Abstract
Several phenotypic differences observed in Parkinson’s disease (PD) patients have been linked to age at onset (AAO). We endeavoured to find out whether these differences are due to the ageing process itself by using a combined dataset of idiopathic PD (n = 430) and healthy controls (HC; n = 556) excluding carriers of known PD-linked genetic mutations in both groups. We found several significant effects of AAO on motor and non-motor symptoms in PD, but when comparing the effects of age on these symptoms with HC (using age at assessment, AAA), only positive associations of AAA with burden of motor symptoms and cognitive impairment were significantly different between PD vs HC. Furthermore, we explored a potential effect of polygenic risk score (PRS) on clinical phenotype and identified a significant inverse correlation of AAO and PRS in PD. No significant association between PRS and severity of clinical symptoms was found. We conclude that the observed non-motor phenotypic differences in PD based on AAO are largely driven by the ageing process itself and not by a specific profile of neurodegeneration linked to AAO in the idiopathic PD patients.
Subject terms: Parkinson's disease, Neurodegeneration, Dementia
Introduction
Although considered as one disease entity, Parkinson’s disease (PD) displays substantial clinical heterogeneity with various phenotypes that translate into different combinations of both motor and non-motor symptoms. To address this heterogeneity, the age at onset (AAO) has been suggested as a key indicator associated with the clinical profile and progression of PD1–3. Previous studies with cross-sectional design have identified later AAO to be related with a stronger motor as well as non-motor impairment suggesting that late AAO is associated with higher progression rate of motor symptoms and cognitive decline. Conversely, early onset PD has been reported to show a specific disease profile with higher rate of motor complications such as early dyskinesia and dystonia4–6. Furthermore, both prospective7 and retrospective studies with autopsy-proven PD8 have shown similar findings, but given the heterogeneity of the study designs and various cut-offs used for categorising AAO, the reproducibility of the findings is limited. Despite reporting multiple AAO-related phenotypic differences, no study so far has endeavoured to integrate the effect of the physiological ageing process. Therefore, the associations between AAO and severity of PD phenotypes require further analysis.
Apart from AAO, the concept of polygenic risk scores (PRS) in sporadic forms of PD has recently been established to assess the complex genetic architecture of PD beyond known rare familial forms of PD with Mendelian inheritance of mutations in disease-causing genes9. Even though PRS were reported to be significantly negatively correlated with AAO10, potential effects of PRS on the disease severity and the phenotypic profile have not yet been explored in detail.
Previous studies focusing on the role of AAO in PD were limited by (i) not addressing the concomitant effect of the physiological ageing process on the clinical phenotype by modelling age-related effects in a healthy control group, (ii) including relatively small numbers of PD patients from highly specific subgroups (e.g. drug naïve), (iii) using different AAO cut-offs across the studies and (iv) lacking a detailed genetic profiling of the study sample to exclude individuals with monogenic forms and variants presenting a genetic risk factor for developing PD. Therefore, our study addresses these issues by combining a mono-centric idiopathic PD dataset and healthy control group (HC) with detailed genetic data with the aim (i) to investigate the effect of AAO on clinical phenotype in idiopathic PD, (ii) to separate the PD-related ageing effect from the natural ageing effect and finally (iii) to explore the effect of the genetic background reflected by PRS on the disease severity in idiopathic PD.
Results
Effect of AAO on clinical outcomes in PD
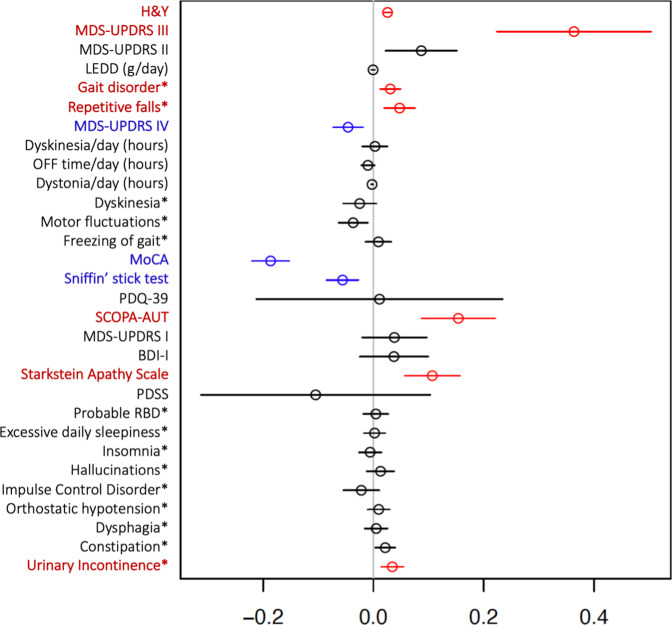
Several traits in PD phenotypic profiles were found in association with AAO. An overview of clinical outcomes, sociodemographic characteristics and comorbidities among participants of the Luxembourg Parkinson’s Study is shown in Tables 1 and 2. As expected, the PD group comprised more males than females (67% vs. 33%) with mean AAO of 61.8 ± 12.0 years and mean disease duration since diagnosis of 5.5 ± 5.5 years. The mean age at assessment (AAA) was 67.3 ± 11.0 years. To investigate the effects of AAO on the clinical outcomes, a multiple regression analysis adjusting for disease duration was performed with results shown in Fig. 1. The overall motor disease severity as reflected by modified H&Y, MDS-UPDRS III, frequency of falls and gait disorder were all significantly positively associated with AAO. With regard to the motor complications of PD, no significant association of AAO was found with total hours of dyskinesia/day, dystonia/day, nor OFF time/day, however, a significant negative association of AAO with the MDS-UPDRS IV total score was identified. Additionally, SCOPA-AUT total score and Starkstein Apathy scale had significant positive associations with AAO indicating that patients with higher AAO experience more non-motor symptoms including urinary incontinence. Cognition as reflected by the MoCA score was significantly negatively associated with AAO showing higher impairment in patients with an older AAO. Similarly, AAO was significantly negatively associated with olfactory dysfunction. All other putative associations were not significantly associated with AAO as shown in Fig. 1.
Table 1.
Overview of sociodemographic characteristics of study dataset including comorbidities and polygenic risk score with p values from Mann–Whitney U test for numerical variables and Fisher’s exact test for binary variables.
| HC n = 556 | PD = 430 | ||||||
|---|---|---|---|---|---|---|---|
| Demographic, PRS and comorbidities | Mean or YES in % | SD or NO/YES | n.a. | Mean or YES in % | SD or NO/YES | n.a. | p value |
| Gender (male)* | 56% | 243/313 | 0 | 67% | 142/288 | 0 | 7.8e−04″ |
| Age at onset (years) | – | – | 556 | 61.84 | 11.99 | 0 | – |
| Age at assessment (years) | 59.61 | 11.78 | 0 | 67.30 | 11.04 | 0 | 6.8e−23″ |
| Disease duration since diagnosis (years) | – | – | 556 | 5.49 | 5.54 | 0 | – |
| Years of education | 14.27 | 3.88 | 5 | 13.09 | 4.10 | 0 | 5.4e−06″ |
| Family history of parkinsonism* | 26% | 408/146 | 2 | 25% | 324/106 | 0 | 5.6e−01 |
| Family history of dementia* | 32% | 373/178 | 5 | 24% | 325/103 | 2 | 5.4e−03′ |
| Polygenic risk score for PD | −0.21 | 0.91 | 6 | 0.16 | 0.94 | 6 | 7.1e−09″ |
| De novo* | – | 0/0 | 556 | 8% | 395/35 | 0 | – |
| Treatment with DBS* | 0% | 556/0 | 0 | 5% | 410/20 | 0 | 4.8e−08″ |
| History or presence of RLS* | 6% | 520/36 | 0 | 9% | 392/38 | 0 | 1.8e−01 |
| Diabetes (type not specified)* | 6% | 523/33 | 0 | 10% | 385/45 | 0 | 1.2e−02′ |
| Arterial hypertension* | 33% | 375/181 | 0 | 44% | 239/191 | 0 | 1.6e−04″ |
| Cardiovascular disease* | 9% | 504/52 | 0 | 21% | 340/90 | 0 | 3.8e−07″ |
| Hypercholesterolemia* | 38% | 347/209 | 0 | 42% | 248/182 | 0 | 1.5e−01 |
| History of stroke* | 3% | 539/17 | 0 | 5% | 410/20 | 0 | 2.4e−01 |
Single and double ticks indicate significance at the 5% level and the Bonferroni-adjusted 5% level respectively. The binary variables are annotated by asterisk.
