Abstract
A glutathione S-transferase (GST) with activity toward 1,2-epoxy-2-methyl-3-butene (isoprene monoxide) and cis-1,2-dichloroepoxyethane was purified from the isoprene-utilizing bacterium Rhodococcus sp. strain AD45. The homodimeric enzyme (two subunits of 27 kDa each) catalyzed the glutathione (GSH)-dependent ring opening of various epoxides. At 5 mM GSH, the enzyme followed Michaelis-Menten kinetics for isoprene monoxide and cis-1,2-dichloroepoxyethane, with Vmax values of 66 and 2.4 μmol min−1 mg of protein−1 and Km values of 0.3 and 0.1 mM for isoprene monoxide and cis-1,2-dichloroepoxyethane, respectively. Activities increased linearly with the GSH concentration up to 25 mM. 1H nuclear magnetic resonance spectroscopy showed that the product of GSH conjugation to isoprene monoxide was 1-hydroxy-2-glutathionyl-2-methyl-3-butene (HGMB). Thus, nucleophilic attack of GSH occurred on the tertiary carbon atom of the epoxide ring. HGMB was further converted by an NAD+-dependent dehydrogenase, and this enzyme was also purified from isoprene-grown cells. The homodimeric enzyme (two subunits of 25 kDa each) showed a high activity for HGMB, whereas simple primary and secondary alcohols were not oxidized. The enzyme catalyzed the sequential oxidation of the alcohol function to the corresponding aldehyde and carboxylic acid and followed Michaelis-Menten kinetics with respect to NAD+ and HGMB. The results suggest that the initial steps in isoprene metabolism are a monooxygenase-catalyzed conversion to isoprene monoxide, a GST-catalyzed conjugation to HGMB, and a dehydrogenase-catalyzed two-step oxidation to 2-glutathionyl-2-methyl-3-butenoic acid.
Mammalian glutathione S-transferases (GSTs) have been studied extensively since they play an important role in the detoxification of a wide range of electrophilic compounds (8, 18). Much less work with bacterial GSTs has been reported, and the available information on these enzymes has recently been reviewed (14, 34). Some bacterial GSTs catalyze hydrolytic or reductive dehalogenation reactions (22, 37). Another enzyme catalyzes a reductive ether bond cleavage and is involved in lignin degradation (23). Surprisingly, the physiological functions of many other bacterial GSTs remain to be established (34). For instance, some GSTs appear to be associated with the metabolism of aromatic compounds, but they are not essential for growth (12).
Recently we reported the presence of a GST in cell extracts of the isoprene-utilizing bacterium Rhodococcus sp. strain AD45 (32). The enzyme was capable of degrading 1,2-dichloroepoxyethanes which occur as toxic products of the cometabolic oxidation of 1,2-dichloroethenes by monooxygenases (15, 25, 30, 31). Cell extracts of strain AD45 catalyzed the glutathione (GSH)-dependent conversion of various epoxides such as the primary oxidation product of isoprene, 1,2-epoxy-2-methyl-3-butene (isoprene monoxide), and cis-1,2-dichloroepoxyethane, indicating that the GST has a broad substrate range (32). To establish the role of the enzyme in isoprene metabolism, we have characterized the protein and investigated the primary reaction product formed after conjugation of GSH to isoprene monoxide. Furthermore, we describe the purification of a highly specific dehydrogenase that catalyzes the NAD+-dependent oxidation of the glutathione-isoprene monoxide conjugate.
MATERIALS AND METHODS
Organisms and growth conditions.
Rhodococcus sp. strain AD45 was grown on isoprene in batch culture or continuous culture, using a mineral medium supplemented with 20 mg of yeast extract per liter as described before (32).
Enzyme assays.
GST activities were assayed at 30°C by following the consumption of substrate by on-line headspace gas chromatography (GC) (32). Reaction mixtures consisted of 50 mM Tris-HCl buffer (pH 8.5) containing 5 mM substrate and 5 mM GSH unless stated otherwise. The dimensionless Henry coefficients that were used for the calculation of activities are 0.001 for cis-1,2-dichloroepoxyethane, 0.007 for epoxypropane, 0.02 for 1,2-epoxyhexane, and 0.02 for 1,2-epoxy-2-methyl-3-butene (32). The dimensionless Henry coefficients of epithiopropane (0.06), 1,2-epoxybutane (0.008), 2,3-epoxybutane (0.01), epifluorohydrin (0.007), epichlorohydrin (0.006), and epibromohydrin (0.004) were determined as described before (30).
The formation of GSH-isoprene monoxide conjugates was monitored by removing samples from a reaction mixture containing 10 mM GSH and 5 mM isoprene monoxide. The samples were quenched by mixing with 1 volume of 1 M formic acid and analyzed for the presence of GSH-isoprene monoxide conjugates by high-pressure liquid chromatography (HPLC).
Dehydrogenase activities were measured in 100 mM glycine-NaOH buffer (pH 10.0) to which 1 mM NAD+ and 7.5 mM GSH-isoprene monoxide conjugate was added from a 0.5 M stock solution, unless stated otherwise. The activity was monitored by following the production of NADH at 340 nm.
Activities are expressed in units per milligram of protein. One unit is defined as the activity that catalyzes the conversion of 1 μmol of substrate per min.
Purification of the GST.
Isoprene-grown cells were harvested from a continuous culture that was operated at a dilution rate of 0.026 h−1 (32) and resuspended in 10 mM Tris-HCl buffer (pH 7.5). All further steps were carried out at 0 to 4°C. Cells were washed twice with this buffer before they were resuspended in 10 mM Tris-HCl buffer containing 1 mM β-mercaptoethanol, 1 mM EDTA, and 3 mM NaN3 (TEMA buffer). After sonication, a cell extract was obtained by centrifugation (40,000 × g, 60 min).
Solid (NH4)2SO4 was added to the extract to 40% saturation. The mixture was gently stirred for 30 min at 0°C and centrifuged (16,000 × g, 20 min). The supernatant was decanted, and solid (NH4)2SO4 was added to 95% saturation and gently stirred for 30 min. The precipitate was collected by centrifugation (16,000 × g, 20 min) and dissolved in TEMA buffer.
The solution was dialyzed against TEMA buffer to remove (NH4)2SO4, and applied to a Resource Q anion-exchange column (6 ml; Pharmacia Biotech, Uppsala, Sweden) that was connected to a model LCC500 fast protein liquid chromatography system (Pharmacia Biotech). The buffer system consisted of TEMA buffer (buffer A) and TEMA buffer with 0.45 M NaCl (buffer B). Retained protein was eluted with a three-step increasing linear gradient: 0 to 15% buffer B in 20 ml, 15 to 40% buffer B in 100 ml, and 40 to 100% buffer B in 10 ml (flow rate, 2 ml min−1; fraction volume, 2 ml). Activity eluted at 90 to 105 mM NaCl, and active fractions were pooled.
