Abstract
Induction of genes expressed from the arabinose PBAD promoter is very rapid and maximal at low arabinose concentrations. We describe here two mutations that interfere with the expression of genes cloned under arabinose control. Both mutations map to the ydeA promoter and stimulate ydeA transcription; overexpression of YdeA from a multicopy plasmid confers the same phenotype. One mutation is a large deletion that creates a more efficient −35 region (ATCACA changed to TTCACA), whereas the other affects the initiation site (TTTT changed to TGTT). The ydeA gene is expressed at extremely low levels in exponentially growing wild-type cells and is not induced by arabinose. Disruption of ydeA has no detectable effect on cell growth. Thus, ydeA appears to be nonessential under usual laboratory growth conditions. The ydeA gene encodes a membrane protein with 12 putative transmembrane segments. YdeA belongs to the largest family of bacterial secondary active transporters, the major facilitator superfamily, which includes antibiotic resistance exporters, Lac permease, and the nonessential AraJ protein. Intracellular accumulation of arabinose is strongly decreased in mutant strains overexpressing YdeA, suggesting that YdeA facilitates arabinose export. Consistent with this interpretation, very high arabinose concentrations can compensate for the negative effect of ydeA transcriptional activation. Our studies (i) indicate that YdeA, when transcriptionally activated, contributes to the control of the arabinose regulon and (ii) demonstrate a new way to modulate the kinetics of induction of cloned genes.
The arabinose regulon of Escherichia coli consists of five operons scattered around the chromosome. AraC is the major transcriptional regulator of the regulon. AraC positively regulates transcription of the four other operons in the presence of arabinose and represses transcription in its absence (34, 35). Transcription of these operons is sensitive to catabolite repression and requires cyclic AMP and the catabolite repressor protein CRP. The interplay of these two transcriptional activators and the positions of their binding sites are slightly different for each promoter (10, 36). The araBAD operon encodes the three enzymes necessary for arabinose metabolism. The araE and araFGH operons encode two transport systems (14, 20, 21). AraE is a low-affinity sugar:proton symporter (23), while the periplasmic binding protein AraF and the two membrane proteins AraG and AraH constitute a high-affinity transport system (14, 15). All mutations which affect growth on arabinose as a carbon source or expression of the araBAD operon map to these eight genes.
A genetic search for arabinose-inducible promoters identified a fifth operon, which maps at 9 min and is now called the araJ operon (10, 22). The araJ gene encodes a nonessential membrane protein of unknown function (31). Disruption of araJ had no visible effect on growth in minimal arabinose medium, whether arabinose uptake was mediated by AraE or AraFGH. Furthermore, the kinetics of PBAD induction were similar in wild-type and ΔaraJ strains, indicating that AraJ is not involved in arabinose regulation. It has been proposed that AraJ can participate in the transport or processing of arabinose polymers, which are abundant nutrients in nature (31). When the sequence of AraJ became available, no homologs were detected in the databases. It is now known that AraJ belongs to a large class of multidrug resistance translocators (7), and in particular to the major facilitator superfamily (MFS), which includes AraE (24, 28, 29). These proteins have 12 transmembrane segments, and a number of them have been shown to export antibiotics and other small molecules (6, 7, 28).
The properties of the arabinose regulon have led to the development of a family of expression plasmids that are extensively used for physiological studies of null mutations in essential genes (9). These vectors encode the positive and negative regulator AraC, and they contain the intergenic control region and the PBAD promoter. A number of features of the arabinose regulon contribute to the versatility of these vectors. Expression in the absence of inducer can be kept to very low levels in the presence of glucose, because of the repressor activity of AraC and the reduced concentration of cyclic AMP, allowing for the cloning of toxic genes. Expression levels can be modulated over a 1,000-fold range, and they are different in rich versus minimal medium. Finally, the kinetics of induction is very rapid, and the kinetics of repression upon removal of arabinose depends on the host Ara phenotype (9).
We have taken advantage of these properties to clone in pBAD24 a chimeric protein in which the signal sequence of a mammalian protein was fused to the mature portion of alkaline phosphatase (AP) (4). This chimeric protein is exported to some extent, but its expression is toxic when the PBAD promoter is fully induced. We have shown that most suppressors of this toxic phenotype map to known sec genes, have a weak Sec phenotype, and selectively slow down export of the toxic protein (4). We report here the characterization of two suppressor mutations that do not directly affect protein export but interfere with induction of the PBAD promoter by arabinose. These experiments led to the characterization of YdeA, a membrane protein that is homologous to AraJ and that interferes with the intracellular accumulation of arabinose.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains used in this study were DHB3 [F− araD139 Δ(ara-leu)7696 Δ(lac)X74 rpsL150 galU galK thi malFΔ3 phoAΔ(PvuII) phoR] (5), SB0 (DHB3/pBAD72K) (4), JCB433 (MC1000 recD1903::mini-Tn10) (32; obtained from J. Bardwell), DB519 (JCB433/pSB13), and AD126 [F′ lacIq Tn5/ΔatpBC ilv::Tn10 malBF13 malT(Con) arg his] (obtained from M. Ehrmann). Plasmid pBAD72K (4) is a derivative of pBAD18 (9) in which the sequence coding for the first 52 amino acids of murine PAI2 (2) was fused to a derivative of TnphoA (8); the chimeric protein is expressed from the arabinose PBAD promoter. A kanamycin resistance (Kanr) cassette (from pUC4Kn; Pharmacia) was inserted downstream of the TnPhoA sequence. Suppressor strains are derivatives of SB0 isolated on NZ plates supplemented with 0.2% arabinose and 40 μg of 5-bromo-4-chloro-3-indolyl phosphate per ml (4). SB1 contains a suppressor of spontaneous origin, whereas SB34 was isolated after UV mutagenesis (26). Strains SB01 and SB034 are derivatives of SB0 in which the suppressor mutations of SB1 and SB34 were cotransduced with the zdf-1::Tn10 transposon (see below). Arabinose uptake was measured in strains DB529, DB530, and DB533, derivatives of DHB3 containing the zdf-1::Tn10 transposon and pBAD24 (9); DB529 contains the suppressor mutation of SB1, DB530 is ydeA+, and DB533 contains the suppressor mutation of SB34. These strains are phenotypically Ara−, although they express AraC from the plasmid.
