Abstract
MobB is a small protein encoded by the broad-host-range plasmid R1162 and required for efficient mobilization of its DNA during conjugation. The protein was shown previously to stabilize the relaxosome, the complex of plasmid DNA and mobilization proteins at the origin of transfer (oriT). We have generated in-frame mobB deletions that specifically inactivate the stabilizing effect of MobB while still allowing a high rate of transfer. Thus, MobB has two genetically distinct functions in transfer. The effect of another deletion, extending into mobA, indicates that both functions require a specific region of MobA protein that is distinct from the nicking-ligating domain. The mobB mutations that specifically affected stability also resulted in poor growth of cells, due to increased transcription from the promoters adjacent to oriT. The effects of the mutations could be suppressed not only by full-length MobB provided in trans, as expected, but also by additional copies of oriT, cloned in pBR322. In addition, in the presence of MobA both the full-length and truncated forms of MobB stimulated recombination between oriT-containing plasmids. We propose a model in which MobB regulates expression of plasmid genes by altering the stability of the relaxosome, in a manner that involves the coupling of plasmid molecules.
Proteins required to process plasmid DNA for conjugal transfer assemble at a unique locus, the origin of transfer (oriT), to form a complex called the relaxosome. For the broad-host-range plasmid R1162, and the nearly identical RSF1010, there are three mobilization (mob) genes (Fig. 1) (4, 7), each encoding a protein important for the activity of the relaxosome. The largest and best characterized of these proteins, MobA, locally disrupts the helical structure of the oriT DNA in the relaxosome and then cleaves one of the strands (31). MobC, a second component of the relaxosome, enhances strand separation at the site of cleavage (32). The linear strand, with MobA covalently attached to the 5′ end (1, 28), is probably unwound from its complement and inserted into a recipient cell in the 5′-to-3′ direction (14). During a late stage in transfer, MobA rejoins the ends of this strand to regenerate a circular molecule.
FIG. 1.
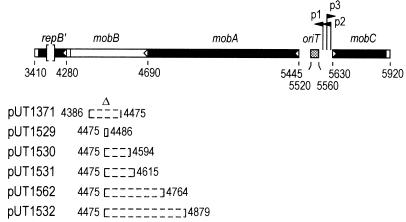
Organization of genes for mobilization and of oriT in plasmid R1162 and locations of different in-frame deletions. The genes mobA and mobB overlap in separate reading frames. The carboxy-terminal domain of mobA is termed repB′ and encodes a primase that is also translated separately (29). Initiation and termination sites for translation are indicated by open triangles and rectangles, respectively. The locations of the promoters p1 to p3 (29) are shown, with the direction of transcription from each indicated by the arrowheads. The base pair coordinates are distances from the unique EcoRI site in R1162.
Reconstitution experiments in vitro have indicated that the third mobilization protein, MobB, stimulates nicking of oriT DNA in the relaxosome (28). In agreement with this, we have found that in vivo MobB stabilizes the assembly of MobA and MobC at oriT (23). This stabilization is shown by the greater proportion of oriT DNA sensitive to oxidation by permanganate, due to strand separation of this DNA within the relaxosome (31). In the absence of MobB, the frequency of mobilization of R1162 decreases 2 to 3 orders of magnitude. The higher transfer frequency in the presence of MobB could simply reflect stabilization of the relaxosome by the protein. We isolated mutations that resulted in unstable relaxosomes but which nevertheless permitted a high level of transfer (23). Although there were fewer complexed molecules, each appeared to be more active in nicking, which could account for the high transfer frequency. However, none of the mutations eliminated the requirement for MobB in transfer. Since mutations could be easily found that compensated for unstable relaxosomes but did not relieve the requirement for MobB, it seemed unlikely that this requirement could be explained solely by the stabilizing effect of the protein.
We have isolated and characterized a set of R1162 derivatives containing in-frame deletions in mobB and overlapping mobA (Fig. 1). The properties of these mutations indicate that stabilization of the relaxosome by MobB is genetically separable from a second role of this protein in transfer and also that a distinct region of MobA is required for MobB activity. In addition, the deletions have revealed that stabilization of the relaxosome can be enhanced by additional copies of oriT in the cell. We propose a model in which coupling of plasmid molecules at oriT, brought about by MobA and MobB, stabilizes the relaxosome.
MATERIALS AND METHODS
Strains and plasmids.
The Escherichia coli K-12 strains used were MV10 (C600 ΔtrpE5) (11) and JW151 (thi endA polA1 T3s), obtained from I. Molineux. The recipient strain in mating experiments was DF1019, a C600 derivative resistant to nalidixic acid (8). M13 derivatives were propagated in RV lacX42 (F′lac) (24), and isolated plaques formed on lawns of JM103 (19).
To construct deletion derivatives of R1162, a CG-to-GC mutation was first introduced in mobB at bp 4468 (Fig. 1) by oligonucleotide-directed mutagenesis (15). The mutation created an Acc65I site and caused a substitution of proline for alanine in MobB. A set of plasmids (Fig. 1) containing deletions that extend from the site of this mutation were constructed by first filling in the Acc65 site with Klenow fragment to create a unique SnaBI site. The XhoI-StuI-XhoI linker CTCGAGGCCTCGAG was introduced at the SnaBI site, and deletion derivatives were generated by digestion with StuI and partial digestion with HaeIII. Only deletions rightward, as shown in Fig. 1, were obtained by this procedure, because the first HaeIII site leftward was in a region encoding a primase essential for plasmid replication (26). The correct reading frame in the resulting plasmids, pUT1529, pUT1530, pUT1531, and pUT1532, was restored by filling in the remaining XhoI site, which also creates a PvuI site. The plasmid pUT1562 was constructed by deletion of DNA from this site to a second PvuI site at bp 4767, created by oligonucleotide mutagenesis. Plasmid pUT1533 is identical to pUT1530 except that it contains a 48-bp deletion that includes all of the oriT DNA but not the adjacent promoters. The plasmid was constructed by exchanging a Bst1107I/EcoO109 fragment from another R1162 derivative containing the deletion (23).
