Abstract
Large numbers of unique recombinant forms (URF) of human immunodeficiency virus (HIV-1) have been found among sexual transmission populations in China. Here, we report a novel second-generation URF of HIV-1 named BD201AQ that was isolated from an HIV-1-positive man who was infected through homosexual transmission in Baoding City, Hebei Province, China. Phylogenetic analysis based on the near-full-length genome (NFLG) sequence indicated that BD201AQ formed a monophyletic branch that did not cluster with other HIV-1 subtypes. Recombination analysis showed that the NFLG of BD201AQ had 12 segments, six CRF07_BC and six CRF01_AE segments, with CRF07_BC as the main framework. These findings indicate that the constant emergence of novel recombinant forms should receive more attention and that more measures should be taken to monitor the molecular epidemiological characteristics of HIV-1 and to prevent the spread of HIV-1 infections.
Human immunodeficiency virus (HIV) is one of the deadliest pathogens worldwide. HIV, which causes acquired immunodeficiency syndrome (AIDS), has been a major global public health threat since the first case was reported in 1981 [1]. The extremely high genetic variability and high-speed replication capacity of HIV have led to a high degree of genetic diversity. Recombinants readily arise when different HIV strains co-circulate in the same area or within a population, and at least 125 circulating recombinant forms (CRFs) and many unique recombinant forms (URFs) have been recorded in the Los Alamos HIV Sequence Database (www.hiv.lanl.gov). The continuous emergence of new recombinant forms has brought new challenges to the monitoring, treatment, and prevention of HIV infections.
The city of Baoding, which is located in central Hebei Province in northern China, is the most populous city in Hebei Province. Baoding has borders with Beijing and Tianjin, and has experienced the second-worst HIV-1 epidemic in Hebei Province. Lu et al. [2] reported that, in 2019, 77.5% of HIV-1-infected individuals in Hebei Province were men who have sex with men (MSM); 49.6% of them were infected with the CRF01_AE strain, and 29.7% were infected with the CRF07_BC strain. In 2020, the Baoding CDC reported data showing that 98.3% of HIV-infected individuals were infected through sexual transmission and that homosexual transmission accounted for 91.7% of the cases [3]. These data indicated that MSM populations are at high risk of HIV infection in Baoding. In a previous study [4], we showed that CRF01_AE (53.0%) and CRF07_BC (26.5%) were the predominant strains among MSM in Baoding. We predicted that co-circulation and dual infections with CRF01_AE and CRF07_BC would contribute to the emergence of second-generation recombinant strains.
In this study, we detected and characterized a second-generation HIV-1 recombinant strain (BD201AQ) derived from the CRF01_AE and CRF07_BC subtypes, isolated from an MSM infected with HIV-1. The BD201AQ-infected man was a Chinese citizen of Han ethnicity who resided in Baoding. He was 28 years old and unmarried, with primary school education, and was diagnosed by the Baoding CDC in November 2020. He was infected with HIV-1 through homosexual transmission. His baseline CD4+T cell count was 333 cells/µl. This study was approved by the Medical Ethics Committee of the Baoding People’s Hospital. Written informed consent was obtained from the subject prior to sample collection.
The near-full-length genome (NFLG) sequence of BD201AQ was amplified because of the discordant subtypes of the gag and pol genes. RNA was extracted using a QIAamp Viral RNA Mini Accessory Set (QIAGEN, Duesseldorf, Germany), and PrimeScript IV 1st Strand cDNA Synthesis Mix (TaKaRa Biotechnology, Dalian, China) was used to reverse transcribe the RNA into 3′ and 5′ half-molecule cDNAs using the primers 1.R3.B3R and 07Rev8. The primer sequences and their locations corresponding to the HIV-1 reference strain HXB2 genome (GenBank no.: K03455) are listed in Table 1. Nested polymerase chain reactions (PCR) was performed using TaKaRa Premix Taq (TaKaRa Biotechnology) to amplify two regions of the NFLG sequence of BD201AQ. The primers used for PCR are listed in Table 1. The same amplification conditions were used in two rounds of amplification of the two regions as follows: 5 min at 94°C, followed by 30 cycles of 30 s at 94 C, 30 s at 60°C, and 6 min at 72℃, and extension for 10 min at 72°C. The amplification products were detected by 1% agarose gel electrophoresis, and those with the expected sizes were purified and sequenced by Beijing Bomaide Gene Technology Co., Ltd. The chromatogram data were assembled using ContigExpress software in the Vector NTI suite v11.5.1 (Invitrogen). The NFLG sequence of BD201AQ is 8971 nucleotides (nt) long, spanning from the gag gene to a region of three long terminal repeats (corresponding to nt 672–9613 in the HXB2 genome). The NFLG sequence was aligned with subtype reference sequences and CRFs in China (https://hiv.lanl.gov/components/sequence/HIV/search/search.html) using MAFFT v7.0 (http://mafft.cbrc.jp/alignment/server/index.html) and adjusted manually using BioEdit (v7.2.5.0). The phylogenetic tree and subregion trees were constructed by the neighbor-joining method, using MEGA 6. Recombination breakpoints were identified by SimPlot (v3.5.1.0) and Bootscan analysis.