Solid (NH4)2SO4 was added to a concentration of 1.5 M, and the protein was applied to a Resource Iso column (1 ml; Pharmacia Biotech). Retained protein was eluted with a 20-ml linear decreasing gradient of 1.5 to 0 M (NH4)2SO4 in buffer A (flow rate, 0.5 ml min−1; fraction volume, 0.5 ml). Activity eluted at 1.0 to 0.85 M (NH4)2SO4. Approximately 25% of the activity that was thus collected eluted as pure protein, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The other 75% was further purified by applying it a second time to the Resource Iso column. The pooled purified enzyme was dialyzed against TEMA buffer and stored at 4°C.
Affinity chromatography with GSH covalently attached with the sulfur atom to agarose via a 12-atom linker (Sigma, St. Louis, Mo.) was carried out as recommended by the manufacturer.
Purification of the dehydrogenase.
In the first step of the purification protocol, cell extract was subjected to anion-exchange chromatography using a DE52 column (diameter, 3 cm; height, 10 cm; Sigma). The protein was eluted with a 500-ml linear gradient of 0 to 0.3 M NaCl in TEMA buffer containing 10% glycerol (flow rate, 0.8 ml min−1; fraction volume, 10 ml). Activity eluted at 0.2 to 0.25 M NaCl. Active fractions were pooled and concentrated by ultrafiltration with an Amicon diaflow membrane (10-kDa exclusion pore) fitted in an Amicon apparatus. Concentrated protein was subjected to gel filtration using a Sephacryl S-300 column (diameter, 0.7 cm; height, 61 cm; Pharmacia) that was equilibrated and eluted with TEMA buffer. Pooled fractions were dialyzed against 20 mM potassium phosphate buffer (pH 7.0) containing 1 mM EDTA, 1 mM β-mercaptoethanol, 1.5 mM NaN3, and 10% glycerol (PEMAG buffer) and subjected to affinity chromatography using 30 ml of Blue Sepharose CL-6B. Protein was eluted with PEMAG buffer (flow rate, 1 ml min−1; fraction volume, 4 ml). Activity was not retained. The active fractions were pooled and dialyzed against TEMA buffer containing 1.5 M (NH4)2SO4 and applied to a Resource Iso column (Pharmacia). Retained protein was eluted with a 20-ml linear decreasing gradient of 1.5 to 0 M (NH4)2SO4 in TEMA buffer (flow rate, 0.5 ml min−1; fraction volume, 0.5 ml). Activity eluted at 1.1 to 0.95 M (NH4)2SO4.
Estimation of molecular mass.
Molecular masses of the native enzymes were estimated by gel filtration on a Superose 12 HR 10/30 column equilibrated with TEMA buffer containing 100 mM NaCl. Immunoglobulin G (160 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and soy bean trypsin inhibitor (20.1 kDa) were used as reference proteins.
Molecular masses of denatured enzymes were determined by SDS-PAGE. Phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), soy bean trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa) were used as reference proteins.
Amino acid analysis and N-terminal amino acid sequence determination.
Determination of amino acid composition and N-terminal amino acid sequence analysis were performed by Eurosequence BV (Groningen, The Netherlands). For N-terminal sequence analysis, approximately 50 μg of pure protein was applied to SDS-PAGE and electroblotted on a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The N-terminal sequence was determined by Edman degradation using an automated sequenator (model 477A; Applied Biosystems). The amino acid composition was determined after the protein was hydrolyzed with 5.7 N HCl at 166°C for 2 h. The hydrolysate was applied to an HP Aminoquant equipped with a Shandon Hypersil OD5 column (2.1 by 200 mm).
Homology search.
The amino acid analysis program at EMBL (Heidelberg, Germany) was used to search the SwissProt database for proteins with a homologous amino acid composition (19). The BLAST program (5) was used to search the SwissProt database for proteins that show sequence similarity with the N-terminal sequences. Multiple sequence alignments were generated with Clustal W version 1.7 (29). Conserved amino acids are indicated according to the following scheme: P, A, G, S, T; Q, N; E, D; R, K; I, L, M, V; F, Y, W; C, H. All used programs were offered as services on the Worldwide Web.
HPLC-MS analysis of GSH conjugates.
GSH-epoxide conjugates present in the supernatants were analyzed by reversed-phase HPLC using a Merck Hitachi L-6200A system equipped with a Lichrosorb 5C18 column (20 by 4.6 mm) or a Lichrosorb 7C18 column (Chrompack, Middelburg, The Netherlands) and a Merck Hitachi L4000 UV detector. For data acquisition, a Kontron PC Integration Pack 3.90 (Kontron Instruments, Milan, Italy) was used. The buffer system consisted of 0.1% trifluoroacetic acid in water (buffer A) and 0.1% trifluoroacetic acid in acetonitrile (buffer B). The following elution protocol was used: 0 to 2 min, 0% buffer B, isocratic; 2 to 32 min, 0 to 20% buffer B, linear gradient; 32 to 36 min, 20 to 100% buffer B, linear gradient (column regeneration). The elution profile was obtained by measuring the absorbance at 214 nm. HPLC with on-line atmospheric pressure ionization mass spectrometry (HPLC-MS) detection was performed as described before (10, 26) at a nozzle voltage of 70 V to detect molecular ions or 170 V to detect collision-induced fragments.
Synthesis of GSH-epoxide conjugates.
Approximately 1 g of GSH-isoprene monoxide conjugate was synthesized in 50 mM sodium carbonate buffer containing 25 mM isoprene monoxide and 20 mM GSH. Carbonate buffers were used to avoid signals in the nuclear magnetic resonance (NMR) analysis arising from buffering compounds. The reaction was started by adding purified GST to a concentration of 0.02 mg ml−1. The mixture was incubated at 30°C for 2 h and filtered with an Amicon diaflow membrane (10-kDa exclusion pore) to remove the enzyme. The filtrate was lyophilized to remove excess isoprene monoxide, and 10% of the sample was dissolved in D2O and subjected to 1H NMR analysis. The remaining 90% was dissolved in Tris-HCl (50 mM, pH 8.0) to a concentration of 0.5 M and used as a substrate for dehydrogenase assays.
