Abstract
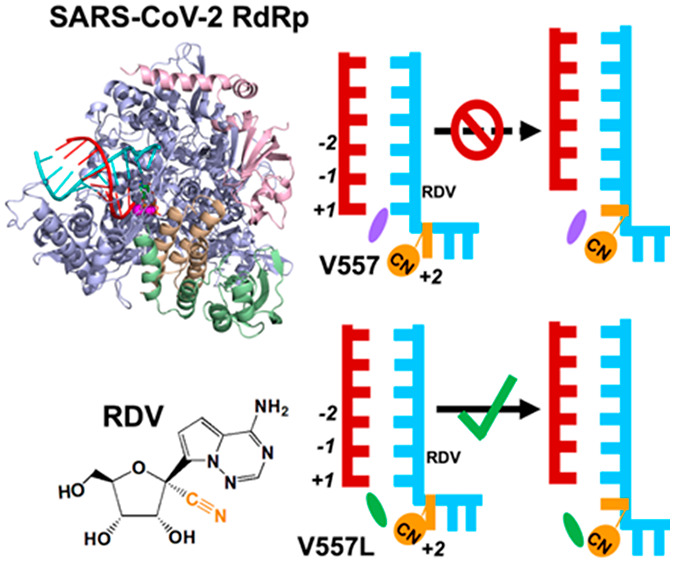
Remdesivir is one nucleotide analogue prodrug capable to terminate RNA synthesis in SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) by two distinct mechanisms. Although the “delayed chain termination” mechanism has been extensively investigated, the “template-dependent” inhibitory mechanism remains elusive. In this study, we have demonstrated that remdesivir embedded in the template strand seldom directly disrupted the complementary NTP incorporation at the active site. Instead, the translocation of remdesivir from the +2 to the +1 site was hindered due to the steric clash with V557. Moreover, we have elucidated the molecular mechanism characterizing the drug resistance upon V557L mutation. Overall, our studies have provided valuable insight into the “template-dependent” inhibitory mechanism exerted by remdesivir on SARS-CoV-2 RdRp and paved venues for an alternative antiviral strategy for the COVID-19 pandemic. As the “template-dependent” inhibition occurs across diverse viral RdRps, our findings may also shed light on a common acting mechanism of inhibitors.
The outbreak of coronavirus disease 2019 (COVID-19) has become a global pandemic, and over 542 million confirmed cases have been reported by June 2022.1 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the novel coronavirus that has caused COVID-19 and can be furtively transmitted among humans.2,3 To curb the health crisis, great efforts have been devoted to exploring the effective treatment for COVID-19.4−13
The core polymerase complex of SARS-CoV-2 has become one promising target for antiviral drug design. It is the minimal scaffold to mediate the RNA synthesis and plays a central role in viral replication and transcription. It is composed of three nonstructural proteins (nsps) (Figure 1A), where nsp12 mainly conducts the RNA synthesis in the active site, while nsp7 and nsp8 serve as cofactors to stimulate RNA-dependent RNA polymerase (RdRp) activity.14−18 Since the outbreak of the pandemic, nucleotide analogues have been widely explored as potential inhibitors to terminate the RNA synthesis in SARS-CoV-2 RdRp.5,7,8,10−12,19
Figure 1.
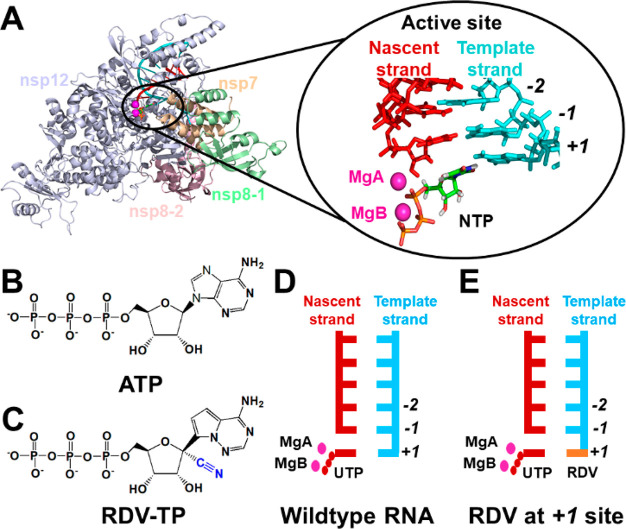
(A) Overall structure of the SARS-CoV-2 RdRp complex is shown on the left, with the catalytically active site amplified on the right. The nascent and template strands are colored in red and cyan, respectively. Nucleoside triphosphate (NTP) is shown in sticks with two Mg2+ ions as magenta spheres. (B) Chemical structures of adenosine triphosphate (ATP). (C) Chemical structure of remdesivir (RDV) in the triphosphate form, with the 1′-cyano group highlighted in blue. (D) Structure diagram of SARS-CoV-2 RdRp with UTP at the active site. (E) Similar to (D) but with RDV (shown in orange) embedded at the +1 site of the template strand.