Tn10 transposons were linked to the suppressor mutations by infection with λ mini-Tet 1098 (37) and repeated cycles of P1 transduction and selection on plates containing arabinose and tetracycline. The closest Tn10 (zdf-1::Tn10) was 75% linked to the suppressor mutations of SB1 and SB34. This transposon was localized after PCR amplification of the chromosomal DNA directly flanking the IS10 sequences (13). The amplified fragment was hybridized to a membrane on which an ordered library of genomic clones had been immobilized (19) (purchased from Takara Shuzo, Tokyo, Japan, through ITC Biotech, Heidelberg, Germany). The probe hybridized to Kohara’s phages 306 and 307, at approximately 35.2 min on the E. coli chromosome.
AP assays.
AP enzymatic activities were measured by determining the rate of p-nitrophenyl phosphate hydrolysis (25). Cultures grown in NZ medium to early log phase were induced with arabinose and collected on ice at the indicated times in the presence of 2 mM iodoacetamide.
RNA isolation, cRNA probes, and hybridizations.
RNAs were isolated from cultures grown in NZ or LB medium to early log phase as described elsewhere (3, 30). The RNAs were digested with RNase-free DNase (Promega), and RNA integrity was verified by gel electrophoresis and Northern blot hybridization. cRNA synthesis, Northern blot hybridization, and RNase protection were performed as described elsewhere (1).
To make the PAI2 probe, a 250-bp KpnI-BamHI fragment from pBAD72K was subcloned into the cognate sites of pBSKS. The plasmid was linearized with SacI and transcribed with T3 RNA polymerase in the presence of 50 to 100 μM unlabeled UTP and 10 to 50 μCi of [32P]UTP. To make the ydeA probe, a 950-bp fragment of SB1 chromosomal DNA was PCR amplified with primers pSB1up (5′GATCACATTCTCAAGACGC) and pSB1don2 (5′GGCATGAGTGGTTGC) and digested with BclI and NsiI; a 387-bp fragment, which is identical in wild-type and SB1 DNAs, was subcloned between the BamHI and PstI sites of pBSKS. The plasmid was linearized with EcoRI and transcribed with T7 RNA polymerase as described above. High-specific-activity probes were synthesized in the presence of 25 μM unlabeled UTP and 50 μCi of [32P]UTP.
Cloning of ydeA and sequence of the suppressor mutations.
The suppressors strains were crossed with F′ strains containing the episomes F500 and F506 (26), in which a Tn10 was introduced by P1 transduction. All Tetr Kanr exconjugants were Arar, indicating that the suppressor mutations in SB1 and SB34 are dominant. DNA from the SB1 suppressor strain was partially digested with Sau3AI and fractionated on a 10 to 40% sucrose gradient, and fragments in the 4- to 10-kbp range were cloned into the BamHI site of pACYC184. The library was screened for multicopy dominant inserts conferring the Arar phenotype to the parental strain (SB0), and four plasmids with overlapping inserts were isolated. A large 12-kbp insert (pSB11) was reduced to a 1.8-kbp insert (pSB13; up to the BspHI site in ydeB) and to a minimal 1.3-kbp insert (pSB16; up to the BsgI site between ydeA and ydeB), both of which conferred the suppressor phenotype of the SB1 strain. DNA sequencing was performed with Sequenase version 2.0 (U.S. Biochemical).
Construction of a ydeA null allele.
Plasmid pDB9722 is derived from pUC19 and contains a 1.3-kbp AvaI-NsiI fragment derived from Kohara phage 304 (19), a Kanr cassette (from pUC4Kn; Pharmacia), and a 1.3-kbp MscI-SalI fragment derived from plasmid pSB11. The left end of the deletion is located between the −35 and −10 regions of the ydeA promoter, and the right end is located 27 bp upstream of the ydeA UAG stop codon. The replacement cassette was excised with SalI and KpnI and electroporated into strains JCB433 and DB519, which express ydeA from pSB13. Kanr recombinants were obtained with both strains, suggesting that ydeA is nonessential. The disruption was verified by Southern blot hybridization with two probes located on either side of the Kanr cassette. The disrupted allele was introduced by P1 transduction with similar frequencies in DHB3 and in DHB3 carrying the ydeA-expressing plasmid pSB13.
Arabinose uptake.