The GSH conjugates with epoxypropane, 1,2-epoxybutane, and 1,2-epoxyhexane were synthesized by using purified GST in a standard enzyme assay mixture containing 5 mM epoxide and 10 mM GSH. After 90% of the epoxide was converted, the sample was lyophilized to remove remaining epoxide. HPLC-MS analysis at 70-V nozzle voltage showed the presence of molecular ions with m/z 366 for the conjugate with epoxypropane, m/z 380 for the conjugate with 1,2-epoxybutane, and m/z 408 for the conjugate with 1,2-epoxyhexane. These values are in agreement with the theoretical values.
NMR analysis of the GSH-isoprene monoxide conjugate.
1H NMR spectra were recorded on a Varian VXR-300 spectrometer (300 MHz) or on a Varian Gemini spectrometer (200 MHz). 1H NMR chemical shifts were determined relative to the internal standard sodium 2,2′,3,3′-tetradeutero-3-trimethylsilyl propionate.
Chemicals.
Organic chemicals used in this study were obtained from Acros Chimica (Geel, Belgium) or Aldrich (Milwaukee, Wis.).
RESULTS
Purification and biochemical properties of the GST.
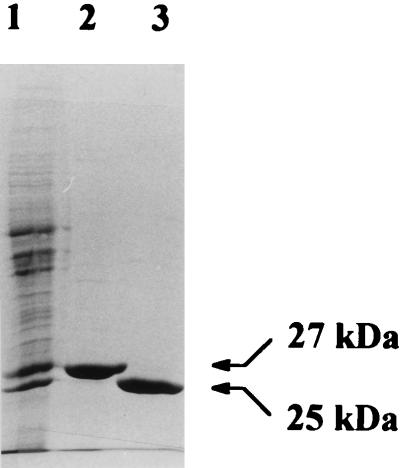
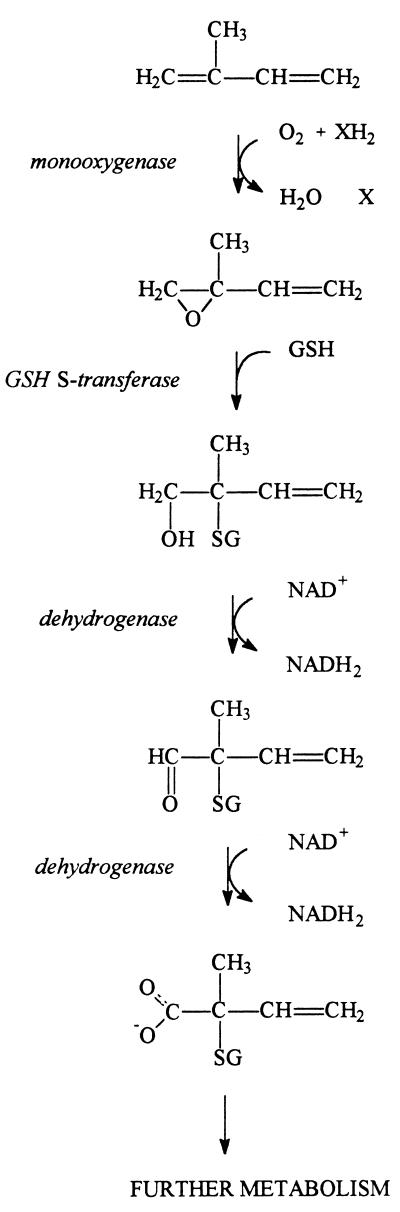
Previously, we observed that only in extracts of isoprene- and isoprene monoxide-grown cells of Rhodococcus sp. strain AD45 could a GST activity with epoxides be detected (32). We purified this enzyme to homogeneity from cells grown on isoprene in continuous culture. No binding or retardation of the protein was observed upon affinity chromatography with GSH-coated agarose. Therefore, the enzyme was purified by (NH4)2SO4 precipitation followed by anion-exchange chromatography and hydrophobic interaction chromatography (Table 1). The purification protocol reproducibly yielded pure protein, as judged by SDS-PAGE (Fig. 1). The protein was purified 13-fold, implying that the GST represents approximately 8% of the total protein in isoprene-grown cells. This was in agreement with the presence of a prominent protein band of the same electrophoretic mobility in cell extracts of isoprene-grown cells (Fig. 1).
TABLE 1.
Purification of the GST and dehydrogenase from Rhodococcus sp. strain AD45
| Step | Total protein (mg) | Total activity (U) | Sp act (U mg of protein−1) | Purification factor | Yield (%) |
|---|---|---|---|---|---|
| GST | |||||
| Cell extract | 188 | 959 | 5.1 | 1 | 100 |
| (NH4)2SO4 | 233 | 932 | 4.0 | 0.8 | 97 |
| Resource Q | 60 | 786 | 13.1 | 2.6 | 82 |
| Resource Iso | 8.9 | 594 | 66.7 | 13.1 | 62 |
| Dehydrogenase | |||||
| Cell extract | 336 | 1,001 | 2.9 | 1 | 100 |
| DE52 | 96 | 932 | 6.3 | 2.2 | 93 |
| Sephacryl S-300 | 42 | 786 | 8.0 | 2.8 | 79 |
| Blue Sepharose | 15 | 198 | 13.3 | 4.6 | 20 |
| Resource Iso | 18.0 | 6.2 |
FIG. 1.
Cell extract of Rhodococcus sp. strain AD45 grown on isoprene in continuous culture (lane 1), purified GST (lane 2), and purified dehydrogenase (lane 3). Each lane contained approximately 20 μg of protein.
Gel filtration indicated that the native protein had a molecular mass of 46 ± 4 kDa. Since the band obtained with SDS-PAGE represented a polypeptide of 27 ± 2 kDa, the enzyme is probably a homodimer (Fig. 1).
The enzyme was not affected by the addition of 1 mM MnCl2, CoCl2, CuSO4, MgSO4, RbCl2, or ZnSO4. Since the activity was also not influenced by the addition of 5 mM EDTA, we concluded that the enzyme does not require divalent cations for optimal activity. Preincubation of the enzyme with N-ethylmaleimide, p-chloromercuribenzoate, or HgCl2, which react with sulfhydryl groups in proteins, did not have a significant effect on activity. Activity was also not influenced by the addition of 1 mM β-mercaptoethanol. The enzyme could be stored at 4 or −20°C for 3 months with less than 20% loss of activity.
The N-terminal amino acid sequence and amino acid composition (Table 2) were determined to investigate whether the enzyme was related to one of the many GSTs that have been characterized. The N-terminal sequence obtained was Met-Ile-Thr - Val - Tyr - Gly - Tyr - Val - Pro - Ala - Trp - Gly - Ile - Pro - Asp - Ile - Ser-Pro-Tyr-Val-Tyr-Lys-Val-?-Asn-Tyr-?-Thr-Phe-Thr-Gly-Ile.
