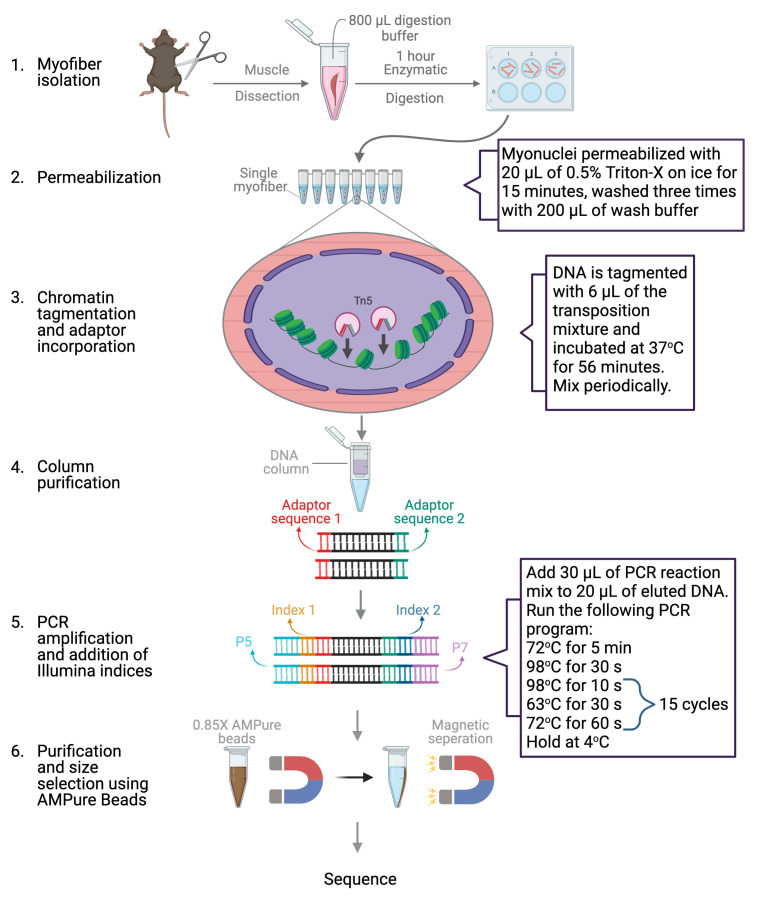
Abstract
Chromatin accessibility is a key determinant of gene expression that can be altered under different physiological and disease conditions. Skeletal muscle is made up of myofibers that are highly plastic and adaptive. Therefore, assessing the genome-wide chromatin state of myofibers under various conditions is very important to gain insight into the epigenetic state of myonuclei. The rigid nature of myofibers, as well as the low number of myonuclei that they contain, have rendered genome-wide studies with myofibers challenging. In recent years, ATAC-Seq from whole muscle and single nucleus ATAC-Seq have been performed. However, these techniques cannot distinguish between different fiber and cell types present in the muscle. In addition, due to the limited depth capacity obtained from single nucleus ATAC-Seq, an extensive comparative analysis cannot be performed. Here, we introduce a protocol where we combine the isolation of a single myofiber with OMNI ATAC-Seq. This protocol allows for genome-wide analysis of accessible chromatin regions of a selected single myofiber at a sufficient depth for comparative analysis under various physiological and disease conditions. This protocol can also allow for a specific myofiber to be selected, such as a regenerating myofiber. In the future, this protocol can help identify global changes in chromatin state under various conditions, as well as between different types of myofibers.
Graphical abstract:
Keywords: Single myofiber , ATAC-Seq , Skeletal Muscle , Epigenetics , Chromatin Accessibility
Background
Skeletal muscle is the largest tissue in the body and it is primarily composed of myofibers ( Buckingham et al. , 2003 ). Myofibers are highly adaptive, and therefore studying changes in myofibers under different stimuli is crucial for understanding how the skeletal muscle adapts and responds to different conditions and diseases ( Deschenes, 2004 ; Wilson et al. , 2012 ). Epigenetics and chromatin accessibility play a key role in the regulation of gene expression and tissue function ( Zhu et al. , 2018 ), making it crucial to study chromatin accessibility of different cell types, including myofibers under various stimuli. ATAC-Seq is the commonly used gold-standard method for genome-wide analysis of accessible chromatin regions, and is based on a hyperactive transposase, Tn5 ( Buenrostro et al. , 2015 ; Corces et al. , 2017 ). The advancements in sequencing and single cell technologies have allowed for ATAC-Seq to be performed at the whole muscle ( Ramachandran et al. , 2019 ) and at the single nucleus levels in recent years (Dos Santos et al. , 2020 ). However, both methods have limitations: whole muscle sequencing does not distinguish between different cell types present in the muscle, and single nucleus ATAC-Seq has lower sequencing depth, restricting its ability for comparative analysis. Therefore, these methods can not assess the chromatin state of only the muscle fibers and can not distinguish different myofiber types. The rigid structure of myofibers ( Janssen et al. , 2000 ; Keire et al. , 2013 ) and the low number of myonuclei that a single myofiber contains (200–300 myonuclei) ( Neal et al. , 2012 ; Cramer et al. , 2020 ), have made it challenging to perform ATAC-Seq on a single myofiber.
