Abstract
Proteomics plays a vital role in biomedical research in the post-genomic era. With the technological revolution and emerging computational and statistic models, proteomic methodology has evolved rapidly in the past decade and shed light on solving complicated biomedical problems. Here, we summarize scientific research and clinical practice of existing and emerging high-throughput proteomics approaches, including mass spectrometry, protein pathway array, next-generation tissue microarrays, single-cell proteomics, single-molecule proteomics, Luminex, Simoa and Olink Proteomics. We also discuss important computational methods and statistical algorithms that can maximize the mining of proteomic data with clinical and/or other ‘omics data. Various principles and precautions are provided for better utilization of these tools. In summary, the advances in high-throughput proteomics will not only help better understand the molecular mechanisms of pathogenesis, but also to identify the signature signaling networks of specific diseases. Thus, modern proteomics have a range of potential applications in basic research, prognostic oncology, precision medicine, and drug discovery.
Proteomics plays a vital role in biomedical research in the post-genomic era. With the technological revolution and emerging computational tools, proteomic methodology has evolved rapidly in the past decade and shed light on solving complicated biomedical problems. Thus, this mini-review summarizes existing and emerging high-throughput proteomics methodologies, including mass spectrometry, protein pathway array, next-generation tissue microarrays, single-cell proteomics, single-molecule proteomics, Luminex, Simoa and OLINK Proteomics.
Introduction
Since the successful completion of the Human Genome Project that mapped the whole human genomes, a massive number of genomic markers have been identified and are being applied to medical sciences1. Many of them have been developed as routine tests in the clinic. However, a significant limitation of genomic or transcriptomic profiling studies is that genomic and transcriptomic data, which only provide indirect measurements of cellular states, may not accurately reflect the corresponding protein changes. These data fail to reveal changes in posttranslational modifications (PTMs), including phosphorylation and protein degradation. Therefore, the genomic data alone cannot bring a full picture of the disease mechanisms with comprehensive understanding2. Nowadays, the Human Proteome Project (https://hupo.org/) has been launched to characterize the entire human proteome by advanced proteomic techniques, which is the next major challenge3. The guidelines on interpreting proteomic data have also been published and recently updated4.
Proteomics, as the combination of proteome experimentation and data analysis, analyzes protein composition, structure, expression, modification status, and the interactions and connections between proteins at an overall level5. It offers complementary information to genomics and transcriptomics. It is also essential for generating a map of the complex, interconnected pathways, networks, and molecular systems, which directly control the major life activities such as cell proliferation, differentiation, senescence, and apoptosis. With the substantial improvement of experimental technology over the past decade6, the proteomics methods have been evolved from conventional methods, such as immunohistochemistry (IHC) staining, western blot, and enzyme-linked immunosorbent assay (ELISA), to high-throughput methods such as tissue microarray (TMA), protein pathway array and mass spectrometry7. Those high-throughput proteomics techniques not only decrease analysis time but also increase the accuracy and depth of proteome coverage. With the advents of bioinformatics and modern multi-analytes “omics” technologies (Supplementary Fig. 1), proteomics holds a great promise for uncovering the molecular mechanisms that underlies diseases towards the discovery of novel biomarkers8 and can be used as specific diagnostic assays, prognostic predictors, and therapeutic targets to enhance personalized medicine further9,10.
In this review, we will discuss the advances in high-throughput proteomic techniques, statistics and algorithms, progress in applying proteomics to disease diagnostics, current challenges and future perspectives.
High-throughput proteomic techniques
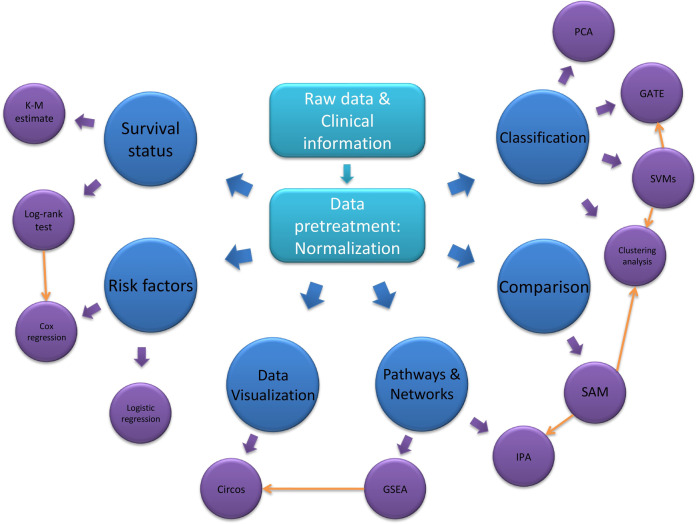
With the rapid development of high-throughput technology6, several new technologies are widely used in proteomics and metabolomics in recent years. Regardless the specific technique, these global proteomic approaches (Fig. 1) can be divided into three phases, namely discovery, network-analysis and clinical proteomics. Discovery is the initial phase to identify the amino acid sequence and unknown protein structure with qualification11. We then in network-analysis phase build the global signaling networks and investigate the relations among the known proteins to explore the potential biomarkers with verification. Finally, in the clinical proteomics phase12, we develop clinical assays related to the productization of the biomarker or panel fitting the clinical flow. The commonly used high-throughput proteomic techniques include mass spectrometry, protein pathway array, next generation tissue microarrays and Luminex and will be discussed in details below.
Fig. 1. The process of proteomics “from bench to bedside”.
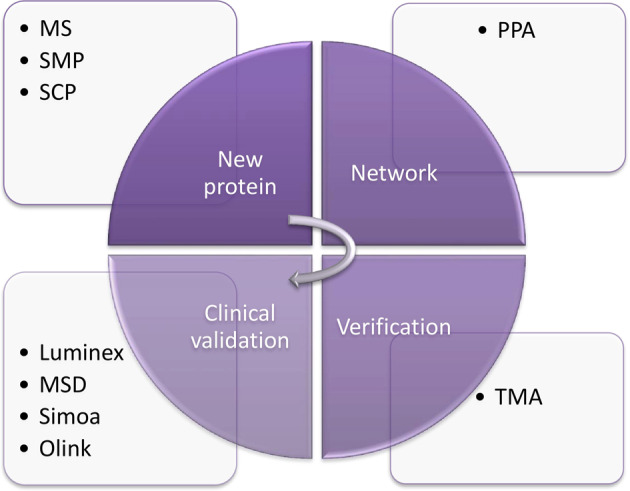
The mass spectrometry (MS)-based methods, single-molecule proteomics (SMP) and single-cell proteomics (SCP) have been widely used to identify and quantify new proteins in the initial discovery stage. Protein pathway array (PPA) is a high-throughput technique to explore the regulation of protein-protein interactions, pathway-pathway interactions, and biological functions to find the position of newly discovered protein in the cell signaling networks. Luminex, Meso-scale Discovery (MSD), Simoa and Olink are effective high-throughput methods for clinical validation after the proteomic markers are verified using tissue microarray (TMA).
Mass spectrometry
Mass spectrometry (MS) has been developed as one of the most essential and popular tools to identify proteins and their isoforms, and quantify posttranslational modifications, either via the fragments directly or the specific proteolytic activity responsible for their formation13–15. The most significant effect of MS is to discover and detect an intact protein or a subset of composite or surrogate peptides as MS-based quantitative proteomics that traditional immunoassays find incredibly challenging or impossible. MS can be combined with multiple separations and pre-fractionation techniques to identify the target protein/peptide and improve identification accuracies and yields16. For example, two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) is based on electrical charge and molecular weight, while liquid chromatography (LC) based on polarity, electrical charge, and protein molecular weight. For an example of 2D-PAGE, mixtures of proteins are separated by the electrical charge as isoelectric point (pI) in the first dimension and further separated by molecular weight in the second dimension on 2-D gels. The protein samples from different resources, which were labeled by different cyanine dyes such as Cy2b, Cy3, and Cy5 as reporter fluorophores, can be processed in the same 2D-PAGE to purify the target protein and enhance the detection accuracy17. After being digitalized 2D-PAGE by fluorescence scanner and the image analysis18, the interesting or significant spots in the gel are cut out and enzymatically digested to peptides for MS as matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) MS analysis where each digest yields a peptide mixture that can be analyzed by bottom-up experiment. Although 2D-PAGE has traditionally been used as a standard procedure for proteomics research, gel-based techniques tend to be labor-intensive and time-consuming, and are therefore not suitable for high-throughput proteomics. By contrast, LC or high-performance liquid chromatography (HPLC) allows continuous separation of thousands of proteins from complex mixtures and can be combined with MS as LC-MS for increased throughput19–21. Among them, Reversed-phase liquid chromatography (RPLC) is the most commonly used LC-based separation platform. It is characterized by the distribution of compounds between a water-containing mobile phase and a relatively nonselective stationary phase and other chromatography formats can be added prior to the RPLC separation to improve the dynamic range of measurement22.