Remdesivir is one representative prodrug of a nucleotide analogue that has demonstrated therapeutic efficacy in the animal model of SARS-CoV-2 RdRp20 and effectively terminated viral RNA synthesis in biochemical experiments through a “delayed termination mechanism”.17,18,21−26 The chemical structure of remdesivir’s active form (RDV-TP) resembles that of natural adenosine triphosphate (ATP) (Figure 1B,C). Previous studies have demonstrated that RDV-TP can efficiently compete with ATP to incorporate into the nascent strand.17,21,27−31 However, after the incorporation of RDV-TP, three nucleoside triphosphates (NTPs) can still be consecutively added into the nascent strand, and the termination occurs when the translocation of remdesivir from the −3 to −4 site is hampered due to the interactions between the 1′-cyano group of remdesivir and the protein residues.17,21,22,24−26 It is worthy to note that such a termination is obvious when the NTP concentration is ∼0.1 μM and only less than 10% of the full-length RNA product can be observed.22 However, when higher concentrations of NTP pools were used, the RNA synthesis arrest caused by the “delayed chain termination” would be overcome, and more than 90% full-length product can be yielded at an NTP concentration as low as 10 μM.21,22 As the intracellular NTP concentrations are in the range of high μM and low mM,32,33 the efficient read-through likely occurs, and the full-length RNA product containing remdesivir can be formed under biologically relevant conditions.21,22
Intriguingly, a recent biochemical experiment has demonstrated that when a copy of RNA strand embedded with remdesivir was used as the template, it would exert the “template-dependent” inhibition, where the UTP incorporation complementary to remdesivir is inhibited.22 In this scenario, remdesivir shows stronger resistance to the increment of NTP concentration, and a concentration of 50–100 μM is required to overcome this obstacle and recover over 90% NTP incorporation efficiency.22 Such a concentration is obviously higher than that required to bypass the “delayed chain termination”, suggesting that the “template-dependent” inhibition is pronounced under biologically relevant conditions. However, the contribution of two mechanisms to the antiviral activity of remdesivir would be highly dependent on the NTP concentrations. Also, when multiple RDV-TPs are incorporated into the nascent strand, the “delayed chain termination” could be amplified. Even so, investigation of “template-dependent” inhibitory mechanism is still one important aspect for gaining a comprehensive understanding about the action of remdesivir on SARS-CoV-2 RdRp. More interestingly, the biochemical experiment has shown such inhibition can be significantly reduced by the point mutation V557L.22 These observations have altogether suggested that the presence of remdesivir at the template strand offers the second opportunity to inhibit the RNA extension. However, the experimental assay alone is insufficient to distinguish whether the inhibition is exerted by directly disrupting the NTP incorporation at the active site or by hindering the translocation. Moreover, it remains elusive how V557L mutation renders the SARS-CoV-2 RdRp drug-resistant to remdesivir at the template strand.
MD simulations have been widely used to understand the acting mechanism of RdRp inhibitors as well as to explore the potential inhibitors since the outbreak of the COVID-19 pandemic.7,12,27,29 In particular, the “delayed chain termination” mechanism exerted by remdesivir has been investigated by MD simulations,24,26 the results of which have not only well-reproduced the experimental observations that the termination occurs at the upstream site of the nascent strand instead of the active site, but also further elucidated the underlying mechanisms. These previous works have suggested that MD simulations are powerful in elucidating the inhibitory mechanisms of nucleotide analogues. Moreover, besides understanding how remdesivir inhibits its complementary UTP incorporation, our current work would also focus on the mechanism about how the V557L mutation gains drug resistance to remdesivir, the prerequisite of which is to explore the conformations of V557L. However, the available experimental structures of SARS-CoV-2 RdRp all possess the wildtype V557, and a static model constructed by manually mutating V557 to leucine may not appropriately estimate the orientation of V557L. In this regard, MD simulations would help to fully sample the conformational space of V557L and explore the orientations of its side chain under a dynamic protein environment, which lay the foundation for the investigation of molecular mechanisms. Therefore, MD simulations serve as a promising tool for the current investigation of the “template-dependent” inhibition.
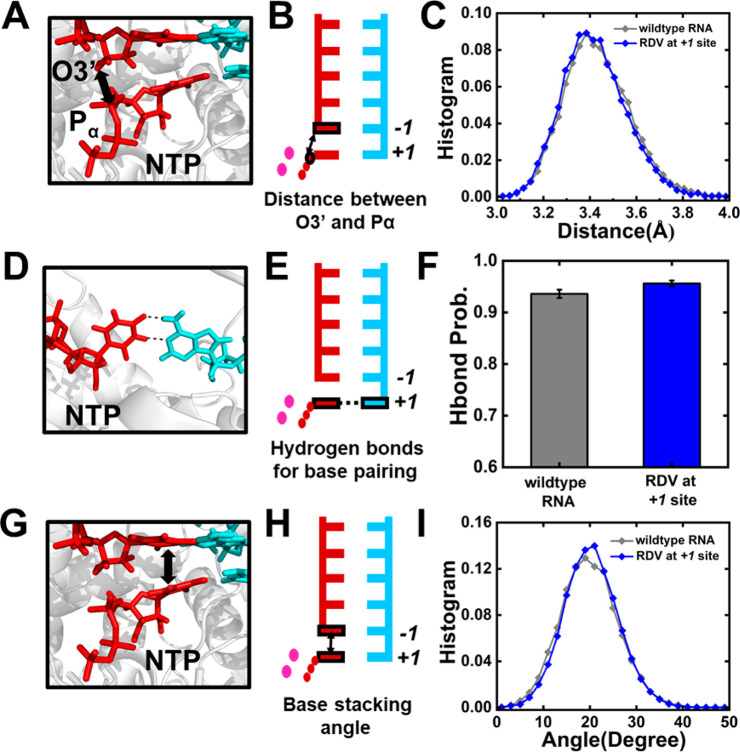
In this study, we have performed MD simulations to elucidate the molecular mechanisms for the “template-dependent” inhibition exerted by remdesivir. We first examined if the NTP incorporation at the active site is directly impaired when remdesivir is at the +1 site (Figure 1D,E). The cryo-EM structure of the SARS-CoV-2 nsp12-nsp7-nsp8 complex (PDB ID: 7BZF(17)) was adopted as our structural basis, and remdesivir was modeled at the +1 site of the template strand with UTP binding at a closed active site (see SI Section 1.2 for details of model construction). A total of five replicas of 50 ns production simulations have been performed to investigate the incorporation capability of UTP opposite to remdesivir (Figure 1E). For comparison, the same amount of MD simulations with UTP opposite to adenosine (“wildtype-RNA” system, Figure 1D) has also been conducted (see SI Section 1.1 for details). A distance between the Pα atom of UTP and the O3′ atom of the 3′-terminal of the nascent strand (Pα–O3′ distance, Figure 2A,B) below 4 Å has been suggested to be necessary for efficient catalysis and formation of a phosphodiester bond.34,35 Consistently, such distance is <4 Å in all the MD conformations with a UTP:A pair at the active site (Figure 2C). Interestingly, when remdesivir is present at the template strand opposite to UTP, all the MD conformations still maintain a Pα–O3′ distance below 4 Å (Figure 2C), which satisfies the prerequisite for the efficient incorporation. The stability of UTP at the active site is also important for ensuring a catalytically active configuration. Accordingly, the base pairing and base stacking stability were examined (Figure 2D–I). Our results have demonstrated that UTP can always form stable base pairing with remdesivir embedded at the +1 site of the template strand (hydrogen bonding probability = 95.6%), resembling that for the UTP:A base pair (hydrogen bonding probability = 93.6%) (Figure 2F). Moreover, UTP can stack well with the 3′-terminal of the nascent strand in both systems, with base-to-base dihedral angles around 19.9° and 20.3° for adenosine and remdesivir, respectively (Figure 2I). To examine the convergence of sampling, we have extended each of the five replicas of 50 ns simulations to 200 ns and found out that the capability of UTP in maintaining the catalytically active conformation at the active site computed using 5 × 100 ns, 5 × 150 ns, or 5 × 200 ns MD simulations is similar to that as observed in 5 × 50 ns simulations (Figure S1). Moreover, we have also computed the root-mean-square deviations (RMSD) of the Cα atoms in the nsp12 along the MD trajectories and found out that the RdRp is stable in our simulations (Figure S2). Altogether, our calculations have suggested that UTP could well maintain a catalytically active configuration opposite to remdesivir at the +1 site of the template strand.
