Abstract
The VanX protein is a d-alanyl-d-alanine (d-Ala-d-Ala) dipeptidase essential for resistance to the glycopeptide antibiotic vancomycin. While this enzymatic activity has been typically associated with vancomycin- and teicoplainin-resistant enterococci, we now report the identification of a d-Ala-d-Ala dipeptidase in the gram-negative species Salmonella enterica. The Salmonella enzyme is only 36% identical to VanX but exhibits a similar substrate specificity: it hydrolyzes d-Ala-d-Ala, dl-Ala-dl-Phe, and d-Ala-Gly but not the tripeptides d-Ala-d-Ala-d-Ala and dl-Ala-dl-Lys-Gly or the dipeptides l-Ala-l-Ala, N-acetyl-d-Ala-d-Ala, and l-Leu-Pro. The Salmonella dipeptidase gene, designated pcgL, appears to have been acquired by horizontal gene transfer because pcgL-hybridizing sequences were not detected in related bacterial species and the G+C content of the pcgL-containing region (41%) is much lower than the overall G+C content of the Salmonella chromosome (52%). In contrast to wild-type Salmonella, a pcgL mutant was unable to use d-Ala-d-Ala as a sole carbon source. The pcgL gene conferred d-Ala-d-Ala dipeptidase activity upon Escherichia coli K-12 but did not allow growth on d-Ala-d-Ala. The PcgL protein localizes to the periplasmic space of Salmonella, suggesting that this dipeptidase participates in peptidoglycan metabolism.
Vancomycin is a glycopeptide antibiotic that inhibits peptidoglycan synthesis by binding to the peptidoglycan precursor UDP-MurNAc-l-Ala-d-Glu-l-Lys-d-Ala-d-Ala at the d-Ala-d-Ala terminus (32). Acquired resistance to glycopeptides in enterococci requires VanX (36), a dipeptidase that cleaves d-Ala-d-Ala and allows the synthesis of d-Ala-d-lactate mediated by the vanH and vanA gene products (6). This decreases the rate of synthesis of the pentapeptide precursor UDP-MurNAc-l-Ala-d-Glu-l-Lys-d-Ala-d-Ala and increases the levels of a pentadepsipeptide precursor, UDP-MurNAc-l-Ala-d-Glu-l-Lys-d-Ala-d-Lac, which exhibits decreased binding to glycopeptide antibiotics (34). Vancomycin resistance also requires the d,d-carboxypeptidase VanY (2) and the regulatory system VanR-VanS, which is responsible for transcriptional control of the vanA, vanH, vanX, and vanY genes (3; see reference 35 for a recent review). While d-Ala-d-Ala dipeptidase activity has been associated typically with vancomycin-resistant enterococci, we now describe the identification of a d-Ala-d-Ala dipeptidase in the gram-negative bacterium Salmonella enterica.
S. enterica is a facultative intracellular pathogen that is responsible for several disease syndromes in humans, including gastroenteritis, typhoid fever, and bacteremia (23). There is a single species in the genus Salmonella which encompasses over 2,300 serotypes that differ in host specificity and the disease condition that they promote in susceptible hosts. During growth within host tissues, Salmonella modifies its peptidoglycan (33) and sheds part of its lipopolysaccharide (LPS) (12). Moreover, Salmonella can introduce modifications in the lipid A portion of its LPS that decrease the ability of the LPS to induce expression of tumor necrosis factor alpha by adherent monocytes (21) and increase resistance to cationic peptide antibiotics (22). While changing the peptidoglycan and the LPS may be a way of preventing activation of the host immune system, the virulence role and molecular basis of these cell surface alterations remain largely unknown.
The PhoP-PhoQ two-component regulatory system governs several aspects of Salmonella virulence, including intramacrophage survival, resistance to antimicrobial peptides, and invasion of epithelial cells (16). The PhoQ protein is a membrane-bound sensor that modifies the transcriptional activity of the PhoP protein in response to the extracytoplasmic levels of Mg2+: PhoP-activated genes are transcriptionally induced during growth in micromolar concentrations of Mg2+ and repressed when bacteria are grown in the presence of millimolar concentrations of Mg2+ (10, 39). Salmonella appears to reside in a low-Mg2+ environment within host tissues because PhoP-activated genes are transcriptionally induced to high levels within host cells (1, 11, 41). The PhoP-PhoQ system controls expression of some 40 different proteins, including two distinct Mg2+ transporters, a UDP-glucose dehydrogenase, and a nonspecific acid phosphatase, as well as proteins required for modification of the lipid A in the LPS (20, 39).