According to different strategies of processing, MS-based methods can be divided into top-down, bottom-up, and shotgun approaches. In the top-down proteomics method, a full-length protein, which can be subsequently fragmented inside the MS and the masses of the fragments be recorded, is directly sent for MS analysis23. By contrast, proteins are enzymatically or chemically digested into peptides that serve as input to the MS equipment in bottom-up proteomics techniques. Moreover, shotgun proteomics is a particular case of bottom-up proteomics where the whole proteins in a complex mixture, such as serum, urine, and cell lysates, are cut into peptides and followed by multidimensional HPLC-MS, which aims to generate a global profile of protein mixtures as genome “shotgun” sequencing24. On the other hand, the separation of peptides prior to MS is not necessarily needed in the bottom-up strategy. Then MS data is matched to identify the target proteins and their associated modifications in the protein sequence database by data-dependent discovery engines25, which can be divided into peptide scoring, protein scoring, and finally protein inference26.
Protein pathway array
Human diseases, especially cancers, are often a complicated biomedical process attributable to complex protein-based signaling network pathway alterations that control cell behaviors, such as apoptosis, invasion, and metastasis27. Uncovering the underlying changes in multidimensional protein signaling networks not only aids in understanding the molecular mechanisms of pathogenesis but also identifies the characteristic signaling network signatures that are unique for the type or stage of the diseases28–32. Measuring many proteins simultaneously is of great importance to the theory of protein-protein interactions (PPIs) in the signaling network, which is a big challenge to conventional immunoassays such as western blot. Therefore, high-throughput proteomic tools were increasingly used for biomarker discovery in basic, translational and clinical research.
Protein pathway array (PPA)27, a gel-based high-throughput platform, employed antibody mixtures to detect antigens in a protein sample which can be extracted from biopsy or tissue. In this approach, microdissection of tumor tissue could be applied to maximize the proportion of the proteins from tumor tissue instead of the surrounding benign tissue27. Then, the immunofluorescence signals of antibody-antigen reactions are converted to numeric data as the value of protein expression by Quantity One (https://www.bio-rad.com/en-us/category/image-lab-software-suite?ID=5291f579-0715-48f4-b3de-766b92222582) from Bio-Rad. The biomarkers and proteomic networks can be explored and trained after data normalization and appropriate statistic modeling. PPA has been applied to many diseases such as essential thrombocythemia33 and papillary thyroid carcinoma34. Its high-throughput protein profiles in a robust quantitative manner provided an advantage over traditional methods.
Next generation tissue microarrays
Immunohistochemistry staining, as one of the traditional and reliable research methods, uses an enzyme-linked chromogenic substrate for detection and requires microscopic examination35. It remains a time-consuming and subjective process and produces a qualitative or semiquantitative assessment of protein expression because of the nonspecific stain or background noise. As technology advanced and high-throughput demand increased over the last decade, the TMA gradually began to be widely used in both research and clinical fields36,37. TMA contains many small representative tissue cores of formalin-fixed paraffin-embedded (FFPE) or frozen blocks from hundreds of different cases assembled in an array fashion on a single histologic slide, and therefore allows a large-scale antibody-based molecular analysis of multiple samples at the same time37. Therefore, it is a practical and valuable tool to confirm and verify new biomarkers generated from PPA or MS proteomics methods. Thus, it is often used in an independent cohort and identifies the location of the target proteins in the cell membrane, cytoplasm, or nucleus. Since digital pathology with multiple smart microscopes has been developed rapidly in the past year, a new approach of TMA, next-generation tissue microarrays (ngTMAs), was recently created35. It allows annotations to be placed directly on the digital slides for a higher accuracy. Two major advantages of ngTMAs are its time-efficiency and high throughput without major compromise on quality38. Due to its improved sensitivity and rapid, large-scale detection capabilities, ngTMAs has become a powerful tool to improve the quality of TMAs used in clinical and translational research39–41 but could be more widely used.
Multiplex bead- or aptamer-based assays
Proteomics plays a critical role in clinical practice, although there are gaps and limitations to translate proteomics from basic molecular research to clinical use. Multiplex bead- or aptamer-based assays have been developed42–45 but have various sensitivity and specificity. Therefore, caution and in-house validation studies must be used before the assay is applied to clinical samples.
Luminex bead-based array system is increasingly used in protein profiling applications in recent years46–50. It makes the detection of proteomic biomarker panel reliable, fast and able to cope with dynamic changes in the variety of clinical practices51. Luminex uses different, flexible fluorescent-labeled beads that are spectrally distinguishable and coated with a different capture antibody or probe to identify the antigen or mutation in samples. It is able to detect up to 500 analytes (FLEXMAP 3D Platform: https://www.luminexcorp.com/flexmap-3d/) in a single sample using a 96-well plate or 384-well plate. For proteomics usage, megaplex microspheres are tagged to allow fluorescent detection and can be used in the development of the multiplex immuno-assays by labeling multiple target antibodies. After microsphere activation and conjugation reactions, a panel of beads-antibody complexes is mixed and incubated with samples to capture protein analytes in the sample. Then the sandwich structure of bead-antibody-antigen complexes is passed and counted by a flow cytometer using different fluorescent of beads. Therefore, the high-throughput Luminex system has a great potential for fast multiplexed analysis of panels of genetic, proteomic, metabolic biomarkers associated with disease diagnosis, prognosis, and therapeutics in patients.
Another widely used platform is Meso-scale Discovery (MSD) assay (https://www.mesoscale.com/) which may be multiplex, single-plex or ultrasensitive. It has been used mostly for cytokine detection in the mice with type 1 diabetes or radiation treatments, and the astrocytes with neuronal networks52–55. It has also been compared with other platforms or detection assays. For human cytokine profiling, the MSD assay is more sensitive than Luminex assay but less specific45, while a recent study shows low or no significant correlations for detecting most of the cytokines (except interleukin 6) among Luminex xMAP®, MSD V-Plex® and Quantikine assays43. A study of 38 epileptic children shows a freeze-thaw cycle results in consistent measurements in 46% (6 of 13) of the analytes using Luminex high-sensitivity assay, 11% (1 of 9) using Luminex standard-sensitivity assay, and in no analytes using MSD assay42. Therefore, the Luminex high-sensitivity assay appears to have better precision than the other 2 assays for epilepsy research. For detecting plasma Alpha-Synuclein in Parkinson’s disease patients, MSD assay has a smaller effect size than Quanterix assay but correlated well with Biolegend56.
One of the widely used bead-based multiplex assays is Simoa® (Single Molecule Array, owned by Quanterix)57. It covers 6 disease areas, is customizable, and includes 109 oncology, 26 neurology, 19 immunology, 13 cardiology and 45 infectious disease assays as of May 2022. Their platform can be used to detect 6 to 10 biomarkers in a single test, and can detect as low as 1 fg/mL of proteins. As a highlight of its performance, Simoa® had the highest sensitivity and precision in a comparison of platforms’ performance in post-traumatic stress disorder and Parkinson’s disease,43 as well as the lowest variation and highest effect size in a 3-platform comparison on Parkinson’s disease56.
Antibodies are the primary detection tool in the bead-based assays but face challenges in high-throughput platforms. To meet the challenge, protein-binding reagents are produced such as slow off-rate aptamer58 and have been commercialized as the SOMAscan® assay. The aptamer-based SOMAscan® assay can assess expression of 1,000 to 9,000 antigens in a single test and has an impressive dynamic range (8 orders of magnitude), a great sensitivity (lower detection limit, 40 fM) and a high precision (median coefficients of variance = ~5%)59,60. In a study on embryonic stem cells, SOMAscan has a higher reproducibility, a higher sensitivity and a larger dynamic range than nano LC-MS/MS and RNA sequencing, but fewer features to detect61. The SOMAscan’s results are overall comparable to those of nano LC-MS/MS and RNA sequencing61. In a study on patients with end-stage renal disease, SOMAscan correlated very well with ELISA in detecting 2 of the 3 targeted proteins, but not in the last one62. However, compared with antibody-based Olink platform, SOMAscan® assay shows a wide range of correlation in assessing protein expression in 2 cohorts of chronic obstructive pulmonary disease and thus should be used with caution63. Moreover, despite the greater coverage and overall good correlation, SOMAscan did not reveal bigger odds ratios of the proteins linked to acute kidney injury than those revealed using one of the immunoassays including MSD (electro-chemiluminescence platform), Access (paramagnetic-chemiluminescence platform) and Unicel (chemiluminescence platform) and Biochip (multiplexed ELISA platform)60.