Figure 2.
Investigation into the NTP incorporation capability in SARS-CoV-2 RdRp by 5 × 50 ns simulations. (A,B) 3D structure (A) and cartoon model (B) showing the distance between the Pα atom of the NTP and the O3′ atom of the 3′-terminal nucleotide of the nascent strand. (C) Histogram of the Pα–O3′ distance for two systems including RdRp with wildtype-RNA (gray) and RDV at the +1 site (blue). (D,E) 3D structure (A) and cartoon model (B) showing the hydrogen bond probability for base pairing at the active site. (F) Hydrogen bonding probability for the two systems. (G,H) 3D structure (G) and cartoon model (H) showing the stacking angle between the base of NTP and the base of the 3′-terminal nucleotide. The stacking angle was defined as the angle formed by the normal vectors of the base of NTP and the base of the 3′-terminal nucleotide of the nascent strand. (I) Histogram of the stacking angles in two systems.
We noticed that a previous study based on structural modeling has suggested that the backbone of the protein residue A558 would clash with the 1′-cyano group of remdesivir at the +1 site, thereby departing from remdesivir and destabilizing the base pairing between UTP and remdesivir.22 In our simulations, we have consistently observed that A558 would move away from the 1′-cyano group of remdesivir (Figures S3 and S4). Specifically, the distance between the backbone oxygen atom of A558 and the C1′ atom of remdesivir is 6.78 ± 0.14 Å, obviously larger than that between A558 and adenosine (4.70 ± 0.23 Å). However, such conformational changes seldom perturb the base pairing stability, as the hydrogen bonding probability of UTP:remdesivir (>90%) is similar to that of the UTP:A pair (Figure 2F), the observation of which is different from that in the previous work based on a static structural model.22 We also noted that the previous study used an alternative cryo-EM structure of SARS-CoV-2 RdRp (PDB ID: 6YYT(16)) as the basis to construct the model. To further examine if the discrepancy of the base pairing stability originated from modeling using different experimental structures, we have constructed two extra models by referring to the previous study. MD simulations have been performed to examine the stability of the active site as well as the position of A558 relative to the template-strand nucleotide (Figure S5, see SI Section 4 for details). We have found out that even though different experimental structures were used for the initial modeling, the UTP:remdesivir base pair at the active site is always well-maintained, and A558 would move away from remdesivir, further consolidating the robustness of our results.
The above observations altogether indicate that the “template-dependent” inhibitory mechanism is not exerted by directly impairing the NTP incorporation at the active site. In this regard, the inhibition could be applied by the second scenario, where the translocation is hampered and the NTP incorporation is inhibited. To examine the propensity of translocation when remdesivir moves from the +2 to +1 site along the template strand, we first evaluated the influence of remdesivir on the stability of both the pre- and post-translocation states by performing MD simulations for SARS-CoV-2 RdRp with remdesivir at the +1 site in the post-translocation state and at the +2 site in the pre-translocation states, respectively (see SI Sections 1.2 and 2.2 for details). SARS-CoV-2 RdRp with wildtype double-stranded RNA (dsRNA) in both states has also been examined for comparison (see SI Sections 1.1 and 2.1 for details). The hydrogen bonding probability between the template and nascent strands was computed to estimate the thermal stability of both states. Our results have demonstrated that when remdesivir is present at the template strand, the base pairs at the −1, −2, and −3 sites are well-maintained in both states (hydrogen bonding probabilities >85%), resembling the scenario observed for the RdRp with wildtype dsRNA (Figure S6). This result has suggested that the translocation of remdesivir from the +2 to +1 site is thermodynamically allowed.