n.a. corresponds to total number of missing values per variable, PD Parkinson’s disease, PRS Polygenic risk score, HC Healthy controls, DBS Deep brain stimulation, SD Standard deviation.
Table 2.
Overview of dataset with clinical variables in healthy control group (HC) and Parkinson’s disease patients (PD) with p values from Mann–Whitney U test for numerical variables and Fisher’s exact test for binary variables.
| HC n = 556 | PD = 430 | ||||||
|---|---|---|---|---|---|---|---|
| Clinical symptoms and scales | Mean or YES in % | SD or NO/YES | n.a. | Mean or YES in % | SD or NO/YES | n.a. | p value |
| H&Y | 0.00 | 0.00 | 2 | 2.24 | 0.81 | 2 | 1.5e−196″ |
| MDS-UPDRS III | 3.45 | 4.76 | 6 | 34.70 | 17.02 | 9 | 3.1e−150″ |
| MDS-UPDRS II | 1.21 | 2.37 | 6 | 11.69 | 8.32 | 8 | 6.2e−126″ |
| LEDD (g/day) | 0.0035 | 0.037 | 0 | 0.53 | 0.42 | 0 | 7.5e−160″ |
| Gait disorder* | 2% | 546/10 | 0 | 57% | 185/245 | 0 | 6.9e−97″ |
| Repetitive falls* | 1% | 552/4 | 0 | 18% | 351/79 | 0 | 1.1e−25″ |
| MDS-UPDRS IV | 0.00 | 0.00 | 4 | 1.88 | 3.52 | 5 | 1.4e−43″ |
| Dyskinesia/day (hours) | 0.00 | 0.00 | 0 | 0.69 | 2.73 | 1 | 1.2e−21″ |
| OFF time/day (hours) | 0.00 | 0.00 | 0 | 0.53 | 1.44 | 2 | 3.3e−34″ |
| Dystonia/day (hours) | 0.00 | 0.00 | 0 | 0.048 | 0.22 | 2 | 6.8e−12″ |
| Dyskinesia* | 0% | 556/0 | 0 | 13% | 375/55 | 0 | 1.9e−21″ |
| Motor fluctuations* | 0% | 556/0 | 0 | 17% | 357/73 | 0 | 1.1e−28″ |
| Freezing of gait* | 0% | 556/0 | 0 | 23% | 331/99 | 0 | 1.6e−39″ |
| MoCA | 27.03 | 2.55 | 3 | 24.28 | 4.41 | 8 | 4.4e−28″ |
| Sniffin’ stick test | 12.86 | 2.39 | 2 | 8.03 | 3.41 | 13 | 1.6e−94″ |
| PDQ-39 | 10.31 | 13.20 | 16 | 39.69 | 26.31 | 39 | 1.3e−80″ |
| SCOPA-AUT | 7.34 | 5.81 | 16 | 14.82 | 8.02 | 21 | 6.9e−53″ |
| MDS-UPDRS I | 4.58 | 4.42 | 9 | 10.47 | 7.04 | 10 | 9.0e−51″ |
| BDI-I | 5.29 | 5.03 | 15 | 9.97 | 7.11 | 23 | 2.3e−30″ |
| Starkstein Apathy Scale | 9.41 | 4.71 | 16 | 13.93 | 5.70 | 26 | 8.2e−35″ |
| PDSS | 122.81 | 19.61 | 13 | 105.17 | 23.85 | 28 | 2.3e−34″ |
| Probable RBD* | 8% | 496/42 | 18 | 24% | 305/95 | 30 | 1.2e−11″ |
| Excessive daily sleepiness* | 3% | 541/15 | 0 | 30% | 299/131 | 0 | 2.4e−36″ |
| Insomnia* | 8% | 514/42 | 0 | 24% | 327/103 | 0 | 9.1e−13″ |
| Hallucinations* | 0% | 554/2 | 0 | 17% | 357/73 | 0 | 1.2e−25″ |
| Impulse Control Disorder* | 0% | 555/1 | 0 | 9% | 392/38 | 0 | 1.8e−13″ |
| Orthostatic hypotension* | 6% | 525/31 | 0 | 27% | 312/118 | 0 | 8.2e−22″ |
| Dysphagia* | 1% | 552/4 | 0 | 25% | 323/107 | 0 | 5.6e−37″ |
| Constipation* | 5% | 528/28 | 0 | 42% | 250/180 | 0 | 3.6e−47″ |
| Urinary Incontinence* | 5% | 530/26 | 0 | 32% | 293/137 | 0 | 3.6e−31″ |
Single and double ticks indicate significance at the 5% level and the Bonferroni-adjusted 5 % level respectively.
Clinical symptoms and scales are described in Supplementary Material. The binary variables are annotated by asterisk.
n.a. (not acquired) corresponds to total number of individuals with missing value, SD Standard deviation.
Fig. 1. Forrest plot with estimated coefficients and corresponding confidence intervals (±1.96 × standard error) for AAO, from linear/logistic regression of numerical/binary outcome on disease duration and AAO.
The colour blue indicates significant negative effects of AAO on the clinical outcome, and the colour red indicates significant positive effects at the Bonferroni-adjusted 5% level. The binary variables are annotated by asterisk. Clinical symptoms and scales are described in Supplementary Material.
Analysing the difference in ageing effect in PD vs HC
When investigating the effects of AAA and AAO on the clinical phenotypes of PD, all associations were found to be comparable in both models (cf. Table 3). The reason is the strong correlation between AAA and AAO (statistically significant Kendall’s tau ρ = 0.73, see Supplementary Fig. 1). To investigate an effect of physiological ageing on the PD phenotypes, we also included the HC group into the regression models. When investigating the ageing-associated effects in PD, we determined a significant positive association in PD between AAA and H&Y, MDS-UPDRS III, frequency of falls and urine incontinence, SCOPA-AUT, Starkstein Apathy Scale as well as significant negative association between AAA and MoCA and Sniffin’ Stick test (cf. Table 3). Similarly in the HC group, we found a significant positive association between AAA and MDS-UPDRS III, SCOPA-AUT, Starkstein Apathy Scale, frequency of urine incontinence and gait disorder as well as significant negative association between AAA and MoCA and Sniffin’ Stick test as demonstrated in Table 4. Surprisingly, after comparing the ageing effect between PD vs HC (i.e. comparing effect of AAA on the clinical variables; see Table 5, column AAA:status), the only significant differences between PD and HC were found for H&Y, MDS-UPDRS III, MDS-UPDRS IV and MoCA indicating that the concomitant ageing process might be the main determinant of the non-motor PD phenotypic differences when studying the isolated effect of age in PD.