Proximity extension assay (Olink)
Proximity extension assay, as one of proximity-dependent ligation assays, is based on oligonucleotide-linked antibody pairs that have slight affinity to each other64–66. When these oligonucleotide-linked antibodies are brought in proximity, the two unique oligonucleotides linked to the antibodies will be extended by a DNA polymerase and amplified exponentially later65. Quantitative real-time PCR is often used to amplify and quantify the oligonucleotides in the sample. Thus, oligonucleotides can serve as a unique surrogate marker of specific antigens which the antibodies recognize. As described in its original reports64,67, the 5 specific assay steps include: 1. Oligonucleotide-linked antibody pairs are added into the sample; 2. The probe pairs bind to the antigen and subsequently the probe oligonucleotides are brought into close proximity; 3. The oligonucleotides form pair-wise binding of matching probe-pairs; 4. The matching probe-pairs are amplified using universal primers. The process is termed as pre-amplification due to the lack of specific primers; 4. The matching probe-pairs are digested using uracil-DNA glycosylase and unbound universal primers are removed; 5. The pre-amplified probe-pairs (DNA templates) are quantified using specific primers and quantitative real-time PCR. Multiplex and 96-muliplex detection methods have been developed and also show very high sensitivity and specificity64,67. The Olink assay has been applied to several clinical fields with great success, including coronavirus disease 2019, traumatic brain injury and renal diseases68–71. It can simultaneously quantify over 3,000 proteins in a miniscule amount of sample (e.g., a few microliters).
Nanopore based single-molecule proteomics
Nanopore has been increasingly used for DNA or RNA sequencing and tried on proteomics72–74. Its early application to proteomics was seen in sequencing peptides of mycobacterium75. Peptide-oligonucleotide conjugates and measurements with nanopore-induced phase-shift sequencing were used and seemed able to sequence short peptides76. Later, addition of helixase was found effective to reduce the reading error rate to 30 rereads per million77. It is also proposed to combine nanopore with other techniques such as fluorescence labeling and protein-fragmentation for better readouts76. The major challenges of nanopore based single-molecule proteomics are low efficiency and lack of sufficient sensitivity for detecting PTM76.
It is noteworthy that the comparisons of these platforms may not be representative of the whole menu of a given technology, and thus should be applicable only to the aforementioned disease-specific areas. For example, the performance of Simoa® on other diseases may not be as good as that on Parkinson’s disease. Thus, caution and in-lab comparison may be warranted.
Statistics and algorithms
Traditional statistics methods, such as Student’s t-test and one-way analysis of variance (ANOVA), have various biases and may be time-consuming to handle big data78. Therefore, new high-throughput approaches or machine learning-based algorithms (Fig. 2) are needed to process big data that are generated from multi-omics79. Machine learning can be divided into supervised learning and unsupervised learning approaches generally80. In terms of supervised learning, it applied a “labeled” training set to train a model and predict a qualitative or quantitative output, such as classification and regression. By contrast, unsupervised learning has an unlabeled output set and enables the algorithms to determine and identify the natural patterns with shared similarities in an unknown dataset, such as cluster analyses80,81. Artificial intelligence and digital pathology are involving rapidly and will play an even more important role in research, pathology and medicine81–83 while some traditional statistic tools remain important such as normalization and batch effect removal.
Fig. 2. The flow chart of data analysis.
Normalization is the most significant step after acquiring the raw data. Data can be analyzed according to specific study design and available clinical information and it can be based on the raw data after normalization or a result from other analyses. For example, the clustering analysis can be performed on raw data or the proteins that have significant changes after SAM.
Normalization
The most common and necessary form of big data pre-processing phase is normalization, which is being used to centralize and rescale all of the data as a whole numerical matrix to improve their numeric stability, overall performance and model fitting78,84. All machine learning-based statistic models, such as distance-based cluster analysis, regression, and principal component analysis, are susceptible to unscaled data distribution. For example, the most commonly used formula of normalization is Z-score, which is also called the standard score85. Z-scoring the data centers the raw data by subtracting the mean (average) of a group of values of expression of genes or proteins first to reduce the influence of an extreme outlier that could affect the mean of a dataset with a small number of samples and then divides each data variable by the standard deviation (SD) to scale the data variable. Furthermore, common housekeeping genes and proteins, including GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and beta-actin, whose expression is considered the same in all samples, can also be used to normalize array data as a house-keeping gene. Besides, another preferred treatment for the ratio data is to log the data but not to Z-score the data.
Significance analysis of microarrays
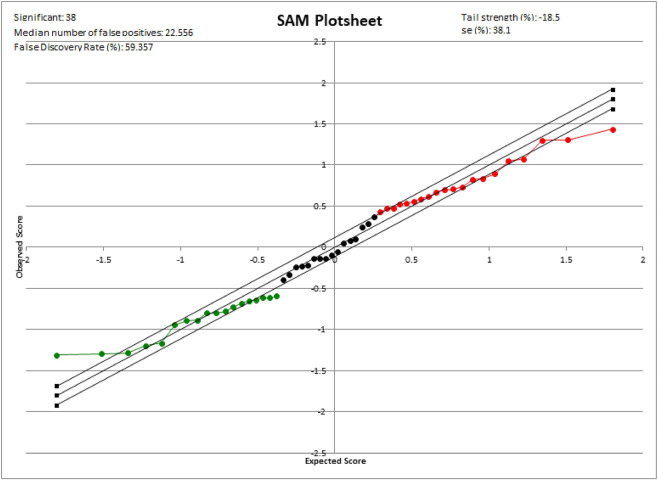
Significance Analysis of Microarrays (SAM) (https://statweb.stanford.edu/~tibs/SAM/) is a supervised learning program for large-scale gene or protein expression data mining developed by the Stanford University Statistics and Biochemistry Labs. SAM, a Microsoft Excel add-in package, is a widely used high-throughput permutation-based approach to identify differentially expressed proteins between sets of samples in abundance proteomics data using modified t-statistics (q-value) which measures the strength of the relationship between protein abundance and disease outcome86. Unlike the regular t-test for small sample size, SAM algorithm is an excellent fit for big data to minimize the number of false positives and negatives by permuting the columns of the protein abundance and automatic imputation of missing data via the nearest neighbor algorithm. Furthermore, one of the SAM’s valuable features is that it gives estimates of the False Discovery Rate using data permutations, which is the proportion of proteins likely to have been identified by chance as being significant (Fig. 3).
Fig. 3. Plotsheet generated by the significance analysis of microarrays: data are presented as a scatter plot of expected (x-axis) vs observed (y-axis) and the solid line indicates the relative difference expression of group.
Red color indicates upgrade and green color indicates downgrade. The data points that exceed a threshold from expected relative differences have significant different.
Clustering and discriminant analyses
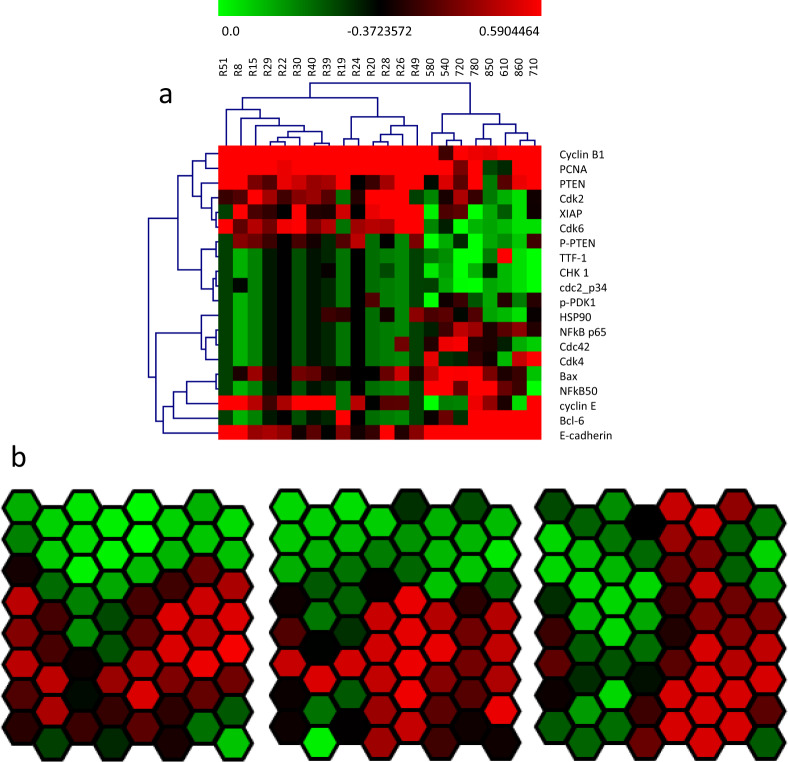
Hierarchical clustering analysis (HCA) has been used to cluster the big data by forming a mathematical model based dendrogram87,88. Several optimized mathematical formula-based models are created to measure the distance between data points, including Manhattan (L1) distance89, Euclidean (L2) distance90, Pearson correlation91 and others92. The Euclidean distance is the most commonly used but is vulnerable to outliers in non-normal distribution data especially, but might be inferior to Pearson correlation in analyzing proteomic data88. Manhattan distance requires the strict normalization. Pearson correlation is a scale-invariant of the similarity measure, etc88,91. It must be noted that the choice of distance measure impacts the performance of HCA88,92 and thus should be decided with caution. Moreover, there are different principles which can be calculated to measure the distance between clusters, such as average distance, minimum distance way and maximum distance ways. The average distance way uses the average of all data points in one cluster to map to the closet one of the other clusters93. Both distance measure and its calculation formula determine which samples and clusters are grouped together. Based on these 2 metrics, the model is optimized to keep the distance between the data points within one cluster as close as possible in the numerical matrix, but keep the distance between the data points in different clusters as far as possible. Besides, clustering results are also affected by both input data and selected variables, such as feature distributions and biomarkers. For example, the clustering results will be significantly different if samples and biomarkers are added, deleted, and/or replaced. Therefore, essential variables (biomarkers), sample selection criteria and study goals should be clearly defined prior to a HCA for robust and reproducible analysis. In addition, HCA can be divided into one way and two-way HCAs. Two-way HCA indicates that the data is clustered using the X-axis (samples) and Y-axis (biomarkers) at the same time (Fig. 4a) comparing with one way, which means either axis clusters the data according to study design.