In this paper, we identify a d-Ala-d-Ala dipeptidase in S. enterica. We establish that this peptidase is encoded by a gene that is specific to Salmonella and regulated by the PhoP-PhoQ regulatory system. We also demonstrate that the Salmonella dipeptidase localizes to the periplasmic space, suggesting a role for this enzyme in peptidoglycan metabolism.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacterial genetic techniques, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. All Salmonella strains are derived from wild-type S. enterica serovar Typhimurium 14028s, except for strain AA3007 which is derived from LT2. Plasmids are derived from pUC19, pBR322, or Mud5005. Bacteria were grown at 37°C in either Luria-Bertani (LB) (29) or N-minimal medium (38) supplemented with 0.1% Casamino Acids, 38 mM glycerol, and different concentrations of MgCl2. Experiments that evaluated the ability of strains to use d-Ala-d-Ala as a sole carbon source were carried out in N-minimal medium supplemented with 0.2% d-Ala-d-Ala, 0.1 mg of thiamine per liter, and MgCl2 (10 mM). Ampicillin and kanamycin were used at 50 μg/ml, and tetracycline was used at 10 μg/ml. Phage P22-mediated transductions were carried out as described before (8). Plasmids were transformed into bacteria either by electroporation with a Bio-Rad apparatus or by chemical transformation using standard procedures.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or sourcea |
|---|---|---|
| S. enterica | ||
| 14028s | Wild type | ATCC |
| MS7953s | phoP7953::Tn10 | 9 |
| EG9331 | pcgL::MudJ | 39 |
| EG10277 | sugR1::MudJ | 4 |
| EG10981 | phoP7953::Tn10 ugtL::lac-kan | This work |
| EG11250 | ugtL::lac-kan | This work |
| E. coli | ||
| DH5α | F′/endA1 hsdr17 (rK− mK+) supE44 thi-1 recA1 gyrA relA1 Δ(lacIzYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | 13 |
| JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14− (McrA−) Δ(lac-proAB) thi gyrA96 endA1 hsdr17 | 43 |
| EG10358 | TK2316Mucts/pEG9129 | This work |
| EG10421 | DH5α/pEG9125 | This work |
| TK2316Mucts | F−hsdR thi thr lacZ(Am) nagA kdp rha Mucts | 15 |
| Plasmids | ||
| pBR322 | reppMB1 Tetr Apr | 5 |
| pEG5005 | pBC0::Mud5005 | 14 |
| pEG9122 | pBR322::pcgL+ | This work |
| pEG9123 | pBR322::pcgL+ | This work |
| pEG9125 | pUC19::pcgL+ ugtL+ | This work |
| pEG9129 | Mud5005::pcgL+ ugtL+ | This work |
| pEG9130 | pUC19::ugtL+ | This work |
| pEG9131 | pUC19::pcgL− | This work |
| pEG9132 | pUC19::pcgL+ ugtL+ | This work |
| pUC19 | reppMB1lacZα Apr | 43 |
ATCC, American Type Culture Collection.
Molecular biological techniques.
The nucleotide sequence of a 3.8-kb region that harbors the pcgL and ugtL genes was determined on both strands by using plasmid pEG9129 as the template and newly synthesized primers. Plasmid pEG9129 DNA was extracted from strain EG10358 and purified by use of the Qiagen plasmid midi kit, and a segment of it was sequenced with the dichlororhodamine dye terminator cycle sequencing kit and an ABI 310 automatic sequencer. DNA sequence analysis and protein sequence alignments were performed with the GeneWorks (IntelliGenetics) and GCG (University of Wisconsin) software packages.
Southern hybridization analysis was carried out on chromosomal DNA that had been digested with EcoRI, size fractionated on a 1% agarose gel, and transferred to a nylon membrane by capillary blotting. To investigate the presence of pcgL-hybridizing sequences, a 782-bp PCR fragment containing the pcgL coding region was generated with primers 858 (5′-GGGTCTCTGCTTAACGG-3′) and 906 (5′-GCGAGGTGTAACATATGG-3′), labeled with [α-32P]dCTP with the Ready to Go kit (Pharmacia Biotech), and used as a probe under previously described conditions (19). To investigate the presence of vanXE. coli (termed ecovanX by Lessard et al. [26])-hybridizing sequences, a 473-bp PCR fragment containing a segment of the vanXE. coli coding region was generated by using primers 744 (5′-AATTGAAATACGCCTGCGCTG-3′) and 745 (5′-GAGCAGAGGGTAACTCGCTGC-3′) and labeled as described above for the pcgL probe. Hybridization experiments with the vanXE. coli probe were carried out under both high- and low-stringency conditions, as previously described (17).
Cloning of the pcgL gene.
We used the in vivo cloning procedure with the mini-Mu replicon Mud5005 (14) to construct a library from wild-type S. enterica serovar Typhimurium. Kanamycin-resistant transductants of strain TK2316Mucts were screened by colony hybridization using as a probe a 202-bp DNA fragment located immediately adjacent to the MudJ transposon in the pcgL::MudJ strain EG9331. The 202-bp fragment was generated by the PCR using primers 633 (5′-GCGTGGGCCAAAGATCCTTCT-3′) and 634 (5′-ACGCAGTACAATTCACCAGTG-3′) and labeled with [α-32P]dCTP with the Ready to Go kit (Pharmacia Biotech). Four hybridizing clones were identified, and one of them, EG10358, harboring plasmid pEG9129 with a 24-kb insert (see Fig. 1), was used for further studies.
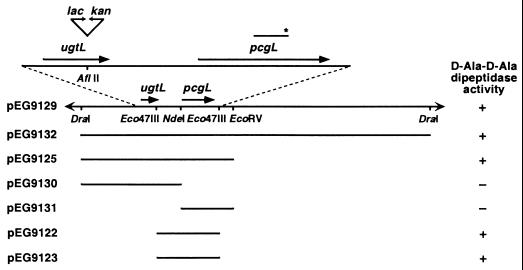
FIG. 1.
Physical and genetic map of pcgL-containing region and localization of pcgL-encoded d-Ala-d-Ala dipeptidase activity. The short horizontal line with an asterisk above indicates the position of the PCR-generated fragment used to screen the wild-type library. Plasmids pEG9122 and pEG9123 are pBR322 derivatives that harbor the same Eco47III fragment in opposite orientations. The pcgL gene is in the same orientation as the tet gene of pBR322 in plasmid pEG9123 and in the opposite orientation in plasmid pEG9122. Plasmid pEG9129 is based on Mud5005, and plasmids pEG9125, pEG9130, pEG9131, and pEG9132 are derived from plasmid pUC19.
Construction of a ugtL mutant.
An 11-kb HindIII fragment from plasmid pEG7125 (18) harboring a promoterless lac operon and kan resistance gene was introduced into the AflII site within the ugtL gene in plasmid pEG9124. (Plasmid pEG9124 was derived from plasmid pEG9125 by digestion with AatII and SmaI followed by religation). Plasmid pFH1, harboring the lac operon in the same orientation as the ugtL gene, was used to transfer the ugtL mutation back to the chromosome as described previously (19). The structure of the ugtL locus in the resulting mutant was confirmed by Southern hybridization analysis using as a probe a 610-bp fragment containing the ugtL gene that was generated by the PCR using primers 758 (5′-GAACACGTCGATTGTCGGCGC-3′) and 759 (5′-ACGATTAGCTGACGGCTTTG-3′).
