Abstract
The expression of CS1 pili by enterotoxigenic strains of Escherichia coli is regulated at the transcriptional level and requires the virulence regulator Rns, a member of the AraC family of regulatory proteins. Rns binds at two separate sites upstream of Pcoo (the promoter of CS1 pilin genes), which were identified in vitro with an MBP::Rns fusion protein in gel mobility and DNase I footprinting assays. At each site, Rns recognizes asymmetric nucleotide sequences in two regions of the major groove and binds along one face of the DNA helix. Both binding sites are required for activation of Pcoo in vivo, because mutagenesis of either site significantly reduced the level of expression from this promoter. Thus, Rns regulates the expression of CS1 pilin genes directly, not via a regulatory cascade. Analysis of Rns-nucleotide interactions at each site suggests that binding sites for Rns and related virulence regulators are not easily identified because they do not bind palindromic or repeated sequences. A strategy to identify asymmetric binding sites is presented and applied to locate potential binding sites upstream of other genes that Rns can activate, including those encoding the CS2 and CFA/I pili of enterotoxigenic E. coli and the global regulator virB of Shigella flexneri.
Pili, which are long proteinaceous rod-like structures extending from the surfaces of bacteria, often serve as adherence factors of bacterial pathogens. Enterotoxigenic strains of Escherichia coli (ETEC), a group of enteric pathogens that cause diarrheal disease in humans and animals, may express one or more of at least 20 antigenically distinct pili (11). These include the CS1 group, consisting of CS1, CS2, CS4, CS14, CS17, CS19, and CFA/I. The amino acid sequences of pili within this group are highly conserved, although these pili differ in their antigenicities and probably in the host receptors to which they bind (35). The ability to adhere to host receptors increases the ability of piliated ETEC to colonize a host and to establish an infection because they are able to resist being rapidly flushed from the gastrointestinal tract (38).
The proteins involved in synthesis of the CS1 group of pili are unrelated to those of other types of pili and therefore constitute a class that differs from the well-known Pap and type IV pilus classes. In addition, all pili within the CS1 group require a regulator for their expression. One such regulator, Rns, activates transcription of the genes encoding CS1 and CS2 pili (3). Activators with significant homology to Rns have also been shown to control expression of ETEC pili unrelated to the CS1 group, including the Pap-related pili CS5 and 987P, which are dependent upon CsvR and FapR, respectively (7, 23). In several other bacterial pathogens, type IV pili are regulated by proteins with homology to Rns. The regulatory proteins for these include PerA (BfpT) of enteropathogenic E. coli, AggR of enteroaggregative E. coli, and ToxT (TcpN) of Vibrio cholerae, which control the expression of bundle forming, AAF/I, and toxin-coregulated pili, respectively (18, 30, 39).
Virulence factors regulated by Rns-like activators are not limited to pili. For example, in addition to the activation of genes encoding the bundle-forming pilus, PerA is also needed for expression of eaeA, encoding the membrane protein intimin, which is required for close contact of enteropathogenic E. coli with host epithelial cells, and the esp genes, which encode secreted proteins that induce signal transduction pathways in host epithelial cells (13, 22). UreR of uropathogenic strains of Proteus mirabilis, Providencia stuartii, and E. coli regulates the expression of an operon for the catalysis of urea to ammonia and carbamate (8). The resulting alkaline environment is thought to enhance the survival and virulence of the uropathogen within the urinary tract. VirF from the genus Yersinia regulates the expression of multiple virulence factors, including secreted Yop proteins encoded by unlinked genes present on the same virulence plasmid that encodes VirF (6). Similarly, VirF of Shigella flexneri regulates plasmid-encoded virulence factors required for invasion and spreading within epithelial cells (9).
Some of these Rns-like virulence regulators are so closely related that they can substitute for one another. Both Rns and CsvR can complement cfaR null mutations for the expression of CFA/I pili in ETEC strains (5, 7). Rns can also complement virF null mutations for the expression of multiple virulence factors in S. flexneri (32). CfaR can complement rns null mutations for expression of CS1 and CS2 pili and aggR null mutations for expression of AAF/I pili (5, 30). Since these regulators are all thought to be DNA binding proteins, the ability of these activators to substitute functionally for each other suggests that they recognize similar DNA binding sites.
Rns and its homologs are related to the AraC family of regulators that includes over 100 members (for a review, see reference 12). Most of the proteins in this family contain 260 to 300 amino acid residues, and most are activators of transcription. Sequence conservation among family members is highest in the carboxy termini, which are known or thought to compose the DNA binding domains of these regulators. The crystal structure of MarA, an AraC family member that regulates the expression of the multiple antibiotic resistance regulon in E. coli (19, 26), reveals that its DNA binding domain carries two helix-turn-helix motifs and that a recognition helix of each motif is placed in the major groove of DNA (34). Rns and other AraC family members probably bind DNA in a similar manner because secondary structure predictions suggest that each has two helix-turn-helix motifs in its carboxy terminus.
The amino terminus of most AraC-type regulators is known or thought to constitute an effector-binding domain for small molecules. The crystal structure of the amino terminus of AraC also reveals that it is involved in protein dimerization (37). With the exception of UreR, which responds to urea, none of the virulence regulators of this family has been found to respond to an effector molecule (12). However, biochemical characterization of these virulence regulators is relatively new compared to that of AraC, and future work may uncover effector molecules for Rns and similar activators. Alternatively, Rns and related regulators may not require effector molecules to function as activators. In this case, the amino termini of these virulence regulators may serve only as dimerization domains.
Some of the activators in the AraC family regulate the expression of genes directly, while others act indirectly through regulatory cascades. For example, VirF of S. flexneri is an indirect regulator, inducing the expression of invasion genes through positive regulation of an unrelated regulator, VirB (1). VirF of Yersinia enterocolitica is a direct activator, binding upstream of promoters for several virulence genes of the yop regulon, including yopE, yopH, virC, and lcrG (41). PerA of enteropathogenic E. coli also acts directly by binding in the vicinity of promoters of genes encoding the bundle-forming pilus and the intimin gene eaeA (39). Although it has been shown that each of these regulators is a DNA binding protein, the exact nucleotides that constitute a binding site are not known and in most cases only the approximate position of the binding site is available.
Because of the ready availability of genome sequence databases, a clearer definition of the binding sites for these and other regulators would facilitate the identification of virulence genes and their expression. Therefore, to advance our understanding of DNA binding site recognition by Rns and related virulence regulators, we characterized Rns-nucleotide interactions in vitro and in vivo. This information was then applied to identify potential Rns binding sites upstream of loci encoding probable or known virulence factors in different enteric bacterial pathogens.