Cultures were grown in M63-glycerol medium supplemented with 18 amino acids to an A600 of 0.2 to 0.4, centrifuged, resuspended at an A600 of 0.6, and induced for 20 min with 2% arabinose. Under these conditions, wild-type and ydeA-expressing strains are induced to the same extent. AraFGH-mediated uptake was measured in strain AD126 containing the araFGH-expressing plasmid pKKATEB (14, 15) and either pSB13 or pACYC184. Cultures grown in LB medium were induced with 5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 30 min. The cells were washed four times with M63, resuspended at an A600 of 0.6, and incubated for 10 min at room temperature. To measure arabinose uptake in the absence of the proton motive force, AD126 cells were fed with 0.2% glucose and preincubated for 30 s with 16 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP); the same volume of dimethyl sulfoxide was added to control cells. [14C]arabinose (250 mCi/mmol; CB-69; CEA-France; a generous gift from R. W. Hogg) was diluted with unlabeled arabinose. At the indicated times after the addition of [14C]arabinose, 200 μl of cells was filtered through nitrocellulose (HAWP; 0.45-μm pore size; Millipore). The filters were immediately washed with 5 ml of M63 and dried, and the radioactivity was measured by liquid scintillation. An unfiltered aliquot was used to determine the radioactivity input and to calculate the intracellular arabinose levels.
RESULTS
Two mutations that interfere with transcriptional activation of the PBAD promoter.
A PAI2-AP chimeric protein was expressed under the control of the arabinose PBAD promoter (4). Induction with arabinose was toxic and prevented colony formation on arabinose plates. A large collection of suppressors of this toxic phenotype have been isolated, and most of them map to one of the secA, secY, and secG genes, which encode components of the protein export machinery (27, 33, 38). These mutants slow down the export kinetics of the chimeric protein upon induction with arabinose (4).
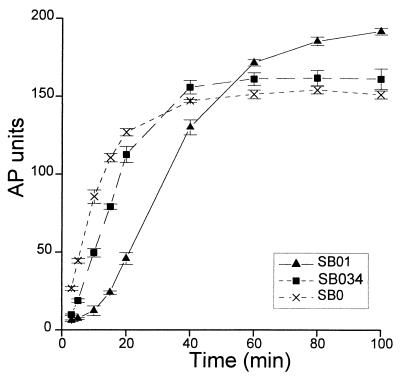
Two strains, SB1 and SB34, contained suppressor mutations that did not map to known sec genes. They were localized between 30 and 45 min on the E. coli chromosome by Hfr mating. A transposon was linked to either of these two mutations, shown to be very closely linked, and mapped near 35 min (see Materials and Methods). These suppressors, like those in sec genes, slow down the accumulation of active AP upon induction, and this effect was more pronounced for strain SB01 than for SB034 (Fig. 1). This effect was particularly evident at early times, and by 60 min all three strains had similar levels of AP activity. The accumulation of active AP integrates transcription, translation, and protein export to the periplasm, and the suppressors could affect any of these processes. The two suppressor strains showed no apparent defect in export of MalE or β-lactamase (data not shown). It has been shown that synthesis of proteins expressed from the PBAD promoter is maximally induced within 2 min upon addition of arabinose (9). In contrast, the synthesis of the PAI2-AP protein was induced much more slowly in the suppressor strains; a similar effect was observed with two unrelated proteins cloned in the same vector (an FtsQ-AP fusion protein and human Bcl-2 [data not shown]).
FIG. 1.
Effects of suppressor mutations on kinetics of PAI2-AP export upon induction with arabinose. The two suppressor mutations of strains SB1 and SB34 were transduced into the parental strain SB0, generating suppressor strains SB01 and SB034, respectively. All strains express the chimeric PAI2-AP protein. Cells grown to an A600 of 0.1 in NZ medium at 37°C were induced with 0.2% arabinose at time zero. AP activity was assayed at the indicated times. Each curve represents the average of three independent cultures; error bars indicate standard deviations.
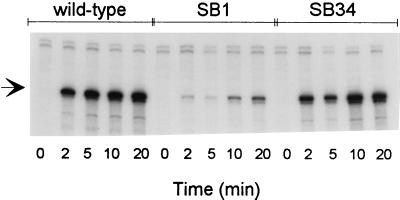
These results suggested that the suppressors could interfere with the activity of the PBAD promoter. To test this hypothesis, we measured the kinetics of PAI2-AP mRNA induction in wild-type and suppressor strains (Fig. 2). Whereas induction of PAI2-AP transcription was maximal within 5 min in the parental strain, it was reduced approximately 2-fold in SB34 and at least 20-fold in SB1. Northern blot hybridizations showed that both suppressors did not affect the size distribution of PAI2-AP transcripts (data not shown). Thus, the decrease in transcriptional induction of the PBAD promoter (Fig. 2) accounts for the reduced kinetics of active AP accumulation (Fig. 1). These results indicated that both suppressors interfere with the transcriptional activation of the arabinose PBAD promoter.
FIG. 2.
Effects of suppressor mutations on kinetics of PAI2-AP transcription upon induction with arabinose. Cells grown to an A600 of 0.1 in NZ medium at 37°C were induced with 0.2% arabinose at time zero. Total RNA was extracted from samples collected at the indicated times. Two micrograms of RNA was hybridized to a 32P-labeled PAI2 cRNA probe (290 nt). After digestion with pancreatic RNase, the hybrids were denatured and electrophoresed in a 6% urea-polyacrylamide gel. The protected cRNA fragment is 193 nt long (arrow); traces of undigested probe are visible in the upper portion of the gel. Lanes 1 to 5, parental strain SB0; lanes 6 to 10, suppressor strain SB1; lanes 11 to 15, suppressor strain SB34.