TABLE 2.
Amino acid composition of the GST of Rhodococcus sp. strain AD45
| Amino acid | No. of residuesa | Amino acid | No. of residues | |
|---|---|---|---|---|
| Asx | 30.3 | Tyr | 9.9 | |
| Glx | 30.8 | Val | 13.3 | |
| Ser | 12.9 | Met | 3.8 | |
| His | 5.0 | Phe | 11.9 | |
| Gly | 23.5 | Ile | 13.4 | |
| Thr | 11.2 | Leu | 21.5 | |
| Ala | 21.9 | Lys | 11.3 | |
| Arg | 12.3 | Pro | 11.5 |
Calculated on the basis of a molecular mass of 27 kDa. Cysteine and tryptophan residues were not determined; hence, the number of the other residues may have been overestimated.
No significant homology was found when the amino acid composition or the N-terminal sequence was compared to those of proteins present in the SwissProt database. However, GSTs typically contain one and often two tyrosine residues between positions 4 and 8 in the N-terminal part of the protein (8, 34). This feature is shared by the enzyme of strain AD45 since it contains tyrosine residues at positions 5 and 7. These tyrosine residues are involved in activation of the sulfhydryl group of bound GSH in all GSTs except the theta class. The latter proteins, to which many bacterial GSTs belong, appear to use a serine residue that is located in the N-terminus of the polypeptide. In the enzyme of strain AD45, this may be the function of the serine at position 17 (8, 35).
Substrate range and kinetics of the GST.
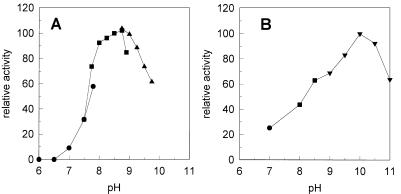
The activities of the GST at different pH values and with various epoxides were measured with on-line GC. Optimal activity was observed at pH 8.5 to 9.0 (Fig. 2). All epoxides tested were substrates for the enzyme, and the relative activities are shown in Table 3. The highest conversion rates were observed with the physiological substrate isoprene monoxide. Other terminal epoxides and epithiopropane were converted at rates of 25 to 37% of that of isoprene monoxide. Another good substrate was the nonterminal epoxide cis-2,3-epoxybutane. Activity with cis-1,2-dichloroepoxyethane was accurately detectable, although the rate of conversion was much lower.
FIG. 2.
Effect of pH on enzyme activity of the GST (A) and the dehydrogenase (B) of Rhodococcus sp. strain AD45. Activity was determined at different pH values in 50 mM potassium phosphate (●), 50 mM Tris-HCl (■), 50 mM sodium carbonate (▴), or 50 mM glycine-NaOH.
TABLE 3.
Substrate specificity of the GST from Rhodococcus sp. strain AD45
| Substrate | % of activitya |
|---|---|
| Isoprene monoxide | 100 |
| Epoxyethane | 37 |
| Epoxypropane | 32 |
| 1,2-Epoxybutane | 26 |
| 1,2-Epoxyhexane | 25 |
| cis-2,3-Epoxybutane | 13 |
| Epithiopropane | 34 |
| Epifluorohydrin | 28 |
| Epichlorohydrin | 31 |
| Epibromohydrin | 25 |
| cis-1,2-Dichloroepoxyethane | 4 |
All activities were determined in 50 mM Tris-HCl buffer (pH 8.5) with 5 mM GSH except for epifluorohydrin, epichlorohydrin, and epibromohydrin, in which case 50 mM Tris-HCl (pH 8.0) was used to reduce the chemical reaction rate between these substrates and GSH. Activities are expressed as percentages of the activity found, at the same pH, with isoprene monoxide.
The enzyme followed Michaelis-Menten kinetics with isoprene monoxide and cis-1,2-dichloroepoxyethane as substrates. At 5 mM GSH, a Vmax of 66 U mg of protein−1 and a Km of 0.1 mM were found with isoprene monoxide as a substrate. With cis-1,2-dichloroepoxyethane, these values were 2.4 U mg of protein−1 and 0.1 mM, respectively. Activity with isoprene monoxide was linearly dependent on the GSH concentration up to 25 mM, above which the nonenzymatic reaction of GSH with isoprene monoxide was too high to allow accurate determination of enzyme-catalyzed reaction rates. From these data, we calculated that the specificity constants (kcat/Km) for GSH are 1.1 × 104 and 4.1 × 102 M−1 s−1 with isoprene monoxide and cis-1,2-dichloroepoxyethane, respectively, as the substrates.
Identification of the reaction product of GSH and isoprene monoxide.
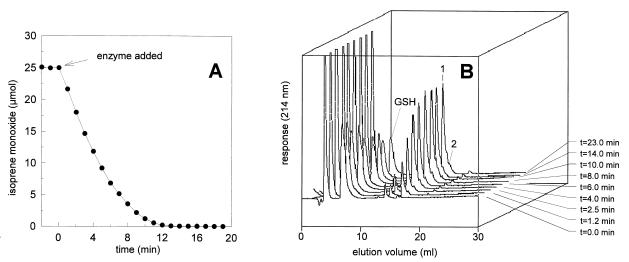
To identify the product formed from isoprene monoxide, we incubated the substrate with twofold excess GSH and analyzed samples by HPLC. The elution profiles showed that the amount of GSH decreased about 50% during conversion, indicating that GSH reacted stoichiometrically with isoprene monoxide (Fig. 3). The major product (compound 1) formed during the degradation of isoprene monoxide eluted at 17 min, and a minor product (compound 2) eluted at 18 min. The ratio between compound 1 and compound 2 was estimated to be 10:1 (Fig. 3 and 6). Analysis with HPLC-MS at 70-V nozzle voltage showed that compound 1 had a molecular ion with m/z 392, which is consistent with the theoretical value for a conjugate of isoprene monoxide and GSH. The presence of such a conjugate was confirmed by the observation of a protonated dimer (m/z 783). At higher nozzle voltage (170 V), collision-induced fragmentation is expected. Indeed, we observed loss of the isoprene moiety from the cysteinyl sulfur, regenerating GSH and producing an ion with m/z 308 (M+ − 84). The other ions observed showed a typical peptide fragmentation pattern (11) from the conjugate generating ions with m/z 392 (M+), 263 (Y"2), 130 (B1), and 76 (Y"1) and for the dealkylated compound with m/z 308 (M+), 291 (Z3), 233 (B2), 179 (Y"2), and 162 (Z2).