The derivation of pUT530 (5), pUT1371, pUT1376, and pUT221 (23) has been described elsewhere. The plasmid pUT1585 was constructed by joining pUT221 and pUT1530 at their unique AflIII sites within R1162 DNA and then screening for spontaneous, second-site recombinants to regenerate a plasmid identical to pUT221 but with the mobB deletion of pUT1530. The plasmid pUT1440 consists of an 802-bp HpaII-EcoO109 fragment of R1162 DNA (coordinates, 5135 to 5936 [Fig. 1]) cloned by replacement of the small ClaI-EcoRV fragment of pBR322 (3).
The plasmid used for measurement of intracellular amounts of specific mRNAs was constructed by first cloning a 2,667-bp ScaI-EcoO109 fragment, containing the entire mob region from R1162, into the EcoRV site of pBR322. The fragment and adjacent DNA were then excised by digestion with SalI and EcoRI and cloned into pLG339 (30) by replacement of the small SalI-EcoRI fragment. The deletion from pUT1530 was introduced by fragment exchange following digestion with BlpI and Bst1107I.
Assaying recombination between plasmids.
MV10 cells containing R1162 or a derivative (see Fig. 6) were transformed with pUT1440, and colonies of cells resistant to ampicillin and streptomycin were obtained by plating. Three unrelated colonies were separately inoculated in 5 ml of broth medium containing antibiotics, and the culture was grown overnight to stationary phase. Plasmid DNA, isolated by the Qiagen miniprep procedure, was used to transform JW151. Transformants were selected by plating them on medium containing streptomycin and, separately, on medium containing ampicillin. The number of colonies in each case was determined after incubation at 37°C for approximately 48 h.
FIG. 6.
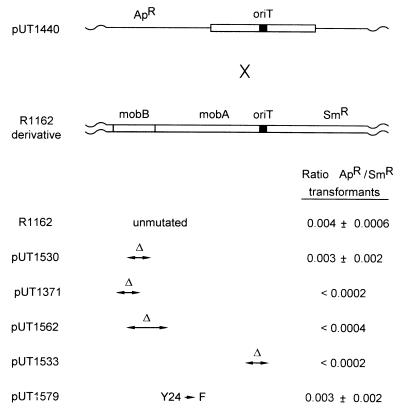
Recombination frequency between pUT1440, a derivative of pBR322 containing a cloned fragment of R1162 mob DNA that includes oriT, and R1162 or a derivative. In each case, the result is the average and standard deviation of three experiments.
Determining relative amounts of specific mRNAs in cells.
Relative amounts of specific mRNAs were determined by hybridization of total RNA, immobilized on a nitrocellulose membrane, with radiolabeled DNA probes. Total cellular RNA from 10 ml of log-phase cells was extracted by a commercially available procedure (Rneasy; Qiagen), and 3 to 5 μg of this preparation was dissolved in 25 mM MgCl2 and digested at 37°C for 1 h with 10 U of DNase I (Sigma). Half of this sample was stored frozen at −70°C; the other half was further digested with 1 μg of DNase-free RNase A under the same conditions. Serial dilutions of the samples were then applied to a nitrocellulose membrane (BA85; Schleicher and Schuell) prepared according to the method of Sambrook et al. (25) in a slot blot apparatus.
Double-stranded, radiolabeled DNA probes were prepared by PCR in a reaction mixture (100 μl) that contained 100 to 200 ng of plasmid DNA; 2 mM MgSO4; a 0.2 μM concentration of each primer; dGTP, dCTP, and dTTP (50 μM each); 45 μM dATP; 10 μl of [α-32P]dATP (3,000 Ci/mmol; 10 μCi/μl); and 1 U of Vent DNA polymerase (New England Biolabs). Unincorporated nucleotides were removed with a spin column (Qiagen). The specific activity of the product was approximately 108 cpm per μg. About 0.5 μg of the probe DNA was diluted in 150 μl of H2O, heated to 100°C for 15 min, and then chilled on ice and added to 15 to 20 ml of hybridization buffer (25) containing the immersed nitrocellulose membrane. After incubation at 42°C for 16 to 20 h, the membrane was washed once at room temperature with 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) and then three times with 0.2× SSC–0.1% SDS at 68°C. The membrane was dried at room temperature. The amounts of hybridized probe were quantified with a phosphorimager; the hybridized DNA was also visualized by autoradiography.
Other procedures.
Bacteria were mated on semisolid medium by a standard procedure (4). The recombination frequency of M13mp9 derivatives containing two directly repeated copies of oriT was determined as previously described (21). DNA in whole cells and cleared lysates (12) was treated with permanganate as described by Zhang and Meyer (31) and Perwez and Meyer (23). DNA prepared from cleared lysates was also used to assay site-specific nicking at oriT by primer extension (23). In this assay, half of the DNA sample was digested with BsmAI, which cleaves the plasmid DNA between oriT and the priming site. The other half was digested with BstZ17, which cleaves distal to oriT in the direction of priming. The samples were then mixed, and the DNA was denatured and annealed to 32P-, end-labeled primer. Strand extension was carried out by thermal cycling with Taq DNA polymerase, as previously described (23), and the sample was applied to a 0.35-mm, 8% polyacrylamide gel. The relative amounts of DNA in the bands after electrophoresis were determined with a phosphorimager. The fraction of nicked molecules was taken as the amount of DNA due to termination at the oriT nick site divided by the amount of DNA resulting from termination at the BsmAI cleavage site.