The kinetics of induction of a FtsQ-AP fusion protein (9) from the PBAD promoter was decreased in the SB1 suppressor strain. In contrast, expression of the same chimeric protein from the PTAC promoter was induced by IPTG with the same kinetics in the parental and SB1 strains (data not shown). Thus, decreased expression of cloned genes in the suppressor strains appeared specific for expression vectors derived from the arabinose regulon.
The suppressor mutations lead to increased expression of YdeA.
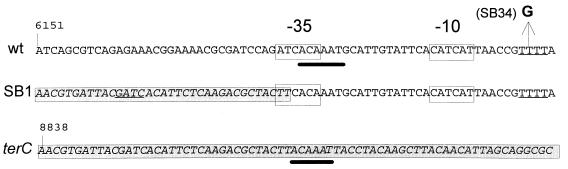
To investigate the mechanism of action of these suppressors, we determined that the suppressor mutations are dominant (see Materials and Methods) and cloned the corresponding mutant genes. A genomic library prepared with DNA of strain SB1 was screened for inserts that suppress the toxicity of the PAI2-AP protein and confer an Arar phenotype. A 1.35-kbp fragment, derived from a large 12-kbp insert, was the minimal restriction fragment able to confer the Arar phenotype. This fragment contains one open reading frame (ORF), that of YdeA (SwissProt accession no. [AN] P31122). The sequence of the insert, isolated from strain SB1, was identical to that of ydeA in the E. coli genome sequence database throughout the coding region. However, the first 23 nucleotides (nt) of the insert were localized 8 kbp upstream of ydeA, suggesting that a large deletion is responsible for the mutant phenotype (Fig. 3). This deletion extends into the −35 region of the ydeA promoter and leads to the replacement of ATCACA by a TTCACA element. This change is expected to increase the activity of the promoter since the mutated element displays a much better match with the −35 consensus sequence (TTGACA). We also failed to detect a mutation in the ydeA coding region of strain SB34 and found a different mutation in the promoter. In this case, the change occurred in the putative transcription initiation site, where the sequence TTTT is changed to TGTT.
FIG. 3.
Nucleotide sequences of mutations in the ydeA promoter. The nucleotide sequence of the wild-type (wt) ydeA promoter region is shown at the top; numbering corresponds to that of database entry ECAE000250. The putative −35 and −10 promoter regions are boxed, and the potential start sites are underlined. The mutation in strain SB34 introduces a G residue in the transcription initiation region. The large deletion in strain SB1 changes the −35 region, which now shows a five-of-six-position match with the TTGACA consensus sequence; the insert of pSB11 starts at the underlined Sau3AI site. The sequence upstream of the deletion is in italics and boxed, as is the wild-type sequence at the beginning of terC, shown at the bottom; numbering corresponds to that of database entry ECAE000249. The ACAAAT repeats (thick lines) upstream of terC (bottom) and ydeA (top) may have been involved in the spontaneous deletion of intervening sequences.
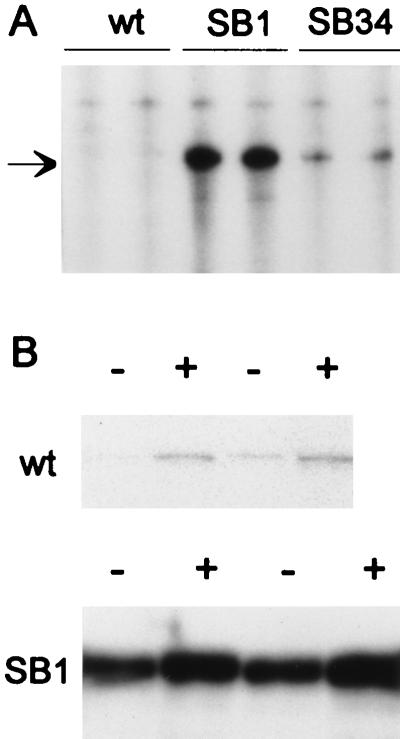
The identification of these two mutations strongly suggested that the suppressors act by increasing ydeA transcription. To test this hypothesis, we analyzed the level of ydeA mRNA by RNase protection in the wild-type and two mutant strains. Large amounts of transcripts were observed with RNA of strain SB1 (40 to 80 pg of mRNA per μg of total cellular RNA); lower levels were found with RNA of SB34. In contrast, only very low levels of ydeA mRNA were detected in RNA of the wild-type strain (Fig. 4A). To determine whether ydeA transcription is under arabinose control, we compared the level of ydeA mRNA in arabinose-induced cells to that in untreated cells (Fig. 4B). In the wild-type strain, the level of ydeA mRNA was only slightly elevated after addition of arabinose. A similar marginal increase was observed in strain SB1. This effect appears minimal compared to the more than 100-fold increase in transcription from the PBAD, PE, PFGH, and PJ promoters upon induction with arabinose (Fig. 2; references 10, 20, and 23). These experiments indicate that the ydeA gene is poorly expressed in wild-type E. coli, at least under exponential growth conditions, and that it is not part of the arabinose regulon.