FIG. 3.
Conversion of isoprene monoxide (25 μmol) and GSH (50 μmol) and formation GSH-isoprene monoxide conjugates (compound 1 and 2) by the GST of Rhodococcus sp. strain AD45. (A) Depletion of isoprene monoxide measured by on-line GC. (B) HPLC elution profiles of samples that were removed from the assay at different times. The major reaction product (compound 1) was identified as HGMB.
FIG. 6.
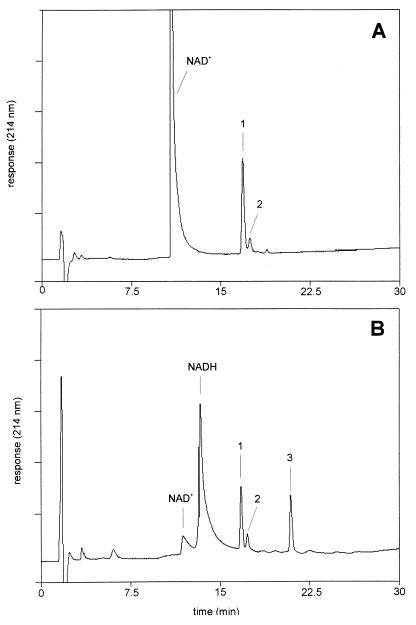
NAD+-dependent HGMB conversion by the dehydrogenase of Rhodococcus sp. strain AD45. HPLC elution profiles of assays without dehydrogenase (A) and with dehydrogenase (70 μg/ml) (B). The assay mixture consisted of 0.4 mM NAD+ and 0.4 mM HGMB in 50 mM glycine-NaOH (pH 10). Activity was monitored by following the generation of NADH at 340 nm, and samples for HPLC-MS analysis were removed at 60 min. Compounds 1 (HGMB) and 2 were identified as conjugates of GSH and isoprene monoxide. Compound 3 was identified as 2-glutathionyl-2-methyl-3-butenoic acid.
For compound 2, analyzed at a nozzle voltage of 70 V, a molecular ion with m/z 392 (M+) was also observed, indicating a GSH conjugate of isoprene monoxide. Results at high ionization energies did not give a clear indication of the structure of this product due to the small amounts present and poor separation at higher column loading. It is likely that compound 2 represents the product formed by substitution of GSH at the less-favored carbon atom in the epoxide ring (see below).
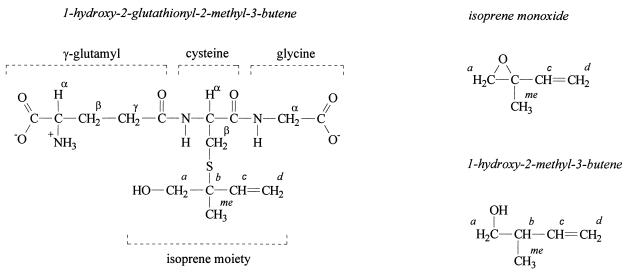
Since no ions arising from fragmentation in the isoprene moiety of compounds 1 and 2 were observed, it was necessary to use NMR to determine whether GSH was linked via a thioether bond to the C-1 or C-2 carbon atom of the isoprene moiety. Thus, the product of the GST-catalyzed reaction of GSH with isoprene monoxide was subjected to 1H NMR analysis, and the spectra were compared with those of related compounds (Fig. 4). Isoprene monoxide is a chiral compound, and since it was used as a racemic mixture, two diastereomeric products would be formed by GSH conjugation. It was found that isoprene oxidation by strain AD45 favors mainly the generation of the (R) enantiomer of isoprene monoxide (enantiomeric excess = 95%) (33). Therefore, we also used isoprene monoxide produced by strain AD45 to synthesize the conjugate with GSH.
FIG. 4.
Structures of compounds subjected to 1H NMR analysis.
With the conjugates formed with racemic isoprene monoxide and with biologically prepared isoprene monoxide, the signals produced by the GSH moiety could easily be identified by comparison with literature values (7, 20, 38) and by comparison with the 1H NMR spectrum of GSH (Table 4). The largest change in chemical shift of the protons of the GSH moiety was observed for the Cys Hβ protons (Table 4), indicating that the GSH moiety is covalently linked via the sulfur atom to the isoprene moiety. Comparison with the 1H NMR spectrum of isoprene monoxide showed that in the conjugates the Ha protons of the isoprene moiety shifted downfield after conjugation with GSH, indicating opening of the epoxide ring. Therefore, the expected reaction product was 1-glutathionyl-2-hydroxy-2-methyl-3-butene or 1-hydroxy-2-glutathionyl-2-methyl-3-butene (HGMB). It was concluded that the GST of strain AD45 catalyzed the nucleophilic attack of GSH on the sterically most hindered carbon atom to yield HGMB for the following reasons. (i) The chemical shifts of the resonances of Ha protons are characteristic for protons on a hydroxyl-substituted carbon atom rather than protons located on a sulfur-substituted carbon atom as in 1-glutathionyl-2-hydroxy-2-methyl-3-butene. (ii) The chemical shift of the resonances of Ha protons were similar in 1-hydroxy-2-methyl-3-butene and HGMB, indicating similar electronic environments.
TABLE 4.
1H NMR spectra of HGMB, GSH, isoprene monoxide, and 1-hydroxy-2-methyl-3-butenea
| H | Chemical shift (intensity, multiplicity)
|
||||
|---|---|---|---|---|---|
| HGMB
|
GSH | Isoprene monoxide | 1-Hydroxy-2-methyl-3-butene | ||
| Racemic | AD45 | ||||
| Glutamate | |||||
| Hα | 3.45 (1, NR) | 3.5 (1, NR) | 3.6 (1, dd) | ||
| Hβ | 1.9 (2, m) | 2.0 (2, m) | 1.9 (2, m) | ||
| Hγ | 2.3 (2, m) | 2.4 (2, m) | 2.3 (2, m) | ||
| Cysteine | |||||
| Hα | 4.35 (1, m) | 4.4 (1, m) | 4.35 (1, d) | ||
| Hβ | 2.65 (1, m)b | 2.70 (1, m) | 2.70 (1, d) | ||
| Hβ′ | 2.80 (1, m) | 2.85 (1, m) | 2.75 (1, d) | ||
| Glycine | |||||
| Hα | 3.55 (1, s)b | 3.6 (2, s)b | 3.6 (2, s) | ||
| Isoprene moiety | |||||
| Ha | 3.451) (NR) | 3.5 (2, NR) | 2.8 (2, dd) | 3.3 (2, dd) | |
| Hb | 2.1 (1, m) | ||||
| Hc | 5.65 (1, dd) | 5.7 (1, dd) | 5.6 (1, m) | 5.6 (1, m) | |
| Hd | 5.05 (1, dd) | 5.1 (2, dd) | 5.3 (2, m) | 4.9 (2, m) | |
| Hme | 1.15 (1.5, s) | 1.2 (0.4, s) | 1.3 (3, s) | 0.8 (3, d) | |
| 1.20 (1.5, s) | 1.25 (2.6, s) | ||||
Spectra were recorded in D2O at room temperature. Chemical shifts were measured relative to sodium 4,4-dimethyl-4-silapentane-1-sulfonate and assigned with 0.05-ppm accuracy. The protons are assigned with reference to Fig. 5. NR, not resolved. Multiplicity of the Hα (Glu), and Ha (isoprene moiety) protons could not be resolved due to overlap of spectral lines.