RESULTS
Two functions of MobB in conjugal mobilization can be distinguished by mutation.
The R1162 derivative pUT1371 (Fig. 1) contains a 162-bp, in-frame deletion in mobB, a mutation that inactivates the gene and causes at least a 100-fold decrease in the mobilization frequency of the plasmid (23) (Table 1). The mutation is complemented when MobB, encoded by the plasmid pUT221 (23), is provided in trans (Table 1). This indicates that the mutation only affects mobB and not the overlapping gene, mobA (Fig. 1).
TABLE 1.
Effect of internal in-frame deletions in mobA and mobB on plasmid mobilization frequency and on generation time of the host
| Plasmid | Mobilization frequencya with:
|
Generation time of hostb (min) with:
|
||
|---|---|---|---|---|
| pACYC184 (−MobB) | pUT221 (+MobB) | pACYC184 (−MobB) | pUT221 (+MobB) | |
| R1162 | 4.3 × 10−3 (1.0) | 3.4 × 10−3 (1.0) | 35 ± 3 | 33 ± 3 |
| pUT1371 | 3.7 × 10−5 (0.009) | 2.0 × 10−3 (0.6) | 81 ± 3 | 40 ± 8 |
| pUT1529 | 6.9 × 10−4 (0.2) | 4.4 × 10−3 (1.3) | 135 ± 10 | 35 ± 12 |
| pUT1530 | 1.3 × 10−3 (0.3) | 3.6 × 10−3 (1.1) | 69 ± 3 | 38 ± 4 |
| pUT1531 | 2.3 × 10−3 (0.5) | 5.1 × 10−3 (1.5) | 115 ± 5 | 71 ± 1 |
| pUT1562 | 6.6 × 10−6 (0.002) | 5.0 × 10−5 (0.01) | 124 ± 4 | 115 ± 5 |
| pUT1532 | <1 × 10−7 | <1 × 10−7 | 102 ± 3 | 119 ± 1 |
| pUT1533 | NT | NT | 81 ± 3 | 78 ± 5 |
| pUT1376 | 3.3 × 10−6 | 4.3 × 10−3 | 121 ± 11 | 159 ± 9 |
Average of two experiments (range, ±25% or less), shown as transconjugants per donor. The mobilization frequency relative to that for R1162 is given in parentheses. NT, not tested.
Average and range of two experiments.
We generated additional in-frame mobB deletions, starting at bp 4475, near the middle of the gene and extending toward the N-terminal coding end (Fig. 1). Three of the resulting plasmids, pUT1529, pUT1530, and pUT1531, contain deletions of 12, 120, and 141 bp, respectively, that are entirely within mobB. These deletions, as well as the one in pUT1371, were all complemented to the same level by pUT221 (Table 1). However, in the absence of complementation, pUT1529, pUT1530, and pUT1531 were each mobilized at a substantially higher frequency than pUT1371. This higher rate of transfer was observed even for pUT1531, the plasmid that contained a deletion removing about one-third of mobB.
The mutation in pUT1371 results not only in a low rate of transfer but also in relaxosomes that are unstable. There is a smaller proportion of nicked molecules in the cell and, as a consequence, an increased yield of supercoiled DNA compared to those with R1162 following alkaline extraction (23) (Fig. 2). Like the low frequency of transfer, this instability was also reversed by providing MobB in trans (Fig. 2). In the same assay, plasmid DNA yields for pUT1529, pUT1530, and pUT1531 were also greater than the yield of R1162, indicating that the relaxosomes of these plasmids were likewise unstable (Fig. 2), and again, the yield was reduced when full-length MobB was present in the cell.
FIG. 2.
DNA yields following alkaline extraction (17) for R1162 and derivatives containing deletions in mobB. The cells also contained pACYC184 (6) or pUT221, a pACYC184 derivative containing mobB (23). Plasmid DNAs were linearized by digestion with EcoRI and displayed by 0.8% agarose gel electrophoresis.
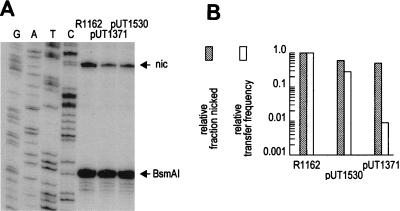
If the effect of MobB on transfer is solely through stabilizing the relaxosome, then the different mobB deletions should have approximately commensurate effects on the proportion of molecules nicked at oriT and on the frequency of mobilization. We measured the fraction of molecules nicked at oriT for pUT1371, pUT1530, and R1162. Plasmid DNA in cleared lysates (12) was treated with SDS and phenol to disrupt the relaxosome, and the proportion of molecules specifically nicked within oriT was determined by primer extension and measurement of radioactivity in DNA bands after gel electrophoresis (Fig. 3A). The proportion of nicked molecules was 0.23 for R1162; this decreased to 0.14 for pUT1530 and 0.12 for pUT1371 (Fig. 3B). Thus, although the deletions in pUT1530 and pUT1371 affected the frequency of plasmid mobilization to different extents (Table 1 and Fig. 3B), they had similar effects on nicking. Moreover, the decrease in transfer frequency and the proportion of nicked molecules were similar for pUT1530, but transfer of pUT1371 was lower than could be accounted for by the level of nicked molecules. We conclude that mutations in mobB can differentially affect the frequency of transfer and the stability of the relaxosome. Mutations within the N-terminal half of MobB destabilize the relaxosome, and this probably accounts in large part for the small but detectable decrease in the transfer frequency of these plasmids (Table 1). The C-terminal region of MobB, conserved in these deletions and inactivated in pUT1371, is important for transfer in a second way, unconnected with relaxosome stability.