Overproduction and purification of the PcgL protein.
The PcgL protein was overproduced in Escherichia coli K-12 DH5α cells harboring the pcgL-containing plasmid pEG9125. Bacterial cells were grown in LB broth, harvested, and dissolved in 50 mM HEPES buffer (pH 7.7). After sonication and centrifugation, the supernatant was precipitated with ammonium sulfate to 80% saturation. After desalting, the protein was purified in a two-step procedure using a high-pressure liquid chromatography apparatus. First, we used an ion-exchange column (Toyopearl SP column) and 200 mM KCl to elute the protein, and then we used a size exclusion column (Waters Protein Pak 300SW). The purity of the PcgL protein was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. The N-terminal sequence of the first 10 residues of the PcgL protein was determined with Edman degradation by Midwest Scientific Laboratory (St. Louis, Mo.).
Enzyme and virulence assays.
Crude extracts from N-minimal medium-grown bacteria were prepared as follows: bacteria were grown in 3 to 5 ml of medium, harvested, washed twice with 50 mM HEPES buffer (pH 7.7), resuspended in 500 μl of the same buffer, and sonicated. The supernatants were used for total protein determination with the bicinchoninic acid protein assay kit (Sigma), with bovine serum albumin as a standard. Periplasmic and cytoplasmic fractions were prepared by osmotic shock as described previously (31). β-Galactosidase and nonspecific acid phosphatase activities were determined as described before (24, 29).
Peptide hydrolysis was determined quantitatively by measuring the release of free amino acids by a modification of the cadmium-ninhydrin method (42). Briefly, substrates (10 mM) were incubated with 10 μg of protein from crude cell extracts or with 1.8, 9, or 18 ng of purified PcgL protein, for 30 min at 37°C in 50 mM HEPES buffer (pH 7.7). Then, 7.5 times the volume of cadmium-ninhydrin solution (0.01 g of ninhydrin/ml of ethanol, 12.5 ml of acetic acid, 1 g of CdCl2/ml of bidistilled water) was added, and the samples were incubated for 5 min at 85°C for color development. The optical density at 505 nm was then determined. Standards of 0.1, 0.5, and 1 mM d-alanine were used, as well as blanks containing 10 mM of the corresponding substrate resuspended in 50 mM HEPES buffer. All substrates were from Sigma.
Macrophage survival assays with the macrophage-like cell line J774 and invasion assays of canine kidney epithelial (MDCK) cells were conducted as described previously (25). Virulence assays were performed with 7- to 8-week-old female BALB/c mice inoculated orally or intraperitoneally with 100 μl of bacteria diluted in phosphate-buffered saline.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank database under accession no. AF120672.
RESULTS
The pcgL gene encodes a novel Salmonella protein.
The Salmonella pcgL locus was originally identified as a PhoP-activated lac gene fusion generated with the promoter probe transposon MudJ (39). To determine the function of the pcgL gene, we isolated the wild-type copy of pcgL by screening a plasmid library by colony hybridization using as a probe a 202-bp DNA segment located immediately adjacent to the MudJ transposon in the pcgL mutant EG9331 (Fig. 1). Sequence analysis of a positive clone revealed that the MudJ had inserted within a novel Salmonella gene encoding a 256-amino-acid protein with a predicted molecular mass of 28,995 Da and an isoelectric point of 6.98.
The PcgL protein is 36% identical over 192 residues to the 202-amino-acid VanX, a d-Ala-d-Ala dipeptidase encoded within the Enterococcus faecium transposon Tn1546 and required for vancomycin resistance (36). The similarity between PcgL and VanX extends over the entire length of these proteins (Fig. 2A). Conserved amino acids include VanX residues His116, Asp123, and His184, which have been implicated in the coordination of Zn2+, and Glu181, which was shown to be the catalytic base (7, 28). The PcgL protein is larger than VanX due to a 20-amino-acid cleavable signal sequence at the N terminus (see below) and to 23 additional residues located between a position equivalent to amino acids 130 and 131 of the VanX protein. The PcgL protein also exhibits similarity to uncharacterized open reading frames from Mycobacterium tuberculosis and Synechocystis sp. and to an E. coli K-12 protein that displays d-Ala-d-Ala dipeptidase activity (26).
FIG. 2.
(A) Alignment of the PcgL protein of S. enterica and the VanX protein of the Enterococcus faecium transposon Tn1546. (B) Alignment of the S. enterica ugtL gene product with a segment from a chitin synthetase from Schizosaccharomyces pombe.
The pcgL gene product is required for d-Ala-d-Ala dipeptidase activity.
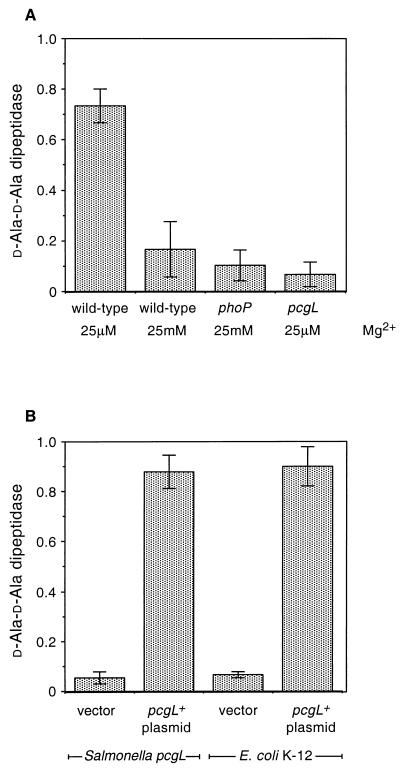
Despite the low-level sequence identity between the PcgL and VanX proteins, we detected high levels of d-Ala-d-Ala dipeptidase activity in crude extracts prepared from wild-type Salmonella grown under conditions that promote expression of PhoP-activated genes (i.e., 25 μM Mg2+) (Fig. 3A). This activity was very much reduced in extracts prepared from wild-type cells grown under conditions that repress transcription of PhoP-activated genes (i.e., 25 mM Mg2+) and from extracts prepared from a phoP mutant, and it was absent from extracts prepared from the pcgL strain (Fig. 3A).
