After nanocomposite deposition, gold chips were washed in PBS by agitation (500 r.p.m.) for 10 min and dried with pressurized air. 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC, ThermoFisher, 22980) and N-Hydroxysuccinimide (NHS, Sigma Aldrich, 130672) were dissolved in 50 mM MES buffer (pH 6.2) at 400 mM and 200 mM, respectively, and deposited on nanocomposite-covered gold chips for 30 min. After surface activation, chips were quickly rinsed with ultra-pure water and dried, and the capture probes were spotted on top of the working electrode area.
Clinical samples and ethics statement
De-identified clinical saliva samples from the Dominican Republic were obtained from Boca Biolistics under their ethical approvals. Because the saliva samples were collected from actively infectious patients, saliva samples were heat-inactivated at 95 °C for 10 min to inactivate SARS-CoV-2 virions present in the samples, which denatured the antibodies present within the samples. RT–qPCR was performed by Boca Biolistics using the Perkin Elmer New Coronavirus nucleic acid detection kit. De-identified clinical plasma samples were obtained from the Crimson Biomaterials Collection Core Facility at Partners Healthcare (currently Mass General Brigham). Additional de-identified clinical plasma and saliva samples were obtained through the Massachusetts Consortium on Pathogen Readiness (MassCPR); these samples had been collected by Prof. Jonathan Li and Prof. Xu Yu. Additional pre-SARS-CoV-2 pandemic samples were obtained from the Walt Laboratory at Brigham and Women’s Hospital. The Institutional Review Boards at the MGH, MGB and Harvard University as well as the Harvard Committee on Microbiological Safety approved the use of the clinical samples in this study. All clinical plasma samples were collected from convalescent patients and were inactivated by heating at 65 °C for 30 min before use to denature any potential SARS-CoV-2 virions that might be present in the samples; this lower temperature denaturation method allows us to retain antigenicity of antibodies present within the samples.
CRISPR-based RNA assay with fluorescent reporter
CRISPR-based assays require the selection of both LAMP isothermal amplification primers and gRNAs to detect the LAMP amplicons. LAMP amplification primers (Supplementary Table 3) were selected after testing a range of LAMP primers, including some from the literature37–41. Cas12a-gRNAs consist of two parts: the handle region (UAAUUUCUACUAAGUGUAGAU) that the Cas protein recognizes and binds, and a user-defined region at the 3’ end of the handle that determines the specificity to the target. Spacer regions were selected following established guidelines20. To synthesize the gRNA (gRNA sequence: UAA UUU CUA CUA AGU GUA GAU GGU GAA ACA UUU GTC ACG CA), synthetic DNA with an upstream T7 promoter sequence (5′ GAAATTAATACGACTCACTATAGGG 3′) was purchased from Integrated DNA Technologies (IDT) and in vitro transcribed using the HiScribe T7 High Yield RNA synthesis kit from New England Biolabs (NEB). Reactions were incubated for 16 h at 37 °C, treated with DNase I (NEB) and purified using the RNA Clean & Concentrator-25 kit (ZymoResearch). gRNA was quantified (ng μl−1) on a Nanodrop 2000 (ThermoFisher).
Simulated SARS-CoV-2 samples were prepared by serially diluting full-length SARS-CoV-2 viral RNA (Twist Biosciences, MT106054.1) in nuclease-free water. Viral RNA extracted from saliva samples was used after purification via the QiAmp viral RNA extraction kit, as explained above. RNA was then amplified by LAMP and further detected by collateral cleavage of the fluorophore-quenched ssDNA RP. Briefly, 5 µl of the diluted genomic DNA, or clinical sample RNA extract was added to 2.5 µl of the 10X primer mix (Supplementary Table 3), 12.5 µl of the LAMP master mix (NEB) and 5 µl water. LAMP mixtures were incubated for 30 min at 65 °C. After LAMP amplification, 4 µl of the amplified LAMP mixture were mixed with 11 µl of nuclease-free water and 5 µl of the CRISPR mixture, which contained 1 µM ssDNA fluorophore-quencher reporter (sequence: 6-FAM/TTATT/IABkFQ), 100 nM Cas and 200 nM gRNA in 10X NEB 2.1 buffer. Reactions were incubated at 37 °C for 20 min and fluorescence kinetics were measured using a BioTek NEO HTS plate reader (BioTek Instruments), with readings every 2 min (excitation 485 nm; emission 528 nm).