Fig. 4. Examples of hierarchical clustering analysis (HCA) and grid analysis of time-series expression (GATE).
a The heatmap of a two-way hierarchical clustering analysis was performed by the Multi Experiment Viewer (MeV) (http://mev.tm4.org/). The color in each square represents a numerical value and the bar is on the top. All samples (x-axis) were clustered into three groups, while all protein markers were clustered into four groups. b Protein markers were cluster by multiple data points using GATE. The multiple data points can be divided by time, dosages, or stages of the disease.
Additionally, there is a particular clustering analysis, called Grid Analysis of Time-series Expression (GATE) (Fig. 4b), to analyze and visualize high-dimensional biomolecular according to time series94. GATE, as an integrated computational software platform, uses a correlation-based clustering algorithm to arrange time series or continuous-time points on a two-dimensional hexagonal array. It dynamically colors individual hexagons according to the expression level of genes or proteins to create animated movies of systems-level molecular regulatory dynamics. Furthermore, GATE allows interactive interrogation of movies against a wide variety of knowledge datasets, such as Protein interaction hubs, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Kinase enrichment analysis (KEA), WikiPathways pathways, etc, to infer potential regulatory control mechanisms from patterns of correlation. Those dynamic protein-protein interactions and clustering are able to allow investigating the continuous changes of cell lines or animals at different time points and dosages of treatment, the snapshots of the disease progression, and the different stages of cancer.
In contrast to clustering analysis that classifies known samples, (predictive) discriminant analysis classifies unknown samples based on what the algorithm learned and built in the training set95. For example, support vector machines (SVMs), which are not data-type dependent, can be applied to linearly separate the numerical or categorical data, and to identify the potential biomarkers as classifiers96. All samples need first to be divided into two groups as the training and validation sets. Then SVMs are trained in the training set for the most optimized algorithm, will be tested it later in the validation set and will finally produce the prediction rate by comparing the predicted with true values. The results will be affected by both input data (samples) and selected variables (also known as features or factors). In light of two data sets, both the training and validation sets must include all of the types of patient samples to cover any related clinical situations, such as stages, grades, histologic classification and complication, to eliminate the false negative and positive in the clinical practice. For example, the SVM algorithm may not recognize any “new” cases that have not been included in the training set, even if it is as simple as common sense for researchers or physicians. Besides, the samples in the training set need strict rule-in and rule-out criteria as well as keep the samples as many and diverse as possible to achieve the most accurate classification. In addition, the validation set can be the same or part of the training dataset as internal validation for retrospective evaluation, which is an option for a small sample size or population. However, an external validation cohort is recommended for prospective evaluation and increases reproducibility, generalizability and scientific rigor of the study. Several issues and problems of discriminant analysis must be noted and avoided during the analysis such as predictive versus descriptive discriminant analyses and linear versus quadratic models97,98. In summary, the main aim of (machine learning-based or not) discriminant analysis is to devise a computationally effective statistic model to classify multiple groups of subjects and identify the potential classifiers with a higher prediction rate.
Kaplan-Meier (K-M) curve and survival analysis
The Kaplan-Meier (K-M) curve is a time-event statistic method to investigate the relationship between the endpoint event and period of time99. It can be used to evaluate survival time, disease recurrence, clinical trial, animal study, etc. For the survival analysis, data can be classified into two types, including complete data and censored data according to the endpoint event. Death and disease recurrence are the most commonly used endpoint events. Complete data is defined as the event occurrence during the experimental period. By contrast, censored data includes the subjects who were lost to follow up or experienced a non-qualified event before the end of the study. The time starting from a defined point (zero time point) to the occurrence of a given event needs to be measured as input data. The higher the censored data ratio is in the study, the less accurate the results generally are. The K-M estimate is the simplest way of computing survival over time. The steep survival curve indicates a low survival rate or shorter survival period. It indicates that there might be confounding factors or effect modification in the cohort which can be determined by stratified analysis and multivariate analysis, if the survival curves of each group cross and inferential analyses show statistical differences.
The two survival curves can be compared statistically by the rudimentary log-rank (Mann–Whitney U) test, which has been widely used, including Breslow and Tarone with different weight functions during computing100. But they, usually as univariable analysis, do not allow to test the effect of the other disease-related variables. By contrast, Cox proportional hazards regression model, which is often used as multivariable analysis, can test the effect of other variables while identifying the independent variables of disease100. For example, biomarkers can be analyzed alone with other risk factors such as age, gender, smoking history, and stage to determine whether it independently affects the prognosis. Therefore, an in-depth and comprehensive survival study of PPIs or microarray is to perform a log-rank test to identify the biomarkers that have statistically significant first and then analyze it with other risk factors together using the Cox regression model. The results of those double analyses can be classified into three categories: (1) Biomarkers have statistically significant in both of the log-rank test and the Cox regression model. It means those biomarkers affect the prognosis as independent factors. (2) Biomarkers have statistically significant only in the log-rank test but not in the Cox regression model. It means those biomarkers are correlated with risk factors as effect modification to impact the disease of interest and may have confounding factors (Cox regression model may reveal them). (3) If biomarkers have statistically significant only in the Cox regression model but not in the log-rank test, bias or study errors such as confounding bias need to be considered in the study. Moreover, the number of cases as complete data should be at least five to ten times greater than the number of variables as multiple secondary endpoints in the Cox regression model to avoid the type I error.
Besides, many popular regression models are being used to analyze the proteomics or microarray-based big data, and their functions are similar to the Cox regression model in varying degrees. For example, the multivariable logistic regression, which is a supervised classification algorithm, is used to model the relationship between a set of continuous, categorical, or dichotomous independent variables and a dichotomous outcome as a dependent variable without time variable55,80,101,102. Cox regression model incorporates time variable but is not able to process the missing values and censored data during a certain amount of time. The advantages of the logistic regression, in which the underlying concept is quite the same as linear regression, are assumed to be a linear association between the features and dependent variable (also known as outcome or label). However, it does not require that the variables normally distributed in the linear discriminant analysis. In addition, the dependent variable is quantitative in the multiple linear regression, rather than a binary outcome in logistic regression.
Principal component analysis
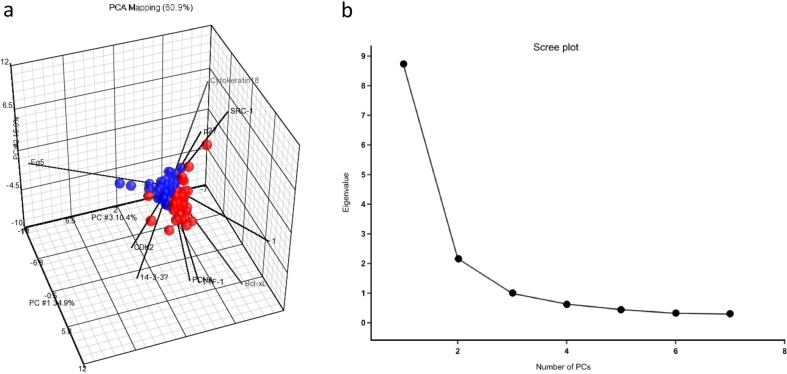
The main objective of principal component analysis (PCA) (Fig. 5a) is to decrease the dimensionality of the big data by creating a set of new variables, called principal components, to represent the majority of the information within the original dataset103. Those new principal components, which may be uncorrelated with each other, are reducing the complexity and noise of the original dataset while minimizing the loss of information. Technically, the number of new principal components can be equal to the number of variables in the original dataset. However, the contribution of new principal components, which represent the proportion of the original data, decrease sequentially (the first principal component accounts for most of the variability of the original dataset, the second subsequent component accounts for as much of the remaining variability as possible, … until the last component). Therefore, only the first few principal components are the most representative, and this trend of progressively decreasing variability of each principal component can be visualized as the scree plot (Fig. 5b). This statistical approach to lower the dimensional representations within a data set through principal components is useful for the classification and the compression of a large dataset or big data.