FIG. 4.
ydeA mRNA levels in wild-type and mutant strains. For each strain, two independent cultures were grown at 37°C to an A600 of 0.4 in NZ medium, and total RNA was isolated. (A) Two micrograms of RNA was hybridized to a 32P-labeled ydeA cRNA probe (450 nt). After digestion with pancreatic RNase, the hybrids were denatured and electrophoresed in a 5% polyacrylamide gel. The protected fragment is 360 nt long (arrow); traces of undigested probe are visible in the upper portion of the gel. The wild-type (wt) strain was SB0. (B) Two cultures of DB530 (ydeA+) and DB529 (SB1 derivative) grown in LB medium were induced for 20 min with 2% arabinose (+) or not induced (−). Ten micrograms of RNA was hybridized to detect the very low levels of ydeA mRNA in wild-type cells. The gel was exposed for 4 days (wild type) and 20 h (SB1).
We have constructed a deletion allele of ydeA (see Materials and Methods). This disruption, which contains a Kanr cassette, could be introduced at similar frequencies in strains expressing ydeA from a plasmid or in strains containing an empty plasmid, indicating that ydeA is a nonessential gene. The deletion strains grew normally at temperatures ranging from 23 to 42°C, in rich as well as in minimal media.
Increased expression of YdeA interferes with the intracellular accumulation of arabinose.
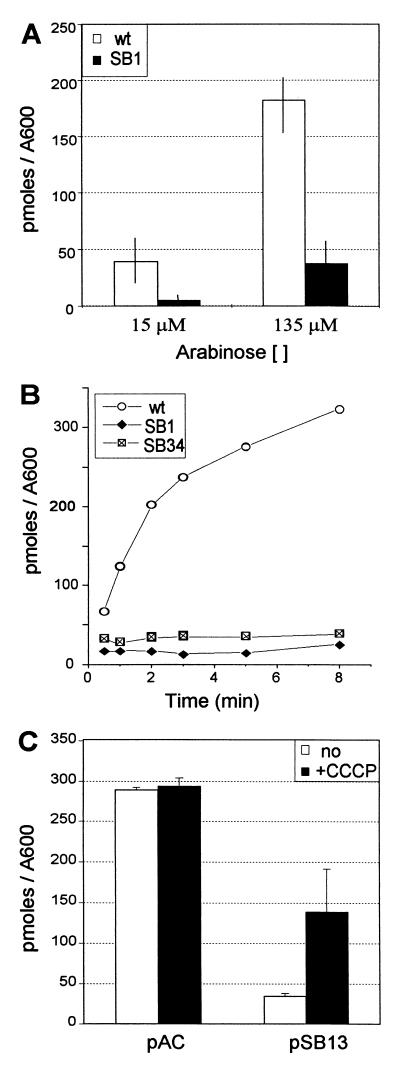
A database search with the YdeA sequence showed that it belongs to the MFS, a large family of integral membrane proteins with 12 transmembrane domains (24, 28, 29). Since increased expression of YdeA interferes with induction of the PBAD promoter by arabinose, it appeared possible that YdeA interferes with the intracellular accumulation of arabinose. To test this hypothesis, we have compared arabinose uptake in the different strains. Induction of the two known arabinose transporters requires the positive regulator AraC (10, 20), which was expressed from an empty pBAD24 plasmid. Cells were first treated with high concentrations of arabinose (see below) to allow induction of the araE and araFGH operons. We then measured arabinose uptake at two concentrations, to assay either mainly the high-affinity AraFGH transport system (Km = 1 to 3 μM) or both the high- and low-affinity (AraE; Km = 60 to 100 μM) systems (Fig. 5A) (15, 21). The amount of arabinose accumulated in cells carrying the SB1 deletion was much lower than that detected in wild-type cells. A similar reduction was observed at both arabinose concentrations, suggesting that YdeA overexpression exerts its effect independently of the arabinose import system. The low amounts of arabinose in strains carrying the SB1 deletion or the SB34 point mutation is probably accounted for by periplasmic binding to AraF (Fig. 5B). The levels of arabinose accumulated in the two mutant strains were too low to directly determine whether YdeA promotes arabinose export or whether it interferes with uptake. The latter hypothesis appears less likely, since YdeA exerts its effect on both the high-affinity and low-affinity transport systems.
FIG. 5.
Effect of the ydeA promoter mutations on arabinose uptake. (A) Two independent cultures of DB529 (SB1 derivative) and wild-type (wt) strain DB530 (ydeA+) were assayed in duplicate for arabinose uptake during a 1-min pulse at the indicated arabinose concentrations. Intracellular arabinose levels (in picomoles/0.2 ml of cells) were normalized to the optical density of the cultures. Error bars indicate ranges. (B) The kinetics of arabinose uptake was measured with two independent cultures of DB529 (SB1 derivative), DB531 (SB34 derivative), and DB530 (ydeA+ [wt]), the average of the two cultures is presented. The arabinose concentration was 62.5 μM. (C) Arabinose uptake by the AraFGH system was assayed in strain AD126 (unc) carrying pKKATEB and either pSB13 or pACYC184 (pAC). Cells were assayed in triplicate for arabinose uptake during a 2-min pulse at 14 μM arabinose. Glucose-fed cells were pretreated for 30 s with 16 μM CCCP (+CCCP) or with the same volume of solvent (no). Intracellular arabinose levels (in picomoles/0.2 ml of cells) were normalized to the optical density of the cultures. Error bars indicate standard deviations.