Singlet Hα (Gly) protons gave two signals due to an increased rotational barrier in the conjugate compared to unmodified GSH.
The methyl group of the isoprene monoxide moiety in HGMB that was synthesized with racemic isoprene monoxide produced two signals with similar intensities (each representing 1.5 protons) and a difference of 0.05 ppm in chemical shift (Table 4). With HGMB synthesized from isoprene monoxide that was produced by strain AD45, the intensity of the signal at 1.25 ppm represented 2.6 protons whereas the signal at 1.20 ppm represented 0.4 proton. This is in agreement with the enantiomeric excess of 70 to 80%, and therefore we concluded that these signals arose from diastereomeric effects.
Purification of the HGMB-dependent dehydrogenase.
Experiments were performed to study the metabolism of HGMB. When NAD+ and HGMB were added to extracts of isoprene-grown cells, rapid formation of NADH was observed. The enzyme catalyzing this activity was purified from isoprene-grown cells that were harvested from a continuous culture (Table 1). The 6.2-fold increase of the specific activity that was observed upon purification indicates that the dehydrogenase represented approximately 16% of the total soluble protein. SDS-PAGE analysis indeed showed that the dehydrogenase was one of the major proteins present in crude extracts of strain AD45 grown on isoprene, although the content may be less than 16% (Fig. 1). The similar intensities of the bands representing the GST and the dehydrogenase indicate that some dehydrogenase was inactivated during the purification procedure, which is in agreement with the low yield.
Optimal activity was observed in a glycine-NaOH buffer at pH 9 to 10 (Fig. 2). No NADH formation was observed when GSH, methanol, ethanol, glycerol, 2-methylbutanol, 3-methylbutanol, 1-hydroxy-3-methyl-3-butene, or 1,2-dihydroxy-2-methyl-3-butene was used as a substrate. We also tested whether the conjugates of GSH with other 1,2-epoxyalkanes would be substrates of the enzyme. When the conjugate of GSH and epoxypropane was added as a substrate, NADH formation was detected at a rate that was eightfold lower than that with HGMB. The conjugates of GSH with 1,2-epoxybutane and 1,2-epoxyhexane were not substrates for the enzyme.
HGMB oxidation followed Michaelis-Menten kinetics for both NAD+ and HGMB. The Km for NAD+ was 0.07 mM at 7.5 mM HGMB. The Km for HGMB was 1.4 mM, and the Vmax was 18 U mg of protein−1.
Analysis of the N-terminal amino acid sequence of the protein by Edman degradation yielded the 23 residues as shown in Fig. 5. Significant homology was found with enzymes of the short-chain dehydrogenase/reductase (SDR) family (Fig. 5), with the highest score for the 3-oxoacyl-acyl carrier protein reductase, an enzyme that is involved in fatty acid biosynthesis and that requires NADP+ for activity. The N-terminal sequence of proteins of the SDR family typically contains a GXXXGXG motif that is involved in the binding of the NAD+ or NADP+ (21). This motif is also present in the enzyme of strain AD45, but the last glycine residue is replaced by an alanine.
FIG. 5.
Alignment of the N-terminal amino acid sequence of the dehydrogenase of Rhodococcus sp. strain AD45 with enzymes belonging to the SDR family. Abbreviations: DH_AD45, dehydrogenase of Rhodococcus sp. strain AD45; DHCA_MOUSE, NADPH-dependent carbonyl reductase from Mus musculus; DHB2_MOUSE, 17 β-hydroxysteroid dehydrogenase 2 from M. musculus; FABG_MYCSM, 3-oxoacyl-acyl carrier protein reductase from Mycobacterium smegmatis; KDUD_BACSU, 2-deoxy-d-gluconate 3-dehydrogenase from Bacillus subtilis.
Identification of the product of the reaction that is catalyzed by the dehydrogenase.
A sample removed from a dehydrogenase assay containing NAD+ and HGMB in a 1:1 molar ratio was analyzed by HPLC-MS (Fig. 6). NADH generation was accompanied by a decrease of HGMB and the generation of a product eluting at 21 min (compound 3). Strikingly, no decrease of compound 2 was observed, emphasizing the high specificity of the enzyme for HGMB (compound 1). Mass spectrometry analysis of compound 3 showed the presence of a molecular ion with m/z 406 and collision-induced fragments with m/z 331 (B2), 303 (A2, weak), 277 (Y"2), 174 (I2), 130 (B1), and 76 (Y"1). Furthermore, a fragment was observed with m/z 259, which is consistent with loss of the carboxyl function in the isoprene moiety from the A2 fragment. From these data, it was concluded that the hydroxyl function in HGMB is oxidized to a carboxyl function to yield 2-glutathionyl-2-methyl-3-butenoic acid.
In the presence of excess HGMB, the formation of a small amount of another product was observed. The compound eluted at 18.5 min and yielded a molecular ion with m/z 390. This is consistent with the molecular ion of the theoretical product of the oxidation of the hydroxyl function in the conjugate to a carbonyl function. From these data we conclude that the dehydrogenase catalyzes the two-step NAD+-dependent oxidation of HGMB. Initially, HGMB is oxidized to 1-oxo-2-glutathionyl-2-methyl-3-butene, which is then oxidized to 2-glutathionyl-2-methyl-3-butenoic acid.
DISCUSSION
A wide range of enzymes, such as hydrolases, reductases, isomerases, metalloglutathione S-transferases, lyases, and carboxylases, are involved in microbial metabolism of epoxides (3, 6, 7, 9, 13, 16, 17, 28, 36). In this paper, we report the purification and characterization of a GST that catalyzes the GSH-dependent metabolism of epoxides in the isoprene-utilizing bacterium Rhodococcus sp. strain AD45. The enzyme had activity with a broad range of epoxides. The best substrate was isoprene monoxide, which is the primary oxidation product of isoprene. The enzyme followed Michaelis-Menten-type kinetics for epoxides but had a very low affinity for GSH, even compared to other bacterial GSTs. With only a few exceptions, all known bacterial GSTs belong to the theta class (34), and the affinity for GSH of these enzymes is often low compared to GSTs of other classes (24).