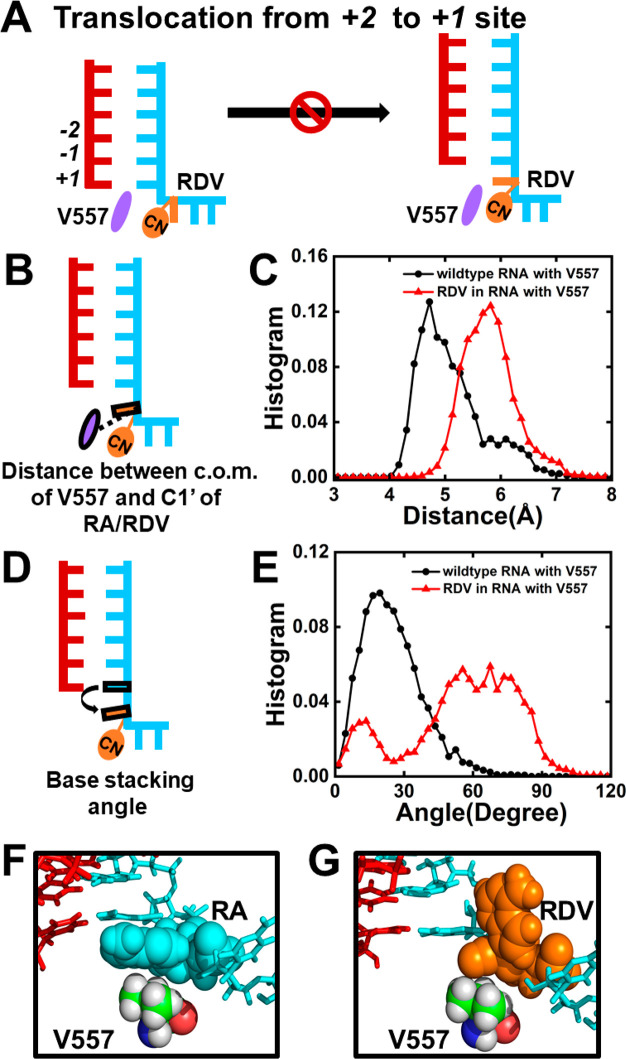
To further examine if the presence of remdesivir would affect the translocation dynamics, a translocation pathway was generated by the Climber algorithm36,37 (Figure S7), and MD simulations were conducted for the translocating intermediates (see SI Section 3 for details). Intriguingly, we have found that the 1′-cyano group of remdesivir would clash with the side chain of V557 when remdesivir moves from the +2 to +1 site (Figure 3A), while such repulsion from V557 has not been observed when adenosine translocates. Specifically, when remdesivir translocates from the +2 to +1 site, its steric clash with V557 would result in a separation distance of 5.87 ± 0.25 Å, larger than that between V557 and adenosine (4.91 ± 0.12 Å) (Figure 3B,C). Moreover, the steric clash would push the base of remdesivir in a tilted configuration with a larger dihedral angle relative to the base of the upstream nucleotide (55.95 ± 5.81°), while the adenosine’s base adopts a more parallel configuration (25.04 ± 1.43°) (Figure 3D–G). Further calculations have validated that the conformation as shown in Figure 3G with an obviously tilted base is a highly populated and representative conformation along the translocation pathway in SARS-CoV-2 RdRp with V557, as K-center clustering based on the conformational similarity has shown that ∼83% of MD conformations fall into the clusters with a mean angle >50° (see SI Section 3.4 for details). Therefore, our simulations have suggested that V557 hinders the translocation of remdesivir from the +2 to +1 site and further inhibits the NTP incorporation against remdesivir at the +1 site. This finding is also consistent with the experimental observation that SARS-CoV-2 RdRp with a single-point V557L mutation would become drug-resistant to remdesivir.22
Figure 3.
Translocation of RDV from the +2 to +1 site is hindered by V557. (A) Diagram showing V557 hampers the translocation of RDV from the +2 to the +1 site, where RDV and V557 are colored in orange and purple, respectively. (B) Cartoon model showing the distance between the center of mass (c.o.m.) of V557 and C1′ of RA/RDV. (C) Histogram of the distance as shown in (B) when RA (black) or RDV (red) translocates. (D) Cartoon model showing the base-stacking angle between RDV/RA and the upstream nucleotide. (E) Histogram of the base-stacking angle for RA (black) or RDV (red) during translocation. (F,G) Typical conformation of RA/RDV with V557 during translocation.
To further understand how the V557L mutation could reduce the inhibitory effect exerted by remdesivir on the transcription of SARS-CoV-2 RdRp,22 we have performed MD simulations with the V577L point mutation for the wildtype-RNA system (see SI Section 1.2 for details). We observed that UTP opposite to adenosine at the +1 site can still maintain the catalytically active conformation upon V557L mutation (Figure S8). Intriguingly, we found that the Val-to-Leu mutation would reorientate its side chain to the direction opposite to the translocation pathway, which is accompanied by the movement of V557L’s backbone (Figure 4A–D). Specifically, the center of mass (c.o.m.) distance between the backbone/side chain of V557L and the base of the upstream nucleotide becomes obviously larger than that for V557 (Figure 4B,D). This variation of the side-chain orientation is observed not only in the wildtype-RNA system (Figure 4B,D), but also when remdesivir is embedded in the template strand (Figure S9). These observations have suggested that remdesivir exerts no extra influence on the side chain orientation of V557L and the bulkier side chain upon Val-to-Leu mutation would spontaneously rotate, resulting in extra space along the translocation pathway.
Figure 4.
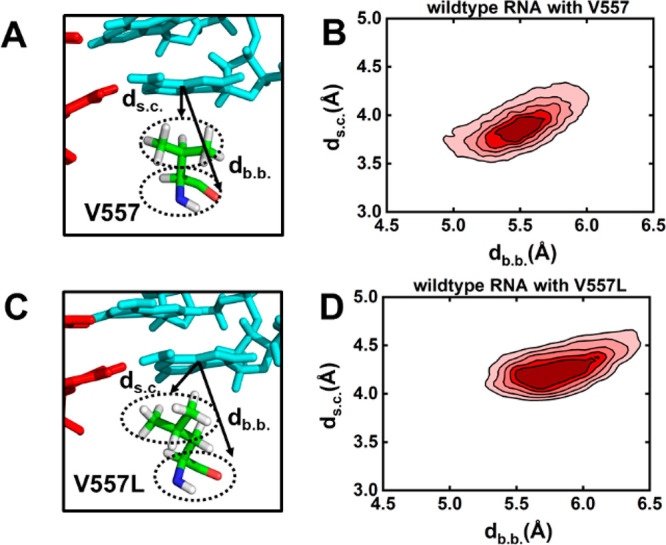
Side chain of V557L is rotated in comparison with that of V557. (A) Representative conformation of V557 with the dashed lines denoting the distance between the base of the nucleotide and the backbone (db.b.)/side-chain (ds.c.) of V557. (B) Density plot of db.b. versus ds.c. for RdRp with wildtype RNA. (C) Similar to (A) but for a V557L mutation. (D) Similar to (B) but for RdRp upon a V557L mutation. In (B) and (D), the simulations of RdRp with UTP binding at the active site were used for the calculations.
