Abstract
Keap1 mutations regulate Nrf2 activity and lead to chemoresistance in cancers. Yet the underlying molecular mechanisms of chemoresistance are poorly explored. By focusing and genotyping head and neck squamous cell carcinoma (HNSCC) that had available pathologic and clinical data, we provide evidence that Keap1 displays frequent alterations (17%) in HNSCC. Functional loss of Keap1 results in significant activation of Nrf2 and promotes cancer cell growth, proliferation, and elevated cancer stem cell (CSCs) self-renewal efficiency and resistance to oxidative stress. Furthermore, decreased Keap1 activity in these cells increased nuclear accumulation of Nrf2 and activation of the Notch pathway, causing enhanced transcriptional alterations of antioxidants, xenobiotic metabolism enzymes, and resistance to chemotherapeutic treatment. Limiting the Nrf2 activity by either Keap1 complementation or by Nrf2 silencing increased the sensitivity to chemotherapy in Keap1-mutated cells and repressed the CSC self-renewal activity. Our findings suggest that Keap1 mutations define a distinct disease phenotype and the Keap1-Nrf2 pathway is one of the leading molecular mechanisms for clinical chemotherapeutic resistance. Targeting this pathway may provide a potential and attractive personalized treatment strategy for overcoming chemotherapeutic resistance conferred by Keap1 mutations.
Subject terms: Oral cancer, Oral cancer
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the utmost global health concern and affects >890,000 patients, and over 450,000 HNSCC-related deaths occur every year [1, 2]. To date, cisplatin (CDDP) and paclitaxel (PTX) based chemotherapy and radiotherapy remain the preferred and effective treatment options for advanced-stage HNSCC patients. Sadly, a proportion of patients do not respond to chemotherapy and develop resistance to treatment. The contributing factors to treatment failures due to therapeutic resistance are linked to cell apoptosis, drug efflux changes, DNA damage repair, and abundance of cancer stem cells (CSCs) [3, 4].
The Cancer Genome Atlas (TCGA) project has profiled an extensive landscape of somatic genomic alterations in HNSCC [5]. This revealed, as expected, that smoking-related HNSCCs have near-universal loss-of-function of TP53 mutations and CDKN2A inactivation with frequent copy number alterations. In addition to these mutations, several other pathway Keap1, and Nrf2- pathway mutations were found in patients with HNSCC.
Inactivation of Kelch-like ECH-associated protein-1 (Keap1) strongly induces NF-E2-related factor 2 (Nrf2), and acquires malignancy in several types of cancer [6]. Nrf2-associated oxidative stress plays a critical role in developing chemoresistance in various cancers such as; lung, breast, colon, and ovarian cancer [7–10]. Recurrent or metastatic HNSCC patients have a poor prognosis with less than 1-year median survival [11]. Platinum-based chemotherapy (cisplatin) has been established as a gold standard systemic agent in HNSCC [12]. However, cisplatin resistance is still a barrier to organ-sparing and the survival of patients with advanced-stage HNSCC. In addition, cisplatin treatment often enhances a fraction of putative head and neck cancer stem cells, which are highly tumorigenic in preclinical models of HNSCC [13]. This small population of CSCs that reside within HNSCC are relatively resistant to chemotherapies and are clinically predicted to contribute to tumor recurrence. Cisplatin-resistant tumor cells consist of a higher proportion of epithelial cell adhesion molecule (EpCAM) and induce the expression of IL-6 and tumorigenic cytokines that contribute to cisplatin-induced stemness [14]. Recently we have shown the Nrf2-associated mechanistic link by which HNSCC cells acquire therapeutic resistance to cisplatin [15]. We have also demonstrated reduced reactive oxygen species (ROS) activity via Nrf2 activation. Nrf2 activation is further concerted with interleukin-6 (IL-6) and p62 [14]. Ongoing evidence has reported that the Keap1-Nrf2 pathway and levels of ROS (reactive oxygen species) contribute to the development of drug resistance and the progression of HNSCC [14]. In addition, recent evidence suggests that Nrf2 and the Notch signaling pathway mutually function by regulating Nrf2 downstream target genes and activating the Notch signaling, suggesting that the Nrf2 and Notch signaling pathway play a critical role in cellular behaviors [16]. However, the effects of somatic alterations in Keap1-Nrf2 on CSCs and chemoresistance have not been deeply explored.