Another unusual feature of the enzyme of strain AD45 is the high pH optimum that coincides with the pKa values of the cysteine residue of GSH in aqueous solution. These data suggest that unlike with most GSTs, the pKa of the cysteine residue is not significantly lowered by interactions with residues in the active site of the enzyme. To our knowledge, the only other GST for which a similarly high pH optimum was observed is FosA. This protein is involved in bacterial fosfomycin (1,2-epoxypropylphosphonic acid) resistance and catalyzes the reaction of GSH and fosfomycin to form 1-glutathionyl-2-hydroxypropylphosphonic acid. However, unlike the GST of strain AD45, which does not need divalent metal ions for activity, FosA is a metalloglutathione S-transferase that is homologous to extradiol dioxygenases and glyoxalase I, rather than a theta class GST (9).
The low affinity of the GST of strain AD45 for GSH and the high pH optimum raised the question of whether GSH is really the physiological cofactor of the enzyme. Previously we showed that in 1,2-epoxyhexane-exposed cells, the conjugate of GSH and 1,2-epoxyhexane accumulated, which indicated that strain AD45 indeed contains an enzyme that covalently couples GSH to epoxides (32). We have checked extracts of isoprene-grown cells of strain AD45 for the presence of other enzymes that have activity for aliphatic epoxides. However, no activity for isoprene monoxide was detected in assays for epoxide hydrolase, epoxide isomerase, or epoxide hydrogenase (32). Furthermore, the GST identified here was one of the two most prominent proteins in isoprene-grown cells of strain AD45.
The GST catalyzes the reaction of GSH and isoprene monoxide to form a stable conjugate. The major product is the conjugate in which the GSH moiety is covalently linked to the tertiary carbon atom of the isoprene monoxide moiety to yield HGMB. This shows that the enzyme catalyzes the nucleophilic attack of GSH to the sterically most hindered carbon atom in the epoxide ring.
Epoxide carboxylation was shown to be a key step in the metabolism of epoxides in both gram-positive and gram-negative epoxypropane-utilizing organisms (3, 4). In these organisms carboxylation is catalyzed by a multiprotein complex that catalyzes epoxide ring opening and the formation of a carbon-carbon bond. Simultaneously, transhydrogenation occurs in which NADPH is oxidized and NAD+ is reduced. Strikingly, 2-methyl-1,2-epoxypropane, which contains a methyl group rather than a hydrogen substituent at the C-2 carbon atom, is a mechanism-based inactivator of epoxide carboxylase activity (2). Reaction of this compound with the carboxylase is thought to result in a covalently modified active site that cannot react further due to the absence of an extractable hydrogen at the C-2 position. Isoprene monoxide does also not contain an extractable hydrogen at C-2, and therefore isoprene monoxide conversion would not be possible by the carboxylation route. In strain AD45, this problem appears to be addressed by a GST-catalyzed conjugation to GSH to yield HGMB. Hence, the GST described here represents a novel type of catabolic epoxide-converting enzyme.
Degradation of HGMB in strain AD45 proceeds by a dehydrogenase that catalyzes the two-step oxidation of the alcohol function to yield 2-glutathionyl-2-methyl-3-butenoic acid with the concomitant reduction of NAD+ to NADH. In contrast to the GST, the dehydrogenase has a very narrow substrate range and seems to be optimized for the oxidation of the HGMB. The toxicity of 1,2-epoxyhexane (32) can now be explained considering the broad substrate range of the GST. The conjugate of 1,2-epoxyhexane and GSH that is formed by this enzyme is not a substrate for the dehydrogenase and will accumulate. Hence, intracellular GSH concentrations will decrease and isoprene monoxide conversion will be inhibited.
The initial steps in the degradation pathway of isoprene in Rhodococcus sp. strain AD45 are summarized in Fig. 7. Degradation starts with oxidation of the methyl-substituted double bond by a monooxygenase to yield 1,2-epoxy-2-methyl-3-butene (32). After conversion by the GST and the two oxidation steps by the dehydrogenase, 2-glutathionyl-2-methyl-3-butenoic acid is generated.
FIG. 7.
Initial steps in isoprene degradation by Rhodococcus sp. strain AD45.
It remains to be elucidated how further degradation proceeds. Currently, we are investigating the genetics of isoprene degradation which may be helpful in understanding the complete metabolic pathway for isoprene.
ACKNOWLEDGMENTS
The work of J.E.T.H.V. was financed by grant IOP91204 from the Dutch IOP Environmental Biotechnology Programme and grant ENV5-CT95-0086 from the EU Environment and Climate Programme.
Piet Wietzes is acknowledged for technical support. C. Margot Jeronimus-Stratingh and Andries P. Bruins (Department of Pharmacy, University of Groningen, Groningen, The Netherlands) are acknowledged for mass spectrometry analysis. Jeffrey Lutje Spelberg is acknowledged for assistance in chiral GC analysis.