FIG. 3.
(A) d-Ala-d-Ala dipeptidase activity of crude extracts from wild-type and pcgL and phoP mutant Salmonella strains grown in N-minimal medium with 25 μM or 25 mM MgCl2. Data correspond to mean values of 20 independent assays. (B) d-Ala-d-Ala dipeptidase activity of S. enterica pcgL mutant strain EG9331 and of E. coli K-12 harboring the pcgL-containing plasmid pEG9122 or the vector pBR322. Data correspond to mean values of three independent assays. d-Ala-d-Ala dipeptidase activity indicates micromoles of d-Ala produced per milligram of protein per minute.
To determine the location of the pcgL gene within plasmid pEG9129, we examined the ability of different subclones to restore d-Ala-d-Ala dipeptidase activity to the pcgL mutant and localized the pcgL gene to the 1.5-kb Eco47III fragment in plasmids pEG9122 and pEG9123, which differ in the relative orientation of the insert (Fig. 1). This fragment includes the complete pcgL open reading frame and could complement the pcgL mutant independent of its orientation, suggesting that it also harbors the regulatory signals necessary for pcgL expression.
Substrate specificity of the PcgL protein.
To ascertain the physiological role of PcgL, we purified the PcgL protein and determined its substrate specificity. The PcgL protein was overproduced in an E. coli K-12 strain harboring the multicopy number plasmid pEG9125 and purified to near homogeneity (E. coli K-12 lacks the pcgL gene and does not express d-Ala-d-Ala dipeptidase activity under the tested growth conditions; see below). Like VanX, the PcgL protein hydrolyzed d-Ala-d-Ala and d-Ala-Gly but not the N-blocked d-Ala-d-Ala species N-acetyl-d-Ala-d-Ala and the tripeptide d-Ala-d-Ala-d-Ala. The dipeptides l-Ala-l-Ala and l-Leu-Pro and the tripeptide dl-Ala-dl-Lys-Gly were not substrates of PcgL either. On the other hand, the PcgL protein hydrolyzed dl-Ala-dl-Phe but not dl-Ala-dl-Val, and this distinguishes it from VanX, which has activity towards both d-Ala-d-Phe and dl-Ala-dl-Val (26, 42). Thus, despite the low-level sequence identity between PcgL and VanX, these proteins have very similar substrate specificities. These results indicate that the pcgL gene encodes a d,d-dipeptidase that uses d-Ala-d-Ala as its preferred substrate.
Subcellular location of the PcgL protein.
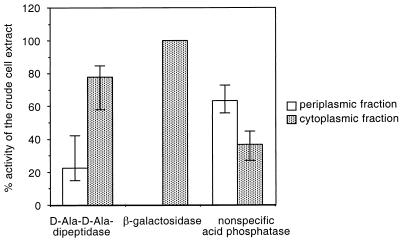
The PSORT protein localization program (30) predicted PcgL to be a periplasmic protein and to have a 20-amino-acid cleavable N-terminal signal sequence. Consistent with the notion that PcgL is a periplasmic protein, the N-terminal sequence of the first 10 residues of the purified PcgL protein was determined by Edman degradation and found to be A-E-N-H-I-D-L-H-Q-P, a perfect match to residues 21 through 30 of the deduced amino acid sequence of the pcgL gene. To further examine the subcellular location of the PcgL protein, we analyzed periplasmic and cytoplasmic fractions from wild-type Salmonella for d-Ala-d-Ala dipeptidase activity (membrane fractions were not tested because the PcgL protein does not have long stretches of hydrophobic residues that could constitute transmembrane domains). We also determined the β-galactosidase and nonspecific acid phosphatase activities of these fractions as prototypical cytoplasmic and periplasmic enzymes, respectively. (Because Salmonella does not harbor the lac operon, these experiments were performed with strain EG10277, which carries the E. coli lac operon.) Despite the predicted periplasmic location of PcgL, only 22.6% of the d-Ala-d-Ala dipeptidase activity localized to the periplasm (Fig. 4). The low recovery of d-Ala-d-Ala dipeptidase activity in the periplasmic space is not unusual for osmotically released enzymes, and similar findings have been reported for the periplasmic proteases DegP and Prc (37, 40). The periplasmic location of PcgL distinguishes it from the cytoplasmic VanX proteins of enterococci and E. coli K-12 and suggests that the Salmonella enzyme acts on d-Ala-d-Ala originating from a peptidoglycan-derived fragment and/or peptidoglycan precursors released into the periplasmic space rather than interacting directly with the cytoplasmic pool of d-Ala-d-Ala.
FIG. 4.
Localization of the d-Ala-d-Ala peptidase activity to different subcellular compartments. The β-galactosidase and nonspecific phosphatase activities were determined as prototypical cytoplasmic and periplasmic enzymes, respectively. Data correspond to mean values of three independent assays.
The pcgL gene is specific to Salmonella.