Here, we introduce a method, single myofiber ATAC-Seq (smfATAC-Seq), where we combine the isolation of a single myofiber with OMNI ATAC-Seq ( Corces et al. , 2017 ) to analyze the genome-wide accessible chromatin regions of a selected single myofiber without the confounding effects of other cell types present in the muscle. Additionally, this method allows for the selection of a specific myofiber, such as a regenerating myofiber, through the patterning of centrally located myonuclei ( Roman and Gomes, 2018 ). Furthermore, it provides sufficient sequencing depth for peak calling and for comparative analysis of myofibers under various physiological and disease conditions. We have successfully used this method to compare the chromatin accessibility between resting and regenerating myofibers as well as between myofibers isolated from a mouse model of Duchenne Muscular Dystrophy (mdx) and wild-type mice. smfATAC-Seq can allow researchers to assess the chromatin state of a single myofiber and compare the epigenetic changes that occur in myofibers under various biological and disease conditions. Therefore, in the future, smfATAC-Seq might help identify novel mechanistic insights on the role of myonuclei in regulating the plasticity of myofibers under various states.
Materials and Reagents
-
Materials
6-well plate (Sarstedt, catalog number: 83.3920)
1.5 mL microtubes (Sarstedt, catalog number: 72.690.300)
200 μL strip tubes (Progene, catalog number: 87-C200-8-TB)
Pipette tips (Sarstedt, catalog numbers: 70.3010 [10 μL], 70.3030 [200 μL], 70.3050 [1,000 μL])
Small and Large Glass Pasteur pipettes (VWR, catalog number: 14672-200)
0.22 μm filter (Ultident, catalog number: 229747)
1 mL syringe with 26G needle (BD Biosciences, catalog number: 309625)
Microplate (Corning, catalog number: 3676)
-
Animals
C57BL/6 mice were acquired from the Jackson Laboratories. The procedure is performed on 4-week-old mice that have gone through intra-muscular cardiotoxin mediated injury on the left hind limb (the procedure has also been successfully performed on 4-week-old uninjured C57BL/6, C57BL/10ScSn- Dmd mdx , and C57BL/10ScSn mice acquired from the Jackson Laboratories). This protocol could be adapted to various strains, but it will be important to adjust the Tn5 dose and incubation time if there is a severe skeletal muscle fiber phenotype.
-
Reagents
For Cardiotoxin-mediated injury
Cardiotoxin (CTX) (Sigma, catalog number: 11061-96-4)
Isoflurane (Fresenius Kabi, catalog number: CP0406V2)
PBS (Wisent, catalog number: 311-425-CL)
Carprofen (Zoetis, RIMADYL)
Ethanol (Commercial Alcohols, catalog number: P016EAAN)
For digestion and isolation of extensor digitorum longus (EDL)- Collagenase from Clostridium histolyctium (Sigma, catalog number: C0130-1G)
- PBS (Wisent, catalog number: 311-425-CL)
- Horse Serum (Wisent, catalog number: 065-250)
- DMEM (Invitrogen, catalog number: 11995073)
- Trypsin (Gibco, catalog number: 15090-046)
- Isoflurane (Fresenius Kabi, catalog number: CP0406V2)
- CO 2 (Praxair, catalog number: GP-700500)
- Digestion Buffer (see Recipes)
- Coating media (see Recipes)
For selection of injured vs. uninjured myofibers
Hoechst (Molecular Probes, catalog number: H1399)
-
PBS (Wisent, catalog number: 311-425-CL)
For lysis and permeabilization of the myofiber
PBS (Wisent, catalog number: 311-425-CL)
Triton X-100 (Sigma, catalog number: T9284)
Permeabilization Buffer (see Recipes)
For transposition
Tagment DNA Buffer and Tn5 transposase (Illumina, catalog number: 20034197)
Tween-20 (Sigma, catalog number: P1379-1L)
Digitonin (Promega, catalog number: G9441)
PBS (Wisent, catalog number: 311-425-CL)
Nuclease free water
Transposition mixture (see Recipes)
For DNA purification
QIAquick PCR Purification Kit (Qiagen, catalog number: 28104)
Ethanol (Commercial Alcohols, catalog number: P016EAAN)
For sequencing-ready library preparation
Q5 High fidelity DNA polymerase (New England Biolabs, catalog number: M0491S)
Deoxynucleotide (dNTP) solution mix (New England Biolabs, catalog number: N0447L)
Nextera XT Index Kit (Illumina, catalog number: FC-131-1001)
Nuclease free water