Table 3.
Multiple regression of clinical outcomes on age at onset (AAO), age at assessment (AAA) and disease duration for Parkinson’s disease group.
| Clinical symptoms and scales | Intercept | Disease duration | AAA | Intercept | Disease duration | AAO | Intercept | AAA | AAO |
|---|---|---|---|---|---|---|---|---|---|
| H&Y | 0.23 | 0.05″ | 0.03″ | 0.23 | 0.07″ | 0.03″ | 0.23 | 0.07″ | −0.05″ |
| MDS-UPDRS III | 5.95 | 0.76″ | 0.36″ | 6.04 | 1.13″ | 0.36″ | 5.98 | 1.13″ | −0.76″ |
| MDS-UPDRS II | 2.38 | 0.63″ | 0.09′ | 2.39 | 0.72″ | 0.09′ | 2.41 | 0.72″ | −0.63″ |
| LEDD (g/day) | 0.37 | 0.04″ | 0.00 | 0.37 | 0.03″ | 0.00 | 0.38 | 0.03″ | −0.04″ |
| Gait disorder* | −2.23 | 0.08″ | 0.03′ | −2.23 | 0.12″ | 0.03′ | −2.22 | 0.12″ | −0.08″ |
| Repetitive falls* | −5.79 | 0.14″ | 0.05″ | −5.74 | 0.19″ | 0.05″ | −5.80 | 0.19″ | −0.15″ |
| MDS-UPDRS IV | 3.43 | 0.28″ | −0.05′ | 3.46 | 0.24″ | −0.05′ | 3.43 | 0.24″ | −0.28″ |
| Dyskinesia/day (hours) | −0.16 | 0.12″ | 0.00 | −0.16 | 0.12″ | 0.00 | −0.15 | 0.12″ | −0.12″ |
| OFF time/day (hours) | 0.87 | 0.06″ | −0.01 | 0.87 | 0.05″ | −0.01 | 0.87 | 0.05″ | −0.06″ |
| Dystonia/day (hours) | 0.15 | 0.01″ | 0.00′ | 0.16 | 0.01″ | 0.00′ | 0.15 | 0.01″ | −0.01″ |
| Dyskinesia* | −1.57 | 0.18″ | −0.02 | −1.55 | 0.15″ | −0.02 | −1.56 | 0.15″ | −0.18″ |
| Motor fluctuations* | −0.35 | 0.18″ | −0.04′ | −0.39 | 0.14″ | −0.04′ | −0.33 | 0.14″ | −0.18″ |
| Freezing of gait* | −2.82 | 0.16″ | 0.01 | −2.87 | 0.17″ | 0.01 | −2.79 | 0.17″ | −0.16″ |
| MoCA | 37.04 | −0.05 | −0.19″ | 37.10 | −0.24″ | −0.19″ | 37.03 | −0.23″ | 0.05 |
| Sniffin’ stick test | 12.40 | −0.11″ | −0.06″ | 12.41 | −0.17″ | −0.06″ | 12.40 | −0.17″ | 0.11″ |
| PDQ-39 | 29.72 | 1.78″ | 0.01 | 29.46 | 1.79″ | 0.01 | 29.87 | 1.77″ | −1.76″ |
| SCOPA-AUT | 2.20 | 0.43″ | 0.15″ | 2.16 | 0.58″ | 0.15″ | 2.23 | 0.58″ | −0.42″ |
| MDS-UPDRS I | 6.05 | 0.35″ | 0.04 | 5.96 | 0.39″ | 0.04 | 6.08 | 0.38″ | −0.35″ |
| BDI-I | 6.14 | 0.24″ | 0.04 | 6.19 | 0.28″ | 0.04 | 6.15 | 0.28″ | −0.24″ |
| Starkstein Apathy Scale | 6.75 | −0.01 | 0.11″ | 6.81 | 0.10 | 0.11″ | 6.74 | 0.10 | 0.00 |
| PDSS | 117.20 | −0.98″ | −0.10 | 117.45 | −1.08″ | −0.10 | 117.11 | −1.06″ | 0.96″ |
| Probable RBD* | −2.13 | 0.11″ | 0.00 | −2.14 | 0.12″ | 0.00 | −2.12 | 0.11″ | −0.11″ |
| Excessive daily sleepiness* | −1.31 | 0.07″ | 0.00 | −1.37 | 0.07″ | 0.00 | −1.29 | 0.07′ | −0.06″ |
| Insomnia* | −1.00 | 0.04′ | −0.01 | −1.01 | 0.04 | −0.01 | −1.00 | 0.04 | −0.04′ |
| Hallucinations* | −3.08 | 0.09″ | 0.01 | −3.06 | 0.10″ | 0.01 | −3.08 | 0.11″ | −0.09″ |
| Impulse Control Disorder* | −1.62 | 0.11″ | −0.02 | −1.64 | 0.09′ | −0.02 | −1.60 | 0.09′ | −0.11″ |
| Orthostatic hypotension* | −1.88 | 0.05′ | 0.01 | −1.92 | 0.06′ | 0.01 | −1.87 | 0.06′ | −0.05′ |
| Dysphagia* | −1.77 | 0.06′ | 0.00 | −1.80 | 0.06′ | 0.01 | −1.76 | 0.06′ | −0.06′ |
| Constipation* | −2.14 | 0.07″ | 0.02′ | −2.19 | 0.10″ | 0.02′ | −2.13 | 0.09″ | −0.07″ |
| Urinary Incontinence* | −3.40 | 0.04′ | 0.04″ | −3.36 | 0.08″ | 0.03″ | −3.41 | 0.08″ | −0.05′ |
Regression coefficients for different outcomes (rows) from three equivalent models with each two out of three features (columns). Single and double ticks indicate significance at the 5% level and the Bonferroni-adjusted 5% level respectively. The bold indicates significant effect where minus value indicates negative significant effect and positive value positive significant effect respectively. The binary variables are annotated by asterisk. Clinical symptoms and scales are described in Supplementary Material.
Table 4.
Simple regression of clinical outcomes with healthy controls.
| Clinical symptoms and scales | Intercept | AAA |
|---|---|---|
| H&Y | 0.00 | 0.00 |
| MDS-UPDRS III | −3.70 | 0.12″ |
| MDS-UPDRS II | −0.21 | 0.02′ |
| LEDD (g/day) | −0.02 | 0.00′ |
| Gait disorder* | −12.56 | 0.13″ |
| Repetitive falls* | −5.14 | 0.00 |
| MDS-UPDRS IV | 0.00 | 0.00 |
| Dyskinesia/day (hours) | 0.00 | 0.00 |
| OFF time/day (hours) | 0.00 | 0.00 |
| Dystonia/day (hours) | 0.00 | 0.00 |
| Dyskinesia* | −26.57 | 0.00 |
| Motor fluctuations* | −26.57 | 0.00 |
| Freezing of gait* | −26.57 | 0.00 |
| MoCA | 29.84 | −0.05″ |
| Sniffin’ stick test | 15.44 | −0.04″ |
| PDQ-39 | 10.68 | −0.01 |
| SCOPA-AUT | 2.53 | 0.08″ |
| MDS-UPDRS I | 3.12 | 0.02 |
| BDI-I | 4.06 | 0.02 |
| Starkstein Apathy Scale | 5.11 | 0.07″ |
| PDSS | 130.37 | −0.13 |
| Probable RBD* | −1.58 | −0.02 |
| Excessive daily sleepiness* | −6.53 | 0.05 |
| Insomnia* | −2.00 | −0.01 |
| Hallucinations* | −3.31 | −0.04 |
| Impulse Control Disorder* | −4.32 | −0.04 |
| Orthostatic hypotension* | −3.92 | 0.02 |
| Dysphagia* | −7.28 | 0.04 |
| Constipation* | −2.37 | −0.01 |
| Urinary Incontinence* | −8.43 | 0.08″ |
Regression coefficients are shown from linear regression of numerical outcome and from logistic regression of binary outcome on age at assessment (AAA). Single and double ticks indicate significance at the 5% level and the Bonferroni adjusted 5% level, the bold indicates significant effect where minus value indicates negative significant effect and positive value positive significant effect respectively. The binary variables are annotated by asterisk. Clinical symptoms and scales are described in Supplementary Material.