MATERIALS AND METHODS
Plasmid and phage constructs.
The Rns expression plasmid pEU2080 was constructed by amplifying the rns gene from plasmid pEU2030 (10) with Pfu DNA polymerase (Stratagene) using primers rnsNcoI (AGGTATAccATGGACTTTAAATACACTGA) and 1201 (AACAGCTATGACCATG). Primer rnsNcoI introduces two base changes, denoted by lowercase letters, immediately before the first codon of the rns gene, producing a NcoI restriction site at the beginning of rns without altering the coding sequence. The expression vector pBAD24 (15) was digested with HindIII and 5′ overhangs were blunted by end filling with the Klenow fragment of DNA polymerase and then digested with NcoI. The 1-kb rns PCR product was then digested with NcoI and cloned between the NcoI and blunted HindIII sites of pBAD24. This arrangement places the expression of Rns under the control of the arabinose-inducible promoter ParaBAD.
Plasmid pEU2082 carries the wild-type DNA fragment of the CS1 promoter Pcoo from −411 to +529 (numbering relative to the transcription start site) from pEU2061 (28) cloned into vector pNEB193 (New England Biolabs). In pEU2082, the 1-kb Pcoo fragment is flanked by restriction sites for BamHI and EcoRI located upstream and downstream of the promoter, respectively. Mutagenic oligonucleotides were used in inverse PCRs on pEU2082, with Pfu DNA polymerase to generate specific point mutations within Rns binding sites upstream of Pcoo. Plasmid pEU2086 carries an A-to-G transition at −45, and plasmid pEU2101 carries a T-to-C transition at −106. Plasmid pEU2086 was used in inverse PCR to generate a double mutant, with a T-to-C change at −106 and an A-to-G change at −45, resulting in plasmid pEU2102.
Reporter plasmids were constructed by cloning the wild-type and mutant Pcoo constructs as 1-kb BamHI-EcoRI fragments into pRS550 (36) digested with BamHI and EcoRI, which are immediately upstream of a promoterless lacZ gene. Reporter plasmids pEU2108, pEU2105, pEU2106, and pEU2107 carry Pcoo constructs cloned from pEU2082, pEU2086, pEU2101, and pEU2102, respectively. λ reporter constructs were generated by homologous recombination in vivo between Pcoo::lacZ reporter plasmids described above and a resident λRS45 prophage (36). λEU2108, λEU2105, λEU2106, and λEU2107 are the products of homologous recombination of λRS45 with pEU2108, pEU2105, pEU2106, and pEU2107, respectively.
Expression and purification of MBP::Rns.
The IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible MBP::Rns expression plasmid, pEU750, was constructed by cloning rns downstream and in frame with malE in vector pMALc2 (New England Biolabs) (28). Strain JM83/pEU750 was grown in Luria-Bertani (LB) medium with 0.2% glucose and 100 μg of ampicillin/ml at 30°C with aeration. The expression of MBP::Rns was induced by addition of IPTG to 300 μM when the culture density reached an optical absorbance at 600 nm of 0.6 to 0.8. The culture was incubated for an additional 2 to 3 h at 30°C, and the cells were pelleted at 4°C and concentrated 100-fold in ice-cold buffer A (10 mM Tris-Cl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol). Cell suspensions were lysed mechanically at 4°C by passage through a French press two to three times. Insoluble material was removed by centrifugation at 18,000 × g for 30 min at 4°C. When necessary to remove residual particulate material, the supernatant was passed through a 0.45-μm-pore-size cellulose acetate syringe tip filter.
MBP::Rns was bound to an amylose resin column equilibrated with buffer A at 4°C and then eluted with 10 mM maltose. Fractions containing MBP::Rns were then applied to a 1-ml heparin column (HiTrap; Pharmacia) equilibrated with buffer A at room temperature. MBP::Rns was eluted from the heparin column in buffer A at 280 mM NaCl and stored at −70°C. The concentration of MBP::Rns was determined by the Bradford method in relation to a standard curve for bovine serum albumin (BSA) without correction for potential differences in dye reactivity between MBP::Rns and BSA.
Preparation of DNA fragments.
DNA for gel mobility, DNase I footprinting, and uracil interference assays was prepared by PCR with 32P end-labeled primers or by incorporation of radiolabeled dATP. The labeled PCR products were separated on nondenaturing acrylamide gels, visualized by autoradiography of the gel, and recovered by crush-soak elution. Eluted DNA was recovered from suspension by binding to Quick Spin PCR columns (Qiagen) and eluted with water.
Gel mobility assay.
Radiolabeled DNA fragments were incubated with MBP::Rns at 37°C for 10 to 30 min in binding buffer (10 mM Tris-Cl [pH 7.4], 50 mM KCl, 1 mM dithiothreitol, 2 ng of poly(dI-dC)/μl, 100 μg of BSA/ml). Glycerol was added to a final concentration of 6.5%, and samples were loaded onto 4 to 6% nondenaturing acrylamide gels with TAE (40 mM Tris-acetate, 1 mM EDTA, pH 8.5) as the gel and running buffer. The gels were run at room temperature, dried, and visualized by exposure to phosphorimager plates.
DNase I protection assay.
DNase I protection assays were conducted as described previously (2) with the following modifications. End-labeled DNA fragments were preincubated with or without MBP::Rns at 37°C for 10 to 30 min in assay buffer (10 mM Tris-Cl [pH 7.4], 50 mM KCl, 1 mM dithiothreitol, 2 ng of poly(dI-dC)/μl, 400 μM MgCl2, 200 μM CaCl2, 100 μg of BSA/ml). DNase I, prepared from lyophilized enzyme (Sigma), was added to 100 ng/ml for 1 min at 37°C and then quenched by addition of 10 volumes of ice-cold precipitation buffer (570 mM NH4OAc, 50 μg of tRNA/ml, 80% ethanol).
Uracil binding interference assay.