Chip functionalization for the CRISPR-based RNA assay with electrochemical reporter
For CRISPR sensors, custom-synthesized amine-terminated peptide nucleic acid (AEEA-ACAACAACAACAACA) where AEEA is an O-linker was obtained from PNAbio. PNA is a synthetic analogue of DNA with a backbone utilizing repeating units of N-(2-aminoethyl) glycine linked through amide bonds. PNA contains the same four nucleotide bases as DNA (adenine, cytosine, guanine and thymine) but are connected through methylene bridges and a carbonyl group to the central amine of a peptide backbone66. Stock PNA was diluted to 20 µM in 50 mM MES buffer and spotted on the working electrode. One electrode was spotted with 1 mg ml−1 BSA as a negative control. The spotted chips were incubated overnight in a humidity chamber. After conjugation, chips were washed and quenched in 1 M ethanolamine dissolved in 10 mM PBS at pH 7.4 for 30 min and blocked with 1% BSA in 10 mM PBS containing 0.05% Tween 20.
CRISPR-based RNA assay with electrochemical reporter
The RP sequence for CRISPR-based EC assays was a ssDNA (sequence: /5Biosg/AT TAT TAT TAT TAT TTG TTG TTG TTG TTG T) conjugated to a biotin linked to polystreptavidin-HRP. Upon Cas12a activation, the ssDNA-biotin RP was cleaved in solution, thus preventing binding to the complementary PNA sequence on the surface. Polystreptavidin-HRP was then added and able to bind to the ssDNA reporter-biotin. The concentration of HRP bound to the electrode was read by HRP-dependent oxidation of precipitating TMB (TMB enhanced one component, Sigma Aldrich, T9455). TMB precipitation forms an insulating, non-soluble layer on the electrode surface. Full-length genomic RNA (Twist Biosciences, MT106054.1) was serially diluted and amplified with 2X LAMP master mix (NEB) for 30 min at 65 °C. Viral RNA was extracted from saliva via purification with the QiAmp viral RNA extraction kit. Similar to the protocol explained above, 5 µl of the viral RNA was added to 2.5 µl of the 10X primer mix (Supplementary Table 3), 12.5 µl of the LAMP master mix (NEB) and 5 µl water. LAMP mixtures were incubated for 30 min at 65 °C. After LAMP amplification, 4 µl of the amplified LAMP product was mixed with 10 µl nuclease-free water and 5 µl CRISPR mix, which contained 4 nM reporter, 100 nM Cas and 200 nM gRNA in 10X NEB 2.1 buffer. Mixtures were incubated for 20 min at 37 °C, during which time the ssDNA biotinylated reporter was cleaved. After that, 15 µl of the LAMP/reporter/Cas mixtures was deposited on the chips for 5 min. Thereafter, the chips were washed and incubated with polystreptavidin-HRP and TMB for 5 min and 1 min, respectively. Final measurement was then performed in PBST using a potentiostat (Autolab PGSTAT128N, Metrohm; VSP, Bio-Logic) by a CV scan with 1 V s−1 scan rate between −0.5 and 0.5 V vs on-chip integrated gold quasi reference electrode. Peak oxidation current was calculated using Nova 1.11 software. Cyclic voltammetry allowed us to measure attomolar concentrations of SARS-CoV-2 target RNA.