Table 5.
Multiple regression model with PD and HC investigating the difference in effect of ageing in HC (AAA) and in PD (AAA:status) adjusted for disease duration.
| Clinical symptoms and scales | Intercept | AAA | Status | Disease duration | AAA:Status |
|---|---|---|---|---|---|
| H&Y | 0.00 | 0.00 | 0.23 | 0.05″ | 0.03″ |
| MDS-UPDRS III | −3.70 | 0.12′ | 9.65 | 0.76″ | 0.24″ |
| MDS-UPDRS II | −0.21 | 0.02 | 2.59 | 0.63″ | 0.06′ |
| LEDD (g/day) | −0.02 | 0.00 | 0.39 | 0.04″ | 0.00 |
| Gait disorder* | −12.56 | 0.13′ | 10.34 | 0.08″ | −0.10′ |
| Repetitive falls* | −5.14 | 0.00 | −0.66 | 0.14″ | 0.04 |
| MDS-UPDRS IV | 0.00 | 0.00 | 3.43 | 0.28″ | −0.05″ |
| Dyskinesia/day (hours) | 0.00 | 0.00 | −0.16 | 0.12″ | 0.00 |
| OFF time/day (hours) | 0.00 | 0.00 | 0.87 | 0.06″ | −0.01 |
| Dystonia/day (hours) | 0.00 | 0.00 | 0.15 | 0.01″ | 0.00′ |
| Dyskinesia* | −20.57 | 0.00 | 18.99 | 0.18″ | −0.02 |
| Motor fluctuations* | −20.57 | 0.00 | 20.22 | 0.18″ | −0.04 |
| Freezing of gait* | −20.57 | 0.00 | 17.75 | 0.16″ | 0.01 |
| MoCA | 29.84 | −0.05″ | 7.20 | −0.05 | −0.14″ |
| Sniffin’ stick test | 15.44 | −0.04″ | −3.03 | −0.11″ | −0.01 |
| PDQ-39 | 10.68 | −0.01 | 19.04 | 1.78″ | 0.01 |
| SCOPA-AUT | 2.53 | 0.08′ | −0.33 | 0.43″ | 0.07 |
| MDS-UPDRS I | 3.12 | 0.02 | 2.94 | 0.35″ | 0.01 |
| BDI-I | 4.06 | 0.02 | 2.09 | 0.24″ | 0.02 |
| Starkstein Apathy Scale | 5.11 | 0.07″ | 1.64 | −0.01 | 0.04 |
| PDSS | 130.37 | −0.13 | −13.17 | −0.98″ | 0.03 |
| Probable RBD* | −1.58 | −0.02 | −0.54 | 0.11″ | 0.02 |
| Excessive daily sleepiness* | −6.53 | 0.05 | 5.22 | 0.07″ | −0.05 |
| Insomnia* | −2.00 | −0.01 | 0.99 | 0.04′ | 0.00 |
| Hallucinations* | −3.31 | −0.04 | 0.23 | 0.09″ | 0.05 |
| Impulse Control Disorder* | −4.32 | −0.04 | 2.70 | 0.11″ | 0.01 |
| Orthostatic hypotension* | −3.92 | 0.02 | 2.04 | 0.05′ | −0.01 |
| Dysphagia* | −7.28 | 0.04 | 5.51 | 0.06′ | −0.03 |
| Constipation* | −2.37 | −0.01 | 0.23 | 0.07″ | 0.03 |
| Urinary Incontinence* | −8.43 | 0.08″ | 5.03 | 0.04′ | −0.05′ |
Regression coefficients are shown for different outcomes (rows). Status takes the value 0 for HC and 1 for PD, the AAA:status is the interaction term of AAA and being PD (status = 1). Single and double ticks indicate significance at the 5% level and the Bonferroni-adjusted 5% level respectively, the bold indicates significant effect where minus value indicates negative significant effect and positive value positive significant effect respectively. The column AAA:status indicates whether the effect of AAA on clinical outcomes differs between PD and HC. The binary variables are annotated by asterisk. Clinical symptoms and scales are described in Supplementary Material.
Correlation between AAO and PRS and its effect on severity of the PD phenotype
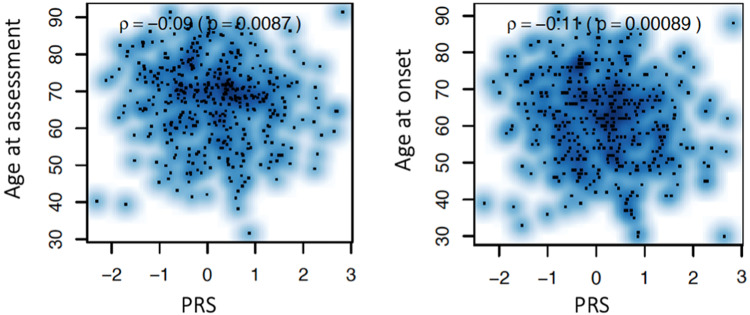
Using a polygenic risk score defined by the imputed genotypic data from the Luxembourg Parkinson’s Study and the summary statistics of 90 single nucleotide polymorphisms (SNP) that were previously identified to be genome-wide significantly associated with PD risk, we identified a significant negative correlation between PRS and AAO as shown in Fig. 2. However, neither Kendall’s tau correlation test for continuous variables nor Mann–Whitney U test for binary variables estimating the effect of PRS on clinical outcomes nor multiple regression models including PRS adjusted for AAA and disease duration showed effects of PRS on the severity of the clinical phenotype as demonstrated in Tables 6 and 7 respectively.
Fig. 2. Pairwise association between age at onset (AAO), age at assessment (AAA) (y-axis) and polygenic risk score (PRS) (x-axis) with Kendall correlation coefficient.
Significant inverse association was determined between AAO and PRS and AAA and PRS indicating the younger the AAO of PD, the higher cummulative burden of small effect size variants (represented by PRS).
Table 6.