The uracil interference assay was done as described previously (33) with the following modifications. One PCR primer was labeled with 32P to generate products that were labeled on only one end. PCR was performed with Taq DNA polymerase and a 1:20 molar ratio of dUTP to dTTP, producing DNA fragments with random substitutions of uracil for thymine. Under these conditions, each Rns binding site carries a maximum of one uracil substitution per strand because the ratio of dUTP to dTTP dictates the substitution frequency and each binding site contains 21 or fewer thymines per strand. Each of the uracil-substituted DNA fragments had only one DNA binding site, and DNA binding conditions were the same as those for gel mobility assays described above. DNA fragments bound by MBP::Rns and DNA fragments not exposed to the protein were recovered from acrylamide gels by crush-soak elution and Quick Spin PCR columns, as described above for the preparation of DNA fragments. The recovered DNA was then treated with uracil-DNA glycosylase (New England Biolabs), an enzyme that hydrolyzes uracil from DNA. The DNA fragments were then treated with piperidine to cleave the phosphodiester backbone at each position lacking a nitrogenous base, and the products were analyzed on denaturing acrylamide gels. DNA fragments from bases −213 to −72 and bases −105 to +83 (numbering relative to the transcription start site of Pcoo) were used to assay binding to the coding strands of site I and site II, respectively. Binding to the noncoding strands of site I and site II was assayed with DNA fragments from bases −213 to −78 and bases −71 to +83, respectively.
Enzymatic assay.
Strains MC4100/pEU745/pEU750 and MC4100/pEU745/pMalc2 were grown to log phase in LB medium with 100 μg of ampicillin/ml and 100 μg of spectinomycin/ml at 37°C and assayed for β-galactosidase activity as described previously (27). MC4100 lysogens carrying Pcoo::lacZ reporter prophage and pEU2080 were grown to log phase at 37°C in LB medium with 0.2% glucose, 100 μg of ampicillin/ml, and 50 μg of kanamycin/ml. Under these conditions, the expression of Rns from pEU2080 was repressed. At time zero, the strains were pelleted, washed once with an equal volume of LB medium, and then diluted fivefold in LB medium with 0.1% arabinose, 100 μg of ampicillin/ml, and 50 μg of kanamycin/ml to induce the expression of Rns from pEU2080. Buffers for cell lysis and β-galactosidase assays were as described previously (27) except that enzymatic reactions were not quenched with Na2CO3. Rather, for each sample the absorbance at 420 nm was continuously monitored for 1 h with an enzyme-linked immunosorbent assay plate reader to develop a kinetic plot. Enzymatic activity was quantitated as the maximum slope of each kinetic plot, Vmax, divided by the optical density of the cell culture at 600 nm. Enzymatic assays were repeated in three separate experiments, and triplicate cultures of each lysogen were assayed within each experiment.
RESULTS
Purification of Rns.
Rns was expressed from an IPTG-inducible Ptac promoter as a 73-kDa fusion protein with the maltose binding protein (MBP) at its amino terminus and was affinity purified on an amylose resin column. Since the fusion protein accounted for only about half of the total protein mass eluted from the column, the eluent was applied to a heparin affinity column with 200 mM NaCl in the column buffer. MBP::Rns eluted from the heparin column at 280 mM NaCl as a single peak. This two-column purification method resulted in a solution containing MBP::Rns that was about 90% pure, as estimated from Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (data not shown).
Cleavage of the 73-kDa fusion protein with protease factor Xa produced two bands on SDS-PAGE. One band ran with an apparent molecular mass of 42 kDa, expected for MBP, and the other ran with an apparent molecular mass of 31 kDa, expected for Rns. The amino-terminal sequence of the 31-kDa protein was found to be AMDFKYTEE. Residues 2 through 8 of this protein were identical to the predicted first seven residues of Rns, and the alanine at position 1 is the result of cloning the rns gene into the expression vector. Thus, factor Xa cleaved the fusion protein at the expected site between MBP and Rns. However, while MBP remained in solution following digestion of the fusion protein with factor Xa, about 50 to 80% of the Rns moiety precipitated. This insolubility is a typical characteristic of regulators within the AraC family and has hampered the analysis of these proteins in vitro (12).
Because of the low solubility of Rns, we wished to use the more soluble fusion protein for in vitro studies. To determine whether the addition of MBP to the amino terminus of Rns affects its activity in vivo, the ability of the fusion protein to activate expression of β-galactosidase from pEU745 was assessed. Plasmid pEU745 carries a Pcoo::lacZ reporter plasmid, and expression of β-galactosidase has been shown to be positively regulated by Rns (29). Plasmid pEU750, which expresses MBP::Rns from the IPTG-inducible Ptac promoter, was compared to pMALc2, the vector in which the fusion protein was cloned, for the ability to activate this reporter plasmid. Both MC410/pEU745/pEU750 and MC4100/pEU745/pMALc2 expressed 600 to 800 Miller units of β-galactosidase in the absence of IPTG induction. However 1 h after the addition of IPTG to 400 μM, expression of β-galactosidase increased to 15,000 Miller units in the strain carrying pEU750 while there was no increase in β-galactosidase in the strain carrying pMALc2. Thus MBP::Rns, like Rns, positively regulates expression from Pcoo, indicating that it is appropriate to use the fusion protein for in vitro analysis of Rns.
Identification of Rns binding sites at Pcoo.
Deletion analysis of a Pcoo::lacZ reporter plasmid in vivo showed that a DNA fragment from bases −411 to +7 (numbering relative to the transcription start site) is sufficient for expression of β-galactosidase to be dependent on Rns (29). A similar Pcoo fragment, from −417 to +83, was used in DNA binding assays with MBP::Rns in vitro. In gel mobility assays, the fusion protein bound to this 500-bp fragment, retarding the mobility of labeled Pcoo DNA (Fig. 1). Protein binding was optimal at 50 mM KCl and inhibited below 10 or above 130 mM KCl (data not shown). A similar trend was observed when NaCl replaced KCl in the binding buffer, but Rns binding was about twofold greater with K+ than with Na+ as the counterion. Extended incubation of protein in solution with DNA at 37°C did not decrease MBP::Rns binding, indicating that the activity of the fusion protein is stable for at least 50 min under these conditions.
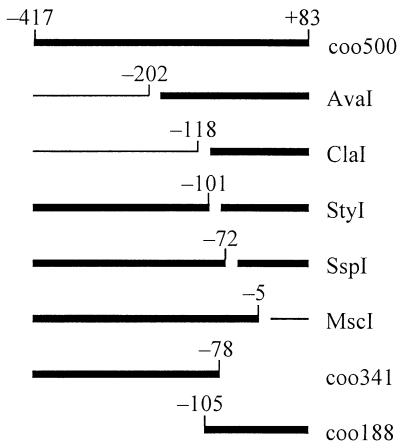
FIG. 1.
Summary of Pcoo DNA fragments bound by MBP::Rns in gel mobility assays. The thick bars indicate DNA fragments whose mobility was retarded by MBP::Rns. The DNA fragment coo500 was digested with the indicated restriction endonucleases, and the gaps within the bars indicate the positions of the restriction sites. The numbering is relative to the transcription start site of the promoter Pcoo. DNA fragments coo500, coo341, and coo188 are PCR products that were not enzymatically digested.