Most members of the MFS are dependent on the proton motive force for substrate transport. To determine whether YdeA requires the proton motive force to interfere with arabinose uptake, we tested whether the effect of YdeA could be prevented by the protonophore CCCP (Fig. 5C). These experiments were performed in unc cells expressing the ATP-dependent AraFGH system from the PTAC promoter. Under these conditions, arabinose uptake was identical in control and CCCP-treated ydeA+ cells. In untreated cells, YdeA overexpression strongly reduced arabinose accumulation. Upon dissipation of the proton motive force, arabinose uptake was increased fourfold, although it did not reach the level measured with ydeA+ cells. These results directly confirm that YdeA interferes with arabinose uptake by the AraFGH transport system and indicate that YdeA is a proton motive force-dependent arabinose export system.
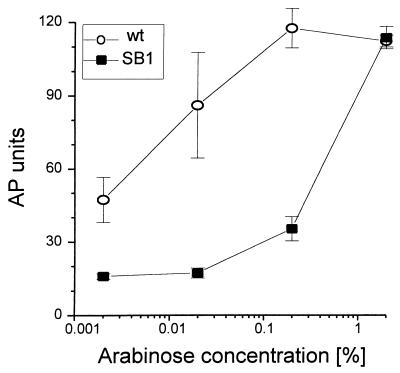
Increased expression of YdeA displaces the dose-response curve of induction by arabinose.
Expression of genes cloned in pBAD vectors can be modulated over a wide range of arabinose concentrations (9). To determine whether it would still be possible to induce fully and rapidly the PBAD promoter in cells overexpressing YdeA, we measured the arabinose dose-response curve in the wild-type and SB1 strains, 20 min after addition of the inducer. In the wild-type strain, the expression of PAI2-AP was already induced with low arabinose concentrations, and it was maximal at 0.2% arabinose (Fig. 6). In contrast, in the SB1 strain, expression of the chimeric protein was very low at arabinose concentrations ranging from 0.002 to 0.2% and reached wild-type levels only when the arabinose concentration was increased to 2%. Similar results were obtained with wild-type cells overexpressing YdeA from plasmid pSB11; in these cells, the level of ydeA mRNA was similar to that found in strain SB1 (data not shown). Thus, increasing the external arabinose concentration can compensate for the overexpression of YdeA and allows rapid and maximal activation of the PBAD promoter.
FIG. 6.
Effect of YdeA on the induction of PAI2-AP by increasing arabinose concentrations. Cells were grown at 37°C in NZ medium to an A600 of 0.4, induced for 20 min with the indicated concentrations of arabinose, and assayed for AP activity. Each curve represents the average of three independent cultures. Error bars represent standard deviations. Both the wild-type (wt; SB0) and suppressor (SB1) strains carry pBAD72K and express the PAI2-AP chimeric protein.
DISCUSSION
The uptake and metabolism of arabinose are believed to involve eight proteins: AraE, a low-affinity transporter; AraF, AraG, and AraH, which constitute the high-affinity transport system; AraB, AraA, and AraD, the three enzymes necessary for arabinose metabolism; and AraC, the major transcriptional regulator of the regulon (34). Arabinose converts AraC from a repressor to an activator, and transcription of the three operons is rapidly and strongly induced (35). We show here that mutational activation of the ydeA promoter, which is essentially silent in wild-type cells, inhibits the transcriptional induction of the PBAD promoter by lowering the intracellular concentration of arabinose. Overexpression of YdeA alone is sufficient for this effect, since the same phenotype was observed in wild-type cells carrying a plasmid with an insert that express only YdeA.
The extremely weak activity of the wild-type ydeA promoter is not surprising. Indeed, the putative −35 and −10 regions display poor matches with the consensus sequences. Furthermore, the initiation site lacks a properly positioned purine residue. Both mutations described here increase the activity of the ydeA promoter. In one case, SB1, a large deletion generated an improved −35 region. In the other case, SB34, a transversion introduced a G residue in the putative initiation site. The deletion provided a stronger stimulation than the initiation site mutation. The higher expression of ydeA in the SB1 strain probably accounts for its stronger effect on the kinetics of induction of genes expressed from the PBAD promoter.
Most genetic studies of promoters have concentrated on the −35 and −10 regions, although initiation has been extensively studied in vitro (references 16 to 18 and references therein). The initiation site mutation of SB34 is almost the exact opposite of a mutation originally described in the phage P22 sar promoter (16). The TGTT-to-TTTT mutation at position +1 in the sar promoter was isolated in a genetic screen for strong promoter mutations and was shown in vitro to cause a defect in promoter clearance. In the ydeA promoter, the TTTT-to-TGTT mutation is at position +2. In strains SB1 and SB34, the ydeA transcription initiation sites appeared indistinguishable at the resolution of the RNase protection assay. Thus, initiation at the SB1 promoter may occur at either the −1 (G) or +1 (T) residue.
The ydeA gene, like many E. coli genes, appears nonessential, at least under the standard laboratory conditions tested. The large 7,856-bp deletion in strain SB1 shows that a number of adjacent genes and DNA elements are also nonessential. This large deletion, which is flanked by two ACAAAT repeats, removes a weak putative clockwise promoter separated by about 1 kbp from the ydeA gene and six ORFs of unknown function. The deletion also removes the hotF locus, a recombinational hot spot, and the terC-terC3-psrA locus, proposed to terminate clockwise replication (12). terC is in fact oriented opposite to what was originally proposed (12), and it is counterclockwise replication that does not require terC. It remains to be determined whether counterclockwise replication terminates only at the terA site (28.78 min) in wild-type cells or whether both sites are used to varying extents. Other large deletions near the terminus have been characterized (11), confirming the low density of essential genes in the E. coli 30- to 35-min map interval.