Serology ELISA assay
ELISA assays were optimized in a 96-well plate format. A volume of 100 μl of 1 μg ml−1 antigens S1 (SinoBiological, 40591-V08H), N (RayBiotech, 130−10760) and S1-RBD (The Native Antigen Company, REC31849) were prepared in a 10 mM PBS buffer at pH 7.4, added to Nunc MaxiSorp ELISA plates (BioLegend, 423501) and immobilized on the plates by overnight incubation at 4 °C. The plates were washed three times with 200 μl PBST, followed by the addition of 250 μl 5% Blotto for 1 h. After washing the plates, 100 μl of the clinical plasma samples diluted in 2.5% Blotto was added and incubated for 1 h at r.t. Plates were further washed and HRP-conjugated anti-human IgA/IgM or biotin-conjugated anti-human IgG detection antibodies were added for 1 h. The secondary antibodies used were: HRP-conjugated anti-human IgM (human IgM mu chain rabbit antibody, 109-4107) or IgA (AffiniPure goat anti-human serum IgA, α-chain specific; Jackson ImmunoResearch, 109-005-011) or biotin-anti-human IgG (AffiniPure goat anti-rabbit IgG, Fc-fragment specific; Jackson ImmunoResearch, 111-005-008). The IgG plate was further mixed with 100 μl streptavidin-HRP (1:200 dilution in 2.5% Blotto) and washed. Turbo TMB (100 μl; ThermoFisher, 34022) was added for 20 min, followed by the addition of 100 μl sulfuric acid (0.1 M H2SO4 in water) to stop the reaction. The absorbance of the plates was immediately read using a microplate reader (BioTek NEO HTS plate reader, BioTek Instruments) at 450 nm.
Electrochemical serology assay
To translate the ELISA assays to EC readouts, S1, N and S1-RBD were diluted to 1 mg ml−1 in the PBS buffer and spotted in three electrodes of the EC chip. An additional electrode was spotted with 1 mg ml−1 BSA as a negative control. The spotted chips were incubated overnight in a humidity chamber. After conjugation, chips were washed and quenched in 1 M ethanolamine (Sigma Aldrich, E9508) dissolved in 10 mM PBS at pH 7.4 for 30 min and blocked with 5% Blotto (Santa Cruz Biotechnology, sc-2324) in 10 mM PBS containing 0.05% Tween 20 (Sigma Aldrich, P9416). The fabricated sensor was then used to detect immunoglobulins from clinical samples. Each sensor was either used to detect IgG, IgM or IgA against the three antigens that were immobilized on the chips. Clinical plasma samples (1.5 μl) were mixed with 13.5 μl 2.5% Blotto and incubated on the chips for 30 min at r.t., followed by a rinsing step. Subsequently, HRP-conjugated anti-human IgM/IgA/biotin-anti-human IgG was added for 30 min at r.t. Polystreptavidin-HRP (1 μg ml−1; ThermoFisher, N200) diluted in 0.1% BSA in PBST was added to chips with IgG for 5 min. The chips were rinsed and precipitating TMB was added for 3 min, followed by a final rinse and EC measurement using a potentiostat by cyclic voltammograms with a scan rate of 1 V s−1 between −0.5 and 0.5 V vs on-chip integrated gold quasi reference electrode. Additional antibodies were screened, including: F(ab’)2 goat anti-human IgG Fc-fragment antibody biotinylated (Bethyl Laboratories, A80-148B), goat anti-human IgG Fc secondary antibody, biotin (ThermoFisher, A18821), purified anti-human IgG Fc antibody (BioLegend, 409302), purified anti-human IgG Fc antibody (BioLegend, 410701) and AffiniPure F(ab’)2 fragment goat anti-human IgG, Fcγ fragment-specific biotin (Jackson ImmunoResearch, 109-006-170).
Multiplexed electrochemical serology and CRISPR-based RNA detection
Multiplexed sensors for both nucleic acid and host antibody detection were prepared by spotting three electrodes of the EC chip with proteins S1, N and S1-RBD, and spotting the amine-terminated peptide nucleic acid (AEEA-ACAACAACAACAACA) reporter on the fourth electrode. The spotted chips were incubated overnight in a humidity chamber. After conjugation, chips were washed and quenched in 1 M ethanolamine (Sigma Aldrich, E9508) dissolved in 10 mM PBS at pH 7.4 for 30 min and blocked with 1% BSA in 10 mM PBS containing 0.05% Tween 20 (Sigma Aldrich, P9416). The fabricated sensor was then used to detect IgGs as well as viral RNA from clinical samples.