PCR reaction mixture (see Recipes)
For library purification and size selection
Ampure XP Beads (Beckman Coulter Life Sciences, catalog number: A63880)
Ethanol (Commercial Alcohols, catalog number: P016EAAN)
Nuclease free water
For quality control of the sequencing libraries
Quant-IT Picogreen dsDNA Assay kit (Invitrogen, catalog number: P7589)
Tris (Hydroxymethyl) Amino Methane (Multicell, catalog number: 600-125-LG)
Glacial acetic acid (Fisherbrand, catalog number: 351272-212)
Na 2 EDTA·2H 2 O (Bioshop, catalog number: EDT 001)
100 bp DNA ladder (GenedireX, catalog number: DM001-R500)
Agarose (Bioshop, catalog number: AGA001.500)
Orange G (Sigma, catalog number: O3756-25G)
Glycerol (Bioshop, catalog number: GLY001.1)
EDTA (Invitrogen, catalog number: AM9260G)
HCl (Honeywell-Fluka, catalog number: 72787)
GelGreen Nucleic Acid Gel Stain (Biotium, catalog number: 41005)
-
Primers (Integrated DNA Technologies, standard desalting, 25 nmole)
VEGFA_TSS_Forward CCGCTGAATAGTCTGCCTTG
VEGFA_TSS_Reverse GAGAAGCGCAGAGGCTTG
Chromosome17qE5_Forward TCATCATGTGTCCTGAAGTTGA
Chromosome17qE5_Reverse GCTTCTCTCCACAGAATTTGC
DNA Loading Dye (see Recipes)
TE Buffer (see Recipes)
TAE Buffer (see Recipes)
Equipment
Pipettes (P20, P200, P1000)
Microscope (Fisher Scientific, inverted microscope, equipped with transmitted light and a 4× objective)
EVOS FLoid Microscope (Life Technologies, catalog number: 4471136)
-
Dissection tools
Fine point high precision forceps (Fisher Scientific, catalog number: 22-327379)
Sharp-pointed dissecting scissors (Fisher Scientific, catalog number: 08-935)
Cell incubator (Thermo Scientific, Forma Series II Water-Jacketed CO2 Incubator, catalog number: 3110)
Heat block (Fisher Scientific, catalog number: 11-718-2)
Centrifuge (Thermo Scientific, Sorval Legend Micro 21R Microcentrifuge, catalog number: 75002447)
Thermocycler (Bio-Rad, C1000 Touch Thermal Cycler, catalog number: 1851148)
Magnetic rack (Thermo Fisher, DynaMag-2 Magnet, catalog number: 12321D)
Microplate reader (BioTek, synergy4)
Power supply (Bio-Rad, PowerPac, catalog number: 1645050)
Bioanalyzer (Agilent, BioAnalyzer 2100)
Gel Imager (LI-COR, Odyssey FC Imaging System)
Procedure
-
Intra-muscular injury of mouse Extensor Digitorum Longus (EDL) muscle (optional)
Note: This step is required if performing ATAC-Seq on regenerating myofibers. If not, you may skip to section B.
Prepare a working solution of 10 µM cardiotoxin (CTX) in PBS; store at -80°C.
-
Perform a subcutaneous injection of 100 µL per 20 g of mouse body weight of Carprofen (4 mg/mL) and wait 20 min.
Note: Carprofen is a non-steroidal anti-inflammatory drug (NSAID) used to manage the pain and inflammation that can be associated with the injury.
Anesthetize the mouse using an institutionally approved method. In this protocol, the mouse was anesthetized with 3% isoflurane in an induction chamber.
Spray the hindlimb that is to be injured with 70% ethanol and inject 50 µL of CTX through the Tibialis Anterior (TA) muscle into the EDL using a 1 mL syringe.
Monitor the mouse for 10 min.
Allow the muscle to regenerate for the desired period of time. We performed this on mouse skeletal muscle 7 days post injury; however, different time points after the injury can be used.
-
Dissection and digestion of Extensor Digitorum Longus (EDL) myofibers from Mus musculus
Prepare a digestion buffer of 1000 U/mL of collagenase from Clostridium histolyctium in unsupplemented DMEM (Recipe 1). Transfer 800 µL of digestion buffer into a 1.5 mL microtube and place it in an incubator at 37°C and 5% CO 2 .
-
Coat a 6-well plate with coating media of 10% Horse Serum (HS) in unsupplemented DMEM (Recipe 2). This will prevent the isolated myofibers from sticking to the plate and becoming difficult to handle.
Notes:
Pure HS or FBS can also be used.
Make sure that the plate is coated for at least 30 min prior to use.
Sacrifice the mouse.
Using a small pair of dissection scissors, remove the skin on the hindlimb and expose the muscles ( Video 1 ).
Dissect the Tibialis Anterior (TA) muscle by cutting the distal tendon, being careful to not cut the adjacent EDL tendon ( Video 1 ).