Kendall correlation coefficient between clinical outcome (row) and polygenic risk score (PRS) for healthy controls (HC) (left) and Parkinson’s disease patients (PD) (right), with annotation by bold indicating significant effect where minus value indicates negative significant effect and positive value positive significant effect respectively (Kendall correlation test).
| Clinical symptoms and scales | HC | PD |
|---|---|---|
| H&Y | 0.0272 | 0.0272 |
| MDS-UPDRS III | −0.0088 | −0.0341 |
| MDS-UPDRS II | −0.0242 | 0.0058 |
| LEDD (g/day) | 0.0091 | −0.0147 |
| Gait disorder* | 0.4070 | −0.0177 |
| Repetitive falls* | 0.6209 | 0.1690 |
| MDS-UPDRS IV | 0.0625 | 0.0625 |
| Dyskinesia/day (hours) | 0.0576 | 0.0576 |
| OFF time/day (hours) | 0.0100 | 0.0100 |
| Dystonia/day (hours) | 0.0491 | 0.0491 |
| Dyskinesia* | – | 0.1426 |
| Motor fluctuations* | – | 0.0864 |
| Freezing of gait* | – | 0.0084 |
| MoCA | 0.0542 | −0.0493′ |
| Sniffin’ stick test | 0.1012 | 0.0576 |
| PDQ-39 | −0.0519 | 0.0386 |
| SCOPA-AUT | −0.0113 | 0.0150 |
| MDS-UPDRS I | −0.0556 | 0.0105 |
| BDI-I | −0.0155 | 0.0290 |
| Starkstein Apathy Scale | −0.0425 | −0.0068 |
| PDSS | −0.0128 | −0.0332 |
| Probable RBD* | −0.1142 | −0.0703 |
| Excessive daily sleepiness* | 0.1638 | −0.1261 |
| Insomnia* | −0.0356 | 0.1387 |
| Hallucinations* | −0.9806 | −0.1225 |
| Impulse Control Disorder* | −2.1177 | −0.0872 |
| Orthostatic hypotension* | 0.0839 | −0.1331 |
| Dysphagia* | −0.6401 | −0.2650′ |
| Constipation* | 0.2701 | −0.0740 |
| Urinary Incontinence* | 0.1011 | 0.0434 |
Single and double ticks indicate significance at the 5% level and the Bonferroni-adjusted 5% level. The binary variables are annotated by asterisk. Clinical symptoms and scales are described in Supplementary Material.
Table 7.
Multiple regression model with coefficients shown from linear regression of numerical outcome and from logistic regression of binary outcome on disease duration, age at assessment (AAA) and polygenic risk score (PRS) in PD group.
| Clinical symptoms and scales | Intercept | Disease duration | AAA | PRS |
|---|---|---|---|---|
| H&Y | 0.17 | 0.05″ | 0.03″ | −0.02 |
| MDS-UPDRS III | 4.44 | 0.76″ | 0.39″ | −0.57 |
| MDS-UPDRS II | 1.81 | 0.63″ | 0.10′ | −0.47 |
| LEDD (g/day) | 0.32 | 0.04″ | 0.00 | 0.01 |
| Gait disorder* | −2.29 | 0.09″ | 0.03′ | −0.04 |
| Repetitive falls* | −5.91 | 0.14″ | 0.05′ | 0.15 |
| MDS-UPDRS IV | 3.37 | 0.28″ | −0.04′ | 0.03 |
| Dyskinesia/day (hours) | −0.36 | 0.12″ | 0.01 | 0.04 |
| OFF time/day (hours) | 0.87 | 0.06″ | −0.01 | −0.04 |
| Dystonia/day (hours) | 0.15 | 0.01″ | 0.00′ | 0.01 |
| Dyskinesia* | −1.62 | 0.18″ | −0.02 | −0.05 |
| Motor fluctuations* | −0.12 | 0.18″ | −0.04′ | −0.12 |
| Freezing of gait* | −2.66 | 0.17″ | 0.01 | −0.13 |
| MoCA | 37.25 | −0.07′ | −0.19″ | 0.40′ |
| Sniffin’ stick test | 12.64 | −0.11″ | −0.06″ | −0.22 |
| PDQ-39 | 30.96 | 1.86″ | −0.01 | −2.98′ |
| SCOPA-AUT | 2.74 | 0.47″ | 0.15″ | −0.98′ |
| MDS-UPDRS I | 6.29 | 0.36″ | 0.03 | −0.84′ |
| BDI-I | 6.81 | 0.27″ | 0.03 | −0.76′ |
| Starkstein Apathy Scale | 6.99 | 0.00 | 0.10″ | −0.18 |
| PDSS | 116.69 | −1.00″ | −0.10 | 0.12 |
| Probable RBD* | −2.07 | 0.12″ | 0.00 | −0.21 |
| Excessive daily sleepiness* | −1.23 | 0.07″ | 0.00 | −0.20 |
| Insomnia* | −1.04 | 0.04′ | −0.01 | 0.11 |
| Hallucinations* | −2.95 | 0.10″ | 0.01 | −0.23 |
| Impulse Control Disorder* | −1.37 | 0.11″ | −0.03 | −0.25 |
| Orthostatic hypotension* | −1.64 | 0.06′ | 0.01 | −0.20 |
| Dysphagia* | −1.77 | 0.07″ | 0.00 | −0.37′ |
| Constipation* | −2.07 | 0.08″ | 0.02′ | −0.13 |
| Urinary Incontinence* | −3.67 | 0.05′ | 0.04″ | 0.06 |
Single and double ticks indicate significance at the 5% level and the Bonferroni adjusted 5% level, the bold indicates significant effect where minus value indicates negative significant effect and positive value positive significant effect respectively. The binary variables are annotated by asterisk. Clinical symptoms and scales are described in Supplementary Material.
Discussion
The presented cross-sectional analysis of PD patients and HC at the baseline clinical visit uses data from one of the largest ongoing observational studies, focusing on PD with demographic and clinical parameters corresponding closely to other recently published large PD datasets11–13. In our study, we have identified several significant associations of different PD-associated motor and non-motor symptoms with AAO using a comprehensive set of clinical assessments. This is in line with previous cross-sectional, retrospective and prospective studies suggesting that later onset PD is associated with a more rapid progression rate of motor symptoms4,11,14,15. Conversely, comparing to the Cardiff community-based PD longitudinal cohort16 and the longitudinal study at the Movement Disorders Clinic Saskatchewan4, both demonstrating higher frequency of dyskinesia, motor fluctuations and dystonia in the younger onset groups vs. older onset groups, we could not identify such associations with AAO. Only an overall burden of motor complications reflected by MDS-UPDRS IV score was significantly negatively associated with AAO in our study. The significant positive association of olfactory dysfunction and significant negative association of cognitive performance with AAO observed in our study correlate with previous findings17,18 and in terms of cognitive impairment it might point to a decreased ability of senescent brain to cope with the pathological neurodegenerative process known as cognitive resilience19. Additionally, another large multi-centric study using the Quebec Parkinson Network (QPN) dataset of over 1000 PD individuals showed comparable results with a positive association between late-onset PD and higher motor burden reflected by H&Y, higher cognitive decline and higher frequency of falls, but differed on significantly higher frequency of constipation and hallucinations late-onset PD (defined as AAO > 50 years) compared to early onset PD11. However, most scales applied in QPN differ from our study and different categorical approaches were used in QPN both for AAO and disease duration, influencing the comparability of results. To summarise our results, the earlier AAO, patients experience a lower level of motor impairment, lower cognitive impairment and less global autonomic dysfunction, apathy and olfactory deficit, but present with more motor complications even after adjusting for disease duration as a main determinant of disease severity.
These phenotypic differences observed in PD based on different AAO were previously not clearly separated from the physiological ageing process and challenged the concept that phenotypic differences are related specifically to the age at which the disease first manifests. This intriguing aspect evolves from the inherent close correlation between the main co-variates (AAA, AAO and disease duration) and thus raises a major methodological concern in most of the cross-sectional studies when aiming at determining the effect of all three co-variates on the clinical outcomes in a single model as discussed by Johnson et al. 200220. Therefore, we tried to disentangle the effect of ageing on the clinical phenotype in the cross-sectional setting by determining the ageing effect in individuals with and without PD. Surprisingly, the effect of ageing (AAA) on clinical outcomes in PD vs HC differed significantly only in motor disease severity (H&Y, MDS-UPDRS III), motor complications (MDS-UPDRS IV) and cognitive performance. These results suggest that the majority of the observed significant non-motor phenotypic differences in PD should be attributed rather to the physiological ageing process itself than age-specific dynamics of PD.