Table 1.
Sequences and locations of the primers used for reverse transcription and nested PCR
| Name | Primer sequence (5ʹ–3ʹ) | Position in HXB2 | |
|---|---|---|---|
| Reverse transcription | 07Rev8 | CCTARTGGGATGTGTACTTCTGAACTT | 5219-5193 |
| 1.R3.B3R |
ACTACTTGAAGCACTCAAGGCAAGCT TTATTG |
9611-9642 | |
| Nested PCR forward | Upper1A | AGTGGCGCCCGAACAGG | 0634-0650 |
| 1.U5.B1F |
CCTTGAGTGCTTCAAGTAGTGTG TGCCCGTCTGT |
0538-0571 | |
| 07For7 |
CAAATTAYAAAAATTCAAAATTTT CGGGTTTATTACAG |
4875-4912 | |
| VIF1 | GGGTTTATTACAGGGACAGCAGAG | 4900-4923 | |
| Nested PCR reverse | 07Rev8 | CCTARTGGGATGTGTACTTCTGAACTT | 5219-5193 |
| Rev11 | ATCATCACCTGCCATCTGTTTTCCAT | 5066-5041 | |
| 2.R3.B6R |
TGAAGCACTCAAGGCAAGCTTTATT GAGGC |
9607-9636 | |
| Low2c | TGAGGCTTAAGCAGTGGGTTCC | 9591-9612 |
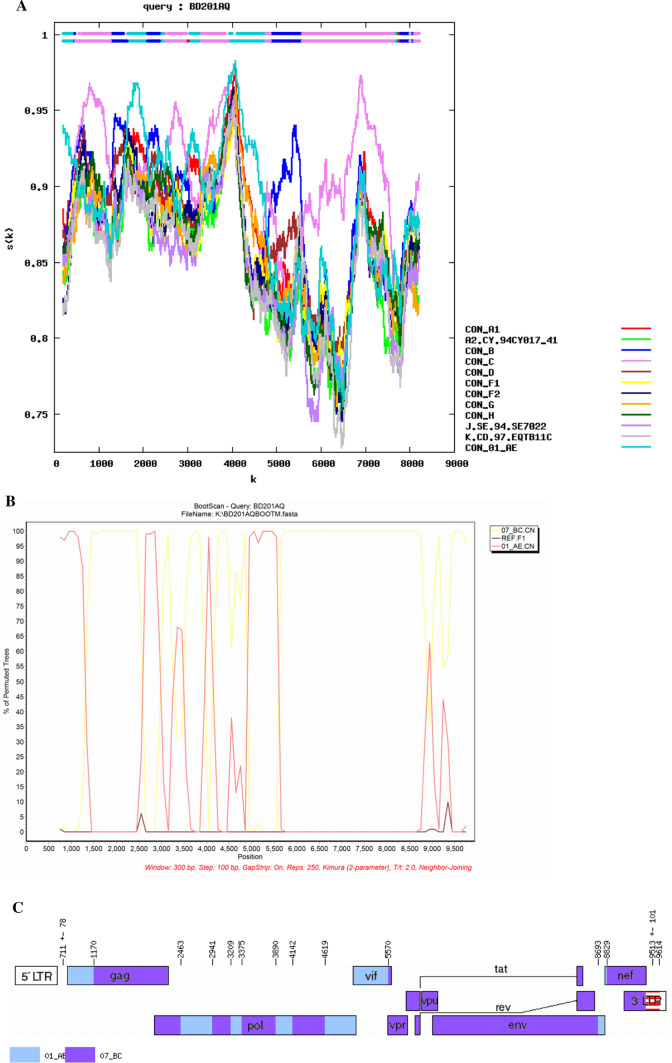
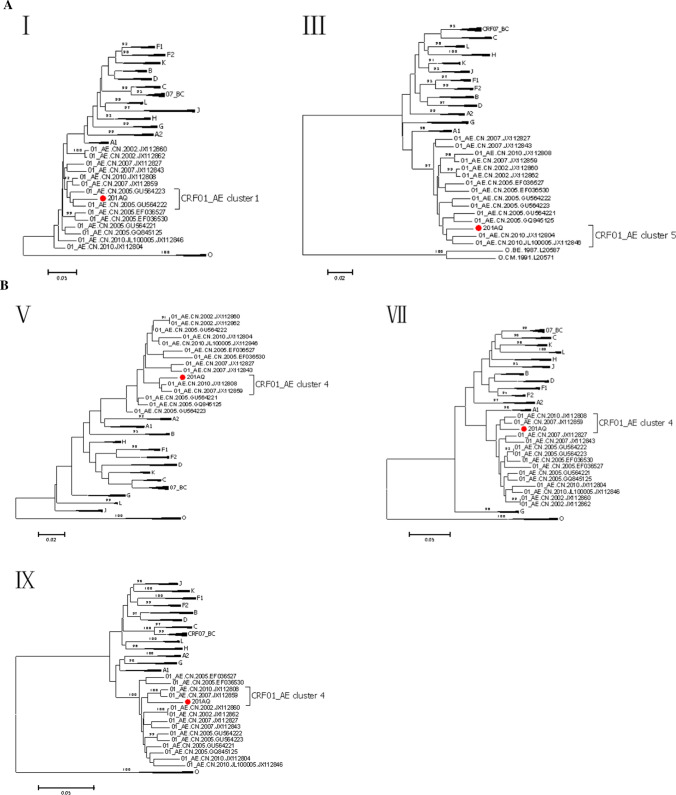
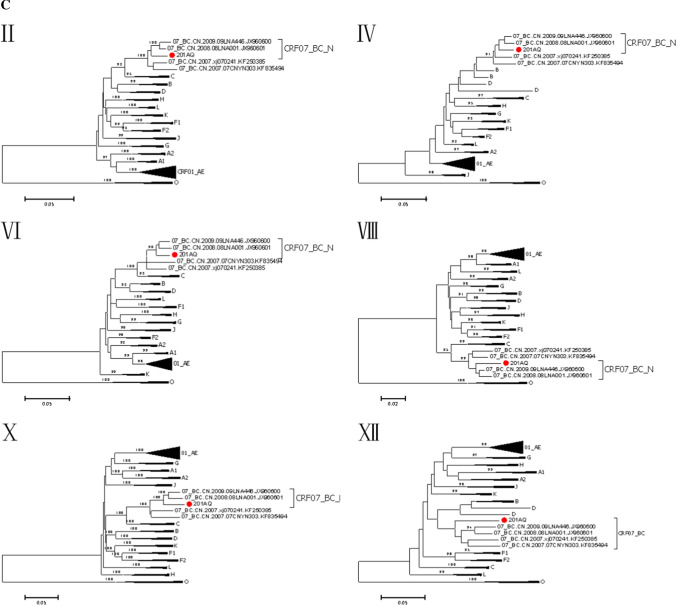
The phylogenetic tree based on the NFLG sequence of BD201AQ (Fig. 1) showed that the BD201AQ sequence was closely related to the CRF01_AE reference sequences but formed a distinct monophyletic branch separately from them. SimPlot analysis indicated that BD201AQ is a recombinant form composed of CRF01_AE and CRF07_BC, with 11 obvious breakpoints (Fig. 2A). Bootscan analysis was carried out to map the exact recombination breakpoints, using CRF01_AE (GenBank nos. KC596062 and KC596061) and CRF07_BC (GenBank nos. BJ070032 and GZ070087) as reference sequences and subtype FI (GenBank nos. AF077336 and AF005494) as an outgroup (Fig. 2B). This analysis indicated that the NFLG sequence of BD201AQ is divided into 12 regions: ICRF01_AE (HXB2, nt 711–1170), IICRF07_BC (HXB2, nt 1171–2463), IIICRF01_AE (HXB2, nt 2464–2941), IVCRF07_BC (HXB2, nt 2942–3209), VCRF01_AE (HXB2, nt 3210–3375), VICRF07_BC (HXB2, nt 3376–3890), VIICRF01_AE (HXB2, nt 3891–4142), VIIICRF07_BC (HXB2, nt 4143–4619), IXCRF01_AE (HXB2, nt 4620–5570), XCRF07_BC (HXB2, nt 5571–8693), XICRF01_AE (HXB2, nt 8694–8829), and XIICRF07_BC (HXB2, nt 8830–9513). A mosaic map of BD201AQ is shown in Fig. 2C. Subregion phylogenetic analysis was used to confirm the genetic origins of each segment. Segments I and III were clustered with CRF01_AE cluster 1, and cluster 5 (Fig. 3A), and segments V, VII, and IX were all clustered with CRF01_AE cluster 4 (Fig. 3B). CRF01_AE cluster 1 strains are prevalent among heterosexuals and injecting drug users (IDUs), and CRF01_AE clusters 4 and 5 are associated with homosexual transmission [5]. Segment XI was too short to construct a neighbor-joining phylogenetic tree. BD201AQ exhibits intra-subtype recombination, which does not rule out the possibility of dual infection. CRF07_BC segments II, IV, VI, VIII, and X were clustered with CRF07_BC_N, which is prevalent among MSM in northern China (Fig. 3C).