FIG. 3.
(A) Proportion of plasmid DNA molecules nicked at oriT for R1162, pUT1371, and pUT1530. The bands on the polyacrylamide gel reflect primer extension with termination at the nick site of oriT (nic) and on an equal amount of template with termination at a BsmAI site proximal to the primer. (B) Relative transfer frequency (obtained from Table 1) and relative fraction of nicked molecules (by quantitative analysis of the bands in the gel), with values set at 1.0 for R1162.
A region of mobA is required for activity of MobB.
We also isolated and characterized two plasmids that contained in-frame deletions extending further from bp 4475, across the beginning of mobB and into mobA (pUT1532 and pUT1562 [Fig. 1]). Because DNA required for initiation of translation of mobB was deleted from these plasmids, we did not expect them to be mobilized at high frequency by R751. The mobilization frequency of pUT1562 was low and similar to that of pUT1371, and conjugal transfer of pUT1532 was not detected (Table 1). However, unlike the plasmids having deletions entirely within mobB, pUT1532 and pUT1562 could not be complemented for transfer by MobB in trans (Table 1). In addition, the high yield of pUT1562 and pUT1532 DNA following alkaline extraction was unaffected when MobB was in the cell (Fig. 2).
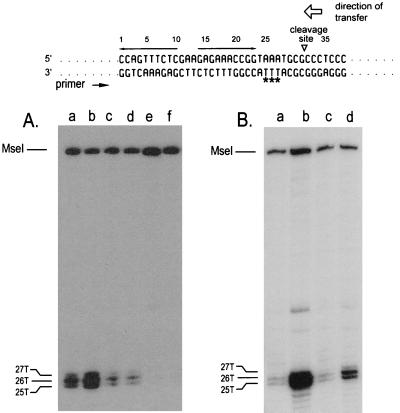
The MobA proteins encoded by pUT1532 and pUT1562 could simply fail to bind to oriT DNA. To test this possibility, we took advantage of the facts that this binding locally disrupts the DNA duplex and the resulting unpaired pyrimidines can be detected by their increased sensitivity to permanganate (31). Strand separation still occurs in the absence of MobB, although because MobB stabilizes the relaxosomes, sensitivity to permanganate is enhanced when this protein is present in the cell (23). We compared the permanganate sensitivity of the oriT DNA in the relaxosomes of pUT1530, pUT1562, and pUT1532. In each case, a cleared lysate of plasmid-containing cells was prepared and exposed to permanganate, and the oxidized bases on the negative strand, the one not transferred, were mapped by primer extension. As reported elsewhere for R1162 (31), three adjacent thymine residues in the oriT DNA of pUT1530 were sensitive to permanganate, and this sensitivity was enhanced by MobB (Fig. 4A, lanes a and b). The sensitive bases are located at bp 25, 26, and 27 in the oriT base sequence shown at the top of the figure. These bases were unreactive in the oriT of pUT1532 (Fig. 4A, lanes e and f), indicating that the MobA encoded by this plasmid binds poorly to oriT DNA or is unstable, thus accounting for the low frequency of mobilization of the plasmid. In contrast, although pUT1562 was mobilized poorly, the bases in the oriT DNA of pUT1562 were still sensitive to oxidation (Fig. 4A, lanes c and d). The sensitivity was about half that detected in the oriT DNA of pUT1530. However, unlike the relaxosome of pUT1530 (or pUT1371 [23]), permanganate sensitivity was not increased when MobB was present in the lysate (Fig. 4A, lane d). Thus, for both stabilization of the relaxosome and mobilization of the plasmid, the region of MobA affected by the deletion in pUT1562 is required for MobB to be active.
FIG. 4.
(A) Permanganate-sensitive bases on the unnicked oriT DNA strand for pUT1530 (lanes a and b), pUT1562 (lanes c and d), and pUT1532 (lanes e and f). The locations of the bases were determined from a sequencing ladder (not shown) generated with the same template and primer. Plasmid DNAs in cleared lysates were treated with permanganate prior to primer extension by PCR (31, 32). The cells also contained either pACYC184 (lanes a, c, and e) or pUT221 (lanes b, d, and f). (B) Permanganate sensitivity of the unnicked oriT DNA strand for pUT1530 isolated from cells also containing pBR322 (lane c) or pUT530 (lane d). Whole cells were exposed to permanganate prior to extraction of the DNA (31). For comparison, the permanganate sensitivity of pUT1530 DNA isolated from cells treated under identical conditions but containing pACYC184 (lane a) or pUT221 (lane b) is also shown. The base sequence of oriT is given at the top of the figure; the permanganate-sensitive bases are identified by asterisks. The inverted repeat (opposing arrows), the site cleaved by MobA in the top (transferred) strand, and the direction of transfer are also indicated.