In this study, we examined whether Nrf2 pathway activation due to Keap1 inactivation plays a role in HNSCC therapeutic resistance and CSC induction. Our results found that Keap1 alterations caused Nrf2 activation concerted with ROS suppression and have led to CSC induction and therapeutic resistance in HNSCC cells. Furthermore, downregulation of Nrf2 activity enhanced chemotherapeutic drug sensitivity in Keap1 mutated HNSCC cells. Our findings suggest that Keap1 mutation status might be helpful for personalized treatment decision-making strategies for patients with HNSCC.
Materials and methods
Cell culture and patient samples
Human SCC9 [17] and Cal33 [18] head and neck cancer cell lines were previously described and purchased from American Type Cell Culture (ATCC, Manassas, VA). All cell lines were authenticated using a short tandem repeat analysis kit (Applied Biosystems, CA). Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin cocktail. Formalin-fixed paraffin-embedded tissues from 24 (n = 24) HNSCC patients treated in Chittagong Medical College Hospital (CMCH), Chittagong, were included in this study after obtaining full patient consent. The study protocol for the collection and use of patient tumor tissues and clinical information was approved by the Institutional Review Board at CMCH (052(1) 04-06-2014) and the King Faisal Specialist Hospital and Research Center (KFSH&RC; RAC# 2210031). We obtained patients’ informed consent following local and international regulations. Tumors were obtained from all consented patients at the time of surgery. Tumors were first minced and enzymatically dissociated with 2 mg/ml of dispase (Roche, USA) and then incubated with 0.25% Trypsin-ethylenediaminetetraacetic acid, passed through a 21-gauge syringe, and filtered through a 23 μm cell filter (Merck Millipore). Cells were either directly cultured in supplemented CSC medium or cryopreserved in 80% FBS and 20% dimethylsulfoxide until further use. The plasmid encoding Keap1 pCMV-Keap1 and pRGBp2 vectors were described previously [19, 20] and purchased from Origene Technologies (Rockville, Maryland, USA). The expression plasmid Keap1 pCMV-Keap1 was transfected into 293FT cells using transfection reagents. Post-transfection supernatants (after 24 h) were collected and used for infection. The medium was then replaced with the complete medium, and cells were grown for 3 days.
Keap1 and Nrf2 DNA sequence analysis
Tumor DNA isolation
Genomic DNA was extracted from a total of 24 (n = 24) formaldehyde-fixed and paraffin-embedded (FFPE) head and neck tumor samples using QIAamp DNA FFPE Tissue Kit (Qiagen). Briefly, 2–3 tumor sections were treated with 1 ml xylene to remove paraffin, tissues were scraped and pooled and the tumor cell pellets were lysed after DNA precipitation, samples were loaded into columns. Following the column wash steps, DNA was eluted with 50 μl elution buffer, quantified, and stored at −20 °C for further analysis.
PCR amplification and Sanger sequencing
The coding regions of Keap1 (ENST00000171111), and Nrf2 (ENST00000397062) genes were PCR amplified using 20 ng DNA-specific primers containing M13 tail sequences in 25 μl reaction volume. Sequences for all the primers are provided in Supplementary Table S1. For tumor samples that did not have a sufficient amount of DNA, nested PCR amplifications were performed to obtain enough products for the Sanger sequencing reaction. High-fidelity Taq polymerase was used for PCR amplification to avoid errors during the amplification reactions. Samples with mutations were verified by repeating the PCR amplification and sequencing steps to rule out any PCR-related artifact. Both M13 forward and reverse primers were used for bidirectional sequencing of the amplified PCR products.
Keap1, Nrf2, Notch1, and Hes1 siRNA transfection and Notch inhibition by DAPT and cell proliferation assay
Keap1 (siGENOME D-001210-01), Nrf2 (siGENOME D-003755-01) SMARTpool, and non-targeting scrambled siRNA sequence (siGENOME D-001210-01) control pool was obtained from Dharmacon and described previously [14]. The ON-TARGETplus pool of Notch1 and Hes1 siRNA was obtained from Thermo Scientific. Transfection was performed in 50% confluent cells using Lipofectamine 200 (Invitrogen) and cultured in reduced serum medium OPTI-MEM following the manufacturer’s instructions. The specific siRNAs were transfected into SCC9 and Cal33 cells in triplicate.