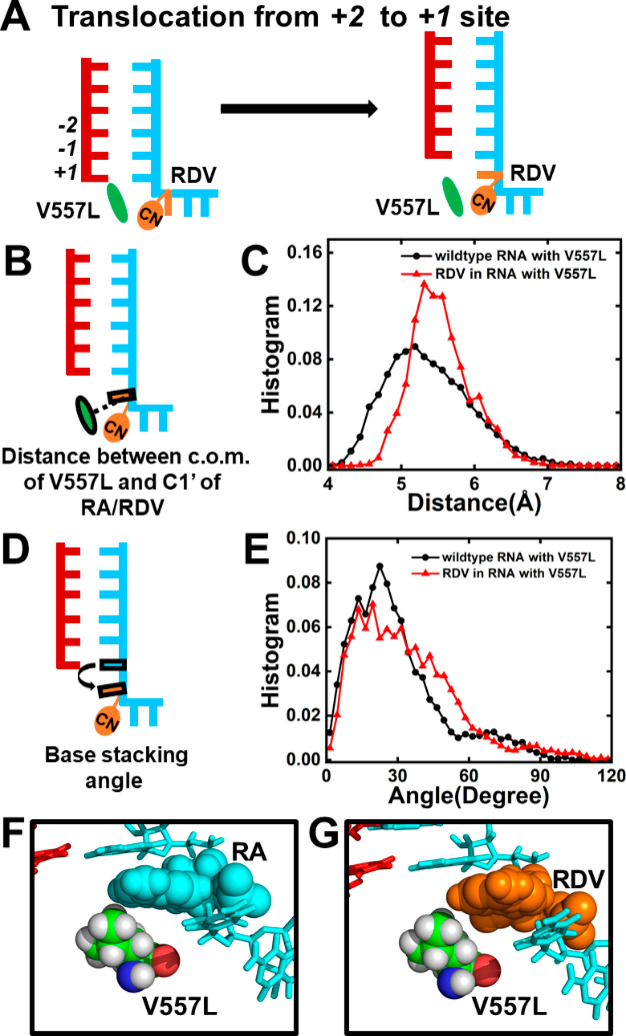
To evaluate the above conjecture, we generated the translocation pathway for SARS-CoV-2 RdRp with the V557L mutation and performed MD simulations with the translocating intermediates (see SI Section 3 for details). We found that the distance between V557L and remdesivir (5.5 ± 0.12 Å) is similar to that between V557L and wildtype adenosine (5.4 ± 0.25 Å) (Figure 5B,C). In addition, remdesivir and adenosine also form similar base-stacking angles relative to the upstream nucleotide, with dihedral angles of 29.72 ± 6.42° and 30.34 ± 5.29°, respectively (Figure 5D–G). Therefore, our results have suggested that the V557L residue would rotate its side chain away from the translocation pathway and thereby relieve the hindrance over the translocation of remdesivir from the +2 to +1 site (Figure 5A). Our calculations have also rationalized the kinetic steady-state experimental data that the UTP incorporation opposite to remdesivir is 5-fold more efficient upon V557L mutation.22 Moreover, our work further consolidates that the steric repulsion from V557 plays the dominant role in hampering the translocation of remdesivir from the +2 to +1 site and thus reduces the incorporation efficiency of UTP complementary to remdesivir at the +1 site.
Figure 5.
V557L mutation relieves the inhibition exerted by RDV. (A) Diagram showing V557L allows the translocation of RDV from the +2 to the +1 site, where RDV and V557L are colored in orange and green, respectively. (B) Cartoon model showing the distance between the c.o.m. of V557L and C1′ of RA/RDV. (C) Histogram of the distance as shown in (B) when RA (black) or RDV (red) translocates. (D) Cartoon model showing the base-stacking angle between RDV/RA and the upstream nucleotide. (E) Histogram of the base-stacking angle for RA (black) or RDV (red) during translocation. (F,G) Typical conformation of RA/RDV with V557L during translocation.