Peel the TA muscle upwards towards the knee using a pair of forceps. Then cut the TA muscle as close to the knee as possible ( Video 1 ).
Using a pair of forceps, detach the biceps femoris muscle from the knee to expose the proximal tendon of the EDL muscle ( Video 1 ).
Cut the distal EDL tendon and gently pull the EDL upwards towards the knee using a pair of forceps. Ensure that there is a bit of tension in the EDL as you are pulling it up without tugging too hard as that may damage the myofibers ( Video 1 ).
Cut the proximal EDL tendon as close to the knee as possible ( Video 1 ).
-
Place the EDL in the 1.5 mL microtube containing the digestion buffer from step B1. Add trypsin to the digestion buffer at a final concentration of 0.25%.
Notes:
Trypsin is added to remove the Muscle Stem Cells (MuSCs) associated with the myofibers.
Make sure to add the trypsin at this step and not at Step B1, as Trypsin can digest the collagenase and render it ineffective.
Incubate the EDL muscle with the digestion buffer containing trypsin at 37°C and 5% CO 2 for 1 h. Invert the tube periodically for mixing.
-
During the incubation period, remove the coating media from the 6-well plate and replace it with 2 mL of PBS 1×. Place the plate into the incubator at 37°C and 5% CO 2 .
Notes:
The coating media can be kept in the fridge and reused for future myofiber isolations.
Make sure that the plate is placed in the incubator for at least 30 min.
-
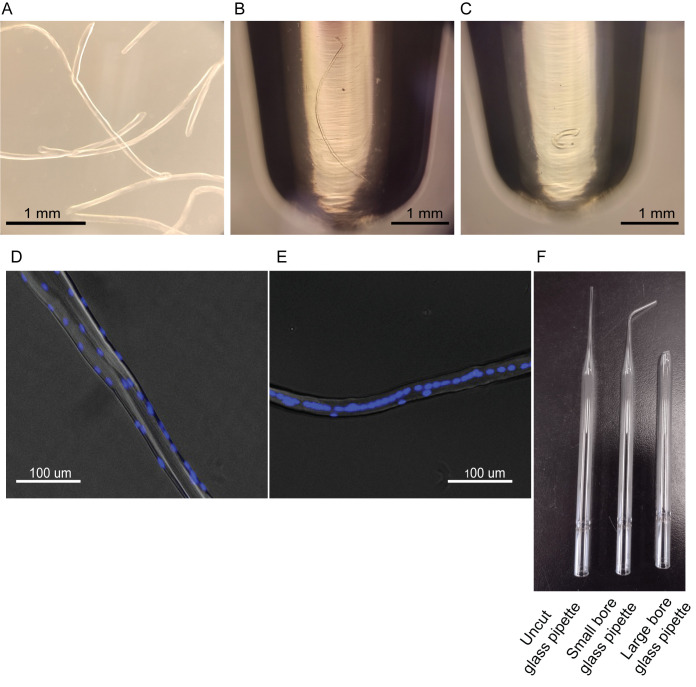
After the 1 h incubation time, transfer the EDL muscle to one of the coated wells of the 6-well plate using a large-bore glass pipette. Coat the large-bore glass pipette with HS prior to use ( Figure 1A ).
Note: Coating is very important because myofibers are very sticky in nature.
Gently pipette the EDL muscle up and down with the large-bore glass pipette coated with HS ( Figure 1F ).
Using a small-bore glass pipette coated with HS, serially transfer a small number of isolated myofibers to the remaining wells of the 6-well plate. This will act as a wash step to remove debris and other cell types.
-
Myofiber selection
-
Add 2 μL of Hoechst (5 mg/mL) into one well of the 6-well plate containing isolated myofibers in 2 mL of PBS 1× (optional).
Note: If you do not wish to select for a specific myofiber such as a regenerating myofiber, you may skip to step 5.
Place the 6-well plate in a 37°C with 5% CO 2 incubator for 5 min.
Remove the 6-well plate from the incubator and place it under the microscope.
-
If the desired myofiber is an injured myofiber, identify the myofibers with centrally located myonuclei along the length of the myofiber, as opposed to the non-regenerating myofibers with no specific myonuclei patterning ( Figure 1D –1E).
Note: During injury, not all the myofibers undergo regeneration simultaneously or to the same degree. However, regenerating myofibers are characterized by the pattern of centrally located myonuclei. Thus, you may select the regenerating myofiber using this hallmark as a marker.
Coat a small-bore glass pipette with HS.
While visualizing the myofibers under the microscope, select the desired myofiber and transfer it into a 200 μL PCR strip tube using the coated small-bore glass pipette.
Visualize the PCR tube under the microscope to ensure that you have selected and transferred a single myofiber into the tube ( Figure 1B ).
-
-
Myofiber lysis and permeabilization
Using a P200 pipette, remove the excess PBS 1× from the PCR tube under the microscope. Make sure not to remove the myofiber.