Measurement of ROS by DCFDA assay
Cells were preincubated with vehicle (control), CB-839 for 48 h and exposed to cisplatin treatment for 24 h. ROS level was measured using the 2’,7-dichlorofluorescein-diacetate DCFH-DA (Sigma; USA). Unless otherwise indicated, cells were treated with Nrf2 siRNA and/or cisplatin for 72 h. Briefly, 1 mL of cell suspension was transferred to a 1.5 mL culture tube and 20 μM of DCFH-DA staining solution was added. Cells were gently mixed and incubated for 45–50 min at 37°C in the dark. Washed the cells and resuspended them in 400 μL of cold PBS. DCFH-DA fluorescence intensity was detected by flow cytometry, using the FITC channel on BD FACSAria flow cytometer (BD BioScience) and data were analyzed with FACSDiva software.
Sphere forming assay
siRNA and scrambled-siRNA treated cells were cultured in six-well ultra-low attachment plates at a density of 1000 cells/well in a growth factor-supplemented CSC medium. The number and size of the spheres were monitored and recorded every 3rd day. Sphere forming efficiency was calculated as the number of actual spheres/number of cells plated × 100.
AlamarBlue cytotoxicity and proliferation assay
Cells were seeded (5000 cells/well) in a 96-well plate in a complete medium. Cells were treated with an increasing concentration of cisplatin alone or a combination of CB-839 for 72 h as indicated concentrations in the figure legends. Cell viability was assessed by AlamarBlue assay using the manufacturer’s instructions. AlamarBlue was added (10% of total volume) and incubated for 4 h in an incubator and fluorescence was measured using the SPECTRAmax Gemini Spectrophotometer (540 nm excitation and 590 nm emission). DRC (Dose-response curve) and GRmetrics packages were used to generate dose–response curves using R-Statistical software (Version 4.0.3). Inhibitory EC50 concentration values were calculated (DRC and GRmetrix package) from the results of cisplatin concentrations in triplicate from three independent experiments.
Quantitative real-time (qRT-PCR)
Total RNA was extracted from fresh tumor tissues, SCC9, and Cal33 cells using RNAeasy Kit (Qiagen) and reversed transcribed. Total RNA was isolated from formalin-fixed, paraffin-embedded tumor tissue sections using RNAeasy FFPE kit (Qiagen) and reverse transcribed. SYBR-Green-1-based RT-PCR amplification was performed in triplicates on the LightCycler-480 (Roche). The primers list is shown in Supplementary Table S2. The relative expression of each gene was analyzed by comparing its expression to that of GAPDH.
Western blotting
Lysed protein was transferred to the PVDF membrane and primary antibodies were added to PVDF membranes in 5% non-fat dry milk in TBS-Tween-20 buffer. Primary antibodies are Nrf2 (Abcam, cat# ab137550, MA, USA) and Keap1 (Abcam, cat# 119403 MA, USA), Notch1 (cat# 14-5785-81) and Hes1 (cat# PA5-28802; Invitrogen, USA), and GAPDH (Santa Cruz, cat# sc32233, CA, USA). HRP-conjugated anti-mouse or anti-rabbit secondary antibodies were used for the detection.
Immunohistochemistry
The following primary antibodies against Nrf2 (Abcam, cat# ab137550, MA, USA), Keap1 (cat# ab119403, Abcam, MA, USA), CD44 (Abcam, cat# ab6124, MA, USA), TP53 (cat# ab238069, Abcam, cat# ab238069, MA, USA) Notch1 (cat# 14-5785-81, Invitrogen, USA), and Hes1(cat# PA5-28802, Invitrogen, USA) were applied on deparaffinized 5-μm thick formalin-fixed tissue sections for overnight. A horseradish peroxidase (HRP)-conjugated secondary antibody was used for the detection. For Nrf2 detection in cells, only nuclear immunostaining was included in this study because only transcriptionally active Nrf2 resides in the nucleus.