In summary, we have elucidated the molecular mechanism of “template-dependent” inhibition exerted by remdesivir on the complementary NTP incorporation in SARS-CoV-2 RdRp by comprehensive analysis based on extensive MD simulations. Our computational results have suggested that the remdesivir at the template strand does not directly abolish the complementary NTP incorporation at the active site. Instead, translocation of remdesivir from the +2 to +1 site is inhibited, and thus the NTP incorporation becomes inefficient. Specifically, the dynamic translocation of remdesivir from the +2 site to the +1 site is kinetically hindered due to the clash between the 1′-cyano group of remdesivir and the side chain of V557, while the V557L mutation tends to reduce the inhibitory effect by rotating its side chain away from the translocation pathway. This finding not only matches with the experimental observation that SARS-CoV-2 RdRp upon V557L mutation would gain drug resistance to remdesivir, but also elucidates an unprecedented mechanism at the molecular level. Although there is no direct experimental evidence to support that the inhibition is due to the translocation rather than the compromised base pairing,22 our prediction can well-distinguish the effect of V557 versus V557L on NTP incorporation as observed in the biochemical experiment, which thus could offer a promising mechanism for “template-dependent” inhibition. We have also noted that the overall fidelity control and the transcriptional efficiency of RNA polymerases could be influenced by several check points,30,38−40 which are more complex than the two scenarios (NTP incorporation and dsRNA translocation) investigated in this study. For example, the presence of remdesivir at the template strand may also affect the initial binding of NTP at the active site and the active site isomerization before catalysis.29,41 However, the most obvious structural discrepancy between remdesivir and the natural adenosine lies in the 1′-cyano substitution, which is neither in direct contact with the complementary NTP when remdesivir is located at the +1 site of the template, nor in proximity to the protein motifs A and C mediating the active site isomerization42,43 or motif F proposed to be involved in NTP loading.41 Therefore, we speculate that the influence of template-dependent remdesivir on these two check points is relatively minor. Instead, we have suggested that the inhibition could be mainly exerted by hampering the translocation of remdesivir from the +2 to +1 site, rendering the active site partially or fully occupied by the 3′-terminal of the nascent strand and unavailable for the arrival of the next NTP. We have also noted that the current investigation of the interpolated and energy-minimized translocation pathway of remdesivir along the template strand is an approximate estimation, while a more accurate description of the translocation dynamics should be achieved by using a more tailor-made method36,44−46 based on expanded sampling. Even so, our proposal about the potential role of V557 in impeding the translocation of remdesivir as well as the influence of V557L in attenuating the inhibition matches well with the experimental observation, which in turn validates that our protocol could still effectively illuminate the “template-dependent” inhibitory mechanism acted by remdesivir in SARS-CoV-2 RdRp. As the “template-dependent” inhibition has been observed across diverse viral RNA polymerases,47−49 our finding may also shed light on the inhibition mechanism acted by nucleotide analogues in other RdRps. Overall, our studies have provided valuable molecular insights into the inhibitory mechanism when remdesivir is embedded in the template strand and underlined the role of 1′-cyano substitution in exerting the inhibitory effect. This offers an alternative strategy for the rational design of antiviral drugs targeted at SARS-CoV-2 RdRp during the synthesis of the second RNA strand for viral transcription and genome synthesis.
Acknowledgments
The authors acknowledge the financial support from the National Key R&D program of China (2021YFA1502300), the National Natural Science Foundation of China (21733007), the Hong Kong Research Grant Council NSFC/RGC Joint Research Scheme (N_HKUST635/20) and the King Abdullah University of Science and Technology (KAUST) Office of Research Administration (ORA) under Award No FCC/1/1976-44-01 and FCC/1/1976-45-01. This research made use of the resources of the Supercomputing Laboratory at King Abdullah University of Science & Technology (KAUST) as well as the computing resources of the X-GPU cluster supported by the Hong Kong Research Grant Council Collaborative Research Fund: C6021-19EF.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpclett.2c01314.
Model construction, MD simulations, and generation of preliminary translocation pathways. Figure S1. Validation of the convergence of the simulations. Figure S2. RMSD of Cα atoms of nsp12 versus time in the five 200 ns MD simulations. Figure S3. Distance between the oxygen atom in the backbone of A558 and the C1′ atom of RDV/adenosine (RA) for RdRp systems with wildtype-RNA (cyan) and RDV at the +1 site (orange). Figure S4. Conformations of A558 along the MD simulations. Figure S5. Examination over the stability of active site using MD simulations initiated from different models. Figure S6. Evaluation of the hydrogen bonding probability between the template and nascent strand at both the pre- (pre-T) and post-translocation (post-T) states. Figure S7. Conformations along the preliminary translocation pathway generated by the Climber algorithm. Figure S8. Estimation of the capability of maintaining the precatalytic conformation for wildtype-RdRp and RdRp with a V557L single-point mutation. Figure S9. Investigation into the orientation of V557/V557L in the system containing RDV in the template strand. Figure S10. The dsRNA conformation at the active site in our model and two cryo-EM structures of SARS-CoV-2 RdRp. Figure S11. Comparison of the conformation before (in steel blue) and after (in green) energy minimization. Figure S12. Comparison of the initial positions of NTP (shown in sticks) and Mg2+ ions (shown in spheres) in our model with those in the two new models. Figure S13. Identifying the translocating intermediates based on the distance between RA and V557/V557L versus the RMSD of dsRNA to the pre-T state during translocation. Figure S14. K-center clustering based on the conformational similarity of the MD conformations initiated from the translocation pathway of remdesivir from the +2 to the +1 site in wildtype RdRp (PDF)
Transparent Peer Review report available (PDF)
The authors declare no competing financial interest.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supplementary Material
References
- WHO . Coronavirus Disease 2019 (COVID-19): Situation Report. Jun 26, 2022.