When considering the effect and role of AAO and age in classification of the respective PD phenotypes, potential underlying genetic determinants need to be considered. It is well known that rare disease-causing mutations in monogenic PD (e.g. in PARKIN, PINK1, SNCA or GBA21–23) have an effect on both AAO and PD phenotype. However, until now only few studies have explored the cumulative effects of common genetic variants with small effect sizes (as defined by PRS) on the clinical phenotype24. Here our results are in line with several recent studies observing no significant association between PRS and cognitive decline, severity of motor symptoms25 or ICD26 in contrast to other longitudinal prospective study27. It is worth noting that our statistical models included individuals without any known PD causing monogenic mutation or genetic risk variant (i.e. PD-associated variants in the GBA gene). Nevertheless, the significance of the PRS effect on clinical outcomes did not change in the models including PD-associated mutation or genetic variant carriers. Together with the significant negative correlation between AAO and PRS (cf. Fig. 2), our findings suggest that PRS may increase the risk to develop PD but might not have an effect on the severity of the disease phenotype. This observation is in favour of the hypothesis that initiation of the disease on one hand and the disease progression rate on the other might be driven by distinct factors.
Besides the mentioned strengths of our study design, several limitations need to be considered. First, the cross-sectional design does not allow for the identification of causal relations between AAO and clinical phenotypes. Second, we cannot consider the Luxembourg Parkinson’s Study as community-based by design, although some clinical indicators (such as mean AAO and male-to-female ratio) correspond closely to several community-based studies28–31. Third, we observe a relatively high frequency of positive family history of parkinsonism in the HC group (26% vs. 25% in PD) as well as high frequency of a family history of dementia in HC (32% vs 24%). We assume that there are two principal reasons why we observe increased frequencies of neurodegenerative diseases in HC group: (i) HC with personal experience with parkinsonism and/or dementia in their family are more aware to support research and (ii) family members of study participants are more inclined to participate in the study. To address these points and eliminate a potential bias, we excluded 1st, 2nd and 3rd degree relatives from our statistical models.
In summary, our study sought to overcome limitations identified in previous studies on the role of AAO in PD by (i) including substantially higher number of PD patients and HCs in the model accounting for the independent effect of ageing, (ii) our study being based on monocentric data collection and including PD patients of all disease stages regardless of the cognitive status, (iii) investigating an idiopathic dataset of PD and PD-related mutation free HC, (iv) refuting the categorisation bias by a priori arbitrary AAO grouping, and finally (v) exploring the effect of PRS on severity of the PD phenotype in a large genotyped sample.
Methods
Study population
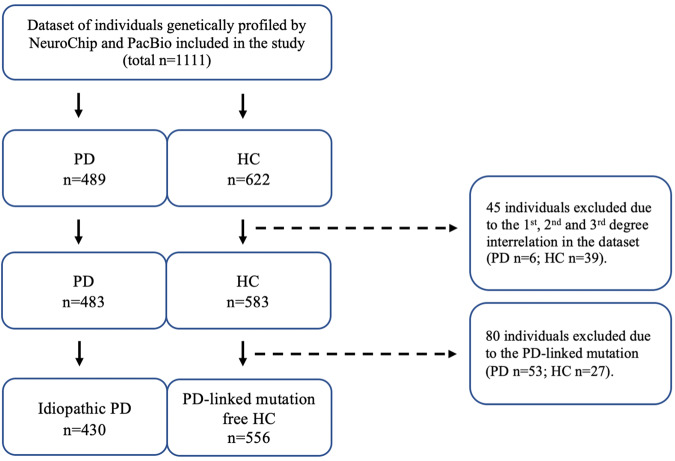
All subjects were recruited from March 2015 until 10th December 2020 in the frame of the nation-wide monocentric observational longitudinal Luxembourg Parkinson’s Study. The diagnosis of PD was based on UKPDSBB diagnostic criteria32. The initial visit dataset of 430 PD patients and 556 HC genetically screened by both NeuroChip and PacBio were analysed after exclusion of 6 PD and 39 HC individuals for 1st, 2nd and 3rd degree relationships and after exclusion of 53 PD carriers and 27 HC carriers of pathogenic PD-associated variants. The overall study design, inclusion and exclusion workflow are illustrated in Fig. 3.
Fig. 3. Description of the study design and study dataset.
PD individuals with Parkinson’s disease, HC healthy control.
All participants taking part in Luxembourg Parkinson’s Study agreed and signed a written informed consent. The study has been approved by the National Ethics Board (CNER Ref: 201407/13). The patients with PD were included regardless of the disease duration, cognitive status, age or disease stage. The HC were partially recruited from the pool of independent observational studies in Luxembourg (ORISCAV-LUX study; EHES-LUX) or were recruited from Luxembourg or the surrounding area of Greater Region based on individual interest not meeting any of the exclusion criteria (presence of a neurodegenerative disorder, active cancer; age under 18 and pregnant women)33.
Clinical assessment and data
A description of the design of the Luxembourg Parkinson’s Study was previously published33. Sociodemographic characteristics and clinical outcomes validated for PD were chosen from the basic clinical assessment battery and listed in Tables 1 and 2. Validated self-administered questionnaires and scales for PD were used. All patients have been evaluated in medication ON state and where applicable, in deep brain stimulation ON state. AAO is defined as age at diagnosis of PD. The clinical symptoms as scales are defined in detail in the Supplementary material.
Missing data statement
The absolute number of missing data per variable are shown in Tables 1 and 2. Given the low proportions of missing values in the outcome variables and 0% of missing values in the co-variates (AAA, AAO and disease duration), we used a pairwise deletion for all statistical models.
Genotyping and quality-control analyses
DNA samples were genotyped using the NeuroChip array (v.1.0 and v1.1; Illumina, San Diego, CA) that was specifically designed to integrate rare and common neurodegenerative disease-related variants34. Quality-control (QC) analysis was performed as follows: samples with call rates < 95% and whose genetically determined sex deviated from reported sex in clinical data were excluded from the analysis, and the filtered variants were checked for cryptic relatedness and excess of heterozygosity. Samples exhibiting excess heterozygosity (F statistic > 0.2) and first-degree relatedness were excluded. Once sample QC was completed, SNPs with Hardy−Weinberg equilibrium P value < 1E−6, and missingness rates >5% were excluded. All samples except for twelve from all individuals entering the analysis after exclusion of the 1st, 2nd and 3rd degree relatives and presence of PD-linked mutation and genetic risk factors passed the QC (424 PD and 550 HC). The data were then imputed using the Haplotype Reference Consortium r1.1 2016 and the Michigan Imputation Server and filtered for imputation quality (RSQ > 0.8)35. Genetic analysis and QC was done using PLINK v1.9. Additionally, all samples underwent targeted sequencing of the GBA locus using single-molecule sequencing on a Sequel II sequencer from Pacific BioScience36. Variants were called with DeepVariant 1.037. PD causing rare variants were defined by the ClinVar classification ‘pathogenic/likely-pathogenic’. All PD causing variants (listed in Supplementary material) identified by any method were Sanger validated and all samples with a validated PD causing variant were excluded from further analysis.