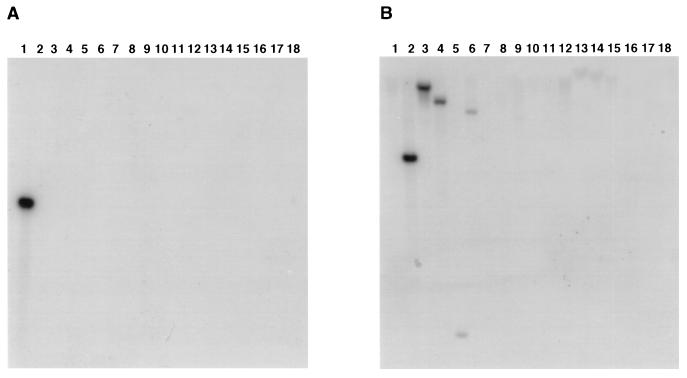
We mapped the pcgL locus to the 37- to 42-min region in the S. enterica serovar Typhimurium chromosome by hybridizing a pcgL-specific probe to an ordered library of the Salmonella genome. We analyzed the DNA sequences flanking the pcgL gene and determined that pcgL is part of a 3.8-kb region with a G+C content of only 41%, which is much lower than the overall G+C content of the Salmonella chromosome (52%). Because unusual G+C contents are often indicative of horizontally acquired sequences, we examined the distribution of the pcgL gene among related bacterial species. We used the pcgL gene as a probe in Southern hybridization experiments carried out under stringent conditions and detected pcgL-hybridizing sequences in S. enterica but not in 14 other microbial species examined (Fig. 5A). Taken together, these data indicate that the pcgL gene is specific to Salmonella and that it was likely incorporated into the Salmonella chromosome by horizontal gene transfer. Acquisition of the pcgL-containing region appears to have occurred early in the evolution of Salmonella because pcgL-hybridizing sequences were detected in all eight subspecies that comprise S. enterica (data not shown).
FIG. 5.
Southern hybridization experiments using pcgL (A) and ecovanX (B) probes were carried out as described in Materials and Methods with DNA from Salmonella enterica serovar Typhimurium (lane 1), E. coli K-12 (lane 2), Shigella flexneri (lane 3), Citrobacter freundii (lane 4), Enterobacter aerogenes (lane 5), Enterobacter cloacae (lane 6), Klebsiella pneumoniae (lane 7), Serratia marcescens (lane 8), Serratia oediferus (lane 9), Proteus mirabilis (lane 10), Proteus vulgaris (lane 11), Erwinia herbicola (lane 12), Yersinia enterocolitica (lane 13), Yersinia pseudotuberculosis (lane 14), Yersinia pestis (lane 15), Pseudomonas aeruginosa (lane 16), Mycobacterium avium (lane 17), and Saccharomyces cerevisiae (lane 18).
Molecular and functional characterization of the pcgL region.
At a position 523 bp upstream from the pcgL gene, we identified a 132-codon open reading frame, encoding a product designated UgtL that exhibits sequence similarity to a chitin synthetase from Schizosaccharomyces pombe (26% identity and 53% similarity over 65 residues to the 859-amino-acid chitin synthetase) (Fig. 2B). Because chitin is the yeast equivalent of bacterial peptidoglycan and d-Ala-d-Ala is produced only for its incorporation into the peptidoglycan, the UgtL protein could function together with the PcgL protein in some aspect of peptidoglycan metabolism. Yet, the region of sequence similarity between UgtL and the chitin synthetase is limited to the predicted transmembrane domains of these putative integral membrane proteins.
To examine the function of the ugtL gene, we constructed a ugtL mutant by introducing a promoterless lac operon and a kanamycin resistance cassette into the chromosomal copy of the ugtL gene (see Materials and Methods). The ugtL mutant was viable and produced wild-type levels of d-Ala-d-Ala dipeptidase activity, consistent with the notion that the ugtL and pcgL genes are not part of the same transcriptional unit and that UgtL is not necessary for d-Ala-d-Ala hydrolysis. Nevertheless, the PcgL and UgtL proteins may participate in the same cellular pathway because transcription of the ugtL gene is regulated by the Mg2+ concentration in the medium and is dependent on a functional PhoP-PhoQ two-component system (Fig. 6). That the ugtL mRNA is likely to be translated is suggested by the presence of an excellent Shine-Dalgarno sequence (AGGA) 9 bp upstream from the ugtL open reading frame.
FIG. 6.
β-Galactosidase activity (in Miller units [29]) from a ugtL-lac transcriptional fusion of Salmonella strains grown in LB or N-terminal medium with 25 μM or 25 mM MgCl2. These results demonstrate that expression of ugtL is regulated by the PhoP-PhoQ system. Data correspond to a single experiment of two independent assays that gave similar results.
The pcgL gene is necessary for growth on d-Ala-d-Ala but dispensable for virulence in mice.
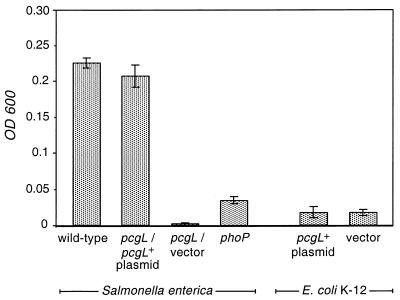
The PcgL-mediated hydrolysis of d-Ala-d-Ala produces d-Ala, an amino acid that can serve as a sole carbon source in E. coli and, presumably, in other enteric species. This raised the possibility of the periplasmic PcgL protein allowing Salmonella to use d-Ala-d-Ala as a sole carbon source, and consistent with this notion, wild-type Salmonella grew on N-minimal liquid medium containing d-Ala-d-Ala (7.5 mM) as a sole carbon source whereas the pcgL mutant did not (Fig. 7). This growth defect is specifically due to the absence of a functional pcgL gene because the pcgL mutant harboring the pcgL+ plasmid grew on d-Ala-d-Ala like wild-type Salmonella did (Fig. 7). The phoP mutant was also defective for growth on d-Ala-d-Ala but not to the same extent as the pcgL strain, and this may reflect the residual level of d-Ala-d-Ala dipeptidase activity exhibited by the phoP mutant. On the other hand, the ugtL mutant grew on d-Ala-d-Ala like wild-type Salmonella did (data not shown), consistent with the notion that the ugtL gene product is not required for the hydrolysis and/or transport of d-Ala-d-Ala.