MobA not only interacts with duplex DNA in the relaxosome but also binds the single oriT DNA strand normally transferred during conjugation and can both cleave this DNA at the site nicked in the relaxosome and ligate the ends (1, 27). This property is reflected in vivo by site-specific recombination between oriTs cloned in M13 single-stranded DNA, when MobA is provided in the cell (21). The reaction can be monitored by infecting plasmid-containing cells with M13mp9 phages (20) having two directly repeated copies of oriT cloned in the lacZ(α) cloning region. These phages form white plaques on medium containing isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). After recombination between the two oriTs, the phages form blue plaques, due to translation through the remaining oriT and production of an altered but active α-complementing fragment. We tested pUT1562, as well as pUT1531, for the ability to support oriT recombination in M13. In both cases, the recombination frequency of the M13 derivative with two directly repeated oriTs was 4% in cells of RV and above the background level of 0.3% in Rec+ cells. These frequencies were unchanged when MobB was present in the cell. Thus, the activity of MobA on single-stranded DNA was unaffected by either MobB or the region of MobA needed for the activity of this protein.
Unstable relaxosomes cause long generation times.
The generation time of MV10 (R1162) cells in broth was approximately 35 min (Table 1). However, cells containing R1162 derivatives with deletions in mobB had substantially longer generation times (Table 1) and formed long filaments (not shown). This was true for pUT1371, in which mobB is completely inactivated, and also for pUT1529, pUT1530, and pUT1531, which are still mobilized at high frequency. For each of these plasmids, MobB in trans restored the characteristics of normal growth (Table 1). The plasmids pUT1562 and pUT1532 also caused long generation times, but in these cases the effect was not complemented by MobB (Table 1). These observations suggested that it was the unstable relaxosomes, resulting from the deletion of either the N-terminal region of MobB or the region of MobA required for MobB activity, that caused the poor growth.
MobA and MobC proteins act together as repressors at oriT (9) and regulate the activity of the adjacent promoters p1, p2, and p3 (29) (Fig. 1). Destabilization of the relaxosome probably decreases the level of repression, resulting in greater synthesis of plasmid proteins, which in turn stresses the cell and causes slower growth. Several other observations are consistent with this interpretation. The plasmid pUT1533 is identical to pUT1530 but contains a deletion that removes oriT but not the adjacent promoters. In contrast to pUT1530, this plasmid resulted in poor growth, whether or not pUT221 was also present (Table 1). Thus, MobB cannot be acting simply as an antidote by binding to MobA and reducing the toxicity of this or some other protein. In addition, oriT must be present in its normal location. The plasmid pUT1376 contains the mobB deletion present in pUT1371, but oriT is at a new position distant from the promoters p1 to p3. Although the oriT in this plasmid was active in mobilization, the cells had a long generation time, whether or not pUT221 was present (Table 1). Finally, point mutations restoring good growth were isolated by serially culturing cells containing pUT1530 in broth. Four independent point mutations were identified, and in each case these mapped in the promoters adjacent to oriT.
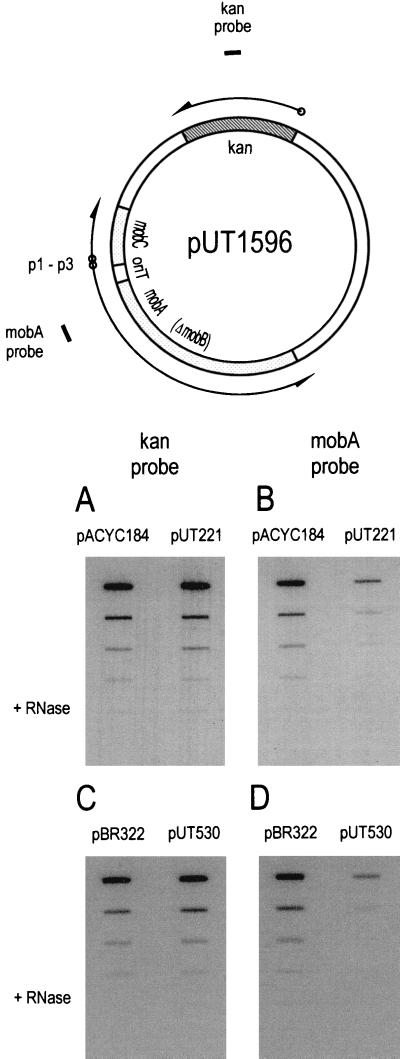
Derepression of transcription by the ΔmobB mutation in pUT1530 was confirmed by hybridization of radiolabeled probes to total cellular RNA. In order to avoid changes in copy number due to changes in transcription from the promoters adjacent to oriT, the mob DNA from pUT1530 was first cloned into the vector pLG339 (30), which has a replicon derived from pSC101. Transcription was measured in cells containing the resulting plasmid (pUT1596 [Fig. 5]) and also either pUT221 (MobB+) or pACYC184 (no MobB present). Two radiolabeled probes were used, one hybridizing to part of the mobA transcript initiated from p1 and p3, and thus regulated at oriT, and another, as an internal control, hybridizing to transcripts of the kanamycin resistance (kan) gene of the vector (Fig. 5). The results (Fig. 5A and B) show that MobB in the cell did not affect transcription of the kan gene but reduced the amount of mobA transcript in the cell. We conclude that when MobB stabilizes the relaxosome, it also increases repression of transcription at oriT.
FIG. 5.
Hybridization of radiolabeled DNA probes to RNA immobilized on a nitrocellulose membrane. An undiluted, DNase I-treated sample and 1:10, 1:20, and 1:40 dilutions were applied in each column of slots. In the bottom slot of each column, three times the amount of undiluted sample was digested with RNase A prior to binding to the membrane. At the top is the reporter plasmid with the approximate locations of the probes for the kan and mobA transcripts.
Multiple copies of oriT in trans suppress poor cell growth and stabilize relaxosomes in strains containing certain defective MobB proteins.