The experiments presented here are compatible with three types of hypotheses concerning the mode of action of YdeA. YdeA could decrease arabinose entry into the cytoplasm, promote arabinose export, or stimulate arabinose degradation. Arabinose uptake experiments, which were performed with uniformly labeled [14C]arabinose, show that the net accumulation is reduced to very low levels. Thus, if YdeA were to activate a catabolic pathway, all C-containing products would have to be rapidly exported from the cell. Furthermore, YdeA shows no homology to known enzymes. Arabinose import is mediated by two transport systems that are structurally and functionally independent (21, 34). AraE is a low-affinity proton:symporter that belongs to the MFS (23, 24), while AraFGH constitutes a high-affinity ATP-dependent transport system related to the ABC primary active transporters (15, 29). We have compared arabinose uptake in SB1 and wild-type cells at a low arabinose concentration, when only the AraFGH system is effective, and at a high arabinose concentration, when both systems contribute to arabinose uptake. Since the effect of YdeA was of the same magnitude, YdeA would have to act on both systems if it were to affect import. Finally, we have shown that YdeA interferes with arabinose uptake in cells that only express the AraFGH system and that this effect is partially abolished by dissipation of the proton motive force. Thus, these results strongly suggest that YdeA promotes arabinose export out of the cytoplasm. Because the amount of arabinose taken up by either SB1 or SB34 cells was so low, it is very difficult to definitively demonstrate, by isotope dilution, that YdeA is an arabinose exporter.
The notion that YdeA promotes arabinose export is supported by sequence comparison with related proteins. YdeA belongs to the very large group of integral membrane proteins with 12 transmembrane segments (24, 28). Within this group, the MFS constitutes one of the largest family, with 64 members in the E. coli genome (29). Approximately one-third of the E. coli members of the MFS have been studied genetically or biochemically, and the remaining are known only as putative proteins. They include sugars transporters (LacY, AraE, etc.), as well as multidrug resistance proteins (EmrB, MdfA, etc.). The closest homologs of YdeA (50% identity) are ORFs in the Helicobacter pylori and Haemophilus influenzae genomes, which most likely correspond to the orthologous genes. Three E. coli proteins show a relatively high homology to YdeA: AraJ, a member of the arabinose regulon (at 9 min, 24% identity; AN P23910) (31), Yicm/f451 (at 83 min, 26% identity; AN P31438), and f389 (at 37 min, 29% identity; AN D90809). The closest YdeA homologue that has been functionally characterized is a chloramphenicol resistance gene of Streptomyces lividans (30% identity; P31141), which exports the drug (6). YdeA overexpression fails to confer resistance to chloramphenicol or tetracycline; it also does not interfere with gene activation by IPTG (a FtsQ-AP fusion protein expressed from PTAC) or maltose (MalE). Thus, the spectrum of molecules that can be exported by YdeA remains to be determined.
Although ydeA appears to be a nonessential gene, the presence of a set of related genes in E. coli, H. influenzae, and H. pylori suggests that they could provide or have provided some evolutionary advantage to these organisms. AraJ has been proposed to facilitate import of arabinose polymers (31). This appears now less likely, since no member of the MFS has been found to transport a substance larger than 1,000 Da (29), which corresponds to six to seven arabinose monomers. Another possibility is that YdeA, when expressed, and/or AraJ could promote the export of arabinose structural analogues that can be imported but not completely metabolized.
Expression vectors based on the arabinose PBAD promoter are highly versatile and have been extensively used (9). Protein levels can be modulated over a wide range by using plasmids with various ori elements, by altering plasmid copy number, and by changing the extracellular arabinose concentration. YdeA overexpression now offers the means to modulate the kinetics of protein expression. For instance, shut off upon arabinose removal, which is slow in ara strains (9), could be accelerated by activation of ydeA gene expression. The most useful advantage is that the kinetics of induction can now be modulated: the effect of protein accumulation could be studied either in a short time frame (in wild-type cells) or over an extended time period (in cells overexpressing YdeA).
ACKNOWLEDGMENTS
We thank J. Beckwith, L.-M. Guzman, and J. Geiselman for helpful discussions, M. Ehrmann, C. Georgopoulos, and K. Khatib for reading the manuscript, J. Deshusses and M. Ehrmann for help with the arabinose uptake experiments, and R. W. Hogg for a generous gift of [14C]arabinose and the pKKATEB plasmid.
This work was supported by grants from the Swiss National Science Foundation and by the Canton de Genève. S.B. was a fellow of the M.D./Ph.D. program of the University of Geneva Medical School and was supported by fellowships from the Dr. Henri-Dubois-Ferrière Dinu Lipatti and Sir Jules Thorn foundations.