Using a P20 pipette, add 20 μL of the permeabilization buffer (Recipe 3) into the tube with the myofiber. Pipette the fiber up and down five times.
Incubate on ice for 15 min ( Figure 1C ).
Remove the permeabilization buffer with a P20 pipette under the microscope while being careful not to remove the myofiber.
Wash the myofiber by adding 200 μL of PBS 1× using a P200 pipette.
Place the tube on ice for 5 min.
Remove the PBS 1× by using a P200 pipette under the microscope.
Repeat steps 5–7 two times.
Note: During the 5 min incubation of the washes, you may start the next step for the preparation of the transposition mixture (Recipe 4).
-
Myofiber transposition by the Tn5 transposase
Prepare the transposition mixture (Recipe 4).
Using a P20 pipette, add 6 μL of the transposition mixture into the tube containing the myofiber. Slowly pipette the myofiber up and down six times.
Place the tube into the heat block set to 37°C for 56 min. Gently shake the tube by flicking it every 5–7 min.
Note: The Tn5 transposase will cut the accessible chromatin regions and add adaptors simultaneously.
-
DNA purification
-
After the transposition, column purify the DNA using the QIAquick PCR Purification Kit according to the manufacturer’s guidelines. Briefly,
Add 70 μL of PB binding buffer into the tube containing the myofiber in the transposition mixture.
Transfer the entire contents of the tube into the column provided by the kit.
Centrifuge the column at 16,000 × g for 1 min.
Discard the flow-through.
Place the column back in the same tube.
Add 700 μL of PE wash buffer into the column.
Centrifuge the column at 16,000 × g for 1 min.
Discard the flow-through. Place the column back in the same tube.
Centrifuge again at 16,000 × g for 1 min.
Discard the flow-through.
Place the column into an empty 1.5 mL microtube.
-
Add 20 μL of elution buffer into the column. Incubate at room temperature for 5 min.
Note: You may place the tube at 37°C for 2 min for better elution.
Centrifuge at 16,000 × g for 1 min.
-
Using a P200 pipette, take out the 20 μL of eluted DNA from the collection tube and add it back into the column for second elution.
Note: Double elution can help increase the yield.
Elute the DNA again by centrifuging at 16,000 × g for 1 min.
Note: This is a stopping point. The eluted DNA can be left at 4°C overnight or at -20°C until library preparation.
-
-
Preparation of sequencing ready libraries
To amplify and incorporate indices for sequencing, prepare the PCR reaction mix (Recipe 5).
-
Add 30 μL of PCR reaction mix to the 20 μL of eluted DNA from the previous step for a final volume of 50 μL.
Note: When preparing the sequencing libraries, make sure that each sample has a different i5 and i7 adaptor combination. In addition, when combining samples in a sequencing lane, make sure that i5 and i7 adaptor combinations for the samples are color balanced for the type of sequencer that is being used (two or four channels). For more information, please visit:
-
Run the PCR program on the thermocycler using the following steps:
72°C for 5 min
98°C for 30 s
98°C for 10 s
63°C for 30 s 15 cycles
72°C for 60 s
Hold at 4°C
Note: This is a stopping point. The libraries can be left at 4°C overnight or at -20°C until library purification.
-
Library purification and size selection
-
Add AMPure XP beads into the PCR tube at a ratio of 0.85× (v/v). Mix well.
Note: This ratio is used specifically to remove fragments that are below the size of 200 bp.
Incubate at room temperature for 8 min ( Figure 2A ).
Place the tube on the magnetic rack. Wait for the solution to become clear ( Figure 2B ).
Discard the supernatant.
Wash the beads with 200 μL of 80% ethanol.
Remove the ethanol from the tube.
Repeat steps H5–H6. Make sure to remove all the ethanol after the second wash.
-
Let the beads dry for 2 min.
Note: Be careful not to over-dry the beads since this leads to lower elution efficiency. However, if the beads do get overdried (a sign of this is the cracking on the beads), do the elution at 37°C for 5–10 min as this can increase the elution efficiency.
Elute the DNA with 15 μL of elution buffer that is provided with the QIAquick PCR Purification Kit. Mix well until beads go into the solution.
-
Incubate at room temperature for at least 5 min.
Note: You may place the tubes at 37°C for 2–5 min for better elution.
Place the tube on the magnetic rack. Wait for the solution to become clear.
-
Keep the supernatant; this is your eluted DNA.
Note: You may proceed to the quality control steps, or you can store at -20°C until sequencing.
-
-
Quantification and quality control of the ATAC-Seq libraries
-
Quantify the libraries by using the Quant-IT Picogreen dsDNA Assay kit ( Table 1 ).
Prepare 1× TE buffer from the 20× TE stock solution that is provided with the kit.
-
Prepare DNA standards by diluting the stock DNA that is provided in 1× TE to the desired concentration.