Notch inhibition and proliferation assay
ON-TARGETplus Pool of siRNA against Notch1 and Hes1 and non-targeting Pool of siRNA (Thermo Scientific) were used to downregulate the expression of Notch1 and Hes1 or used as a control in the experiments. Cells were seeded in 96-well plates in a complete medium and allowed to grow up to 70% confluence. Cells were transfected with siRNA using Lipofectamine RNAiMAX Reagent (Invitrogen). AlamarBlue was added (10% of total volume) and incubated for 4 h in an incubator and fluorescence was measured using the SPECTRAmax Gemini Spectrophotometer (540 nm excitation and 590 nm emission). Notch1 was inhibited with DAPT, a gamma-secretase inhibitor.
Computational analysis of TCGA datasets
We downloaded the head and neck cancer RNA-Seq data set (Illumina HiSeq 2000) from the Cancer Genomic Atlas (TCGA, https://portal.gdc.cancer.gov), and analyzed using R-statistical software (Version 4.0.3).
Statistical analysis
Experiments were performed in triplicates where necessary and results were presented as mean ± SEM. For independent data with two specimens, a two-tailed t test for equal variance, or one-way ANOVA Tukey post hoc comparison for three or more groups were applied. For all statistical analysis we used “R” statistical software (version 4.0.3), and for graphs “ggplot2” packages in “R”. Kaplan–Meier survival curves were generated and analyzed using “R” packages “survival” and “survminer”. The significance was calculated using Log-rank and Mantel-Cox test. Dose–response was analyzed using a DRC (Dose–response curve) and GRmetrix packages in “R” statistical software (version 4.0.3). The significance was defined based on P < 0.05.
Results
Keap1 and Nrf2 mutations predict shorter overall survival in patients with advanced HNSCC
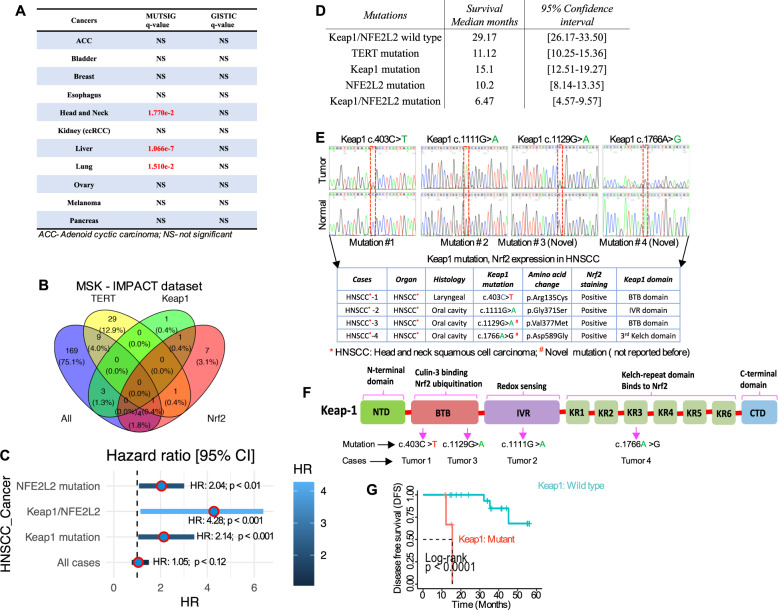
Keap1 mutations and the resulting Nrf2 activations have been reported in many cancers [5, 21]. As an approach to exploring the tumor-associated Keap1 alterations, resulting in Nrf2 activation and chemotherapeutic resistance through CSC induction, we first investigated the presence of genomic alterations of Keap1 in a large panel of 21 distinctive cancers sequenced by The Cancer Genomic Atlas consortium (TCGA) and recently developed mutations significance method (MutSigCV), which provides a statistical metric to identify driver candidates in cancer with respect to the gene nucleotide length and the background mutations rate of each type of cancer analyzed [5, 22]. This analysis revealed that the Keap1 mutations occurred in several cancers, including head and neck cancer (Fig. 1A; Suppl. Fig. 1A). Memorial-Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) is a platform for archiving a hybridization capture-based next-generation sequencing panel that detects protein-coding mutations and copy number alterations (CNAs) and selects promoter mutations and structural rearrangements in more than 410 cancer-associated genes [23, 24]. We explored a cohort of 186 sequentially profiled HNSCC patients for tumor-specific somatic mutation in Keap1 only (n = 1), Nrf2 only (n = 7), or both (n = 1) (Fig. 1B). Additionally, we included a third mutation in our analysis, TERT (n = 40), since TERT mutations often co-occur with Nrf2 (n = 1), but not with Keap1 (n = 0). In this cohort, we observed a marked increase in the hazard ratio (HR 4.28, p < 0.001) and a significant decrease in median survival from 29.17 months in patients with Keap1 and or Nrf2 wild-type (WT) to 15.1, 10.2, and 6.47-months patients harboring either Keap1 and or Nrf2 alone or double mutations (Fig. 1C, D). In a multivariate analysis, Keap1 and Nrf2 double mutations significantly (p < 0.001) predicted overall poor survival (Fig. 1D). In the TCGA data set, Keap1 gene alterations are mostly missense mutations that occur in Kelch or BTB domains of Keap1 (Suppl. Fig. 1B), thereby impeding Keap1 protein interaction with Nrf2. On the other hand, Nrf2 mutations are mostly missense and occur within the first 100 amino acids that contain the Neh2 domain. That includes the two degrons bound by the Keap1 and thus likely impede Keap1-mediated Nrf2 degradation [25] (Suppl. Fig. 1C).