Polygenic risk score (PRS)
We generated PRSs with PRSice-2 under default settings. PRSs for each individual were calculated using the imputed genotype data from Luxembourg Parkinson’s Study as a target sample. The base GWAS data used to determine PRS for PD was the summary statistics of the 90 SNPs that were previously found to be genome-wide significantly associated with PD risk38. The criteria for linkage disequilibrium (LD) clumping of SNPs were pairwise LD r2 < 0.1 within the 250 kb window. Briefly, PRSs were calculated by summing the weighted effects of GWAS PD risk genetic variants present in the target samples, with a possible proxy of R2 > 0.9, meeting p value thresholds ranging from 5e−08 to 0.5. The values of PRS were Z-normalised.
Statistical analysis
Firstly, we performed an intergroup comparison (PD vs HC) of sociodemographic and clinical characteristics as well as polygenic risk score and comorbidities with the Mann−Whitney U test for numerical variables and Fisher’s exact test for binary variables (Tables 1 and 2). Secondly, we used multiple regression models (linear and logistic) to identify effects of AAO (as a numerical variable) on numerical or binary clinical outcomes accounting for disease duration (Fig. 1). Subsequently, we performed a multiple regression model for both HC and PD (Table 5) to examine whether the effect of ageing (AAA) on clinical outcomes differs between HC and PD adjusted for disease duration. For this, we included the main effects of the continuous variable AAA and the binary variable status (HC: status = 0, PD: status = 1), their interaction effect (HC: status*AAA = 0, PD: status*AAA > 0), and the main effect of the continuous variable disease duration (HC: duration = 0, PD: duration > 0). To investigate the role of PRS in PD, a pairwise association analysis with Kendall’s tau correlation test between PRS and AAO and AAA was performed (Fig. 2). Furthermore, we performed a Kendall correlation test between PRS and clinical outcome for PD and HC respectively (Table 6). As a last step, we employed a multiple regression model including PRS adjusting for AAA and disease duration, to investigate the effect of PRS on the clinical phenotype in PD (Table 7). At all instances, the significance at the 5% level and the Bonferroni-adjusted 5% level was set.
Supplementary information
Acknowledgements
We would like to give special thanks to all participants in the Luxembourg Parkinson’s Study. Furthermore, we acknowledge the joint effort of the NCER-PD consortium members generally contributing to the Luxembourg Parkinson’s Study as listed below. The work presented here was supported within the framework of the National Centre of Excellence in Research on Parkinson’s Disease (NCER-PD) funded by the Luxembourg National Research Fund (FNR/NCER13/BM/11264123), the PEARL program (FNR/P13/6682797 to R.K.), MotaSYN (12719684 to R.K.), MAMaSyn (to R.K.), MiRisk‐PD (C17/BM/11676395 to R.K., E.G., P.M.), the FNR/DFG Core INTER (ProtectMove, FNR11250962 to P.M.), and the PARK-QC DTU (PRIDE17/12244779/PARK-QC to R.K. and S.P.), FNR PD-Strat (INTER/11651464 to EG), DIGIPD (ERAPERMED 2020-314 to EG).
Author contributions
L.P.: Conceived, organised, and executed the research project; co-executed the statistical analysis and interpretation of results; wrote the manuscript; substantially participated in data collection, data exportation and data curation. R.K.: Conceived, organised, and co-executed the research project; participated in interpretation of results; critically revised the manuscript. A.R.: Executed the statistical analysis and interpretation, critically revised the manuscript, substantially participated in data curation. E.G.: Co-executed the research project; participated in interpretation of results; critically revised the manuscript. P.M.: Executed the genetic analysis; participated in interpretation of results; critically revised the manuscript. S.P.: Contributed genetic data; co-executed the research project; critically revised the manuscript. Z.L.: Executed the genetic analysis; participated in interpretation of results; critically revised the manuscript.
Data availability
The dataset for this manuscript is not publicly available as it is linked to the Luxembourg Parkinson’s Study and its internal regulations. Any requests for accessing the dataset can be directed to request.ncer-pd@uni.lu.
Code availability
The code for the statistical models is available at: 10.17881/hr67-ba06.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Change history
9/2/2022
A Correction to this paper has been published: 10.1038/s41531-022-00378-9
Contributor Information
L. Pavelka, Email: lukas.pavelka@uni.lu
R. Krüger, Email: rejko.krueger@uni.lu
on behalf of the NCER-PD Consortium:
Geeta Acharya, Gloria Aguayo, Myriam Alexandre, Muhammad Ali, Dominic Allen, Wim Ammerlann, Rudi Balling, Michele Bassis, Katy Beaumont, Regina Becker, Camille Bellora, Guy Berchem, Daniela Berg, Alexandre Bisdorff, Kathrin Brockmann, Jessica Calmes, Lorieza Castillo, Gessica Contesotto, Nico Diederich, Rene Dondelinger, Daniela Esteves, Guy Fagherazzi, Jean-Yves Ferrand, Manon Gantenbein, Thomas Gasser, Piotr Gawron, Soumyabrata Ghosh, Enrico Glaab, Clarissa Gomes, Elisa Gómez De Lope, Nikolai Goncharenko, Jérôme Graas, Mariella Graziano, Valentin Groues, Anne Grünewald, Wei Gu, Gaël Hammot, Anne-Marie Hanff, Linda Hansen, Maxime Hansen, Michael Heneka, Estelle Henry, Sylvia Herbrink, Eve Herenne, Sascha Herzinger, Michael Heymann, Michele Hu, Alexander Hundt, Nadine Jacoby, Jacek Jaroslaw Lebioda, Yohan Jaroz, Quentin Klopfenstein, Rejko Krüger, Pauline Lambert, Zied Landoulsi, Roseline Lentz, Inga Liepelt, Robert Liszka, Laura Longhino, Victoria Lorentz, Paula Cristina Lupu, Clare Mackay, Walter Maetzler, Katrin Marcus, Guilherme Marques, Tainá Marques, Patrick May, Deborah Mcintyre, Chouaib Mediouni, Francoise Meisch, Myriam Menster, Maura Minelli, Michel Mittelbronn, Brit Mollenhauer, Kathleen Mommaerts, Carlos Moreno, Serge Moudio, Friedrich Mühlschlegel, Romain Nati, Ulf Nehrbass, Sarah Nickels, Beatrice Nicolai, Jean-Paul Nicolay, Wolfgang Oertel, Marek Ostaszewski, Sinthuja Pachchek, Claire Pauly, Laure Pauly, Lukas Pavelka, Magali Perquin, Roslina Ramos Lima, Armin Rauschenberger, Rajesh Rawal, Dheeraj Reddy Bobbili, Eduardo Rosales, Isabel Rosety, Kirsten Rump, Estelle Sandt, Venkata Satagopam, Marc Schlesser, Margaux Schmitt, Sabine Schmitz, Reinhard Schneider, Jens Schwamborn, Amir Sharify, Ekaterina Soboleva, Kate Sokolowska, Olivier Terwindt, Hermann Thien, Elodie Thiry, Rebecca Ting Jiin Loo, Christophe Trefois, Johanna Trouet, Olena Tsurkalenko, Michel Vaillant, Mesele Valenti, Liliana Vilas Boas, Maharshi Vyas, Richard Wade-Martins, and Paul Wilmes
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-022-00342-7.