FIG. 7.
Growth of wild-type and phoP and pcgL mutant Salmonella strains harboring either the pcgL-containing plasmid pEG9122 or the vector pBR322 and of E. coli K-12 harboring the pcgL-containing plasmid pEG9122 or the vector pBR322 in N-minimal medium with d-Ala-d-Ala as a sole carbon source. The final optical density at 600 nm (OD 600) of the bacterial cultures was determined after 24 h of incubation. Similar results were obtained after 48 h of incubation except that the phoP mutant Salmonella strain grew better than the E. coli K-12 strains. Data correspond to mean values of two independent experiments.
As described above, the pcgL gene is specific to Salmonella and requires the PhoP-PhoQ virulence regulatory system for expression (39). This raised the possibility that PcgL may be necessary for Salmonella pathogenesis and/or may be responsible for phenotypes associated with mutations in the phoP locus. However, the pcgL mutant was as virulent as wild-type Salmonella when inoculated into BALB/c mice by either the oral or intraperitoneal route. Likewise, the pcgL mutant displayed wild-type levels of invasion into the epithelial cell line MDCK, survival within the macrophage-like cell line J774, and growth in low-Mg2+ defined medium.
The pcgL gene confers d-Ala-d-Ala dipeptidase activity upon E. coli K-12.
The E. coli K-12 genome (GenBank accession no. D90789) harbors an open reading frame coding for a 193-amino-acid protein that exhibits sequence similarity to the enterococcal VanX protein (40% identity over 141 residues). However, several lines of evidence argue against this open reading frame, vanXE. coli (designated ecovanX by Lessard et al. [26]), encoding the functional homolog of the Salmonella PcgL protein: (i) VanXE. coli is only 40% identical to PcgL, whereas homologous proteins in E. coli and Salmonella typically exhibit >85% sequence identity; (ii) unlike PcgL, VanXE. coli does not appear to have a signal sequence and is likely a cytoplasmic protein; (iii) no d-Ala-d-Ala dipeptidase activity was detected in extracts prepared from E. coli K-12 when cells were grown under conditions that promote expression of the Salmonella pcgL gene (Fig. 7); and (iv) sequences hybridizing to the vanXE. coli gene were detected in Shigella flexneri, Citrobacter freundii, Enterobacter aerogenes, and Enterobacter cloacae (Fig. 5B), bacterial species lacking pcgL-hybridizing sequences (Fig. 5A). Cumulatively, these data demonstrate that the vanXE. coli and pcgL genes have different phylogenetic distributions and suggest that these products play different physiological roles.
To gain further insight into the function of the Salmonella pcgL gene, we investigated the behavior of an E. coli K-12 strain harboring the pcgL-containing plasmid pEG9122. As expected, the E. coli strain exhibited d-Ala-d-Ala dipeptidase activity (Fig. 3B). Moreover, this activity was regulated by the PhoP protein since reduced d-Ala-d-Ala dipeptidase levels were present in extracts prepared from an isogenic phoP mutant of E. coli K-12 (data not shown). On the other hand, the E. coli K-12 strain harboring the pcgL+ plasmid could not grow on d-Ala-d-Ala (Fig. 7). Because the PcgL protein was normally exported to the periplasmic space in E. coli K-12 (see above), these data indicate that, in addition to pcgL, other genes are necessary for E. coli K-12 to use d-Ala-d-Ala as a sole carbon source.
DISCUSSION
d-Ala-d-Ala dipeptidase activity was first described by Reynolds and coworkers in vancomycin-resistant enterococci (36). In these organisms, the VanX protein cleaves d-Ala-d-Ala, which decreases the cytoplasmic pool of this dipeptide and allows the incorporation of d-Ala-d-lactate into peptidoglycan precursors. The resulting pentadepsipeptide precursor, UDP-MurNAc-l-Ala-d-Glu-l-Lys-d-Ala-d-Lac, exhibits lower binding to glycopeptide antibiotics than the normal pentapeptide precursor, UDP-MurNAc-l-Ala-d-Glu-l-Lys-d-Ala-d-Ala, and this accounts for vancomycin resistance (35).
The glycopeptide antibiotic-producing organisms Streptomyces toyocaensis NRRL 15009 and Amycolatopsis orientalis C329.2 contain proteins that exhibit 61 to 64% sequence identity to the VanX protein of vancomycin-resistant enterococci (27). However, Streptomyces toyocaensis NRRL 15009 and A. orientalis C329.2 do not appear to have been the source of glycopeptide resistance genes in vancomycin-resistant enterococci because the G+C contents of the resistance gene clusters in Streptomyces toyocaensis NRRL 15009 and A. orientalis C329.2 are 65.3 and 63.6%, respectively, much higher than those of vancomycin-resistant enterococci (27).
Unexpectedly, VanX-like proteins have been recently identified in three gram-negative species (i.e., E. coli K-12 and Synechocystis sp. during genome projects and S. enterica in the present study), and d-Ala-d-Ala dipeptidase activity has been demonstrated for the purified Salmonella PcgL protein (this work) and for maltose-binding protein fusion derivatives of the E. coli K-12 and Synechocystis VanX homologs (26). Because the outer membrane of gram-negative bacteria prevents access of vancomycin to its target, these VanX-like proteins must have roles other than mediation of vancomycin resistance. Then, what is the physiological function(s) of d-Ala-d-Ala dipeptidases in gram-negative bacteria?
A periplasmic d-Ala-d-Ala dipeptidase in S. enterica.
We have purified the PcgL protein of S. enterica serovar Typhimurium, a d-Ala-d-Ala dipeptidase that exhibits a substrate specificity similar to that displayed by the enterococcal VanX proteins, with which it has only 36% sequence identity. The Salmonella protein localizes to the periplasmic space (Fig. 4), and this distinguishes it from the enterococcal VanX proteins, which are cytosolic enzymes. Thus, the PcgL protein is likely to act on d-Ala-d-Ala released from pentapeptide precursors and/or peptidoglycan-derived fragments rather than directly control the cytoplasmic pool of d-Ala-d-Ala.