Fig. 1. Keap1/NFE2L2 (Nrf2) mutations predict shorter overall survival in patients with HNSCC.
A Alterations of the Keap1 gene in major cancer types from TCGA database. B Venn diagram indicates the number of patients with head and neck cancer in the MISK-IMPACT database that is wild-type for the mutation (green), mutant for TERT (yellow), mutant for Keap1(light blue), and mutant for Nrf2 (purple) (n = 186). C Multivariate Cox regression analysis for each indicated variable was performed. D The risk ratio of overall survival corresponds to each indicated variable. Nrf2 (P < 0.01), Keap1/Nrf2 (p < 0.001) and Keap1 (p < 0.001) mutations are independently identified as significant covariate for overall survival. The table indicates the overall survival across each group with a 95% confidence interval. E Electropherogram depicting Keap1 mutation sequence analysis for HNSCC. The top part shows the detection of the Keap1 mutations identified in HNSCC patients’ tumors and the bottom part shows the non-cancerous normal individuals’ Keap1 sequence. The table below shows the details of each patient and amino acid changes and corresponding Nrf2 positivity. F Schematic diagram of conserved domain showing the structure of Keap1 protein and the location of each mutation within Keap1 protein. NTD N-terminal domain (amino acids), BTB broad complex-Tramtrack-Bric-a-brack, IVR intervening regions, KR Kelch repeat. G Kaplan–Meier disease-free survival analysis curve of Keap1 wild-type and Keap1 mutant HNSCC patients (n = 24; 4-Keap1 mutant and 20- Keap1 wild-type patient) (Log-rank p < 0.0001).
To explore the role of Keap1 mutations in HNSCC pathogenesis, and given the size of the available clinical samples, we begin by examining the Keap1 mutations in our samples (n = 24). We amplified and sequenced all five protein-coding exons of the Keap1 from 24 HNSCC surgical samples. We found 4 (17%) Keap1 mutations and 2 out of 4 mutations had novel pathogenic somatic Keap1 mutations (c.403 C > T and c.1129 G > A), while the remaining two mutations (c.1112 G > A and c.1766A > G) had likely pathogenic and benign germline in nature (Fig. 1E). Intriguingly, tumors with Keap1 mutations showed positive Nrf2 expression (Fig. 1E; the table below). These mutations reside in the functionally important Keap1 protein domain, such as BTB, IVR, and KR regions, governing Nrf2 ubiquitination, redox sensing, and Nrf2 binding sites (Fig. 1F). The significance of mutations was further analyzed by in silico predictors (Suppl. Table 3). We also detected five separate synonymous germline variants in various frequencies (Suppl. Table 4). The most notable variant is Keap1 c.1815G > A and is highly enriched in the HNSCC population compared to previously reported global healthy population frequencies. Prognostic analysis of patient tumors carrying Keap1 mutations revealed a significant correlation with poor DFS (p < 0.0001 by Log-rank analysis; Fig. 1G). Due to frequent alteration of Keap1 and TP53 [26] in HNSCC, we were interested in whether Keap1 alterations showed association with TP53 molecular alterations. As expected, frequent TP53 overexpression (12/24, 50.0%) was detected in our cohort. Interestingly, Keap1 alterations were detected exclusively in the TP53-overexpressed HNSCC tumors (Suppl. Table 5).