Fig. 1.
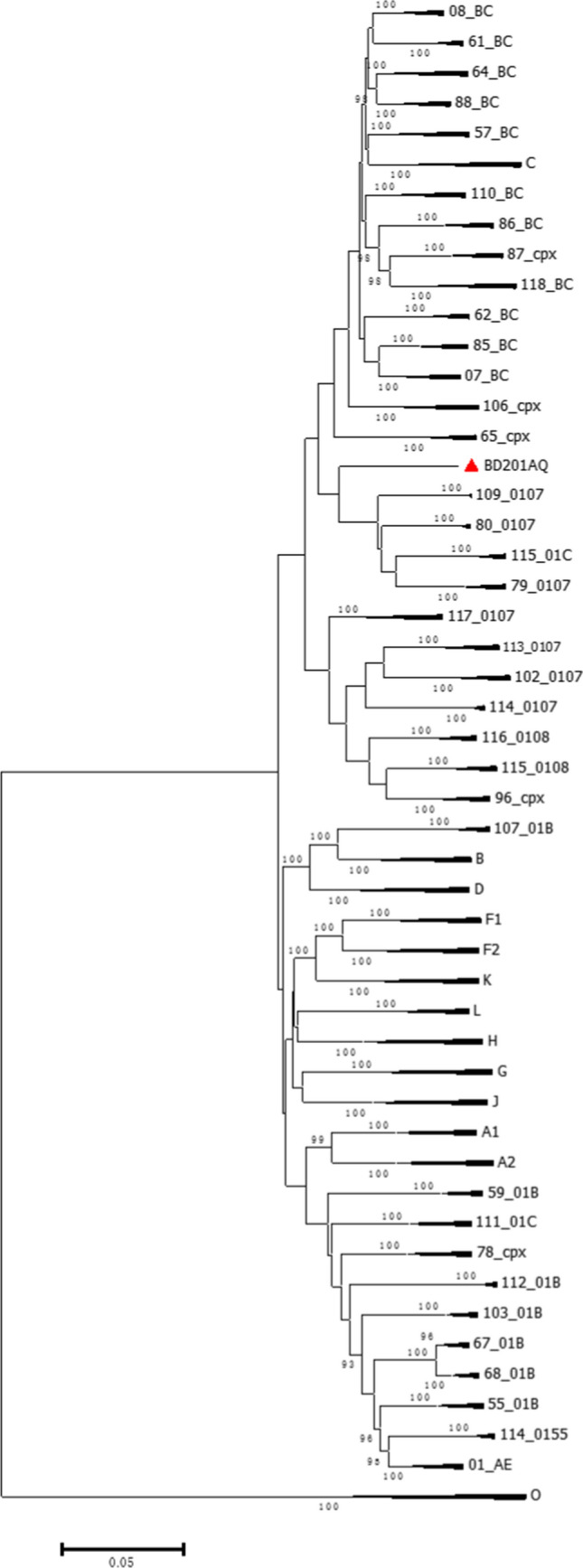
Phylogenetic tree of the near-full-length genome sequence of BD201AQ (8971 bp, ▲) and the sequences of other HIV-1 strains. The tree was constructed by the neighbor-joining method, using MEGA 6. The stability of each node was assessed by bootstrap tests with 1000 replicates. Bootstrap values ≥ 90% are shown at the corresponding nodes. The scale bar indicates 5% genetic distance.
Fig. 2.