Fig. 5. Principal component analysis (PCA).
a The PCA mapping was performed using the Partek Genomics Suite (Partek, St. Louis, MO) (https://www.partek.com/partek-genomics-suite/). Patients with different survival status (red represents dead and blue represents alive) were separated by eight proteins. The first principal component is plotted on the X-axis and captures 34.9% of the variance. The second principal component is plotted on the Y-axis and achieves 15.6 % of the variance. b The scree plot represents the contribution of each principal component in PCA, and each principal component’s contribution decreases sequentially.
Ingenuity pathway analysis, gene-set enrichment analysis and circos
Many analytical methods combined with online databases to analyze proteomics and microarray data, and are more suitable for discovering clinical significance rather than in-depth statistical analysis. Three commonly used computational tools are described and may be useful for some studies.
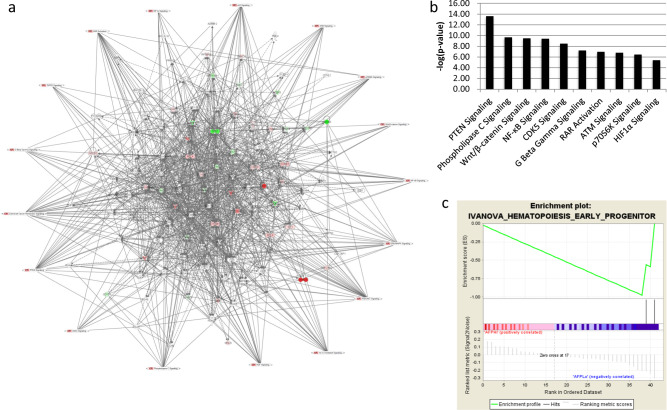
Ingenuity Pathway Analysis (IPA) was a web-based software application for causal analysis using expression datasets104. It is now owned by Qiagen with >109,000 expression datasets and 8.5 million findings (https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/). It can generate hypothetical molecular interactions to understand cellular processes based on knowledge databases such as Biomolecular Interaction Network Database (BIND) database, Biological General Repository for Interaction Datasets (BioGRID) database, Cognia database, DIP database (Database of Interacting Proteins), IntAct database, Molecular INTeraction database (MINT), Munich Information Center for Protein Sequences (MIPS) database, QIAGEN’s Ingenuity Knowledge Base, etc. Therefore, IPA can simultaneously visualize and analyze cross-database data of genomics, proteomics, and metabolomics data for signaling networks and canonical pathways from integrated various omics formats. The 2-dimensional signaling network offers a landscape survey of multi-omics (Fig. 6a) in which all upgraded and downgraded genes or proteins are visualized and connected or linked based on the latest database, and can be labeled by either function or pathway. The principle of ranking canonical pathways activity contains the research-based changes of each molecule such as the fold changes from PPA or microarray and the database-based the importance of each molecular in each canonical pathway, which is calculated with the Fisher’s exact test as the negative log of this p value (Fig. 6b).
Fig. 6. Examples of ingenuity pathway analysis (IPA) and gene-set enrichment analysis (GSEA).
a The signaling networks generated by database-based Ingenuity Pathway Analysis (IPA). The up- and downregulated proteins are represented by molecules in red and green color, respectively. The pathways were labeled outside of the network. b The top canonical pathways that were most significant to the dataset were identified by the IPA. The score assigned to each pathway was presented in –log (p value) using Fisher’s exact test. c The enrichment plot generated by the database-based gene set enrichment analysis (GSEA). The bar in the middle of the figure was labeled in red and blue from left to right, which means risk factor and protective factor separately. The enriched gene set is the IVANOVA_HEMATOPOIESIS_EARLY_PROGENITOR (https://www.gsea-msigdb.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_EARLY_PROGENITOR), which is a protective factor in this figure.
Gene set enrichment analysis (GSEA) (https://www.gsea-msigdb.org/gsea/index.jsp) is another computational method that provides pathway enrichment tools to help interpret datasets105. This approach focuses on cumulative changes in the expression of multiple genes as a gene set, which shares similar biological function, chromosomal location, or regulation, instead of an individual gene to identify pathways106. One similar web-based GSEA tool is Enrich for analyzing human and mouse data107 and modEnrich for analyzing fish, fly, worm and yeast data108. The most significant advantage of this GSEA method is that it can catch some pathways, in which several genes change in a small amount but in a coordinated way (Fig. 6c). The results reflect many of the complexities of co-regulation and modular expression by enrichment score (ES), corresponding to a weighted Kolmogorov–Smirnov-like statistic.
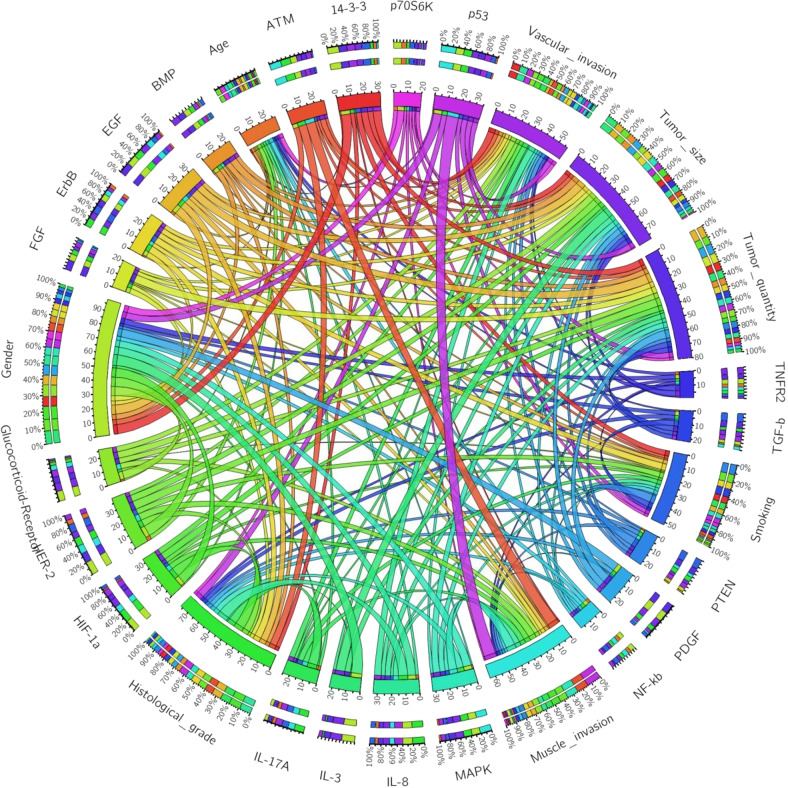
Additionally, Circos (http://circos.ca/) is a software package for visualizing omics-based data and information in a circular layout109. It has an online version (http://circos.ca/circos_online) which however was nonfunctional as of January 2022. Circos plot can be created for exploring relationships and contributions between canonical pathways and clinical clinicopathological characteristics or risk factors (Fig. 7). Each signaling pathway and clinicopathological category are assigned with a unique color in the figure, and the arcs depict the correlation between the clinicopathological categories and signaling pathways. It not only represents the rank of activity of each canonical pathway in the disease but also illustrates the status of activation of the signaling network in each clinicopathological category. The larger the circumference of the arc, the more active the canonical pathway or the more significant the influence of this clinicopathological category on the signaling network. The area of each colored ribbon delineates the proportion of the signaling pathway that contributes to a particular clinicopathological category.
Fig. 7. Circos plot: Among all eight clinicopathological categories, the gender occupied the most significant proportion of the distribution, suggesting that it is the clinical factor that has the most impact on the signaling network.
Among 20 canonical pathways altered in the disease, the HER-2 and p53 are affected most, suggested that they play essential roles in the pathogenesis of the disease.
Single cell proteomics
Single cell proteomics is an emerging technique focused on single cells. It will compete and complement single cell transcriptomics for understanding single-cell biology in the near future. Single cell proteomics recently became a reality when advanced technology showed that peptides in a single cell could be efficiently delivered to the MS instruments110,111. These single-cell MS methodologies can be broadly divided into cell-free and multiplex methods, the latter of which allows proteomic analyses of multiple cells at the same time. The SCoPE2 and Scp are the R-packages for analyzing multiplex single cell proteomic data112,113, while the SCeptre is their counterpart implemented in Python114. Some general proteomic pipelines may also be used to process single-cell proteomic data. They include computational quality control tools115 and a single pipeline (MSnbase) for data processing and visualization116,117.