Note: You may prepare DNA standards of 10 ng/μL, 5 ng/μL, 2 ng/μL, 1.5 ng/μL, 1.0 ng/μL, 0.75 ng/μL, 0.5 ng/μL, 0.25 ng/μL, 0.1 ng/μL, and 0.05 ng/μL.
-
Pipette 5 µL of TE 1× buffer into each well of a microplate. Calculate the number of wells needed depending on the number of samples you have. Usually, each standard and sample is run in triplicates.
Note: Due to the small concentration of DNA following smf-ATAC-Seq , you may run the samples in duplicates instead in order to not waste the samples.
Add 1 μL of the sample and the standards to the corresponding wells containing 5 μL of 1× TE buffer.
-
Thaw the stock Picogreen dye solution and mix well. Then make a 1:200 dilution of the stock Picogreen in 1× TE buffer.
Note: Make sure to not expose the Picogreen Dye to direct light for long periods of time.
-
Pipette 5 μL of the diluted Picogreen solution into each well. Quantify the amount of DNA in each sample by using a microplate reader.
Note: We usually get a final concentration of 3–7 ng/μL ( Table 1 ).
-
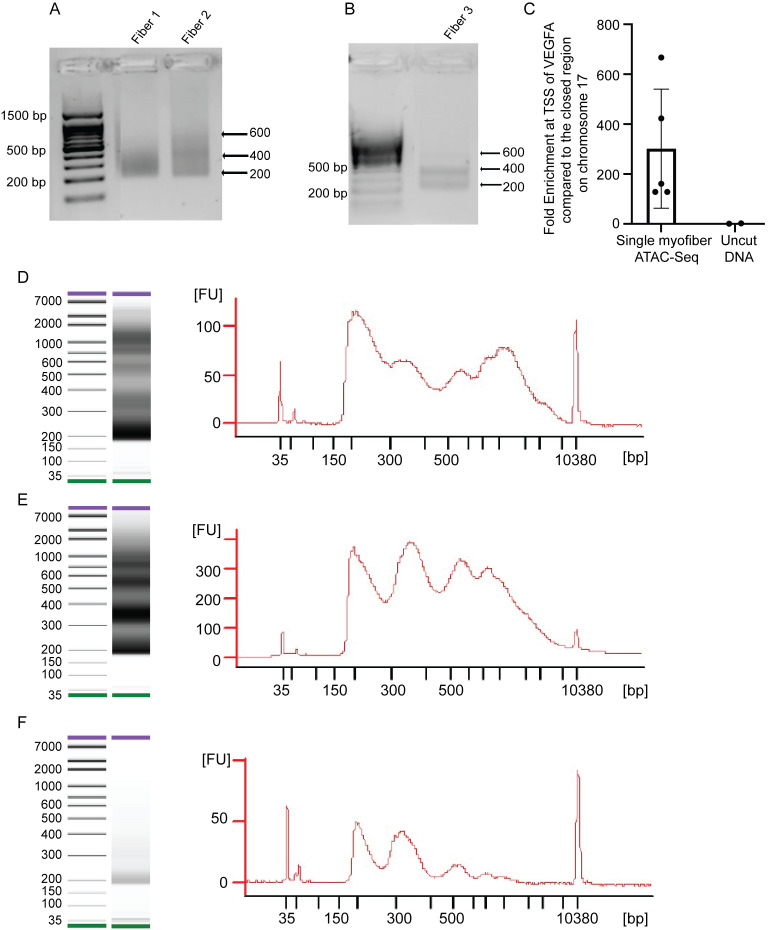
To verify the library size, run 11 ng of the sample on 1.25% agarose gel with dsGreen (1:10000 dilution) ( Figure 3A –3B).
Notes:
The minimum amount is 11 ng; depending on the amount of DNA, you may run up to 20 ng of the sample on the gel.
Make sure to run only 3 μL of the DNA ladder that is diluted 1:2. This will ensure that the DNA ladder does not wash out the signal from the samples.
-
To check the enrichment for open regions of chromatin, you may run a qPCR for marker genes in the myofibers ( Figure 3C ).
Note: Since myofibers express VEGFA ( Lazure et al., 2020 ), we use the Transcription Start Site (TSS) of VEGFA to check for enrichment. You may use the TSS of other expressed genes in the myofibers. As a negative control, we use a closed region on chromosome 17. We have also included the fold enrichment for the input DNA, which corresponds to the myofibers that were not transposed with the Tn5, called uncut DNA. In a successful ATAC-Seq, you will observe much higher fold enrichments in your sample for your marker gene compared to the enrichment in the Uncut DNA sample.
-
Run the sequencing libraries on a bioanalyzer ( Figure 3D –3F)
Video 1. Dissection of the EDL muscle.

The procedure in the video was approved by McGill University Animal Care Committee (UACC) under the protocol #7512.
Figure 1. Myofiber isolation and selection.