- Chen N.; Zhou M.; Dong X.; Qu J.; Gong F.; Han Y.; Qiu Y.; Wang J.; Liu Y.; Wei Y.; et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: a Descriptive Study. Lancet. 2020, 395, 507–513. 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N.; Zhang D.; Wang W.; Li X.; Yang B.; Song J.; Zhao X.; Huang B.; Shi W.; Lu R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroker A.; Tirzite M. Repurposed Pharmacological Agents for the Potential Treatment of COVID-19: a Literature Review. Respir. Res. 2021, 22, 304. 10.1186/s12931-021-01885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L.; Qiang T.; Liang C.; Ren X.; Jia M.; Zhang J.; Li J.; Wan M.; YuWen X.; Li H.; et al. RNA-dependent RNA Polymerase (RdRp) Inhibitors: The Current Landscape and Repurposing for the COVID-19 Pandemic. Eur. J. Med. Chem. 2021, 213, 113201. 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong C. H. T.; Mu J.; Li S.; Fang Y.; Liu Q.; Zhang X.; Kam H.; Lee S. M. Y.; Chen Y.; Deng F.; et al. Reviving Chloroquine for Anti-SARS-CoV-2 Treatment with Cucurbit[7]uril-based Supramolecular Formulation. Chin. Chem. Lett. 2021, 32, 3019–3022. 10.1016/j.cclet.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C.; Goonetilleke E. C.; Unarta I. C.; Huang X. Incorporation Efficiency and Inhibition Mechanism of 2′-substituted Nucleotide Analogs Against SARS-CoV-2 RNA-dependent RNA Polymerase. Phys. Chem. Chem. Phys. 2021, 23, 20117–20128. 10.1039/D1CP03049C. [DOI] [PubMed] [Google Scholar]
- Jockusch S.; Tao C.; Li X.; Anderson T. K.; Chien M.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. A Library of Nucleotide Analogues Terminate RNA Synthesis Catalyzed by Polymerases of Coronaviruses that Cause SARS and COVID-19. Antiviral Res. 2020, 180, 104857. 10.1016/j.antiviral.2020.104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M.; Anderson T. K.; Jockusch S.; Tao C.; Li X.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert M.; Bera S. C.; van Nies P.; Kirchdoerfer R. N.; Shannon A.; Le T. T.; Meng X.; Xia H.; Wood J. M.; Harris L. D.; et al. Inhibition of SARS-CoV-2 Polymerase by Nucleotide Analogs from a Single-molecule Perspective. Elife. 2021, 10, e70968. 10.7554/eLife.70968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. A.; Dangerfield T. Mechanisms of Inhibition of Viral RNA Replication by Nucleotide Analogs. Enzymes. 2021, 49, 39–62. 10.1016/bs.enz.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zhang D.; Gao X.; Wang X.; Zhang L. 2′- and 3′-Ribose Modifications of Nucleotide Analogues Establish the Structural Basis to Inhibit the Viral Replication of SARS-CoV-2. J. Phys. Chem. Lett. 2022, 13, 4111–4118. 10.1021/acs.jpclett.2c00087. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Wang T.; Chen Z.; Jin L.; Wu Z.; Yan J.; Zhao X.; Cai L.; Deng Y.; Guo Y.; et al. The Point-of-care-testing of Nucleic Acids by Chip, Cartridge and Paper Sensors. Chin. Chem. Lett. 2021, 32, 3675–3686. 10.1016/j.cclet.2021.06.025. [DOI] [Google Scholar]
- Kirchdoerfer R. N.; Ward A. B. Structure of the SARS-CoV Nsp12 Polymerase Bound to Nsp7 and Nsp8 Co-factors. Nat. Commun. 2019, 10, 2342. 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Yan L.; Huang Y.; Liu F.; Zhao Y.; Cao L.; Wang T.; Sun Q.; Ming Z.; Zhang L.; et al. Structure of the RNA-dependent RNA Polymerase from COVID-19 Virus. Science. 2020, 368, 779–782. 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H. S.; Kokic G.; Farnung L.; Dienemann C.; Tegunov D.; Cramer P. Structure of Replicating SARS-CoV-2 Polymerase. Nature. 2020, 584, 154–156. 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Wu J.; Wang H.; Gao Y.; Liu Q.; Mu A.; Ji W.; Yan L.; Zhu Y.; Zhu C.; et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell. 2020, 182, 417–428. 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.; Mao C.; Luan X.; Shen D.-D.; Shen Q.; Su H.; Wang X.; Zhou F.; Zhao W.; Gao M.; et al. Structural Basis for Inhibition of the RNA-dependent RNA Polymerase from SARS-CoV-2 by Remdesivir. Science. 2020, 368, 1499–1504. 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M.; Anderson T. K.; Jockusch S.; Tao C.; Li X.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A. J.; George A. S.; Schafer A.; Leist S. R.; Gralinksi L. E.; Dinnon K. H.; Yount B. L.; Agostini M. L.; Stevens L. J.; Chappell J. D.; et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020, 32, 107940. 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C. J.; Tchesnokov E. P.; Woolner E.; Perry J. K.; Feng J. Y.; Porter D. P.; Gotte M. Remdesivir is a Direct-acting Antiviral that Inhibits RNA-dependent RNA polymerase from Severe Acute Respiratory Syndrome Coronavirus 2 with High Potency. J. Biol. Chem. 2020, 295, 6785–6797. 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E. P.; Gordon C. J.; Woolner E.; Kocinkova D.; Perry J. K.; Feng J. Y.; Porter D. P.; Gotte M. Template-dependent Inhibition of Coronavirus RNA-dependent RNA Polymerase by Remdesivir Reveals a Second Mechanism of Action. J. Biol. Chem. 2020, 295, 16156–16165. 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J. P. K.; Dangerfield T. L.; Taylor D. W.; Johnson K. A. Remdesivir is a Delayed Translocation Inhibitor of SARS-CoV-2 Replication. Mol. Cell 2021, 81, 1548–1552. 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Zhang D.; Wang X.; Yuan C.; Li Y.; Jia X.; Gao X.; Yen H.-L.; Cheung P. P.-H.; Huang X. 1′-Ribose Cyano Substitution Allows Remdesivir to Effectively Inhibit Nucleotide Addition and Proofreading During SARS-CoV-2 Viral RNA Replication. Phys. Chem. Chem. Phys. 2021, 23, 5852–5863. 10.1039/D0CP05948J. [DOI] [PubMed] [Google Scholar]