REFERENCES
- 1.Allen J R, Ensign S A. Carboxylation of epoxides to β-ketoacids in cell extracts of Xanthobacter strain Py2. J Bacteriol. 1996;178:1469–1472. doi: 10.1128/jb.178.5.1469-1472.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J R, Ensign S A. Characterization of three protein components required for functional reconstitution of the epoxide carboxylase multienzyme complex from Xanthobacter strain Py2. J Bacteriol. 1997;179:3110–3115. doi: 10.1128/jb.179.10.3110-3115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen J R, Ensign S A. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J Biol Chem. 1997;272:32121–32128. doi: 10.1074/jbc.272.51.32121. [DOI] [PubMed] [Google Scholar]
- 4.Allen J R, Ensign S A. Identification and characterization of epoxide carboxylase activity in cell extracts of Nocardia corallina B276. J Bacteriol. 1998;180:2072–2078. doi: 10.1128/jb.180.8.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Arca P, Hardisson C, Suarez J E. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob Agents Chemother. 1990;34:844–848. doi: 10.1128/aac.34.5.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arca P, Rico M, Brana A F, Villar C J, Hardisson C, Suarez J E. Formation of an adduct between fosfomycin and glutathione: a new mechanism of antibiotic resistance in bacteria. Antimicrob Agents Chemother. 1988;32:1552–1556. doi: 10.1128/aac.32.10.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong R N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem Res Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 9.Bernat A B, Laughlin L T, Armstrong R N. Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry. 1997;36:3050–3055. doi: 10.1021/bi963172a. [DOI] [PubMed] [Google Scholar]
- 10.Bruins A P, Covey T R, Henion J D. Ion spray interface for combined liquid chromatography/atmospheric pressure ionization mass spectrometry. Anal Chem. 1987;59:2642–2646. [Google Scholar]
- 11.Carr S A, Hemling M E, Bean M F, Roberts G D. Integration of mass spectrometry in analytical biotechnology. Anal Chem. 1991;63:2802–2824. doi: 10.1021/ac00024a003. [DOI] [PubMed] [Google Scholar]
- 12.Daubaras D L, Hershberger C D, Kitano K, Chakrabarty A M. Sequence analysis of a gene cluster involved in metabolism of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1995;61:1279–1289. doi: 10.1128/aem.61.4.1279-1289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Bont J A M, Harder W. Metabolism of ethylene by Mycobacterium E20. FEMS Microbiol Lett. 1978;3:89–93. [Google Scholar]
- 14.Field J A, Thurman E M. Glutathione conjugation and contaminant transformation. Environ Sci Technol. 1996;30:1413–1418. [Google Scholar]
- 15.Fox B G, Borneman J G, Wackett L P, Lipscomb J D. Haloalkene oxidation by the soluble methane monooxygenase from Methylosinus trichosporium OB3b: mechanistic and environmental implications. Biochemistry. 1990;29:6419–6427. doi: 10.1021/bi00479a013. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths E T, Harries P C, Jeffcoat R, Trudgill P W. Purification and properties of α-pinene oxide lyase from Nocardia sp. strain P18.3. J Bacteriol. 1987;169:4972–4979. doi: 10.1128/jb.169.11.4980-4983.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmans S, Smits J P, van der Werf M J, Volkering F, De Bont J A M. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter 124X. Appl Environ Microbiol. 1989;55:2850–2855. doi: 10.1128/aem.55.11.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes J D, Pulford D J. The glutathione S-transferase super-gene family and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 19.Hobohm U, Houthaeve T, Sander C. Amino acid analysis and protein database compositional search as a rapid and inexpensive method to identify proteins. Anal Biochem. 1994;222:202–209. doi: 10.1006/abio.1994.1474. [DOI] [PubMed] [Google Scholar]
- 20.Huckerby T N, Tudor A J, Dawber J G. Acid-base studies of glutathione (l-γ-glutamyl-l-cysteinyl-l-glycine) by one- and two-dimensional nuclear magnetic resonance spectroscopy. J. Chem. Soc. Perkin Trans. 2. 1985. pp. 759–763. [Google Scholar]
- 21.Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 22.Leisinger T, Bader R, Hermann R, Schmid-Appert M, Vuilleumier S. Microbes, enzymes and genes involved in dichloromethane utilization. Biodegradation. 1994;5:237–248. doi: 10.1007/BF00696462. [DOI] [PubMed] [Google Scholar]
- 23.Masai E, Katayama Y, Kubota S, Kawai S, Yamasaki M, Morohoshi N. A bacterial enzyme degrading the model lignin compound-etherase is a member of the glutathione S-transferase superfamily. FEBS Lett. 1993;323:135–140. doi: 10.1016/0014-5793(93)81465-c. [DOI] [PubMed] [Google Scholar]
- 24.Meyer D J. Significance of an unusually low Km for glutathione in glutathione S-transferases of the α, μ, and π classes. Xenobiotica. 1993;23:823–834. doi: 10.3109/00498259309059411. [DOI] [PubMed] [Google Scholar]
- 25.Oldenhuis R, Oedzes J Y, van der Waarde J J, Janssen D B. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol. 1991;57:7–14. doi: 10.1128/aem.57.1.7-14.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pries F, Kingma J, Krooshof G H, Jeronimus-Stratingh C M, Bruins A P, Janssen D B. Histidine 289 is essential for hydrolysis in the alkyl-enzyme intermediate of haloalkane dehalogenase. J Biol Chem. 1995;270:10405–10411. doi: 10.1074/jbc.270.18.10405. [DOI] [PubMed] [Google Scholar]
- 27.Rink R, Fennema M, Smids M, Dehmel U, Janssen D B. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- 28.Shirai K, Hisatsuka K. Production of β-phenetyl alcohol from styrene by Pseudomonas 305-STR-1-4. Agric Biol Chem. 1979;43:1399–1406. [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Hylckama Vlieg J E T, de Koning W, Janssen D B. Transformation kinetics of chlorinated ethenes by Methylosinus trichosporium OB3b and detection of unstable epoxides by on-line gas chromatography. Appl Environ Microbiol. 1996;62:3304–3312. doi: 10.1128/aem.62.9.3304-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Hylckama Vlieg J E T, de Koning W, Janssen D B. Effect of chlorinated ethene conversion on viability and activity of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1997;63:4961–4964. doi: 10.1128/aem.63.12.4961-4964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Hylckama Vlieg J E T, Kingma J, van den Wijngaard A J, Janssen D B. A glutathione S-transferase with activity toward cis-1,2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl Environ Microbiol. 1998;64:2800–2805. doi: 10.1128/aem.64.8.2800-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Hylckama Vlieg, J. E. T., H. Leemhuis, J. Lutje Spelberg, and D. B. Janssen. Unpublished data.
- 34.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vuilleumier S, Leisinger T. Protein engineering studies of dichloromethane dehalogenase/glutathione S-transferase from Methylophilus sp. strain DM11. Ser12 but not Tyr6 is required for enzyme activity. Eur J Biochem. 1996;239:410–417. doi: 10.1111/j.1432-1033.1996.0410u.x. [DOI] [PubMed] [Google Scholar]
- 36.Weijers C A G M, de Haan A, de Bont J A M. Microbial production and metabolism of epoxides. Microb Sci. 1988;5:156–159. [PubMed] [Google Scholar]
- 37.Xun L, Topp E, Orser C S. Purification and characterization of a tetrachloro-p-hydroquinone reductive dehalogenase from a Flavobacterium sp. J Bacteriol. 1992;174:8003–8007. doi: 10.1128/jb.174.24.8003-8007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.York M J, Beilharz G R, Kuchel P W. Conformation of reduced glutathione in aqueous solution by 1H and 13C N.M.R. Int J Peptide Protein Res. 1987;29:638–646. doi: 10.1111/j.1399-3011.1987.tb02294.x. [DOI] [PubMed] [Google Scholar]