Multiplexed chips were used to detect viral RNA as well as IgG against the three antigens that were immobilized on the EC sensor chips. Negative control saliva (RT–qPCR negative) was heat-inactivated and spiked with plasma at a ratio of 1:20 to simulate IgG concentrations in saliva. Two experiments were done in parallel on each chip, as follows: (1) 15 µl of plasma-spiked saliva was used for the serology assays as explained above. Briefly, 0.75 µl of the plasma sample was mixed with 14.25 µl of control saliva and incubated on the chips for 30 min at r.t., followed by a rinsing step. Subsequently, biotin-anti-human IgG was added for 30 min at r.t. Chips were then rinsed. (2) In parallel, RNA extracted from RT–qPCR positive and negative clinical samples was amplified by LAMP for 30 min at 65 °C as explained above. Then, 4 µl of the amplified LAMP product was mixed with the CRISPR mix, which contained the biotinylated RP, and incubated for 20 min at 37 °C. A volume of 15 µl of the LAMP/reporter/Cas mixtures was deposited on the chips after the chips had been exposed to SARS-CoV-2 IgG. Thereafter, the chips were washed and incubated with polystreptavidin-HRP and TMB for 1 min. Final measurement was then performed in PBST using a potentiostat (Autolab PGSTAT128N, Metrohm; VSP, Bio-Logic) by a CV scan with 1 V s−1 scan rate between −0.5 and 0.5 V vs on-chip integrated gold quasi reference electrode. Peak oxidation current was calculated using Nova 1.11 software. Cyclic voltammetry allowed us to measure both the presence of IgG antibodies as well as attomolar concentrations of SARS-CoV-2 target RNA.
Microfluidic chip for multiplexed electrochemical serology and CRISPR-based RNA detection
A microfluidic chip for a point-of-care diagnostic that integrated the multiplexed chip was designed using Autocad software and printed on a Formlabs Form 3B 3D SLA printer in grey resin (Version 4; Formlabs RS-F2-GPGR-04). The layer thickness chosen for the 3D printing parameters was 50 µm. The chip was cleaned in an isopropanol bath for 20 min, then cured in a heated UV chamber for 1 h at 60 °C (Form wash and Form cure, Formlabs). The chip was designed with chambers for saliva, LAMP reagents and CRISPR reagents, as well as a reaction chamber lined with a PES membrane and a waste port. The PES membrane (Millipore, GPWP04700) was laser cut using Epilog Fusion edge 24 laser cutter to fit within the serpentine reaction chamber. The chambers were designed to be connected to the reaction chamber through channels that are closed when under vacuum. Additionally, the chip had a channel and waste port that allow for the integration of the EC sensor chip. The chip was sealed using a clear delayed tack adhesive tape (3M 9795R) to prevent evaporation. Two high-power resistors were used as heating elements for the sample preparation chamber and reaction chamber (Digikey, 355018RJT) and placed in a custom-printed backing to ensure that accurate and repeatable heating was provided. A full list of parts along with cost analysis can be found in Supplementary Table 4. The voltages used for each temperature are provided in Supplementary Fig. 5 and were generated using an Agilent E3631A DC power supply. A complete view of the microfluidic chip can be seen in Fig. 1 and Supplementary Fig. 5. A small DC pump (Takasago, RP-Q series) connected to an Arduino Uno was used for the pumping. The workflow for the multiplexed EC assay on the microfluidic chip is as follows: (1) The microfluidic chip was sealed with clear delayed tack adhesive tape. (2) Saliva from patient samples (BioIVT) was inactivated at 95 °C for 5 min and mixed with 10% (by volume) proteinase K (NEB P8107S) that was diluted 1:10 in nuclease-free water. (3) 450 µl of the saliva mixture was added into the sample preparation chamber and the chamber was heated to 55 °C for 15 min, followed by 95 °C for 5 min. (4) The sample was pumped over the PES membrane within the reaction chamber at a speed of 50–100 µl min−1. (5) The reaction chamber was heated to 95 °C for 3 min. (6) 25 ul of LAMP reaction (12.5 µl of NEB 2X WarmStart Mastermix, 10 µl nuclease-free water and 2.5 µl of 10x LAMP primer mix) was pumped into the reaction chamber and incubated at 65 °C for 30 min. (7) The heat was decreased to 37 °C and 22 µl of the Cas mix (15 µl nuclease-free water and 7 µl CRISPR mix, which contained 4 nM RP, 100 nM Cas and 200 nM gRNA in 10X NEB 2.1 buffer) was pumped into the reaction chamber and incubated for 30 min. (8) The LAMP/CRISPR reaction was pumped over the EC sensor chip at 4 µl min−1 for 5 min, followed by 20 µl PBST at 15 µl min−1. (9) 20 µl of saliva sample spiked with NIBSC antibody calibrant mixed with anti-IgG detection antibody linked with biotin was flowed through the EC sensor at 4 µl min−1 for 5 min. (10) The EC sensor chip was washed with 20 µl PBST at 15 µl min−1, followed by addition of 20 µl polystreptavidin-HRP at 6 µl min−1. (11) The EC sensor chip was washed with 20 µl PBST at 15 µl min−1 and 20 µl TMB at 8 µl min−1. (12) The EC sensor chip was washed with 30 µl PBST at 15 µl min−1. (13) The EC sensor chip was read on a potentiostat and the cyclic voltammogram was generated.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary figures and tables.