The plasmid pUT530 (5) consists of a copy of the R1162 oriT cloned into the vector pBR322. When this plasmid was introduced into cells containing pUT1529 or pUT530, the resulting strains became healthy and grew with short generation times (Table 2). This effect depended on the cloned oriT in pUT530, because suppression by pBR322 was not observed (data not shown). Thus, copies of oriT in trans suppressed the poor growth phenotype caused by the mobB deletion in pUT1530. A copy of oriT in pUT1530 was also required, since the poor growth of cells containing pUT1533, a derivative of pUT1530 lacking oriT, was unaffected by pUT530 (Table 2).
TABLE 2.
Effect of cloned copies of oriT on inhibition of cell growth by R1162 derivatives
| R1162 derivative present | Generation time (min) of strain containing complementing plasmida
|
|
|---|---|---|
| None | pUT530 | |
| pUT1530 | 80 ± 4 | 35 ± 4 |
| pUT1529 | 82 ± 6 | 40 ± 2 |
| pUT1371 | 81 ± 3 | 95 ± 5 |
| pUT1533 | 78 ± 3 | 74 ± 4 |
| pUT1371 + pUT1585 | NT | 35 ± 4 |
| pUT1371 + pACYC184 | NT | 87 ± 3 |
| pUT1562 + pUT1585 | NT | 91 ± 6 |
| pUT1562 + pACYC184 | NT | 104 ± 4 |
Average and range for two experiments. NT, not tested.
Plasmid pUT530 did not suppress the poor growth of cells containing pUT1371, in contrast to those containing pUT1529 and pUT1530 (Table 2). This could mean that suppression required MobB and that the necessary activity was conserved on the MobB fragments encoded by pUT1529 and pUT1530 but not pUT1371. To test this, we constructed the plasmid pUT1585, analogous to pUT221 but with the mobB deletion of pUT1530. When cells contained this plasmid as well as pUT1371, the introduction of pUT530 resulted in healthy cells (Table 2). There was no suppression when the cells contained the parental vector pACYC184 instead of pUT1585 (Table 2). In contrast, pUT530 was nonsuppressing when introduced into cells that contained pUT1585, but with pUT1562 instead of pUT1371 (Table 2). Thus, suppression required the region of MobB conserved in pUT1530 (and pUT1529) and also the domain of MobA required for MobB activity.
Cells containing pUT1530 grow poorly because the R1162 promoters adjacent to oriT are partially derepressed; the cells become healthy when full repression is restored by providing MobB in trans to stabilize the relaxosomes. Do copies of oriT in trans suppress poor growth by a similar mechanism? We asked first whether pUT530, like MobB, increased the permanganate sensitivity of the oriT DNA in pUT1530. In order to minimize disruption of any macromolecular complexes in the cytoplasm, we treated whole cells rather than cleared lysates with permanganate (31). Sensitive bases on the nonnicked strand were again identified by primer extension. As before, full-length MobB, encoded by pUT221, enhanced strand separation within oriT (Fig. 4B, lanes a and b). In addition, we found that copies of oriT cloned in pUT530 likewise increased the permanganate sensitivity of the oriT DNA of pUT1530 (Fig. 4B, lanes c and d).
Extra copies of oriT also repressed transcription initiated from the promoters adjacent to oriT. As before with MobB, we measured the relative amounts of the kan and mobA transcripts for plasmid pUT1596, but this time in cells containing either extra copies of oriT (pUT530) or only the vector pBR322. The results (Fig. 5C and D) indicate that although pUT530 had no effect on the amount of kan transcript, it repressed transcription of mobA. Moreover, the levels of repression, determined by comparing the relative amounts of bound radioactive probe at the same dilution of RNA, were similar for MobB and oriT. We estimate that the amount of mobA transcript decreased about 12-fold when MobB was present and 5- to 6-fold in the presence of the additional copies of oriT.
The relaxosome is recombinogenic at oriT by a mechanism that requires MobB.
Additional copies of oriT in the cell can act in trans, in a way that depends on MobA and MobB, to repress transcription from neighboring promoters. If repression came about by a direct interaction between plasmid molecules, then it might increase the frequency of their recombination. To test this, we used the plasmid pUT1440, which is similar to pUT530 but contains a larger, 802-bp fragment of cloned R1162 mob DNA. This DNA includes oriT and adjacent parts of mobA and mobC. The plasmid was introduced by transformation into MV10 cells containing R1162 or a derivative, and the DNA was then extracted and used to transform JW151 (polA1) for resistance to ampicillin and, separately, for resistance to streptomycin. Because the replicon of pUT1440 is inactive in JW151, ampicillin-resistant colonies were primarily the result of recombination that had taken place between pUT1440 and R1162 prior to extraction of the DNA. The ratio of ampicillin-resistant transformants to streptomycin-resistant transformants was thus a measure of the recombination frequency in MV10.
The recombination frequency between pUT1440 and R1162 was 0.004, and this frequency was little changed when pUT1530 instead of R1162 was in the cell (Fig. 6). In contrast, recombination between pUT1440 and either pUT1371 or pUT1562, plasmids which do not encode an active MobB, was undetectable under our assay conditions. In addition, no recombination was observed when oriT was deleted from pUT1530 (pUT1533 [Fig. 6]). These results suggested that MobB promotes an interaction between plasmid DNA molecules at oriT and that this interaction influences the rate of recombination. However, it was also possible that a greater overall level of nicking in the relaxosomes of R1162 and pUT1530 was resulting in a substrate more favorable for recombination. The plasmid pUT1579 is identical to R1162 but contains a mutation causing a substitution of phenylalanine for tyrosine-24, the active nucleophile in strand cleavage (27). The protein forms a normal complex as assayed by sensitivity of the DNA to permanganate, but nicking is undetectable in vivo (33). Nevertheless, R1162 and pUT1579 recombined with pUT1440 at essentially the same frequency (Fig. 6). We conclude that the relaxosome enhances plasmid recombination by a mechanism that does not depend on nicking at oriT.