- Kokic G.; Hillen H. S.; Tegunov D.; Dienemann C.; Seitz F.; Schmitzova J.; Farnung L.; Siewert A.; Hobartner C.; Cramer P. Mechanism of SARS-CoV-2 Polymerase Stalling by Remdesivir. Nat. Commun. 2021, 12, 279. 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylehn F.; Menendez C. A.; Perez-Lemus G. R.; Alvarado W.; de Pablo J. J. Modeling the Binding Mechanism of Remdesivir, Favilavir, and Ribavirin to SARS-CoV-2 RNA-Dependent RNA Polymerase. ACS Cent. Sci. 2021, 7, 164–174. 10.1021/acscentsci.0c01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Zhou R. Structural Basis of the Potential Binding Mechanism of Remdesivir to SARS-CoV-2 RNA-Dependent RNA Polymerase. J. Phys. Chem. B 2020, 124, 6955–6962. 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- Dangerfield T. L.; Huang N. Z.; Johnson K. A. Remdesivir is Effective in Combating COVID-19 because It is a Better Substrate than ATP for the Viral RNA-Dependent RNA Polymerase. iScience. 2020, 23, 101849. 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero M. E.; Long C.; La Rocco D.; Keerthi A. M.; Xu D.; Yu J. Probing Remdesivir Nucleotide Analogue Insertion to SARS-CoV-2 RNA Dependent RNA Polymerase in Viral Replication. Mol. Syst. Des. Eng. 2021, 6, 888–902. 10.1039/D1ME00088H. [DOI] [Google Scholar]
- Long C.; Romero M. E.; La Rocco D.; Yu J. Dissecting Nucleotide Selectivity in Viral RNA Polymerases. Comput. Struct. Biotec. 2021, 19, 3339–3348. 10.1016/j.csbj.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulgi S.; Jani V.; V N M. U.; Sonavane U.; Joshi R. Structural Insight into the Binding Interactions of NTPs and Nucleotide Analogues to RNA Dependent RNA Polymerase of SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 6, 1–15. 10.1080/07391102.2021.1894985. [DOI] [PubMed] [Google Scholar]
- Traut T. W. Physiological Concentrations of Purines and Pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Kennedy E. M.; Gavegnano C.; Nguyen L.; Slater R.; Lucas A.; Fromentin E.; Schinazi R. F.; Kim B. Ribonucleoside Triphosphates as Substrate of Human Immunodeficiency Virus Type 1 Reverse Transcriptase in Human Macrophages. J. Biol. Chem. 2010, 285, 39380–39391. 10.1074/jbc.M110.178582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan R. Coupling of Fast and Slow Modes in the Reaction Pathway of the Minimal Hammerhead Ribozyme Cleavage. Biophys. J. 2007, 93, 2391–2399. 10.1529/biophysj.107.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgrignani J.; Magistrato A. The Structural Role of Mg2+ Ions in a Class I RNA Polymerase Ribozyme: a Molecular Simulation Study. J. Phys. Chem. B 2012, 116, 2259–2268. 10.1021/jp206475d. [DOI] [PubMed] [Google Scholar]
- Silva D. A.; Weiss D. R.; Pardo Avila F.; Da L. T.; Levitt M.; Wang D.; Huang X. Millisecond Dynamics of RNA Polymerase II Translocation at Atomic Resolution. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 7665–7670. 10.1073/pnas.1315751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. R.; Levitt M. Can Morphing Methods Predict Intermediate Structures?. J. Mol. Biol. 2009, 385, 665–674. 10.1016/j.jmb.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydow J. F.; Cramer P. RNA Polymerase Fidelity and Transcriptional Proofreading. Curr. Opin. Struct. Biol. 2009, 19, 732–739. 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Yu J. Efficient Fidelity Control by Stepwise Nucleotide Selection in Polymerase Elongation. Mol. Based Math. Biol. 2014, 2, 141–160. [Google Scholar]
- Yuzenkova Y.; Bochkareva A.; Tadigotla V. R.; Roghanian M.; Zorov S.; Severinov K.; Zenkin N. Stepwise Mechanism for Transcription Fidelity. BMC Biol. 2010, 8, 54. 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Ouirane K.; Boulard Y.; Bressanelli S. The Hepatitis C virus RNA-dependent RNA Polymerase Directs Incoming Nucleotides to Its Active Site through Magnesium-dependent Dynamics within Its F Motif. J. Biol. Chem. 2019, 294, 7573–7587. 10.1074/jbc.RA118.005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung J.; Lin Y. S.; Yang Y. H.; Chou Y. L.; Shu L. H.; Cheng Y. C.; Liu H. T.; Wu C. Y. The Potential Chemical Structure of Anti-SARS-CoV-2 RNA-dependent RNA Polymerase. J. Med. Virol. 2020, 92, 693–697. 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P.; Peersen O. B. Structural Basis for Active Site Closure by the Poliovirus RNA-dependent RNA Polymerase. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 22505–22510. 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Pardo-Avila F.; Unarta I. C.; Cheung P.; Wang G.; Wang D.; Huang X. Elucidation of the Dynamics of Transcription Elongation by RNA Polymerase II using Kinetic Network Models. Acc. Chem. Res. 2016, 49, 687–694. 10.1021/acs.accounts.5b00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Cao S.; Zhu L.; Huang X. Constructing Markov State Models to Elucidate the Functional Conformational Changes of Complex Biomolecules. WIREs Comput. Mol. Sci. 2017, 8, e1343. [Google Scholar]
- Zhu L.; Sheong F. K.; Cao S.; Liu S.; Unarta I. C.; Huang X. TAPS: A Traveling-Salesman Based Automated Path Searching Method for Functional Conformational Changes of Biological Macromolecules. J. Chem. Phys. 2019, 150, 124105. 10.1063/1.5082633. [DOI] [PubMed] [Google Scholar]
- Gordon C. J.; Lee H. W.; Tchesnokov E. P.; Perry J. K.; Feng J. Y.; Bilello J. P.; Porter D. P.; Gotte M. Efficient Incorporation and Template-dependent Polymerase Inhibition are Major Determinants for the Broad-spectrum Antiviral Activity of Remdesivir. J. Biol. Chem. 2022, 298, 101529. 10.1016/j.jbc.2021.101529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee W. C.; Aldern K. A.; Hostetler K. Y.; Evans D. H. Cidofovir and (S)-9-[3-hydroxy-(2-phosphonomethoxy)propyl]adenine are Highly Effective Inhibitors of Vaccinia Virus DNA Polymerase when Incorporated into the Template Strand. Antimicrob. Agents Chemother. 2008, 52, 586–597. 10.1128/AAC.01172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D.; Castro C.; Hong Z.; Cameron C. E. Hepatitis C Virus RNA-dependent RNA Polymerase (NS5B) as a Mediator of the Antiviral Activity of Ribavirin. J. Biol. Chem. 2001, 276, 46094–46098. 10.1074/jbc.C100349200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.