References
- 1.Elizabeth Qian, Yue Huang. Subtyping of Parkinson’s disease - where are we up to? Aging Dis. 2019;10:1130–1139. doi: 10.14336/AD.2019.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodríguez-Violante M, Cervantes-Arriaga A, Fahn S, Tolosa E. Two-hundred years later: Is Parkinson’s disease a single defined entity? Rev. Invest Clin. 2017;69:308–313. doi: 10.24875/RIC.17002291. [DOI] [PubMed] [Google Scholar]
- 3.Krüger R, et al. Classification of advanced stages of Parkinson’s disease: translation into stratified treatments. J. Neural Transm. 2017;124:1015–1027. doi: 10.1007/s00702-017-1707-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagano G, Ferrara N, Brooks DJ, Pavese N. Age at onset and Parkinson disease phenotype. Neurology. 2016;86:1400–1407. doi: 10.1212/WNL.0000000000002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jellinger KA. Parkinson disease with old-age onset. Arch. Neurol. 2003;60:1814–1815. doi: 10.1001/archneur.60.12.1814. [DOI] [PubMed] [Google Scholar]
- 6.Zhou MZ, et al. The association between non-motor symptoms in Parkinson’s disease and age at onset. Clin. Neurol. Neurosurg. 2013;115:2103–2107. doi: 10.1016/j.clineuro.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson LW, Rajput AH, Rajput A. Early-onset vs. Late-onset Parkinson’s disease: a clinical-pathological study. Can. J. Neurol. Sci. 2016;43:113–119. doi: 10.1017/cjn.2015.244. [DOI] [PubMed] [Google Scholar]
- 8.Jellinger KA. Very old onset parkinsonism: a clinical-pathological study. Parkinsonism Relat. Disord. 2018;57:39–43. doi: 10.1016/j.parkreldis.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Ohnmacht J, May P, Sinkkonen L, Krüger R. Missing heritability in Parkinson’s disease: the emerging role of non-coding genetic variation. J. Neural Transm. 2020;127:729–748. doi: 10.1007/s00702-020-02184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escott-Price V, International Parkinson’s Disease Genomics Consortium. Nalls MA, et al. Polygenic risk of Parkinson disease is correlated with disease age at onset. Ann. Neurol. 2015;77:582–591. doi: 10.1002/ana.24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan-Or Z, et al. The Quebec Parkinson network: a researcher-patient matching platform and multimodal biorepository. J. Parkinsons Dis. 2020;10:301–313. doi: 10.3233/JPD-191775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos García D, et al. COPPADIS-2015 (COhort of Patients with PArkinson’s DIsease in Spain, 2015): an ongoing global Parkinson’s disease project about disease progression with more than 1000 subjects included. Results from the baseline evaluation. Eur. J. Neurol. 2019;26:1399–1407. doi: 10.1111/ene.14008. [DOI] [PubMed] [Google Scholar]
- 13.Marek K, et al. The Parkinson’s progression markers initiative (PPMI) - establishing a PD biomarker cohort. Ann. Clin. Transl. Neurol. 2018;5:1460–1477. doi: 10.1002/acn3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diederich NJ, Moore CG, Leurgans SE, Chmura TA, Goetz CG. Parkinson disease with old-age onset: a comparative study with subjects with middle-age onset. Arch. Neurol. 2003;60:529–533. doi: 10.1001/archneur.60.4.529. [DOI] [PubMed] [Google Scholar]
- 15.Wickremaratchi MM, et al. The motor phenotype of Parkinson’s disease in relation to age at onset. Mov. Disord. 2011;26:457–463. doi: 10.1002/mds.23469. [DOI] [PubMed] [Google Scholar]
- 16.Mehanna R, Moore S, Hou JG, Sarwar AI, Lai EC. Comparing clinical features of young onset, middle onset and late onset Parkinson’s disease. Parkinsonism Relat. Disord. 2014;20:530–534. doi: 10.1016/j.parkreldis.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Chase BA, Markopoulou K. Olfactory dysfunction in familial and sporadic Parkinson’s Disease. Front Neurol. 2020;11:447. doi: 10.3389/fneur.2020.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim R, Shin JH, Park S, Kim HJ, Jeon B. Longitudinal evolution of non-motor symptoms according to age at onset in early Parkinson’s disease. J. Neurol. Sci. 2020;418:117157. doi: 10.1016/j.jns.2020.117157. [DOI] [PubMed] [Google Scholar]
- 19.Pettigrew C, Soldan A. Defining cognitive reserve and implications for cognitive aging. Curr. Neurol. Neurosci. Rep. 2019;19:1. doi: 10.1007/s11910-019-0917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SB, Meltzer LJ. Disentangling the effects of current age, onset age, and disease duration: parent and child attitudes toward diabetes as an exemplar. J. Pediatr. Psychol. 2002;27:77–86. doi: 10.1093/jpepsy/27.1.77. [DOI] [PubMed] [Google Scholar]
- 21.Blauwendraat C, et al. Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia [published correction appears in Brain] Brain. 2020;143:234–248. doi: 10.1093/brain/awz350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yahalom G, et al. Age at onset of Parkinson’s disease among Ashkenazi Jewish patients: contribution of environmental factors, LRRK2 p.G2019S and GBA p.N370S mutations. J. Parkinsons Dis. 2020;10:1123–1132. doi: 10.3233/JPD-191829. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez DG, Reed X, Singleton AB. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 2016;139:59–74. doi: 10.1111/jnc.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G, et al. Genome-wide survival study identifies a novel synaptic locus and polygenic score for cognitive progression in Parkinson’s disease. Nat. Genet. 2021;53:787–793. doi: 10.1038/s41588-021-00847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ihle J, et al. DIGPD study group; Parkinson’s disease polygenic risk score is not associated with impulse control disorders: a longitudinal study. Parkinsonism Relat. Disord. 2020;75:30–33. doi: 10.1016/j.parkreldis.2020.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Paul KC, Schulz J, Bronstein JM, Lill CM, Ritz BR. Association of polygenic risk score with cognitive decline and motor progression in Parkinson disease. JAMA Neurol. 2018;75:360–366. doi: 10.1001/jamaneurol.2017.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alves G, et al. Incidence of Parkinson’s disease in Norway: the Norwegian ParkWest study. J. Neurol. Neurosurg. Psychiatry. 2009;80:851–857. doi: 10.1136/jnnp.2008.168211. [DOI] [PubMed] [Google Scholar]
- 29.Granieri E, et al. Parkinson’s disease in Ferrara, Italy, 1967 through 1987. Arch. Neurol. 1991;48:854–857. doi: 10.1001/archneur.1991.00530200096026. [DOI] [PubMed] [Google Scholar]
- 30.Winter Y, et al. Incidence of Parkinson’s disease and atypical parkinsonism: Russian population-based study. Mov. Disord. 2010;25:349–356. doi: 10.1002/mds.22966. [DOI] [PubMed] [Google Scholar]
- 31.Savica R, Grossardt BR, Rocca WA, Bower JH. Parkinson disease with and without Dementia: a prevalence study and future projections. Mov. Disord. 2018;33:537–543. doi: 10.1002/mds.27277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litvan I, et al. SIC Task Force appraisal of clinical diagnostic criteria for parkinsonian disorders. Mov. Disord. 2003;18:467–486. doi: 10.1002/mds.10459. [DOI] [PubMed] [Google Scholar]
- 33.Hipp G, et al. The Luxembourg Parkinson’s study: a comprehensive approach for stratification and early diagnosis. Front Aging Neurosci. 2018;10:326. doi: 10.3389/fnagi.2018.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blauwendraat C, et al. NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol. Aging. 2017;57:247.e9–247.e13. doi: 10.1016/j.neurobiolaging.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das S, et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korlach Jonas, et al. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 2010;472:431–455. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- 37.Poplin R, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 38.Nalls MA, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset for this manuscript is not publicly available as it is linked to the Luxembourg Parkinson’s Study and its internal regulations. Any requests for accessing the dataset can be directed to request.ncer-pd@uni.lu.
The code for the statistical models is available at: 10.17881/hr67-ba06.