DISCUSSION
We show here that two effects of MobB, stabilization of the relaxosome and enhancement of the frequency of transfer, can be genetically distinguished by mutations within the gene. The mobB deletions in pUT1529, pUT1530, and pUT1531 destabilize the relaxosome but have only a small effect on the frequency of mobilization (Table 1). These deletions map in the region of the gene encoding the amino-terminal half of the protein.
Deletion of the region of mobA adjacent to mobB results in a relaxosome that is no longer responsive to MobB, and as a result mobilization decreases to the level observed when MobB is effectively absent (pUT1562 [Table 1]). However, this deletion has a smaller effect on the DNA-processing reactions carried out by MobA: cleavage and ligation of single-stranded DNA, as monitored by phage recombination, or the separation of DNA strands within the relaxosome. We interpret these results to mean that the region of mobA adjacent to mobB encodes a domain of the protein required for recognition of MobB. When this region is absent, MobA becomes blind to the presence of MobB, and the frequency of mobilization becomes similar to that of pUT1371. R1162 and pSC101, a plasmid which is unrelated to R1162 overall, have oriTs with very similar base sequences (16). In addition, pSC101 encodes a mobilization protein with tracts of amino acids identical to those in MobA. The similarities between the two proteins map at the amino-terminal end of MobA, throughout the DNA-binding region, but do not extend into the domain required for recognition of MobB. Interestingly, there is no protein encoded by pSC101 that can be identified as MobB-like on the basis of sequence similarities. Thus, MobB and a cognate site within MobA might represent a particular adaptation within the IncQ plasmid group.
Relaxosomes stabilized by MobB cause maximal repression of transcription from the promoters adjacent to oriT. When the relaxosomes are unstable, partial derepression presumably leads to cells that form filaments and grow with long generation times. Since derepression affects the expression of not only the mob genes but also the replication genes downstream from mobA (9), it is not clear whether overexpression of a particular gene or overall plasmid gene expression is the basis for the poor growth. The nicking domain of MobA is not solely responsible, since a deletion that eliminates this region does not restore normal growth (pUT1532 [Table 1]). In any case, the effect of derepression is probably amplified by an increase in plasmid copy number, due to the increase in expression of the replication genes (10, 13).
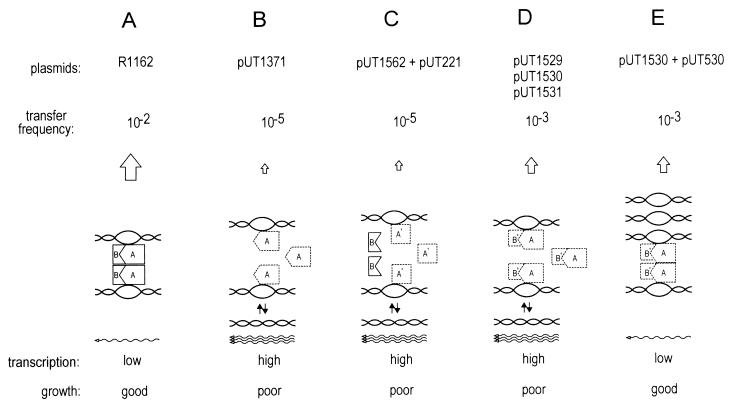
The destabilizing effect of the mobB mutations in pUT1529 and pUT1530 could be overcome by additional copies of oriT in trans. This suppression required the remaining fragment of MobB as well as MobA protein with its recognition domain for MobB. An increased frequency of recombination between oriT-containing plasmid molecules showed identical requirements. From these observations, we propose that MobA and MobB link the oriTs on different plasmid molecules by means of a protein bridge and that this structure is required for stable relaxosomes and, consequently, for optimal repression of plasmid promoters and healthy growth of cells (Fig. 7A). As a result, inactivation of MobB (in the plasmid pUT1371) results in both a low level of transfer and high levels of transcription, with the cells growing poorly (Fig. 7B). The same effects are observed when the region of mobA adjacent to mobB is deleted (Fig. 7C), because MobB is then no longer able to recognize MobA. The mutations in pUT1529, pUT1530, and pUT1531 partially impair the ability of MobB to stabilize the relaxosomes, so that transcription is high and cells again grow poorly (Fig. 7D). However, an increased dosage of oriT, provided by pUT530, partially offsets this instability by driving the oriT interaction toward the coupled complex, thereby decreasing gene repression (Fig. 7E).
FIG. 7.
Model describing the interactions of MobA and MobB with oriT DNA and the effects of these interactions on transfer, transcription and cell growth.
These molecular interactions proposed in Fig. 7 are reminiscent of the coupling or “handcuffing” which is thought to be part of the mechanism of replication control for plasmids RK2 and R6K and phage P1 (2, 18, 22). In these cases, excess iterons bind the plasmid-specific initiation protein, freezing it at the origin of replication and preventing a new round of replication. For R1162, control of replication would be more indirect. Copy number is determined by the level in the cell of the plasmid-specific replication protein RepC (10, 13), and enhanced repression at oriT reduces the overall level of transcription through the gene for this protein (9).