Recombination breakpoints in the near-full-length genome sequence of BD201AQ. A Similarity distance analysis of BD201AQ performed using the Recombinant Identification Program v3.0 (http://hiv-web.lanl.gov), which is one of the HIV Database tools, with window size of 300 bp. B Bootscan analysis of BD201AQ conducted with window size of 300 bp and step size of 90 bp along with the reference sequences of CRF01_AE (red line), CRF07_BC (yellow line), and subtype F1 (black line). C Mosaic map of BD201AQ created using the Recombinant HIV-1 Drawing Tool (http://www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html), which is one of the HIV Database tools.
Fig. 3.
Subregion phylogenetic trees of the different segments of BD201AQ (●) and the sequences of other HIV-1 strains. The trees were constructed by the neighbor-joining method with 1000 bootstrap replications, using MEGA 6. Bootstrap values ≥ 90% are shown at the corresponding nodes. The scale bars indicate a genetic distance of 5%. Segment XI of BD201AQ was too short to construct a neighbor-joining phylogenetic tree.
In recent years, countless URFs derived from CRF01_AE and CRF07_BC have been reported in Hebei, Beijing, and Tianjin [6–9]. Among the four CRF01_AE/CRF07_BC recombinants reported in Hebei Province, two used CRF01_AE as the backbone to insert four CRF07_BC fragments, and two used CRF07_BC as the backbone to insert three and four CRF01_AE fragments, respectively. The CRF01_AE fragment is from the CRF01_AE cluster 4 strain in the Chinese MSM population [6, 7]. Jiao et al. [8] reported that the Beijing BJ2015EU16 strain includes two CRF07_BC and one CRF01_AE fragments, with CRF07_BC as the backbone and two recombination breakpoints in the vpu and env genes. Zhou et al. [9] reported that two recombinants in Tianjin consisted of the CRF01_AE backbone and part of the CRF07_BC sequence, with four breakpoints in the gag, pol, vif, and tat genes. It can be inferred that the BD201AQ recombinant identified in this study is different from the recombinants reported previously because not only are there breakpoints in the gag, pol, vif, env, and nef regions, but there are also more breakpoints (i.e., 12).
In conclusion, we reported a novel HIV-1 recombinant strain (BD201AQ) isolated from an MSM individual in the city of Baoding. Phylogenetic analysis of BD201AQ showed that recombinant fragments within the CRF01_AE subtype could be traced back to clusters 1, 4, and 5 of CRF01_AE strains. The CRF07_BC fragments were derived from HIV-1 genotypes circulating among Chinese MSM. With CRF01_AE and CRF07_BC emerging as the dominant subtypes in China, the recombination diversity of the recently identified lineage of new 0107 unique recombinant forms of HIV-1 may be unimaginably high, threatening populations across the country. In China, CRF07_BC_N, CRF01_AE cluster 4 and CRF01_AE cluster 5 are prevalent mainly among MSM populations [10], and the high mobility, condom-free sex, and multiple sexual partners of MSM populations have led to the emergence of HIV-1 URFs in MSM populations [11–15]. HIV-1 recombination is a major way in which the virus evolves rapidly, increasing the fitness of the virus and helping the virus rapidly acquire drug resistance or escape the surveillance of the immune system. Frequent recombination also makes HIV-1 molecular diagnosis, treatment, and vaccine development more difficult. The recombination analysis of BD201AQ showed that the recombination breakpoints are different from those of the 0107 recombinant forms reported in Hebei Province. The emergence of new complex recombinant forms has increased the diversity of HIV-1 isolates circulating in Baoding, suggesting the need for further molecular monitoring of HIV-1 diversity in the region to better respond to the HIV-1 epidemic.
Acknowledgments
The authors thank all staff involved in this study. We thank Margaret Biswas, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
Author contributions
XY: manuscript writing, data analysis. HZ: manuscript correction, sample information collection. WA: manuscript correction, laboratory procedure, experimental operation. WS: experimental operation. JZ: sample information collection. XL: sequence assembly, data analysis. YL: Experiment design, data analysis.
Funding
This study was supported by the Science and Technology Planning Project of Baoding prefecture (grant number 2041ZF150).
Data availability
The nucleotide sequence of the near-full-length genome of BD201AQ has been deposited in the GenBank database under accession number OL401523.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This study was approved by the Medical Ethics Committee of the Baoding People’s Hospital (protocol number 2019–03).
Consent for publication
Written informed consent was signed by the participant before sample collection. Using a blood sample for further related studies was also permitted.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuegang Yang, Huiying Zhu and Weina An have contributed equally to this work.