To map binding sites for MBP::Rns within the 500-bp Pcoo fragment, the DNA was digested with a series of restriction endonucleases and the restriction fragments were used in gel mobility assays (Fig. 1). Assays with ClaI- and MscI-digested DNA fragments showed that MBP::Rns does not bind upstream of base −118 or downstream of −5. Digestion of the 500-bp Pcoo fragment with StyI produced two DNA fragments, both of which were bound by MBP::Rns. Similarly, the mobilities of both SspI fragments were retarded. In separate gel mobility assays, MBP::Rns also retarded the mobility of DNA fragments from bases −417 to −78 and from bases −105 to +83 that were synthesized by PCR. These findings revealed that there are at least two Rns binding sites within the 500-bp Pcoo fragment and that they are separated by at least 29 bp, the distance between the StyI and SspI recognition sites.
DNase I footprinting of Rns.
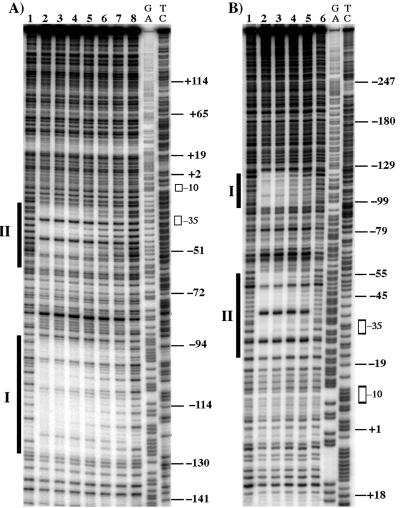
DNase I footprinting was used to precisely define the location of Rns binding sites within Pcoo. In agreement with gel mobility assays that showed MBP::Rns has at least two binding sites, two discrete MBP::Rns footprints were found upstream of Pcoo (Fig. 2). Binding site I begins 30 bp upstream of site II, extending from bases −93 to −129. The promoter-proximal site (binding site II) begins at base −63 and overlaps the promoter −35 hexamer, extending into the spacer region to −23 (Fig. 3).
FIG. 2.
DNase I footprints of MBP::Rns bound to Pcoo. The vertical bars indicate the positions of Rns binding sites I and II. The open rectangles show the positions of the promoter −10 and −35 hexamers. The numbering is relative to the transcription start site. (A) MBP::Rns bound to the coding strand of Pcoo DNA. Lanes 1 and 8 are without MBP::Rns; lanes 2 through 7 contain 300, 200, 133, 89, 59, and 40 nM MBP::Rns. (B) MBP::Rns bound to the noncoding strand of Pcoo DNA. Lanes 1 and 6 are without MBP::Rns; lanes 2 through 5 contain 200, 133, 89, and 60 nM MBP::Rns. The lanes labeled GA and TC contain Maxam-Gilbert sequence ladders.
FIG. 3.
Summary of DNase I protection and uracil interference assays. The nucleotides within Pcoo that were protected from DNase I by MBP::Rns binding are shaded. Uracil substitutions that interfere with MBP::Rns binding are indicated by the letter U. The transcription start site is indicated by an arrow.
Titration of MBP::Rns into DNase I footprinting reactions demonstrated that the sites are saturated at equivalent concentrations of protein, suggesting that the affinities of Rns for both sites are similar. This was confirmed by binding site competition in gel mobility assays. MBP::Rns binding to a radiolabeled DNA fragment carrying only binding site I was inhibited by an equivalent concentration of cold competitor DNA fragment carrying either binding site I or II (data not shown).
Identification of thymine nucleotides recognized by Rns.
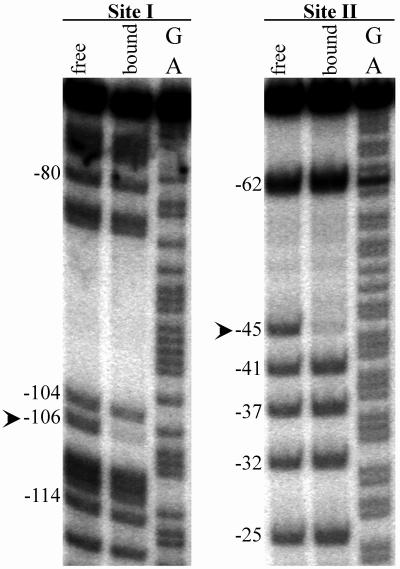
The uracil interference assay was used to identify specific protein-nucleotide interactions required for MBP::Rns binding to better understand how this regulator recognizes each DNA binding site. The experimental identification of these nucleotides was necessary for two reasons. First, although nucleotides contacted by a DNA binding protein are typically conserved at each binding site, no single alignment of Rns binding sites I and II could be found that produced an obvious consensus sequence. Second, only a small subset of nucleotides within a DNase I footprint are actually contacted by a DNA binding protein because steric hinderance limits access of DNase I to DNA. The uracil interference assay identifies thymine C5-methyl groups required for Rns binding because uracil lacks this group. In this assay, a population of DNA fragments is generated with approximately one random uracil-for-thymine substitution per binding site. DNA fragments with substitutions that do not interfere with MBP::Rns binding are separated from the total population by recovering MBP::Rns-DNA complexes from acrylamide gels. The locations of uracil substitutions within these bound fragments are compared to those within the total population of DNA fragments by cleaving the phosphodiester backbone at each uracil substitution and separating the products on denaturing acrylamide gels (Fig. 4).
FIG. 4.
Uracil interference assay of MBP::Rns binding to coding strand of site I and noncoding strand of site II. The phosphodiester backbones of DNA fragments were specifically cleaved at each position where uracil was substituted, and the products were separated on a denaturing acrylamide gel. The lanes labeled “free” contain the total population of DNA fragments in which thymines have been randomly substituted by uracils. The lanes labeled “bound” contain the subpopulation of fragments bound by MBP::Rns in gel mobility assays. The lanes labeled GA contain Maxam-Gilbert sequence ladders. The arrowheads indicate uracil substitutions that prevent MBP::Rns binding. The numbering is relative to the transcription start site of Pcoo.
Sites I and II were assayed individually, and within each site three thymine C5-methyl groups were identified that are essential for MBP::Rns binding. At binding site I, substitution of U for T at base −106 on the coding strand interfered with MBP::Rns binding (Fig. 4), and interference also occurred when U was substituted for T at −113 and −115 on the noncoding strand (data not shown). At site II, interference of binding was observed following substitution of U for T at base −45 on the noncoding strand (Fig. 4) and at positions −36 and −38 on the coding strand (data not shown).