(A) Typical isolated EDL myofibers in a 6-well plate. (B) Live individual myofiber in a 0.2 mL microtube. (C) Single myofiber in a 0.2 mL microtube post permeabilization. (D) Hoechst staining of an uninjured myofiber. (E) Hoechst staining of a regenerating myofiber, displaying the hallmark of centrally located myonuclei. (F) Glass pipettes used in the procedure. Small-bore glass pipette can be obtained by flame polishing the uncut glass pipette on a Bunsen burner. Large-bore glass pipette can be obtained by cutting a glass pipette and then flame polishing on the Bunsen burner.
Figure 2. Size selection and purification of ATAC-Seq libraries by AMPure XP beads.
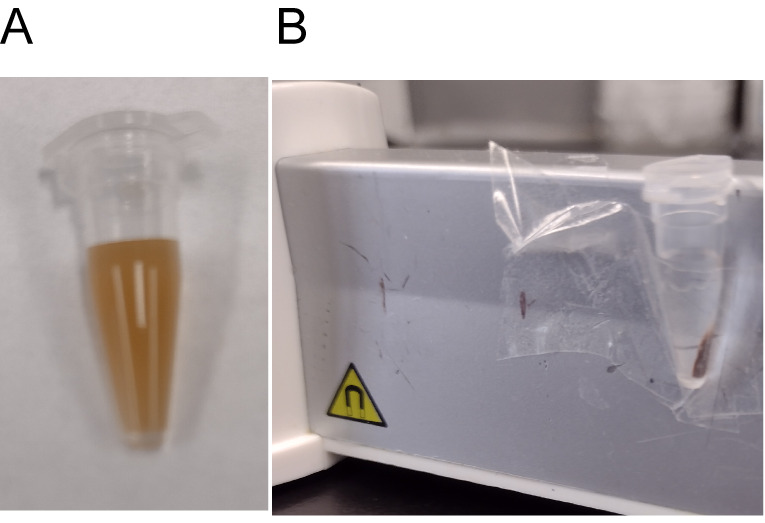
(A) AMPure beads in solution. (B) AMPure beads separated from the solution on the magnetic rack.
Table 1. Representative final concentrations of single myofiber ATAC-Seq libraries .
| Sample | Concentration (ng/μL) |
| Fiber 1 | 3.59 |
| Fiber 2 | 4.61 |
| Fiber 3 | 3.76 |
| Fiber 4 | 6.97 |
| Fiber 5 | 3.39 |
| Fiber 6 | 5.66 |
| Fiber 7 | 4.08 |
Figure 3. Quality control of smfATAC-Seq libraries.
(A–B) Representative pictures of smfATAC-seq libraries after size selection, visualized on an agarose gel. (C) qPCR for the TSS of VEGFA compared with a negative control region of Chromosome 17 qE5 for the smfATAC-Seq libraries (n = 5 biological replicates. Error bars = ± SD). (D–F) Examples of bioanalyzer profiles of smfATAC-Seq.
Data analysis
Follow the standard Encode ATAC-Seq pipeline for the analysis. For more information, visit: https://www.encodeproject.org/atac-seq/ .
Recipes
-
Digestion buffer
Dissolve powdered collagenase from Clostridium histolyctium at a final concentration of 1,000 U/mL.
Add trypsin to the buffer at a final concentration of 0.25% once the muscle has been placed in the digestion buffer; this will prevent the trypsin from digesting the collagenase beforehand.
Note: Always prepare the digestion buffer fresh, immediately before sacrificing the mouse.
-
Coating media
Dilute HS into un-supplemented DMEM at a final concentration of 10% (V/V).
-
Permeabilization buffer
Prepare a stock solution of 10% Triton X-100 in ddH 2 O.
Prepare a working solution of permeabilization buffer from the 10% stock solution to a final concentration of 0.5% Triton X-100 in ddH 2 O.
-
Transposition mixture
Prepare a transposition mixture for six fibers in a total of 40 μL.
Add 20 μL of Tagment DNA Buffer (TD buffer)
Add 13.3 μL of PBS 1×
Add 4.61 μL of Nuclease free H 2 O
Add Digitonin for a final concentration of 0.02%
Add Tween-20 for a final concentration of 0.2%
Add 1.39 μL of Tn5
Note: Add the Tn5 enzyme at the end.
-
PCR reaction mix (1 reaction, 30 μL)
10 μL of Q5 Buffer
10 μL of Q5 enhancer
1 μL of dNTP’s
2.5 μL of i7 index
2.5 μL of i5 index
3.5 μL of nuclease-free water
0.5 μL of Q5 High Fidelity DNA polymerase
-
DNA loading dye
50% TE Buffer (see Recipe 7)
50% Glycerol
Orange G
-
TE buffer
1 mL of Tris-HCl pH 8.0, 1M
200 μL of EDTA pH 8.0, 0.5 M
Complete to 100 mL with dH 2 O
-
TAE buffer
242 g of Tris base
57.1 mL of Glacial acetic acid
37.2 g of anhydrous Na 2 EDTA
Complete to 1 L total volume with dH 2 O
Acknowledgments
This work was funded by a discovery grant from the Natural Sciences and Engineering Research Council (NSERC) to VDS.