References
- 1.Gottlieb MS, Schroff R, Schanker HM, et al. Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N Engl J Med. 1981;305(24):1425–1431. doi: 10.1056/NEJM198112103052401. [DOI] [PubMed] [Google Scholar]
- 2.Lu X, Zhang J, Wang Y, et al. Large transmission clusters of HIV-1 main genotypes among HIV-1 individuals before antiretroviral therapy in the Hebei Province, China. AIDS Res Hum Retroviruses. 2020;36(5):427–433. doi: 10.1089/AID.2019.0199. [DOI] [PubMed] [Google Scholar]
- 3.Shi P, Wang X, Fan W, et al. Pre-treatment drug resistance analysis of HIV-1 infected patients in Baoding City, 2019–2020. Chin J Dermatovenereol. 2021;35(09):50–55. doi: 10.13735/j.cjdv.1001-7089.202103052. [DOI] [Google Scholar]
- 4.Shi P, Chen Z, Meng J, et al. Molecular transmission networks and pre-treatment drug resistance among individuals with acute HIV-1 infection in Baoding, China. PLoS ONE. 2021;16(12):e0260670. doi: 10.1371/journal.pone.0260670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An M, Han X, Zhao B, et al. Cross-continental dispersal of major HIV-1 CRF01_AE clusters in China. Front Microbiol. 2020;11:61. doi: 10.3389/fmicb.2020.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han L, Li H, Wang L, et al. Near full-length genomic characterization of two novel HIV-1 unique recombinant forms (CRF01_AE/CRF07_BC) among men who have sex with men in Shijiazhuang City, Hebei Province, China. AIDS Res Hum Retroviruses. 2021;37(12):978–984. doi: 10.1089/AID.2021.0100. [DOI] [PubMed] [Google Scholar]
- 7.Xing Y, Guo Y, Wang L, et al. Identification of two novel HIV-1 second-generation recombinant forms (CRF01_AE/CRF07_BC) in Hebei, China. AIDS Res Hum Retroviruses. 2021;37(12):967–972. doi: 10.1089/AID.2021.0057. [DOI] [PubMed] [Google Scholar]
- 8.Jiao Y, Wang Y, Kong D, et al. Characterization of a new HIV-1 CRF01_AE/CRF07_BC recombinant virus form among men who have sex with men in Beijing, China. AIDS Res Hum Retroviruses. 2018;34(6):550–554. doi: 10.1089/AID.2018.0049. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Z, Ma P, Feng Y, et al. Characterization of a new HIV-1 CRF01_AE/CRF07_BC recombinant virus in Tianjin, China. AIDS Res Hum Retroviruses. 2018;34(8):705–708. doi: 10.1089/AID.2018.0077. [DOI] [PubMed] [Google Scholar]
- 10.Ge Z, Feng Y, Zhang H, et al. HIV-1 CRF07_BC transmission dynamics in China: two decades of national molecular surveillance. Emerg Microbes Infect. 2021;10(1):1919–1930. doi: 10.1080/22221751.2021.1978822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan Y, Zhu W, Hu F, et al. Genetic characterization of a novel HIV-1 CRF01_AE/CRF07_BC recombinant form among men who have sex with men in Guangdong, China. Arch Virol. 2018;163(11):3093–3097. doi: 10.1007/s00705-018-3928-1. [DOI] [PubMed] [Google Scholar]
- 12.Yao T, Wu J, Zhang Y, et al. Near full-length genomic characterization of a novel unique recombinant (CRF01_AE/CRF07_BC) in Fuyang City of China. AIDS Res Hum Retroviruses. 2020;36(6):527–532. doi: 10.1089/AID.2020.0002. [DOI] [PubMed] [Google Scholar]
- 13.Yin Y, Zhou Y, Lu J, et al. First detection of a cluster novel HIV-1 second-generation recombinant (CRF01_AE/CRF07_BC) among men who have sex with men in Nanjing, Eastern China. Intervirology. 2021;64(2):81–87. doi: 10.1159/000512135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang XH, Yu G, Zheng C, et al. Genomic characterization of a new CRF01_AE/CRF07_BC case from a MSM patient in Guangdong, China. J Med Virol. 2021;93(11):6383–6387. doi: 10.1002/jmv.26799. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Chen L, Chaillon A, et al. The dynamics of the HIV epidemic among men who have sex with men (MSM) from 2005 to 2012 in Shenzhen, China. Sci Rep. 2016;29(6):28703. doi: 10.1038/srep28703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequence of the near-full-length genome of BD201AQ has been deposited in the GenBank database under accession number OL401523.