In conclusion, during the last decade, proteomic technology and research has advanced tremendously. The increasing ability of high-throughput proteomics methods have generated real-time and in-depth datasets. The effective data mining technologies also significantly helped with the pursuit of novel and useful biomarkers, which are essential for disease early-detection and treatment. With the breakthrough of computing power and the rise of artificial intelligence, the role of proteomics has been further expanded. The highly advanced statistic/computational models enable proteomics to be integrated into multi-omics. Under this new trend, proteomics data analysis will be revolutionized for a bigger blueprint with a large amount of clinical and health-related data. It is an exciting time for proteomics developing into an essential new discipline and integrated with other disciplines. Although proteomics has to face emerging challenges during this process, it will move toward more in-depth single-cell biology and individualized precision medicine to boost both basic research and clinical practice to another level.
Supplementary information
Author contributions
M.C. and L.Z. drafted the manuscript. All authors reviewed, discussed and edited the manuscript.
Funding
This work is in part supported by National Science Foundation (IIS-2128307 to L.Z.) and Cancer Prevention & Research Institute of Texas (RR180061 to C.C.). The funders have no role in writing this work or the decision to submit it for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chao Cheng, Email: chao.cheng@bcm.edu.
Lanjing Zhang, Email: lanjing.zhang@rutgers.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41374-022-00830-7.
References
- 1.Collins FS, McKusick VA. Implications of the Human Genome Project for medical science. JAMA. 2001;285:540–544. doi: 10.1001/jama.285.5.540. [DOI] [PubMed] [Google Scholar]
- 2.Wang K, Huang C, Nice E. Recent advances in proteomics: towards the human proteome. Biomed Chromatogr. 2014;28:848–857. doi: 10.1002/bmc.3157. [DOI] [PubMed] [Google Scholar]
- 3.Mathivanan S. Integrated Bioinformatics Analysis of the Publicly Available Protein Data Shows Evidence for 96% of the Human Proteome. J Proteomics Bioinform. 2014;7:41–49. doi: 10.4172/jpb.1000301. [DOI] [Google Scholar]
- 4.Deutsch EW, Lane L, Overall CM, Bandeira N, Baker MS, Pineau C, et al. Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 3.0. J Proteome Res. 2019;18:4108–4116. doi: 10.1021/acs.jproteome.9b00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 6.Ren AH, Diamandis EP, Kulasingam V. Uncovering the Depths of the Human Proteome: Antibody-based Technologies for Ultrasensitive Multiplexed Protein Detection and Quantification. Mol Cell Proteomics. 2021;20:100155. doi: 10.1016/j.mcpro.2021.100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH. Proteomics: Technologies and Their Applications. J Chromatogr Sci. 2017;55:182–196. doi: 10.1093/chromsci/bmw167. [DOI] [PubMed] [Google Scholar]
- 8.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 9.Huang W, Zhan D, Li Y, Zheng N, Wei X, Bai B, et al. Proteomics provides individualized options of precision medicine for patients with gastric cancer. Sci China Life Sci. 2021;64:1199–1211. doi: 10.1007/s11427-021-1966-4. [DOI] [PubMed] [Google Scholar]
- 10.Forler S, Klein O, Klose J. Individualized proteomics. J Proteomics. 2014;107:56–61. doi: 10.1016/j.jprot.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Uzozie AC, Aebersold R. Advancing translational research and precision medicine with targeted proteomics. J Proteomics. 2018;189:1–10. doi: 10.1016/j.jprot.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 12.Parker CE, Borchers CH. The Special Issue: Clinical Proteomics for Precision Medicine. Proteomics Clin Appl12, (2018). [DOI] [PubMed]
- 13.Macklin A, Khan S, Kislinger T. Recent advances in mass spectrometry based clinical proteomics: applications to cancer research. Clin Proteomics. 2020;17:17. doi: 10.1186/s12014-020-09283-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Wang W, Chen J. Recent progress in mass spectrometry proteomics for biomedical research. Sci China Life Sci. 2017;60:1093–1113. doi: 10.1007/s11427-017-9175-2. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 16.Peng L, Cantor DI, Huang C, Wang K, Baker MS, Nice EC. Tissue and plasma proteomics for early stage cancer detection. Mol Omics. 2018;14:405–423. doi: 10.1039/C8MO00126J. [DOI] [PubMed] [Google Scholar]
- 17.Morak M, Schmidinger H, Krempl P, Rechberger G, Kollroser M, Birner-Gruenberger R, et al. Differential activity-based gel electrophoresis for comparative analysis of lipolytic and esterolytic activities. J Lipid Res. 2009;50:1281–1292. doi: 10.1194/jlr.M800566-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva TS, Richard N, Dias JP, Rodrigues PM. Data visualization and feature selection methods in gel-based proteomics. Curr Protein Pept Sci. 2014;15:4–22. doi: 10.2174/1389203715666140221112334. [DOI] [PubMed] [Google Scholar]
- 19.Corbett JR, Robinson DE, Patrie SM. Robustness and Ruggedness of Isoelectric Focusing and Superficially Porous Liquid Chromatography with Fourier Transform Mass Spectrometry. J Am Soc Mass Spectrom. 2021;32:346–354. doi: 10.1021/jasms.0c00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cupp-Sutton KA, Wu S. High-throughput quantitative top-down proteomics. Mol Omics. 2020;16:91–99. doi: 10.1039/C9MO00154A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirzaei H, Carrasco M. Modern Proteomics-Sample Preparation, Analysis and Practical Applications: Springer, 2016.
- 22.Zhang Z, Wu S, Stenoien DL, Pasa-Tolic L. High-throughput proteomics. Annu Rev Anal Chem (Palo Alto Calif) 2014;7:427–454. doi: 10.1146/annurev-anchem-071213-020216. [DOI] [PubMed] [Google Scholar]
- 23.Cai W, Tucholski TM, Gregorich ZR, Ge Y. Top-down Proteomics: Technology Advancements and Applications to Heart Diseases. Expert Rev Proteomics. 2016;13:717–730. doi: 10.1080/14789450.2016.1209414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sechi S. Quantitative Proteomics by Mass Spectrometry. Humana Press; 2017. [Google Scholar]
- 25.Vidova V, Spacil Z. A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition. Anal Chim Acta. 2017;964:7–23. doi: 10.1016/j.aca.2017.01.059. [DOI] [PubMed] [Google Scholar]
- 26.Matthiesen R, Bunkenborg J. Introduction to Mass Spectrometry-Based Proteomics. Methods Mol Biol. 2020;2051:1–58. doi: 10.1007/978-1-4939-9744-2_1. [DOI] [PubMed] [Google Scholar]
- 27.Zhang DY, Ye F, Gao L, Liu X, Zhao X, Che Y, et al. Proteomics, pathway array and signaling network-based medicine in cancer. Cell Div. 2009;4:20. doi: 10.1186/1747-1028-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerin CL, Guyonnet L, Goudot G, Revets D, Konstantinou M, Chipont A, et al. Multidimensional Proteomic Approach of Endothelial Progenitors Demonstrate Expression of KDR Restricted to CD19 Cells. Stem Cell Rev Rep. 2021;17:639–651. doi: 10.1007/s12015-020-10062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Cao Y, Zhao J, Fang Y, Liu N, Zhang Y. Multidimensional Proteomics Identifies Declines in Protein Homeostasis and Mitochondria as Early Signals for Normal Aging and Age-associated Disease in Drosophila. Mol Cell Proteomics. 2019;18:2078–2088. doi: 10.1074/mcp.RA119.001621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadi SA, Waters WR, Palmer M, Lyashchenko KP, Sreevatsan S. Development of a Multidimensional Proteomic Approach to Detect Circulating Immune Complexes in Cattle Experimentally Infected With Mycobacterium bovis. Front Vet Sci. 2018;5:141. doi: 10.3389/fvets.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai M, Liang L, Chen J, Qiu N, Ge S, Ji S, et al. Multidimensional Proteomics Reveals a Role of UHRF2 in the Regulation of Epithelial-Mesenchymal Transition (EMT) Mol Cell Proteomics. 2016;15:2263–2278. doi: 10.1074/mcp.M115.057448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buhimschi IA, Zhao G, Rosenberg VA, Abdel-Razeq S, Thung S, Buhimschi CS. Multidimensional proteomics analysis of amniotic fluid to provide insight into the mechanisms of idiopathic preterm birth. PLoS One. 2008;3:e2049. doi: 10.1371/journal.pone.0002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hui W, Ye F, Zhang W, Liu C, Cui M, Li W, et al. Aberrant expression of signaling proteins in essential thrombocythemia. Ann Hematol. 2013;92:1229–1238. doi: 10.1007/s00277-013-1755-8. [DOI] [PubMed] [Google Scholar]