This method has been successfully used to analyze the chromatin state of mouse single myofibers under injury and disease conditions ( Sahinyan et al. , 2022 ).
Competing interests
The authors declare no competing interests.
Ethics
All procedures that were performed on animals were approved by the McGill University Animal Care Committee (UACC) under the protocol #7512, valid through July/1/2021 – July/1/2022.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Buckingham M. , Bajard L. , Chang T. , Daubas P. , Hadchouel J. , Meilhac S. , Montarras D. , Rocancourt D. and Relaix F. ( 2003 . ). The formation of skeletal muscle: from somite to limb . J Anat 202 ( 1 ): 59 - 68 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buenrostro J. D. , Wu B. , Chang H. Y. and Greenleaf W. J. ( 2015 . ). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide . Curr Protoc Mol Biol 109 : 21 29 21-21 29 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corces M. R. , Trevino A. E. , Hamilton E. G. , Greenside P. G. , Sinnott-Armstrong N. A. , Vesuna S. , Satpathy A. T. , Rubin A. J. , Montine K. S. , Wu B. , et al. .( 2017 . ). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues . Nat Methods 14 ( 10 ): 959 - 962 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cramer A. A. W. , Prasad V. , Eftestol E. , Song T. , Hansson K. A. , Dugdale H. F. , Sadayappan S. , Ochala J. , Gundersen K. and Millay D. P. ( 2020 . ). Nuclear numbers in syncytial muscle fibers promote size but limit the development of larger myonuclear domains . Nat Commun 11 ( 1 ): 6287 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deschenes M. R. ( 2004 . ). Effects of aging on muscle fibre type and size . Sports Med 34 ( 12 ): 809 - 824 . [DOI] [PubMed] [Google Scholar]
- 6. Dos Santos M. , Backer S. , Saintpierre B. , Izac B. , Andrieu M. , Letourneur F. , Relaix F. , Sotiropoulos A. and Maire P. ( 2020 . ). Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers . Nat Commun 11 ( 1 ): 5102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janssen I. , Heymsfield S. B. , Wang Z. M. and Ross R. ( 2000 . ). Skeletal muscle mass and distribution in 468 men and women aged 18-88 yr . J Appl Physiol(1985) 89 ( 1 ): 81 - 88 . [DOI] [PubMed] [Google Scholar]
- 8. Keire P. , Shearer A. , Shefer G. and Yablonka-Reuveni Z. ( 2013 . ). Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells . Methods Mol Biol 946 : 431 - 468 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lazure F. , Blackburn D. M. , Corchado A. H. , Sahinyan K. , Karam N. , Sharanek A. , Nguyen D. , Lepper C. , Najafabadi H. S. , Perkins T. J. , et al. .( 2020 . ). Myf6/MRF4 is a myogenic niche regulator required for the maintenance of the muscle stem cell pool . EMBO Rep 21 ( 12 ): e49499 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neal A. , Boldrin L. and Morgan J. E. ( 2012 . ). The satellite cell in male and female, developing and adult mouse muscle: distinct stem cells for growth and regeneration . PLoS One 7 ( 5 ): e37950 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramachandran K. , Senagolage M. D. , Sommars M. A. , Futtner C. R. , Omura Y. , Allred A. L. and Barish G. D. ( 2019 . ). Dynamic enhancers control skeletal muscle identity and reprogramming . PLoS Biol 17 ( 10 ): e3000467 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roman W. and Gomes E. R. ( 2018 . ). Nuclear positioning in skeletal muscle . Semin Cell Dev Biol 82 : 51 - 56 . [DOI] [PubMed] [Google Scholar]
- 13. Sahinyan K. , Blackburn M D. and Soleimani D V. ( 2022 . ). Application of ATAC-Seq for genome-wide analysis of the chromatin state at single myofiber resolution . eLife 11 : e72792 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilson J. M. , Loenneke J. P. , Jo E. , Wilson G. J. , Zourdos M. C. and Kim J. S. ( 2012 . ). The effects of endurance, strength, and power training on muscle fiber type shifting . J Strength Cond Res 26 ( 6 ): 1724 - 1729 . [DOI] [PubMed] [Google Scholar]
- 15. Zhu F. , Farnung L. , Kaasinen E. , Sahu B. , Yin Y. , Wei B. , Dodonova S. O. , Nitta K. R. , Morgunova E. , Taipale M. , et al. .( 2018 . ). The interaction landscape between transcription factors and the nucleosome . Nature 562 ( 7725 ): 76 - 81 . [DOI] [PMC free article] [PubMed] [Google Scholar]