Analysis of Rns binding sites in vivo.
The effects of mutations within each Rns binding site were assayed in vivo from a Pcoo-::lacZ reporter prophage to determine if either site is required for positive regulation of Pcoo (see Materials and Methods). For each binding site, one thymine identified as critical for Rns binding by the uracil interference assay was changed to cytosine. In total, four reporter phages were constructed, each carrying a 1-kb Pcoo fragment from bases −411 to +529: one carrying the wild-type promoter, one with a T-to-C transition at base −45 on the noncoding strand, one with a T-to-C transition at base −106 on the coding strand, and one with both transitions. For these β-galactosidase assays, the expression of Rns was placed under the control of the arabinose-inducible promoter ParaBAD in plasmid pEU2080 and repressed with glucose because Pcoo::lacZ reporter plasmids are unstable when activated by Rns expressed from its own promoter (29).
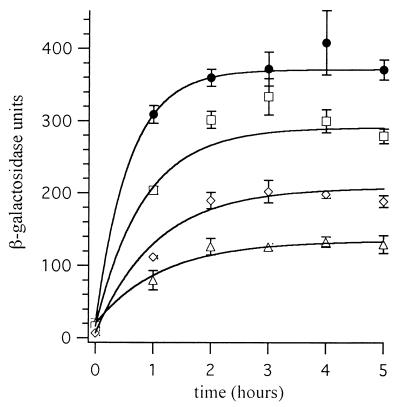
In the presence of glucose without arabinose, the expression of β-galactosidase from all four reporter prophages was only 7 to 20 U. The expression of β-galactosidase from all four reporter constructs increased when the expression of Rns was induced by the removal of glucose and the addition of arabinose (Fig. 5). In all cases the increased expression of β-galactosidase was Rns dependent, because expression did not increase in strains without the Rns expression vector pEU2080 (data not shown). However, constructs carrying mutations within Rns binding sites expressed less β-galactosidase than the wild-type construct. Two hours after the expression of Rns was induced, the expression of β-galactosidase increased 18-fold from that of wild-type Pcoo and remained at this high level throughout the assay. The single mutation within binding site I, T to C at −106, decreased Rns-dependent expression of β-galactosidase 24% ± 3% compared to that of the wild type. The mutation of binding site II, T to C at −45, decreased β-galactosidase expression 43% ± 6%. The effects of these point mutations were additive, since the construct carrying both point mutations expressed 64% ± 4% less β-galactosidase than wild-type Pcoo. These results show that the thymine nucleotides that MBP::Rns interacts with in vitro are also important for Rns activation of Pcoo in vivo.
FIG. 5.
Rns regulation of Pcoo in vivo. The expression of β-galactosidase from wild-type and mutant Pcoo constructs was assayed after the induction of Rns expression by removal of glucose and addition of arabinose to the growth medium at time zero. Solid circles, wild-type Pcoo; squares, T to C at −106; diamonds, T to C at −45; triangles, T to C at −106 and T to C at −45. Each point is the mean (± standard deviation) of three independent cultures.
DISCUSSION
Rns binds at two sites upstream of Pcoo.
Previous deletion analysis of Pcoo showed that a DNA fragment containing bases −417 to +7 was sufficient for Rns-dependent expression from this promoter (29). However, the minimum promoter region required for Rns regulation of Pcoo was not determined because further deletions upstream of Pcoo also destroyed the promoter. Furthermore, the in vivo analysis of Pcoo regulation did not address the question of whether Rns regulates this promoter directly or indirectly. To address these issues, a MBP::Rns fusion protein was purified and studied in vitro after it was shown that this fusion did not alter activity of Rns in vivo. The fusion protein was used in DNase I footprinting and gel mobility assays with DNA fragments of Pcoo. Both assays revealed that MBP::Rns binds to two sites within the −417 to +7 Pcoo fragment. These sites are centered at −112 (site I) and −44 (site II) (Fig. 3). DNase I footprinting of additional downstream sequence to base +237 revealed no other Rns binding sites (data not shown).
DNase I footprinting demonstrated that MBP::Rns can occupy sites I and II simultaneously (Fig. 2). The 31 bp between these sites remain accessible to DNase I cleavage even at the highest concentration of MBP::Rns assayed (500 nM). MBP::Rns can also bind to either site independently of the other. In gel mobility assays, MBP::Rns bound to DNA fragments carrying only site I or site II. The affinities of MBP::Rns for the sites are nearly equivalent because DNA fragments carrying either site I or site II competed equally well for MBP::Rns binding to site I (data not shown).
Both Rns binding sites are required for full expression from Pcoo.
Mutations were introduced into each Rns binding site to determine if either or both are required for Rns regulation of Pcoo in vivo. At each site one thymine, shown by the uracil interference assay to be recognized by MBP::Rns, was changed to a cytosine. Mutation of site I, T−106C, reduced Rns-dependent expression from Pcoo by 24% compared to that from the wild-type promoter. Mutation of site II, T−45C, had a more dramatic effect: expression was reduced by 43%. Thus, while the two binding sites are not equivalent in their contributions to activation, they are both required for full expression from Pcoo.
The more severe effect of the alteration of site II is expected because activator binding sites that are close to the promoter usually have a greater influence on transcription than more distal sites (14). When bound at site II, Rns would be in close proximity to RNA polymerase (RNAP) bound at Pcoo. The DNase I footprint of Rns overlaps the −35 hexamer of Pcoo and extends into the promoter spacer, and the uracil interference assay revealed that Rns interacts with two thymines at −38 and −36. From this position, surface residues of Rns, like those of many activators, could readily form intimate contacts with RNAP. Other regulators homologous to Rns that have binding sites that overlap or are near the promoter −35 hexamer include VirF of S. flexneri (40) and Y. enterocolitica (41), XylS of Pseudomonas putida (21), and AraC of E. coli at araBAD (24) and araF (16).
The double mutation that substituted a C for a T at both Rns binding sites reduced expression from the Pcoo::lacZ reporter prophage by 64% (Fig. 5). Although reduced, this level of expression is Rns dependent because it is fivefold higher than expression from the same construct in the absence of Rns. This Rns dependency indicates that the point mutations introduced at each site diminish but do not abolish Rns binding. This differs from our in vitro analysis, which found that Rns could not bind to either site in which a U had replaced the critical T. The apparent discrepancy between the in vivo and in vitro results might be because cytosine was used to replace thymine in vivo while uracil was used in vitro. The discrepancy may also be the result of differences between in vitro binding conditions and in vivo conditions. For example, MBP::Rns was used for in vitro binding assays while Rns was used for in vivo assays. Also, both binding sites I and II were present on the same DNA fragment in vivo while the effect of uracil substitutions was assayed with DNA fragments carrying each site individually in vitro.