- 34.Huang K, Cui M, Ye F, Li Y, Zhang D. Global profiling of the signaling network of papillary thyroid carcinoma. Life Sci. 2016;147:9–14. doi: 10.1016/j.lfs.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Zlobec I, Suter G, Perren A, Lugli A. A next-generation tissue microarray (ngTMA) protocol for biomarker studies. J Vis Exp, 51893 (2014). [DOI] [PMC free article] [PubMed]
- 36.Kim MK, Ye F, Wang D, Cui M, Ward SC, Warner RR, et al. Differential Protein Expression in Small Intestinal Neuroendocrine Tumors and Liver Metastases. Pancreas. 2016;45:528–532. doi: 10.1097/MPA.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jawhar NM. Tissue Microarray: A rapidly evolving diagnostic and research tool. Ann Saudi Med. 2009;29:123–127. doi: 10.4103/0256-4947.51806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zlobec I, Koelzer VH, Dawson H, Perren A, Lugli A. Next-generation tissue microarray (ngTMA) increases the quality of biomarker studies: an example using CD3, CD8, and CD45RO in the tumor microenvironment of six different solid tumor types. J Transl Med. 2013;11:104. doi: 10.1186/1479-5876-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zysset D, Montani M, Spalinger J, Schibli S, Zlobec I, Mueller C, et al. Molecular and Histological Profiling Reveals an Innate-Shaped Immune Microenvironment in Solitary Juvenile Polyps. Clin Transl Gastroenterol. 2021;12:e00361. doi: 10.14309/ctg.0000000000000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen HG, Lundstrom O, Blank A, Dawson H, Lugli A, Anisimova M, et al. Image-based assessment of extracellular mucin-to-tumor area predicts consensus molecular subtypes (CMS) in colorectal cancer. Mod Pathol, (2021). [DOI] [PMC free article] [PubMed]
- 41.Zahnd S, Braga-Lagache S, Buchs N, Lugli A, Dawson H, Heller M, et al. A Digital Pathology-Based Shotgun-Proteomics Approach to Biomarker Discovery in Colorectal Cancer. J Pathol Inform. 2019;10:40. doi: 10.4103/jpi.jpi_65_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Numis AL, Fox CH, Lowenstein DJ, Norris PJ, Di Germanio C. Comparison of multiplex cytokine assays in a pediatric cohort with epilepsy. Heliyon. 2021;7:e06445. doi: 10.1016/j.heliyon.2021.e06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasseter HC, Provost AC, Chaby LE, Daskalakis NP, Haas M, Jeromin A. Cross-platform comparison of highly sensitive immunoassay technologies for cytokine markers: Platform performance in post-traumatic stress disorder and Parkinson’s disease. Cytokine X. 2020;2:100027. doi: 10.1016/j.cytox.2020.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim SY, Lee JH, Welsh SJ, Ahn SB, Breen E, Khan A, et al. Evaluation of two high-throughput proteomic technologies for plasma biomarker discovery in immunotherapy-treated melanoma patients. Biomark Res. 2017;5:32. doi: 10.1186/s40364-017-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods. 2009;340:55–64. doi: 10.1016/j.jim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Pan J, Zheng QZ, Li Y, Yu LL, Wu QW, Zheng JY, et al. Discovery and Validation of a Serologic Autoantibody Panel for Early Diagnosis of Esophageal Squamous Cell Carcinoma. Cancer Epidemiol Biomarkers Prev. 2019;28:1454–1460. doi: 10.1158/1055-9965.EPI-18-1269. [DOI] [PubMed] [Google Scholar]
- 47.Cui L, Shu C, Liu Z, Tong W, Cui M, Wei C, et al. The expression of serum sEGFR, sFlt-1, sEndoglin and PLGF in preeclampsia. Pregnancy Hypertens. 2018;13:127–132. doi: 10.1016/j.preghy.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Cui L, Shu C, Liu Z, Tong W, Cui M, Wei C, et al. Serum protein marker panel for predicting preeclampsia. Pregnancy Hypertens. 2018;14:279–285. doi: 10.1016/j.preghy.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 49.Tong W, Ye F, He L, Cui L, Cui M, Hu Y, et al. Serum biomarker panels for diagnosis of gastric cancer. Onco Targets Ther. 2016;9:2455–2463. doi: 10.2147/OTT.S86139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taniuchi M, Verweij JJ, Noor Z, Sobuz SU, Lieshout L, Petri WA, Jr., et al. High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites. Am J Trop Med Hyg. 2011;84:332–337. doi: 10.4269/ajtmh.2011.10-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson RJ, Greening DW. Serum/plasma proteomics: methods and protocols. New York, NY, USA: Humana Press; 2017. [Google Scholar]
- 52.Lantoine J, Proces A, Villers A, Halliez S, Buee L, Ris L, et al. Inflammatory Molecules Released by Mechanically Injured Astrocytes Trigger Presynaptic Loss in Cortical Neuronal Networks. ACS Chem Neurosci. 2021;12:3885–3897. doi: 10.1021/acschemneuro.1c00488. [DOI] [PubMed] [Google Scholar]
- 53.Jiang Z, Kamerud J, You Z, Basak S, Seletskaia E, Steeno GS, et al. Feasibility of singlicate-based analysis in bridging ADA assay on Meso-Scale Discovery platform: comparison with duplicate analysis. Bioanalysis. 2021;13:1123–1134. doi: 10.4155/bio-2021-0095. [DOI] [PubMed] [Google Scholar]
- 54.Jia R, Chen YX, Du YR, Hu BR. Meso-scale Discovery Assay Detects the Changes of Plasma Cytokine Levels in Mice after Low or High LET Ionizing Irradiation. Biomed Environ Sci. 2021;34:540–551. doi: 10.3967/bes2021.074. [DOI] [PubMed] [Google Scholar]
- 55.Sivakumaran D, Ritz C, Gjoen JE, Vaz M, Selvam S, Ottenhoff THM, et al. Host Blood RNA Transcript and Protein Signatures for Sputum-Independent Diagnostics of Tuberculosis in Adults. Front Immunol. 2020;11:626049. doi: 10.3389/fimmu.2020.626049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Youssef P, Kim WS, Halliday GM, Lewis SJG, Dzamko N. Comparison of Different Platform Immunoassays for the Measurement of Plasma Alpha-Synuclein in Parkinson’s Disease Patients. J Parkinsons Dis. 2021;11:1761–1772. doi: 10.3233/JPD-212694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quanterix. Quanterix: Publications And Posters. Vol. 2022, 2022.
- 58.Lollo B, Steele F, Gold L. Beyond antibodies: new affinity reagents to unlock the proteome. Proteomics. 2014;14:638–644. doi: 10.1002/pmic.201300187. [DOI] [PubMed] [Google Scholar]
- 59.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, et al. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol Ther Nucleic Acids. 2014;3:e201. doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu RX, Thiessen-Philbrook HR, Vasan RS, Coresh J, Ganz P, Bonventre JV, et al. Comparison of proteomic methods in evaluating biomarker-AKI associations in cardiac surgery patients. Transl Res. 2021;238:49–62. doi: 10.1016/j.trsl.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Billing AM, Ben Hamidane H, Bhagwat AM, Cotton RJ, Dib SS, Kumar P, et al. Complementarity of SOMAscan to LC-MS/MS and RNA-seq for quantitative profiling of human embryonic and mesenchymal stem cells. J Proteomics. 2017;150:86–97. doi: 10.1016/j.jprot.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 62.Han Z, Xiao Z, Kalantar-Zadeh K, Moradi H, Shafi T, Waikar SS, et al. Validation of a Novel Modified Aptamer-Based Array Proteomic Platform in Patients with End-Stage Renal Disease. Diagnostics (Basel)8, (2018). [DOI] [PMC free article] [PubMed]
- 63.Raffield LM, Dang H, Pratte KA, Jacobson S, Gillenwater LA, Ampleford E, et al. Comparison of Proteomic Assessment Methods in Multiple Cohort Studies. Proteomics. 2020;20:e1900278. doi: 10.1002/pmic.201900278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fredriksson S, Dixon W, Ji H, Koong AC, Mindrinos M, Davis RW. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat Methods. 2007;4:327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 65.Gullberg M, Gustafsdottir SM, Schallmeiner E, Jarvius J, Bjarnegard M, Betsholtz C, et al. Cytokine detection by antibody-based proximity ligation. Proc Natl Acad Sci U S A. 2004;101:8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 67.Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fraser DD, Chen M, Ren A, Miller MR, Martin C, Daley M, et al. Novel severe traumatic brain injury blood outcome biomarkers identified with proximity extension assay. Clin Chem Lab Med. 2021;59:1662–1669. doi: 10.1515/cclm-2021-0103. [DOI] [PubMed] [Google Scholar]