REFERENCES
- 1.Belin D. The use of RNA probes for the analysis of gene expression. Mol Biotechnol. 1997;7:153–163. doi: 10.1007/BF02761751. [DOI] [PubMed] [Google Scholar]
- 2.Belin D, Bost S, Vassalli J D, Strub K. A two-step recognition of signal sequences determines the translocation efficiency of proteins. EMBO J. 1996;15:468–478. [PMC free article] [PubMed] [Google Scholar]
- 3.Belin D, Hedgpeth J, Selzer G B, Epstein R H. Temperature-sensitive mutation in the initiation codon of the rIIB gene of bacteriophage T4. Proc Natl Acad Sci USA. 1979;76:700–704. doi: 10.1073/pnas.76.2.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost S, Belin D. A new genetic selection identifies essential residues in SecG, a component of the Escherichia coli protein export machinery. EMBO J. 1995;14:4412–4421. doi: 10.1002/j.1460-2075.1995.tb00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dittrich W, Betzler M, Schrempf H. An amplifiable and deletable chloramphenicol-resistance determinant of Streptomyces lividans 1326 encodes a putative transmembrane protein. Mol Microbiol. 1991;5:2789–2797. doi: 10.1111/j.1365-2958.1991.tb01987.x. [DOI] [PubMed] [Google Scholar]
- 7.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. . (Erratum, 179:5654.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrmann M, Boyd D, Beckwith J. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc Natl Acad Sci USA. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrickson W, Stoner C, Schleif R. Characterisation of the Escherichia coli araFGH and araJ promoters. J Mol Biol. 1990;215:497–510. doi: 10.1016/S0022-2836(05)80163-9. [DOI] [PubMed] [Google Scholar]
- 11.Henson J M, Kopp B, Kuempel P L. Deletion of 60 kilobase pairs of DNA from the terC region of the chromosome of Escherichia coli. Mol Gen Genet. 1984;193:263–268. doi: 10.1007/BF00330678. [DOI] [PubMed] [Google Scholar]
- 12.Hidaka M, Akiyama M, Horiuchi T. A consensus sequence of the three DNA replication terminus sites on the E. coli chromosome is highly homologous to the terR sites of the R6K plasmid. Cell. 1988;55:467–475. doi: 10.1016/0092-8674(88)90033-5. [DOI] [PubMed] [Google Scholar]
- 13.Higashitani A, Higashitani N, Yasuda S, Horiuchi K. A general and fast method for mapping mutations on the Escherichia coli chromosome. Nucleic Acids Res. 1994;22:2426–2427. doi: 10.1093/nar/22.12.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horazdovsky B F, Hogg R W. High-affinity l-arabinose transport operon. J Mol Biol. 1987;197:27–35. doi: 10.1016/0022-2836(87)90606-1. [DOI] [PubMed] [Google Scholar]
- 15.Horazdovsky B F, Hogg R W. Genetic reconstitution of the high affinity l-arabinose transport system. J Bacteriol. 1989;171:3053–3059. doi: 10.1128/jb.171.6.3053-3059.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacques J-P, Susskind M M. Pseudo-templated transcription by Escherichia coli RNA polymerase at a mutant promoter. Genes Dev. 1990;4:1801–1810. doi: 10.1101/gad.4.10.1801. [DOI] [PubMed] [Google Scholar]
- 17.Jin D J. Slippage synthesis at the galP2 promoter of Escherichia coli and its regulation by UTP concentration and cAMP:cAMP receptor protein. J Biol Chem. 1994;269:17221–17227. [PubMed] [Google Scholar]
- 18.Jin D J. A mutant RNA polymerase reveals a kinetic mechanism for the switch between nonproductive stuttering synthesis and productive initiation during promoter clearance. J Biol Chem. 1996;271:11659–11667. [PubMed] [Google Scholar]
- 19.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;31:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 20.Kolodrubetz D, Schleif R. Regulation of the l-arabinose transport operons in Escherichia coli. J Mol Biol. 1981;151:215–227. doi: 10.1016/0022-2836(81)90512-x. [DOI] [PubMed] [Google Scholar]
- 21.Kolodrubetz D, Schleif R. L-arabinose transport systems in Escherichia coli K-12. J Bacteriol. 1981;148:472–479. doi: 10.1128/jb.148.2.472-479.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosiba B E, Schleif R. Arabinose inducible promoter from Escherichia coli, its cloning from chromosomal DNA, identification as the araFG promoter and sequence. J Mol Biol. 1982;156:53–66. doi: 10.1016/0022-2836(82)90458-2. [DOI] [PubMed] [Google Scholar]
- 23.Maiden M C J, Jones-Mortimer M C, Henderson P J F. The cloning, DNA sequence, and overexpression of the gene araE coding for the arabinose-proton symport in Escherichia coli K12. J Biol Chem. 1988;263:8003–8010. [PubMed] [Google Scholar]
- 24.Marger M D, Saier M H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 25.Michaelis S, Inouye H, Oliver D, Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 27.Nishiyama K, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsen I T, Sliwinski M K, Saier M H., Jr Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J Mol Biol. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 30.Pogliano J, Lynch A S, Belin D, Lin E C, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 31.Reeder T, Schleif R. Mapping, sequence and apparent lack of function of araJ, a gene of the Escherichia coli arabinose regulon. J Bacteriol. 1991;173:7765–7771. doi: 10.1128/jb.173.24.7765-7771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell C B, Thaler D S, Dahlquist F W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schatz P J, Beckwith J. Genetic analysis of protein export in Escherichia coli. Annu Rev Genet. 1990;24:215–248. doi: 10.1146/annurev.ge.24.120190.001243. [DOI] [PubMed] [Google Scholar]
- 34.Schleif R. The l-arabinose operon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1473–1480. [Google Scholar]
- 35.Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–203. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 36.Stoner C, Schleif R. The araE low affinity l-arabinose transport promoter. J Mol Biol. 1983;171:369–381. doi: 10.1016/0022-2836(83)90035-9. [DOI] [PubMed] [Google Scholar]
- 37.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 38.Wickner W, Driessen A J, Hartl F-U. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem. 1991;60:101–124. doi: 10.1146/annurev.bi.60.070191.000533. [DOI] [PubMed] [Google Scholar]