The pcgL gene appears to have been incorporated into the Salmonella genome by horizontal gene transfer because pcgL-hybridizing sequences were not detected in related bacterial species (Fig. 5A) and the G+C content of the pcgL-containing region is only 41%, very different from that of the rest of the Salmonella chromosome (52%). This suggests that the pcgL locus endows Salmonella with unique abilities not present in related enteric species. The identification of an open reading frame closely linked to pcgL whose product exhibits similarity to a chitin synthetase from Schizosaccharomyces pombe and is also regulated by the PhoP-PhoQ system suggests that the pcgL region participates in some aspect of peptidoglycan metabolism. While no significant differences in peptidoglycan structure were detected between wild-type and pcgL strains grown in laboratory media (our unpublished results), these findings are consistent with the notion that the PcgL protein acts on a substrate (i.e., d-Ala-d-Ala) liberated by an unidentified endopeptidase cleaving between diaminopimelic acid and d-Ala.
While there are several potential functions for the Salmonella PcgL protein, we are now entertaining three possibilities: nutrient acquisition, virulence, and resistance to a toxic compound. In contrast to wild-type Salmonella, the pcgL mutant was unable to use d-Ala-d-Ala as a sole carbon source, suggesting that the PcgL protein may play a role in nutrient acquisition. d-Ala-d-Ala could originate from peptidoglycan-derived fragments of dying microorganisms or, as suggested by Lessard and colleagues for E. coli K-12 (26), as a result of peptidoglycan turnover in the same microorganism. However, it is presently unknown whether, in its natural environment, Salmonella encounters levels of d-Ala-d-Ala which are high enough to sustain bacterial growth.
While the pcgL gene is Salmonella specific (Fig. 5A) and transcriptionally controlled by the PhoP-PhoQ virulence regulatory system (39), a pcgL mutant retained its ability to cause a lethal infection in BALB/c mice. However, our experiments do not rule out the possibility that the PcgL protein may be required for virulence in other animal species known to be natural hosts for Salmonella or for other aspects of the pathogen-host interaction such as chronic infection. Finally, the periplasmic location of the PcgL protein suggests that it may play a defensive role, mediating the detoxification of a noxious compound that Salmonella may encounter in soil or water.
A cytoplasmic d-Ala-d-Ala dipeptidase in E. coli K-12.
E. coli K-12 harbors a protein, VanXE. coli that is 40% identical to the enterococcal VanX and exhibits d-Ala-d-Ala dipeptidase activity (26). Yet, the VanXE. coli and PcgL proteins are not homologs because they exhibit low-level sequence identity, localize to different subcellular compartments, and are encoded by genes with different phylogenetic distributions (Fig. 5). Moreover, transcription of pcgL is governed by the PhoP-PhoQ regulatory system (39) and does not require RpoS (our unpublished results), the alternative sigma factor controlling transcription of several genes expressed during stationary phase. This is in contrast to vanXE. coli, which is transcriptionally regulated by RpoS (26). Finally, whereas wild-type Salmonella could use d-Ala-d-Ala as a sole carbon source, wild-type E. coli K-12 did not (Fig. 7) unless the vanXE. coli gene was artificially transcribed from the lac promoter or in derivatives expressing the enterococcal vanX gene (26).
The vanXE. coli gene is immediately adjacent to an operon encoding proteins exhibiting similarity to dipeptide permeases (26). Interestingly, transcription of both vanXE. coli and the putative permease genes is under RpoS control. And growth of E. coli on d-Ala-d-Ala was enhanced when the putative dipeptide permease was coexpressed with vanXE. coli. This led Lessard and coworkers to hypothesize that d-Ala-d-Ala generated as a result of peptidoglycan turnover may be transported by this putative peptide transporter into the cytosol, where it would be cleaved by the VanXE. coli protein into d-Ala, providing a source of carbon and energy during stationary phase (26). However, two findings argue against this hypothesis: first, as stated above, wild-type E. coli K-12 cannot grow on d-Ala-d-Ala (26) (Fig. 7). And second, very small amounts of d-Ala-d-Ala are generated from peptidoglycan turnover to sustain bacterial growth.
Conclusions.
The PcgL and VanXE. coli proteins are likely to play different physiological roles in their respective organisms. Yet, these proteins exhibit certain features in common. (i) These enzymes are d-Ala-d-Ala dipeptidases and not carboxypeptidases. Thus, they presumably act on d-Ala-d-Ala generated by equivalent periplasmic endopeptidases cleaving between diaminopimelic acid and d-Ala on peptidoglycan that was not cross-linked and/or on peptidoglycan precursors. And (ii), pcgL and vanXE. coli exhibit a limited phylogenetic distribution (Fig. 5), raising the possibility that these genes may have been incorporated into the Salmonella and E. coli K-12 genomes as a result of horizontal gene transfer. In the case of the pcgL gene, this view is supported by the usually low G+C content of the pcgL-containing region. On the other hand, the G+C content and codon usage of vanXE. coli are similar to those of ancestral E. coli K-12 genes, suggesting that if vanXE. coli was acquired by lateral gene transfer, it must have originated from an organism with a G+C content and a codon usage similar to those of E. coli K-12. Further experiments will be required to determine the specific role that these enzymes play in Salmonella and E. coli K-12.
ACKNOWLEDGMENTS
We thank Felix Solomon and John Burd for technical assistance in this project.