- 69.Fraser DD, Cepinskas G, Patterson EK, Slessarev M, Martin C, Daley M, et al. Novel Outcome Biomarkers Identified With Targeted Proteomic Analyses of Plasma From Critically Ill Coronavirus Disease 2019 Patients. Crit Care Explor. 2020;2:e0189. doi: 10.1097/CCE.0000000000000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel H, Ashton NJ, Dobson RJB, Andersson LM, Yilmaz A, Blennow K, et al. Proteomic blood profiling in mild, severe and critical COVID-19 patients. Sci Rep. 2021;11:6357. doi: 10.1038/s41598-021-85877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carlsson AC, Ingelsson E, Sundstrom J, Carrero JJ, Gustafsson S, Feldreich T, et al. Use of Proteomics To Investigate Kidney Function Decline over 5 Years. Clin J Am Soc Nephrol. 2017;12:1226–1235. doi: 10.2215/CJN.08780816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayer SF, Cao C, Dal, Peraro M. Biological nanopores for single-molecule sensing. iScience. 2022;25:104145. doi: 10.1016/j.isci.2022.104145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oppenheim S, Cao X, Rueppel O, Krongdang S, Phokasem P, DeSalle R, et al. Whole Genome Sequencing and Assembly of the Asian Honey Bee Apis dorsata. Genome Biol Evol. 2020;12:3677–3683. doi: 10.1093/gbe/evz277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou A, Lin T, Xing J. Evaluating nanopore sequencing data processing pipelines for structural variation identification. Genome Biol. 2019;20:237. doi: 10.1186/s13059-019-1858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan S, Zhang J, Wang Y, Guo W, Zhang S, Liu Y, et al. Single Molecule Ratcheting Motion of Peptides in a Mycobacterium smegmatis Porin A (MspA) Nanopore. Nano Lett. 2021;21:6703–6710. doi: 10.1021/acs.nanolett.1c02371. [DOI] [PubMed] [Google Scholar]
- 76.Boskovic F, Keyser UF. Toward single-molecule proteomics. Science. 2021;374:1443–1444. doi: 10.1126/science.abn0001. [DOI] [PubMed] [Google Scholar]
- 77.Brinkerhoff H, Kang ASW, Liu J, Aksimentiev A, Dekker C. Multiple rereads of single proteins at single-amino acid resolution using nanopores. Science. 2021;374:1509–1513. doi: 10.1126/science.abl4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu J, Lin Y, Yang M, Zhang L. Statistics and pitfalls of trend analysis in cancer research: a review focused on statistical packages. J Cancer. 2020;11:2957–2961. doi: 10.7150/jca.43521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krempel R, Kulkarni P, Yim A, Lang U, Habermann B, Frommolt P. Integrative analysis and machine learning on cancer genomics data using the Cancer Systems Biology Database (CancerSysDB) BMC Bioinformatics. 2018;19:156. doi: 10.1186/s12859-018-2157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rashidi HH, Tran NK, Betts EV, Howell LP, Green R. Artificial Intelligence and Machine Learning in Pathology: The Present Landscape of Supervised Methods. Acad Pathol. 2019;6:2374289519873088. doi: 10.1177/2374289519873088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022;35:23–32. doi: 10.1038/s41379-021-00919-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.La Porta CAM, Zapperi S. Explaining the dynamics of tumor aggressiveness: At the crossroads between biology, artificial intelligence and complex systems. Semin Cancer Biol. 2018;53:42–47. doi: 10.1016/j.semcancer.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 84.Smail-Tabbone M, Rance B. Section Editors for the IYSoB, Translational I. Contributions from the 2019 Literature on Bioinformatics and Translational Informatics. Yearb Med Inform. 2020;29:188–192. doi: 10.1055/s-0040-1702002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stechow, von, L. Cancer Systems Biology: Methods and Protocols. (Humana Press: New York, NY, USA, 2016..
- 86.Korenberg MJ. Microarray Data Analysis: Methods and Applications: Springer Science & Business Media, 2007.
- 87.Ward JH., Jr Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 88.Meunier B, Dumas E, Piec I, Bechet D, Hebraud M, Hocquette JF. Assessment of hierarchical clustering methodologies for proteomic data mining. J Proteome Res. 2007;6:358–366. doi: 10.1021/pr060343h. [DOI] [PubMed] [Google Scholar]
- 89.Virmani AK, Tsou JA, Siegmund KD, Shen LY, Long TI, Laird PW, et al. Hierarchical clustering of lung cancer cell lines using DNA methylation markers. Cancer Epidemiol Biomarkers Prev. 2002;11:291–297. [PubMed] [Google Scholar]
- 90.Draisma HH, Reijmers TH, Meulman JJ, van der Greef J, Hankemeier T, Boomsma DI. Hierarchical clustering analysis of blood plasma lipidomics profiles from mono- and dizygotic twin families. Eur J Hum Genet. 2013;21:95–101. doi: 10.1038/ejhg.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw46, (2012). [PMC free article] [PubMed]
- 92.Vagni M, Giordano N, Balestra G, Rosati S. Comparison of different similarity measures in hierarchical clustering. 2021 IEEE International Symposium on Medical Measurements and Applications (MeMeA), 2021, p. 1-6.
- 93.Cai Y, Sun Y. ESPRIT-Tree: hierarchical clustering analysis of millions of 16S rRNA pyrosequences in quasilinear computational time. Nucleic Acids Res. 2011;39:e95. doi: 10.1093/nar/gkr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.MacArthur BD, Lachmann A, Lemischka IR, Ma’ayan A. GATE: software for the analysis and visualization of high-dimensional time series expression data. Bioinformatics. 2010;26:143–144. doi: 10.1093/bioinformatics/btp628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lachenbruch PA. An almost unbiased method of obtaining confidence intervals for the probability of misclassification in discriminant analysis. Biometrics. 1967;23:639–645. doi: 10.2307/2528418. [DOI] [PubMed] [Google Scholar]
- 96.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface15, (2018). [DOI] [PMC free article] [PubMed]
- 97.Huberty CJ. Discriminant analysis: Issues and problems. In: Lovric M, editor. International Encyclopedia of Statistical Science. Berlin, Heidelberg. Berlin, Heidelberg: Springer Berlin Heidelberg; 2011. pp. 390–392.. [Google Scholar]
- 98.Huberty CJ. Issues in the use and interpretation of discriminant analysis. Psychological Bulletin. 1984;95:156–171. doi: 10.1037/0033-2909.95.1.156. [DOI] [Google Scholar]
- 99.Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1:274–278. doi: 10.4103/0974-7788.76794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Swinscow TDV, Campbell MJ. Statistics at square one: Bmj London, 2002.
- 101.Roher AE, Maarouf CL, Sue LI, Hu Y, Wilson J, Beach TG. Proteomics-derived cerebrospinal fluid markers of autopsy-confirmed Alzheimer’s disease. Biomarkers. 2009;14:493–501. doi: 10.3109/13547500903108423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–1304. doi: 10.1093/clinchem/48.8.1296. [DOI] [PubMed] [Google Scholar]
- 103.Keerthikumar S, Mathivanan S. Proteome Bioinformatics. New York, NY, USA: Humana Press; 2017. [Google Scholar]
- 104.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zaim SR, Li Q, Schissler AG, Lussier YA. Emergence of pathway-level composite biomarkers from converging gene set signals of heterogeneous transcriptomic responses. Pac Symp Biocomput. 2018;23:484–495. [PMC free article] [PubMed] [Google Scholar]
- 107.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kuleshov MV, Diaz JEL, Flamholz ZN, Keenan AB, Lachmann A, Wojciechowicz ML, et al. modEnrichr: a suite of gene set enrichment analysis tools for model organisms. Nucleic Acids Res. 2019;47:W183–W190. doi: 10.1093/nar/gkz347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Slavov N. Scaling Up Single-Cell Proteomics. Mol Cell Proteomics. 2022;21:100179. doi: 10.1016/j.mcpro.2021.100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vistain LF, Tay S. Single-Cell Proteomics. Trends Biochem Sci. 2021;46:661–672. doi: 10.1016/j.tibs.2021.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petelski AA, Emmott E, Leduc A, Huffman RG, Specht H, Perlman DH, et al. Multiplexed single-cell proteomics using SCoPE2. Nat Protoc. 2021;16:5398–5425. doi: 10.1038/s41596-021-00616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cheung TK, Lee CY, Bayer FP, McCoy A, Kuster B, Rose CM. Defining the carrier proteome limit for single-cell proteomics. Nat Methods. 2021;18:76–83. doi: 10.1038/s41592-020-01002-5. [DOI] [PubMed] [Google Scholar]
- 114.Schoof EM, Furtwangler B, Uresin N, Rapin N, Savickas S, Gentil C, et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat Commun. 2021;12:3341. doi: 10.1038/s41467-021-23667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bittremieux W, Valkenborg D, Martens L, Laukens K. Computational quality control tools for mass spectrometry proteomics. Proteomics17, (2017). [DOI] [PubMed]
- 116.Gatto L, Gibb S, Rainer J. MSnbase, Efficient and Elegant R-Based Processing and Visualization of Raw Mass Spectrometry Data. J Proteome Res. 2021;20:1063–1069. doi: 10.1021/acs.jproteome.0c00313. [DOI] [PubMed] [Google Scholar]
- 117.Gatto L, Breckels LM, Naake T, Gibb S. Visualization of proteomics data using R and bioconductor. Proteomics. 2015;15:1375–1389. doi: 10.1002/pmic.201400392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.