Rns interactions with its target DNA are typical of an AraC family member.
At both binding sites I and II, Rns interacts with the three thymine C5-methyl groups which were shown by the uracil interference assay to be required for MBP::Rns binding in vitro. At each site, two thymines are separated by an intervening adenosine and the third is 7 nucleotides 5′ to the conserved TAT on the opposite strand (Fig. 3 and 6). The spatial distribution of these three thymine C5-methyl groups places them in two adjacent regions of the major groove, indicating that Rns binds in the major groove of the DNA helix at two locations within a single binding site (Fig. 6). Like Rns, other regulators within the AraC family have also been shown to bind in the major groove of the DNA helix. Methylation of particular guanine N7s in adjacent major groove regions interferes with AraC and XylS binding to their respective sites (17, 21, 25). More definitive is the crystal structure of a MarA-DNA complex, which shows that this AraC family member binds within two adjacent regions of the major groove (34). MarA does not make minor groove contacts, although this possibility cannot be excluded for Rns.
FIG. 6.
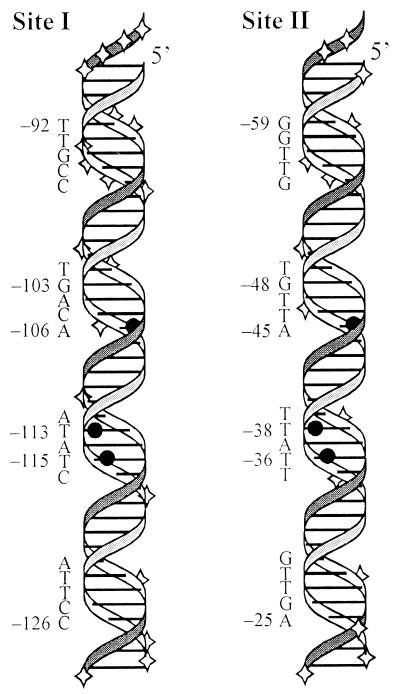
Three-dimensional representation of Rns binding sites I and II. The positions within each binding site that remain accessible or become hypersensitive to DNase I cleavage upon MBP::Rns binding are indicated by diamonds. Within each binding site the three thymine C5-methyl groups that MBP::Rns has hydrophobic interactions with are shown by solid circles. To align these thymines so that they appear in the same orientation in the figure, the sequence of binding site I has been inverted. The numbering is relative to the transcription start site of Pcoo.
The pattern of DNase I cleavage and protection when Rns is bound to either site I or site II suggests that it binds along one face of the DNA helix, leaving the other face exposed. For three helical turns the phosphodiester backbone of one face of the DNA helix is fully protected between and flanking the two major groove regions contacted by Rns. Within the same three helical turns several positions along the opposite face are cleaved by DNase I (Fig. 6). Footprinting, binding interference, and structural studies of other AraC family members have led to similar conclusions. For example, ethylation of phosphates along one face of the helix interferes with AraC binding while ethylation of those on the opposite face does not (17, 25). Similarly, the phosphodiester backbone is protected from cleavage by hydroxyl radicals along only one face of the DNA helix when XylS and VirF are bound to their respective sites (21, 41). The crystal structure of MarA bound to DNA reveals that this family member also binds exclusively along one face of the DNA helix (34).
The three thymine C5-methyl groups with which Rns has hydrophobic interactions are arranged asymmetrically across two regions of the major groove (Fig. 6). Similarly, the three guanine N7s contacted by AraC at site araI1 are arranged asymmetrically across two major groove regions. It has been shown that only one monomer of an AraC dimer binds within this site, contacting two guanines in one region of the major groove and a third guanine in the adjacent major groove region (17). Systematic substitution of every base pair within site araI1 demonstrated that the nucleotides critical for AraC binding lie solely within the adjacent major groove regions and that these critical nucleotides are different in each major groove region (31). As in site araI1, the nucleotides of the two major groove regions in which Rns binds are not the same, so it is probable that each major groove region is contacted by a different DNA binding domain of Rns. These domains may be the two predicted helix-turn-helix motifs in the carboxy terminus of Rns. This hypothesis is supported by the crystal structure of a DNA binding domain from an AraC family member. When bound to its target DNA, the crystal structure of MarA reveals that its two helix-turn-helix motifs place a recognition helix within adjacent regions of the DNA major groove (34). These recognition helices are not identical, and each contacts a unique set of nucleotides.
In summary, our experimental analysis of Rns binding, the nucleotide sequence of each binding site, and the predicted structural features of Rns suggest that Rns interacts with its target DNA like other AraC family members. Rns binds along one face of the DNA helix, forming contacts in two adjacent regions of the major groove. These contacts are different in adjacent major groove regions, and the nucleotides are not conserved between regions. It seems likely that Rns uses both of the predicted helix-turn-helix motifs in its carboxy terminus to contact these different sets of nucleotides. Thus, an asymmetric Rns monomer is probably responsible for all of the contacts at each binding site. The asymmetry of the binding protein is reflected in the sequence asymmetry of each binding site. Because they are asymmetric, these binding sites cannot be identified by simple searches for nucleotide palindromes or repeats. With this new understanding, it appears likely that previous predictions of Rns binding sites, and those of homologous virulence regulators, which were based upon identification of symmetric nucleotide sequences, are probably incorrect.
Identification of potential Rns binding sites.
In this report, we have shown that Rns regulates the expression of CS1 pilin genes directly by binding to two sites upstream of Pcoo and that these sites are asymmetric. The sequence of Rns is homologous to that of CfaR, which activates expression of the genes needed for synthesis of CFA/I pili, cfaABCE, in some ETEC strains (5). Rns is also closely related to VirF, which activates expression of VirB, which in turn regulates other virulence factors of S. flexneri (9). Homology among these virulence regulators is particularly high in the carboxy termini, which contain the two helix-turn-helix motifs probably responsible for specific protein-nucleotide contacts. Because the DNA binding domains of Rns, VirF, and CfaR are conserved, we expect that these regulators recognize similar DNA binding sites. Experimental evidence supports this prediction. Rns can substitute for both CfaR and VirF, and CfaR can substitute for Rns (4, 32). These observations suggest that nucleotide sequences similar to Rns binding sites I and II should be present upstream of loci directly regulated by CfaR and VirF. One or more Rns binding sites should also be found upstream of genes encoding the CS2 pilus, cotBACD, because expression of this pilus is also positively regulated by Rns in some ETEC strains (3).