This work was supported, in part, by grant GM54900 from the National Institutes of Health and grant CRG971613 from the North Atlantic Treaty Organization. F.H. was supported by a Erwin-Schroedinger fellowship from the Austrian Science Fund. E.A.G. is an Associate Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alpuche-Aranda C M, Swanson J A, Loomis W P, Miller S I. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci USA. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Depardieu F, Snaith H, Reynolds P, Courvalin P. Contribution of VanY d,d-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38:1899–1903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanc-Potard A-B, Solomon F, Kayser J, Groisman E A. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol. 1999;181:998–1004. doi: 10.1128/jb.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 6.Bugg T D H, Wright G D, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 7.Bussiere D E, Pratt S D, Katz L, Severin J M, Holzman T, Park C H. The structure of VanX reveals a novel amino-dipeptidase involved in mediating transposon-based vancomycin resistance. Mol Cell. 1998;2:75–84. doi: 10.1016/s1097-2765(00)80115-x. [DOI] [PubMed] [Google Scholar]
- 8.Davis R W, Bolstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 9.Fields P I, Groisman E A, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 10.García Véscovi E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 11.Garcia del Portillo F, Foster J W, Maguire M E, Finlay B B. Characterization of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol Microbiol. 1992;6:3289–3297. doi: 10.1111/j.1365-2958.1992.tb02197.x. [DOI] [PubMed] [Google Scholar]
- 12.Garcia del Portillo F, Stein M A, Finlay B. Release of lipopolysaccharide from intracellular compartments containing Salmonella typhimurium to vesicles of the host epithelial cell. Infect Immun. 1997;65:24–34. doi: 10.1128/iai.65.1.24-34.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant S G N, Jesee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groisman E A, Casadaban M J. Mini-Mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusion. J Bacteriol. 1986;168:357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groisman E A, Casadaban M J. Cloning of genes from members of the family Enterobacteriaceae with mini-Mu bacteriophage containing plasmid replicons. J Bacteriol. 1987;169:687–693. doi: 10.1128/jb.169.2.687-693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman E A, Heffron F. Regulation of Salmonella virulence by two-component regulatory systems. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 319–332. [Google Scholar]
- 17.Groisman E A, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12:3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groisman E A, Parra C A, Salcedo M, Lipps C J, Heffron F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11939–11943. doi: 10.1073/pnas.89.24.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groisman E A, Sturmoski M A, Solomon F, Lin R, Ochman H. Molecular, functional, and evolutionary analysis of sequences specific to Salmonella. Proc Natl Acad Sci USA. 1993;90:1033–1037. doi: 10.1073/pnas.90.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunn J S, Belden W J, Miller S I. Identification of phoP-phoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb Pathog. 1998;25:77–90. doi: 10.1006/mpat.1998.0217. [DOI] [PubMed] [Google Scholar]
- 21.Guo L, Lim K B, Gunn J S, Bainbridge B, Darveau R P, Hackett M, Miller S I. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 22.Guo L, Lim K B, Poduje C M, Daniel M, Gunn J S, Hackett M, Miller S I. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 23.Jones B D, Falkow S. Salmonellosis: host immune responses and bacterial virulence determinants. Annu Rev Immunol. 1996;14:533–561. doi: 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- 24.Kier L D, Weppelman R M, Ames B N. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J Bacteriol. 1979;138:155–161. doi: 10.1128/jb.138.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessard I A D, Pratt S D, McCafferty D G, Bussiere D E, Hutchins C, Wanner B L, Katz L, Walsh C T. Homologs of the vancomycin resistance d-Ala-d-Ala dipeptidase VanX in Streptomyces toyocaensis, Escherichia coli and Synechocystis: attributes of catalytic efficiency, stereoselectivity and regulation with implications for function. Chem Biol. 1998;5:489–504. doi: 10.1016/s1074-5521(98)90005-9. [DOI] [PubMed] [Google Scholar]
- 27.Marshall C G, Lessard I A D, Park I-S, Wright G D. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother. 1998;42:2215–2220. doi: 10.1128/aac.42.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCafferty D G, Lessard I A D, Walsh C T. Mutational analysis of potential zinc-binding residues in the active site of the enterococcal d-Ala-d-Ala dipeptidase VanX. Biochemistry. 1997;36:10498–10505. doi: 10.1021/bi970543u. [DOI] [PubMed] [Google Scholar]
- 29.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 30.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 31.Neu H C, Heppel L A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 32.Nieto M, Perkins H R. Modification of the acyl-d-alanyl-d-alanine terminus affecting complex-formation with vancomycin. Biochem J. 1971;123:789–803. doi: 10.1042/bj1230789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintela J C, de Pedro M A, Zöllner P, Allmaier G, Garcia del Portillo F. Peptidoglycan structure of Salmonella typhimurium growing within cultured mammalian cells. Mol Microbiol. 1997;23:693–704. doi: 10.1046/j.1365-2958.1997.2561621.x. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds P E. Structure, biochemistry, and mechanism of action of glycopeptide antibiotics. Eur J Microbiol Infect Dis. 1989;8:943–950. doi: 10.1007/BF01967563. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds P E. Control of peptidoglycan synthesis in vancomycin-resistant enterococci: d,d-peptidases and d,d-carboxypeptidases. Cell Mol Life Sci. 1998;54:325–331. doi: 10.1007/s000180050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds P E, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol Microbiol. 1994;13:1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 37.Silber K R, Keiler K C, Sauer R T. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci USA. 1992;89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snavely M D, Gravina S A, Cheung T-B T, Miller C G, Maguire M E. Magnesium transport in Salmonella typhimurium: regulation of mgtA and mgtB expression. J Biol Chem. 1991;266:824–829. [PubMed] [Google Scholar]
- 39.Soncini F C, García Véscovi E, Solomon F, Groisman E A. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swamy K H S, Goldberg A L. Subcellular distribution of various proteases in Escherichia coli. J Bacteriol. 1982;149:1027–1033. doi: 10.1128/jb.149.3.1027-1033.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2010. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 42.Wu Z, Wright G D, Walsh C T. Overexpression, purification, and characterization of VanX, a d-,d-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1995;34:2455–2463. doi: 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]