Acknowledgements
We thank T. G. Hitron, R. A. Lee and N. Weckman for helpful discussions and advice. This work was supported by the Wyss Institute for Biologically Inspired Engineering at Harvard University and the Paul G. Allen Frontiers Group. J.R. was funded through the UK Natural Environment Research Council (NERC) GW4 FRESH CDT. H.d.P. was supported by the Harvard University Center for AIDS Research (CFAR), an NIH-funded programme (P30 AI060354), which is supported by the following NIH co-funding and participating Institutes and Centers: NIAID, NCI, NICHD, NIDCR, NHLBI, NIDA, NIMH, NIA, NIDDK, NINR, NIMHD, FIC and OAR. M.Y. acknowledges Fonds de recherche du Québec nature et technologie (FRQNT) postdoctoral fellowship no. 260284. The MGH/MassCPR COVID biorepository was supported by a gift from E. Schwartz, by the Mark and Lisa Schwartz Foundation, the Massachusetts Consortium for Pathogen Readiness, and the Ragon Institute of MGH, MIT and Harvard.
Source data
Source data for Figs. 2–4.
Author contributions
D.N., J.R., S.S.T., P.J., H.d.P., M.Y., N.D. and H.S. conceived the study under the guidance of J.A.P., P.E., J.J.C. and D.E.I. Experiments were performed and validated by D.N., J.R., S.S.T., P.J. and H.d.P. H.d.P. and P.J. formulated the idea, organized the experiments and managed the project. M.Y., N.D., D.R.W., G.A., J.Z.L. and X.G.Y. contributed to the collection and characterization of clinical serum and saliva samples. All authors contributed to manuscript preparation and editing.
Peer review
Peer review information
Nature Biomedical Engineering thanks Dipanjan Pan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
All data needed to evaluate the findings can be found in the paper and its Supplementary Information. The raw and analysed datasets generated during the study are available from the corresponding authors on request. Source data for Figs. 2–4 are provided with this paper.
Competing interests
H.d.P., P.J., J.J.C. and D.E.I. are inventors on patents describing the CRISPR-EC sensing technology. P.J. and D.E.I. are listed as inventors on patents describing the EC sensor platform. The EC sensor platform technology has been licensed to Antisoma Therapeutics Inc. for infectious disease and cancer diagnostics and to StataDX Inc. for cardiovascular, neurological and kidney disease diagnostics. P.J. and D.E.I. hold equity in StataDx and D.E.I. is a board member. J.J.C. and D.R.W. are co-founders and directors of Sherlock Biosciences. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Devora Najjar, Joshua Rainbow, Sanjay Sharma Timilsina, Pawan Jolly, Helena de Puig.
Contributor Information
James J. Collins, Email: jimjc@mit.edu
Donald E. Ingber, Email: don.ingber@wyss.harvard.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41551-022-00919-w.