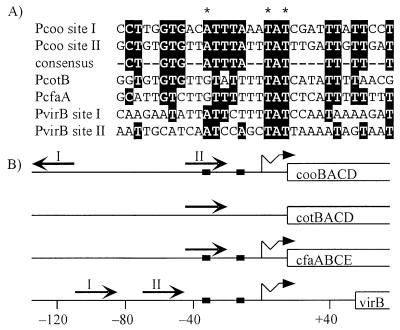
These predictions were tested by searching upstream of cfaA, virB, and cotB for nucleotide sequences that are similar to a Rns binding site consensus sequence. This consensus was developed by using our analysis of specific Rns-nucleotide interactions. The uracil interference assay showed that Rns forms hydrophobic interactions with three thymine C5-methyl groups at site I and site II. These thymine triads are the key to orienting and aligning site I to site II because Rns specifically interacts with each thymine of a triad and the spatial arrangement of the three thymines within a triad is identical at both sites (Fig. 6). However, the orientation of the two triads with respect to each other is inverted. Therefore the coding strand of site I was aligned to the noncoding strand of site II so that the triads were aligned and oriented in the same direction. When aligned in this manner, sites I and II have a consensus of 18 identical nucleotides (Fig. 7).
FIG. 7.
Similar sequences are found upstream of other genes that Rns regulates. (A) A consensus sequence was developed from the alignment of the experimentally identified Rns binding sites upstream of Pcoo, sites I and II. Sequences upstream of cotB, cfaA, and virB were searched for sequences similar to the consensus. Nucleotides identical to the consensus are shaded. The asterisks indicate the positions of thymines within Pcoo sites I and II with which Rns has hydrophobic interactions on the noncoding or coding strand. (B) The locations and orientations of known and potential Rns binding sites are shown by straight arrows. The numbering is relative to the transcription start sites of Pcoo (29), Pcfa (20), and PvirB (40), which are shown by wavy arrows. The transcription start site of Pcot has not been determined. The solid boxes show the locations of promoter −35 and −10 hexamers. The open-ended boxes represent open reading frames.
One potential Rns binding site that is 67% identical to the consensus is located 38 bp upstream of the cotB open reading frame (Fig. 7A). This distance is the same as that between site II and cooB (Fig. 7B). Similarly, a potential binding site was found 38 bp upstream of cfaA. This site is 78% identical to the consensus and, like binding site II, it overlaps the promoter −35 hexamer. The sites upstream of cotB and cfaA conserve only two of the three thymines with which Rns has hydrophobic interactions. At both of these sites, a cytosine replaces the third thymine on the noncoding strand, like the mutations we have introduced into binding sites I and II, which reduce but do not abolish Rns regulation of Pcoo. Potential binding sites upstream of virB conserve all three contacted thymines, but these sites are only 50 and 56% identical to the Rns binding consensus.
The variance of these potential Rns binding sites from the consensus sequence suggests that Rns might activate expression of virB and the CS2 and CFA/I pilin genes less efficiently than CS1 genes because Rns may bind less tightly to these divergent sites than to sites closer to the consensus. In complementation studies of cfaR mutants this appears to be true (5). However, in those and other complementation studies, the concentration of the virulence regulator is an unknown variable (7, 32). Therefore the differences in expression levels may result either from one regulator binding less effectively than another or from a lower concentration of one regulator versus another. It is also likely that the 18-bp Rns consensus sequence we have defined includes nucleotides that are not contacted by Rns. Supporting this is the observation that only 9 bp of the 17-bp AraC binding site araI1 are critical for AraC binding (31). Additionally, the crystal structure of MarA reveals that it interacts with only 12 bp of a 22-bp double-stranded oligonucleotide (34).
The orientations of these potential binding sites relative to a promoter may be as important as their locations and sequence conservation. As discussed above, Rns binding sites are asymmetric, and it is probable that a single monomer occupies a binding site. Because a Rns monomer lacks internal symmetry, the orientation of the binding site will dictate which surface of Rns will be proximal to RNAP. If surface interactions between Rns and RNAP are important for activation, as they are for many activators, an incorrectly oriented binding site may not allow activation to occur because the critical protein surface is not presented to RNAP. Each of the potential binding sites we have identified is in the same orientation as Rns binding site II relative to the promoter it regulates (Fig. 7B). Thus, when bound at these sites, Rns would present the same surface to RNAP as it does at Pcoo.
The potential Rns binding sites we have identified are within regions previously shown to be required for positive regulation by CfaR and VirF. Deletion analysis of the region upstream of cfaA showed that sequences downstream of base −77 were sufficient for CfaR-dependent expression of cfaA, and the potential Rns binding site we found lies downstream of base −58 (Fig. 7B) (20). Upstream deletions of virB to base −116 or −110 did not decrease VirF regulation of this promoter (Fig. 7B). However, deletion of sequences upstream of −100 decreased VirF-dependent expression of virB dramatically (40). This deletion removes part of one site that we have identified as a potential Rns binding site. Also, the in vitro DNase I footprint of VirF extends from base −117 to −17, covering both potential Rns binding sites. Thus, sites we have identified as potential Rns binding sites upstream of cfaA and virB may also serve as the binding sites for CfaR and VirF, respectively.
Like Rns, all regulators within the AraC family probably recognize asymmetric nucleotide sequences. This complicates the identification of their binding sites, even in cases where several binding sites are known, because these binding sites are not distinguished by repeating or palindromic features. The deduction of a consensus sequence from asymmetric sites presents a challenging puzzle to the investigator because these sites must be placed in both the proper register and the proper orientation. In many of these cases, as for Rns, the only key to the puzzle may be physical mapping of nucleotide contacts. These contacts can then be used to set the register and orientation of binding sites so that a consensus binding site can be developed with confidence. We have used this strategy to develop a consensus binding site that predicts the location of binding sites for Rns. It seems likely that this consensus is also recognized by VirF and CfaR because some of these sites are within regions at which CfaR (20) or VirF (40) is known to act, and these regulators can substitute for one another (5, 7, 32). Additionally, the virulence regulators AggR and CsvR may also recognize this consensus because they, like CfaR and VirF, are homologous to Rns.
The experimental identification of additional Rns binding sites will further refine the consensus binding sequence and increase the confidence of binding site predictions for Rns and homologous virulence regulators. With the ever-increasing availability of genomic sequences, the ability to identify binding sites for Rns and related virulence regulators within nucleotide databases will provide a useful tool, facilitating the identification of genes that may play an important role in bacterial pathogenesis.