Although lacking the amino-terminal region, the MobB proteins encoded by pUT1529, pUT1530, and pUT1531 are still very active for transfer. MobB might also be required for the R1162 relaxosome to recognize the conjugal apparatus of the mobilizing, self-transmissible plasmid. MobB could act as a bridge, or it could modify the conformation of MobA to better fit the conjugal machinery. Thus, MobB could mediate interactions between relaxosomes and also between the relaxosome and a docking site for mobilization. Experiments to obtain physical evidence for these interactions are now under way.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of Health (GM37462).
We thank S. R. Kushner for providing a strain containing pLG339.
REFERENCES
- 1.Bhattacharjee M K, Meyer R J. A segment of a plasmid gene required for conjugal transfer encodes a site-specific, single-strand DNA endonuclease and ligase. Nucleic Acids Res. 1991;19:1129–1137. doi: 10.1093/nar/19.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasina A, Kittell B, Toukdarian A, Helinski D. Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc Natl Acad Sci USA. 1996;93:3559–3564. doi: 10.1073/pnas.93.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of amplifiable multicopy DNA cloning vehicles. II. A multiple cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 4.Brasch M A, Meyer R J. Genetic organization of plasmid R1162 DNA involved in conjugative mobilization. J Bacteriol. 1986;167:703–710. doi: 10.1128/jb.167.2.703-710.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasch M A, Meyer R J. A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J Mol Biol. 1987;198:361–369. doi: 10.1016/0022-2836(87)90286-5. [DOI] [PubMed] [Google Scholar]
- 6.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derbyshire K, Hatfull G, Willetts N. Mobilization of the non-conjugative plasmid RSF1010: a genetic and DNA sequence analysis of the mobilization region. Mol Gen Genet. 1987;206:161–168. doi: 10.1007/BF00326552. [DOI] [PubMed] [Google Scholar]
- 8.Figurski D, Meyer R, Miller D, Helinski D R. Generation in vitro of deletions in the broad host range plasmid RK2 using phage Mu insertions and a restriction endonuclease. Gene. 1976;1:107–119. doi: 10.1016/0378-1119(76)90010-x. [DOI] [PubMed] [Google Scholar]
- 9.Frey J, Bagdasarian M M, Bagdasarian M. Replication and copy number control of the broad host-range plasmid RSF1010. Gene. 1992;113:101–106. doi: 10.1016/0378-1119(92)90675-f. [DOI] [PubMed] [Google Scholar]
- 10.Haring V, Scholz P, Scherzinger E, Frey J, Derbyshire K, Hatfull G, Willetts N, Bagdasarian M. Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proc Natl Acad Sci USA. 1985;82:6090–6094. doi: 10.1073/pnas.82.18.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershfield V, Boyer H W, Yanofsky C, Lovett M A, Helinski D R. Plasmid ColE1 as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci USA. 1974;71:3455–3459. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz L, Kingsbury D T, Helinski D R. Stimulation of cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973;114:577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Meyer R J. Copy number of the broad host-range plasmid R1162 is determined by the amounts of essential plasmid-encoded proteins. J Mol Biol. 1985;185:755–767. doi: 10.1016/0022-2836(85)90060-9. [DOI] [PubMed] [Google Scholar]
- 14.Kim K, Meyer R J. Unidirectional transfer of broad host-range plasmid R1162 during conjugative mobilization. Evidence for genetically distinct events at oriT. J Mol Biol. 1989;208:501–505. doi: 10.1016/0022-2836(89)90513-5. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 17.Marko M A, Chipperfield R, Birnboim H C. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982;121:382–387. doi: 10.1016/0003-2697(82)90497-3. [DOI] [PubMed] [Google Scholar]
- 18.McEachern M J, Bott M A, Tooker P A, Helinski D R. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci USA. 1989;86:7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messing J, Crea R, Seeburg P H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messing J, Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digested restriction fragments. Gene. 1982;19:269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- 21.Meyer R. Site-specific recombination at oriT of plasmid R1162 in the absence of conjugative transfer. J Bacteriol. 1989;171:799–806. doi: 10.1128/jb.171.2.799-806.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal S, Chattoraj D. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J Bacteriol. 1988;170:3554–3560. doi: 10.1128/jb.170.8.3554-3560.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perwez T, Meyer R. MobB protein stimulates nicking at the R1162 origin of transfer by increasing the proportion of complexed plasmid DNA. J Bacteriol. 1996;178:5762–5767. doi: 10.1128/jb.178.19.5762-5767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rotman G, Cooney R, Malamy M. Cloning of the pif region of the F sex factor and identification of a pif protein product. J Bacteriol. 1983;155:254–264. doi: 10.1128/jb.155.1.254-264.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Scherzinger E, Bagdasarian M M, Scholz P, Lurz R, Ruckert B, Bagdasarian M. Replication of the broad host range plasmid RSF1010: requirement for three plasmid-encoded proteins. Proc Natl Acad Sci USA. 1984;81:654–658. doi: 10.1073/pnas.81.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherzinger E, Kruft V, Otto S. Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 1993;217:929–938. doi: 10.1111/j.1432-1033.1993.tb18323.x. [DOI] [PubMed] [Google Scholar]
- 28.Scherzinger E, Lurz R, Otto S, Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 30.Stoker N, Fairweather N, Spratt B. Versatile low-copy-number vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Meyer R J. Localized denaturation of oriT DNA within relaxosomes of the broad host-range plasmid R1162. Mol Microbiol. 1995;17:727–735. doi: 10.1111/j.1365-2958.1995.mmi_17040727.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Meyer R. The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer. Mol Microbiol. 1997;25:509–516. doi: 10.1046/j.1365-2958.1997.4861849.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, S., and R. J. Meyer. Unpublished observations.