References
- 1.Isho B, et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Science. Immunology. 2020;5:eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfel R, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 3.Pan Y, Zhang DT, Yang P, Poon LLM, Wang QY. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou LR, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, et al. Humoral immune response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaebler C, et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Elslande J, et al. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin. Microbiol. Infect. 2020;26:1557.e1–1557.e7. doi: 10.1016/j.cmi.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dispinseri S, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat. Commun. 2021;12:2670. doi: 10.1038/s41467-021-22958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsson AC, et al. Comparison of six commercially available SARS-CoV-2 antibody assays - choice of assay depends on intended use. Int. J. Infect. Dis. 2021;103:381–388. doi: 10.1016/j.ijid.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenwick C, et al. Changes in SARS-CoV-2 spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies. J. Virol. 2021;95:e01828–20. doi: 10.1128/JVI.01828-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manisty C, et al. Time series analysis and mechanistic modelling of heterogeneity and sero-reversion in antibody responses to mild SARS-CoV-2 infection. EBioMedicine. 2021;65:103259. doi: 10.1016/j.ebiom.2021.103259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer B, Drosten C, Muller MA. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert PB, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mak GCK, et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JS, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gootenberg JS, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gootenberg JS, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee RA, et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl Acad. Sci. USA. 2020;117:25722–25731. doi: 10.1073/pnas.2010196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaminski MM, et al. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat. Biomed. Eng. 2020;4:601–609. doi: 10.1038/s41551-020-0546-5. [DOI] [PubMed] [Google Scholar]
- 20.Gayet RV, et al. Creating CRISPR-responsive smart materials for diagnostics and programmable cargo release. Nat. Protoc. 2020;15:3030–3063. doi: 10.1038/s41596-020-0367-8. [DOI] [PubMed] [Google Scholar]
- 21.English MA, et al. Programmable CRISPR-responsive smart materials. Science. 2019;365:780–785. doi: 10.1126/science.aaw5122. [DOI] [PubMed] [Google Scholar]
- 22.Zamani M, et al. Electrochemical strategy for low-cost viral detection. ACS Cent. Sci. 2021;7:963–972. doi: 10.1021/acscentsci.1c00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, et al. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens. Bioelectron. 2021;179:113073. doi: 10.1016/j.bios.2021.113073. [DOI] [PubMed] [Google Scholar]
- 24.Hajian R, et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Jin T, Dai YF, Liu CC. Surpassing the detection limit and accuracy of the electrochemical DNA sensor through the application of CRISPR Cas systems. Biosens. Bioelectron. 2020;155:112100. doi: 10.1016/j.bios.2020.112100. [DOI] [PubMed] [Google Scholar]
- 26.Bruch R, et al. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 2019;31:1905311. doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 27.de Puig H, Bosch I, Collins JJ, Gehrke L. Point-of-care devices to detect Zika and other emerging viruses. Annu. Rev. Biomed. Eng. 2020;22:371–386. doi: 10.1146/annurev-bioeng-060418-052240. [DOI] [PubMed] [Google Scholar]
- 28.Liu R, et al. Analysis of adjunctive serological detection to nucleic acid test for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection diagnosis. Int. Immunopharmacol. 2020;86:106746. doi: 10.1016/j.intimp.2020.106746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torrente-Rodriguez RM, et al. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyllie AL, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott, I. M. et al. Simply saliva: stability of SARS-CoV-2 detection negates the need for expensive collection devices. Preprint at medRxiv10.1101/2020.08.03.20165233 (2020).