ACKNOWLEDGMENTS
We thank Annette Woodring for assistance with enzymatic assays, Denise Murphree for construction of the MBP::Rns expression vector pEU750, and Robert Simons for kindly providing vectors pRS550 and λRS45. Amino-terminal sequencing was done by the Emory Microchemical Facility.
This work was supported by Public Health Service grant AI24870 from the NIAID.
REFERENCES
- 1.Adler B, Sasakawa C, Tobe T, Makino S, Komatsu K, Yoshikawa M. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol. 1989;3:627–635. doi: 10.1111/j.1365-2958.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 2.Brenowitz M, Senear D F, Shea M A, Ackers G K. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 3.Caron J, Coffield L M, Scott J R. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc Natl Acad Sci USA. 1989;86:963–967. doi: 10.1073/pnas.86.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caron J, Maneval D R, Kaper J B, Scott J R. Association of rns homologs with colonization factor antigens in clinical Escherichia coli isolates. Infect Immun. 1990;58:3442–3444. doi: 10.1128/iai.58.10.3442-3444.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caron J, Scott J R. A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect Immun. 1990;58:874–878. doi: 10.1128/iai.58.4.874-878.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis G, Sluiters C, Lambert de Rouvroit C, Michiels T. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haan L A M, Willshaw G A, Van Der Zeijst B A M, Gaastra W. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O167:H5. FEMS Microbiol Lett. 1991;83:341–346. doi: 10.1111/j.1574-6968.1991.tb04487.x. [DOI] [PubMed] [Google Scholar]
- 8.D’Orazio S E, Collins C M. The plasmid-encoded urease gene cluster of the family Enterobacteriaceae is positively regulated by UreR, a member of the AraC family of transcriptional activators. J Bacteriol. 1993;175:3459–3467. doi: 10.1128/jb.175.11.3459-3467.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman C J, Porter M E. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol Microbiol. 1998;29:677–684. doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 10.Froehlich B J, Husmann L, Caron J, Scott J R. Regulation of rns, a positive regulatory factor of pili of enterotoxigenic Escherichia coli. J Bacteriol. 1994;176:5385–5392. doi: 10.1128/jb.176.17.5385-5392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaastra W, Svennerholm A-M. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4:444–452. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 12.Gallegos M T, Schleif R, Bairoch A, Hofmann K, Ramos J L. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory element. In: Niedhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 15.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickson W, Flaherty C, Molz L. Sequence elements in the Escherichia coli araFGH promoter. J Bacteriol. 1992;174:6862–6871. doi: 10.1128/jb.174.21.6862-6871.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson W, Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc Natl Acad Sci USA. 1985;82:3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins D E, Nazareno E, DiRita V J. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jair K-W, Martin R G, Rosner J L, Fujita N, Ishihama A, Wolf R E J. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordi B J A M, Dagberg B, de Haan L A M, Hamers A M, van der Zeijst B A M, Gaastra W, Uhlin B E. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992;11:2627–2632. doi: 10.1002/j.1460-2075.1992.tb05328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaldalu N, Mandel T, Ustav M. TOL plasmid transcription factor XylS binds specifically to the Pm operator sequence. Mol Microbiol. 1996;20:569–579. doi: 10.1046/j.1365-2958.1996.5381060.x. [DOI] [PubMed] [Google Scholar]
- 22.Kenny B, Finlay B B. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc Natl Acad Sci USA. 1995;15:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klaasen P, De Graaf F K. Characterization of FapR, a positive regulator of expression of the 987P operon in enterotoxigenic Escherichia coli. Mol Microbiol. 1990;4:1779–1783. doi: 10.1111/j.1365-2958.1990.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee N L, Gielow W O, Wallace R G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD in the l-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, Flaherty C, Hendrickson W. AraC protein contacts asymmetric sites in Escherichia coli araFGH promoter. J Biol Chem. 1992;267:24848–24857. [PubMed] [Google Scholar]
- 26.Martin R G, Jair K-W, Wolf R E J, Rosner J L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Murphree D. Ph.D. dissertation. Atlanta, Ga: Emory University; 1997. [Google Scholar]
- 29.Murphree D, Froehlich B, Scott J R. Transcriptional control of genes encoding CS pili: negative regulation by a silencer and positive regulation by Rns. J Bacteriol. 1997;179:5736–5743. doi: 10.1128/jb.179.18.5736-5743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nataro J P, Yikang D, Yingkang D, Walker K. AggR, a transcriptional activator of aggregative adherence fimbria 1 expression in enteroaggregative Escherichia coli. J Bacteriol. 1994;176:4691–4699. doi: 10.1128/jb.176.15.4691-4699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niland P, Huhne R, Muller-Hill B. How AraC interacts specifically with its target DNAs. J Mol Microbiol. 1996;264:667–674. doi: 10.1006/jmbi.1996.0668. [DOI] [PubMed] [Google Scholar]
- 32.Porter M E, Smith S G, Dorman C J. Two highly related regulatory proteins, Shigella flexneri VirF and enterotoxigenic Escherichia coli Rns, have common and distinct regulatory properties. FEMS Microbiol Lett. 1998;162:303–309. doi: 10.1111/j.1574-6968.1998.tb13013.x. [DOI] [PubMed] [Google Scholar]
- 33.Pu W T, Struhl K. Uracil interference, a rapid and general method for defining protein-DNA interactions involving the 5-methyl group of thymines: the GCN4-DNA complex. Nucleic Acids Res. 1992;25:771–775. doi: 10.1093/nar/20.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee S, Martin R G, Rosner J L, Davies D R. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakellaris H, Scott J R. New tools in an old trade: CS1 pilus morphogenesis. Mol Microbiol. 1998;30:681–687. doi: 10.1046/j.1365-2958.1998.01088.x. [DOI] [PubMed] [Google Scholar]
- 36.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 37.Soisson S M, MacDougall-Shackleton B, Schelif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Science. 1997;276:421–425. doi: 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- 38.Svennerholm A M, Wenneras C, Holmgren J, McConnell M M, Rowe B. Roles of different coli surface antigens of colonization factor antigen II in colonization by and protective immunogenicity of enterotoxigenic Escherichia coli in rabbits. Infect Immun. 1990;58:341–346. doi: 10.1128/iai.58.2.341-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tobe T, Schoolnik G K, Sohel I, Bustamante V H, Puente J L. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 40.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wattiau P, Cornelis G R. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J Bacteriol. 1994;176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]