- 32.Lalli MA, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. 2021;67:415–424. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linnes JC, Rodriguez NM, Liu L, Klapperich CM. Polyethersulfone improves isothermal nucleic acid amplification compared to current paper-based diagnostics. Biomed. Microdevices. 2016;18:1–12. doi: 10.1007/s10544-016-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlappi TS, McCalla SE, Schoepp NG, Ismagilov RF. Flow-through capture and in situ amplification can enable rapid detection of a few single molecules of nucleic acids from several milliliters of solution. Anal. Chem. 2016;88:7647–7653. doi: 10.1021/acs.analchem.6b01485. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez NM, et al. Paper-based RNA extraction, in situ isothermal amplification, and lateral flow detection for low-cost, rapid diagnosis of influenza A (H1N1) from clinical specimens. Anal. Chem. 2015;87:7872–7879. doi: 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Puig H, et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 2021;7:eabh2944. doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl Acad. Sci. USA. 2020;117:24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu RF, et al. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol. Sin. 2020;35:499. doi: 10.1007/s12250-020-00223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohaim MA, et al. Artificial intelligence-assisted loop mediated isothermal amplification (AI-LAMP) for rapid detection of SARS-CoV-2. Viruses. 2020;12:972. doi: 10.3390/v12090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nawattanapaiboon K, et al. Colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) as a visual diagnostic platform for the detection of the emerging coronavirus SARS-CoV-2. Analyst. 2021;146:471–477. doi: 10.1039/D0AN01775B. [DOI] [PubMed] [Google Scholar]
- 41.Yan C, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel - Manufacturer Instructions/Package Insert (FDA, accessed 15 January 2021); https://www.fda.gov/media/134922/download
- 43.Zupancic U, Jolly P, Estrela P, Moschou D, Ingber DE. Graphene enabled low-noise surface chemistry for multiplexed sepsis biomarker detection in whole blood. Adv. Funct. Mater. 2021;31:2010638. doi: 10.1002/adfm.202010638. [DOI] [Google Scholar]
- 44.Ter-Ovanesyan D, et al. Ultrasensitive measurement of both SARS-CoV-2 RNA and antibodies from saliva. Anal. Chem. 2021;93:5365–5370. doi: 10.1021/acs.analchem.1c00515. [DOI] [PubMed] [Google Scholar]
- 45.Satarker S, Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51:482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Premkumar L, et al. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amanat F, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timilsina, S. S. et al. Rapid antifouling nanocomposite coating enables highly sensitive multiplexed electrochemical detection of myocardial infarction and concussion markers. Preprint at medRxiv10.1101/2021.06.13.21258856 (2021).
- 50.del Rio JS, Henry OYF, Jolly P, Ingber DE. An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 2019;14:1143–1149. doi: 10.1038/s41565-019-0566-z. [DOI] [PubMed] [Google Scholar]
- 51.Sterlin D, et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zohar T, et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yurkovetskiy L, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calistri P, et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int. J. Infect. Dis. 2021;105:753–755. doi: 10.1016/j.ijid.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fajnzylber J, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Kampen JJA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golubchik, T. et al. Early analysis of a potential link between viral load and the N501Y mutation in the SARS-COV-2 spike protein. Preprint at medRxiv10.1101/2021.01.12.20249080 (2021).
- 58.Dörschug A, et al. Comparative assessment of sera from individuals after S-gene RNA-based SARS-CoV-2 vaccination with spike-protein-based and nucleocapsid-based serological assays. Diagnostics. 2021;11:426. doi: 10.3390/diagnostics11030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ndaye AN, Hoxha A, Madinga J. Challenges in interpreting SARS-CoV-2 serological results in African countries. Lancet Glob. Health. 2021;9:E597. doi: 10.1016/S2214-109X(21)00060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaffer DeRoo S, Pudalov NJ, Fu LY. Planning for a COVID-19 vaccination program. JAMA. 2020;323:2458–2459. doi: 10.1001/jama.2020.8711. [DOI] [PubMed] [Google Scholar]
- 61.Broughton JP, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rauch JN, et al. A scalable, easy-to-deploy protocol for Cas13-based detection of SARS-CoV-2 genetic material. J. Clin. Microbiol. 2021;59:e02402–e02420. doi: 10.1128/JCM.02402-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arizti-Sanz J, et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020;11:5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang LQ, et al. Rapid and ultrasensitive electromechanical detection of ions, biomolecules and SARS-CoV-2 RNA in unamplified samples. Nat. Biomed. Eng. 2022;6:276–285. doi: 10.1038/s41551-021-00833-7. [DOI] [PubMed] [Google Scholar]
- 65.Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J. Clin. Microbiol. 2021;59:e02881–02820. doi: 10.1128/JCM.02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jolly P, Rainbow J, Regoutz A, Estrela P, Moschou D. A PNA-based lab-on-PCB diagnostic platform for rapid and high sensitivity DNA quantification. Biosens. Bioelectron. 2019;123:244–250. doi: 10.1016/j.bios.2018.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.
Data Availability Statement
All data needed to evaluate the findings can be found in the paper and its Supplementary Information. The raw and analysed datasets generated during the study are available from the corresponding authors on request. Source data for Figs. 2–4 are provided with this paper.