Keap1 mRNA expression and concurrent Keap1 and Nrf2 mutations in Nrf2 immunopositive HNSCC tumors
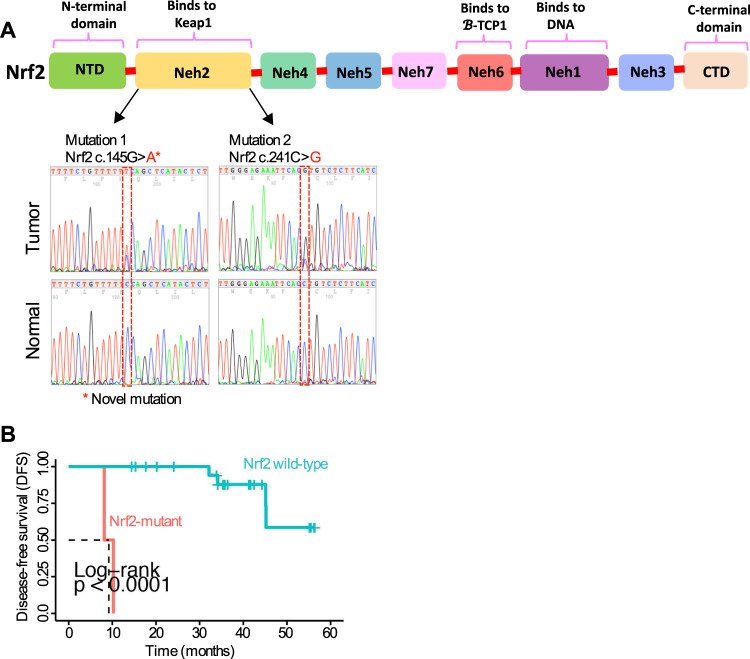
Keap1 is an essential regulator of Nrf2 functions, and the role of Keap1 in regulating Nrf2 signaling in cancers has been reported previously [27]. To evaluate Keap1 expression and concurrent mutations of Keap1 and Nrf2 in Nrf2 immunopositive tumors, we first analyzed Keap1 mRNA expression by qRT-PCR, followed by Nrf2 sequence analysis in Nrf2 immunopositive tumors (n = 24). As shown in Supplementary Table S6, 4 tumors with positive nuclear Nrf2 staining had absent Keap1 transcript expression, with the remaining 19 tumors being positive for Keap1 transcript. Notably, although, one tumor had absent Keap1 transcript expression (HNSCC-17, oral cavity, Suppl. Table 6) but harbored no Keap1 mutations and was also positive for Nrf2 protein expression. We then sequenced the Nrf2 gene from all tumor samples that had positive Nrf2 staining (Fig. 2A). Somatic Nrf2 mutations were found only in 2 tumors (c.145 G > A and c.241 G > C) including a novel Nrf2 mutation (c.145 G > A) in the Neh2 domain where Keap1 binds and with high cytoplasmic Keap1 expression (Fig. 2A; Suppl. Table 3). In our samples, no tumors harbored both Keap1 and Nrf2 mutations concurrently, confirming the MISK-IMPACT results in Fig. 1B. Given the smaller size of the available clinical samples, however, prognostic analysis of HNSCC carrying a Nrf2 mutation revealed a significant correlation with poor DFS (~ 10 months) (p < 0.0001 by Log-rank analysis; Fig. 2B, Suppl. Fig. 2).
Fig. 2. Keap1 mRNA expression and concurrent Keap1/Nrf2 mutations in Nrf2 immunopositive HNSCC tumors.
A Electropherogram depicting Nrf2 mutation sequence analysis for head and neck cancer. The top part shows the locations of each mutation within the Nrf2 protein. The bottom part shows the detection of Nrf2 mutation identified in HNSCC patients’ tumors in non-cancerous normal individuals in the Nrf2 sequence. B Kaplan–Meier disease-free survival analysis curve of Nrf2 wild-type and mutant HNSCC patients (n = 24, 2-Nrf2 mutant and 22 Nrf2 wild-type patient) (Log-rank p < 0.0001).
The biological effect of Keap1 mutations and effects of glutaminase inhibitor CB-839 in chemosensitizing Keap1 mutant cells
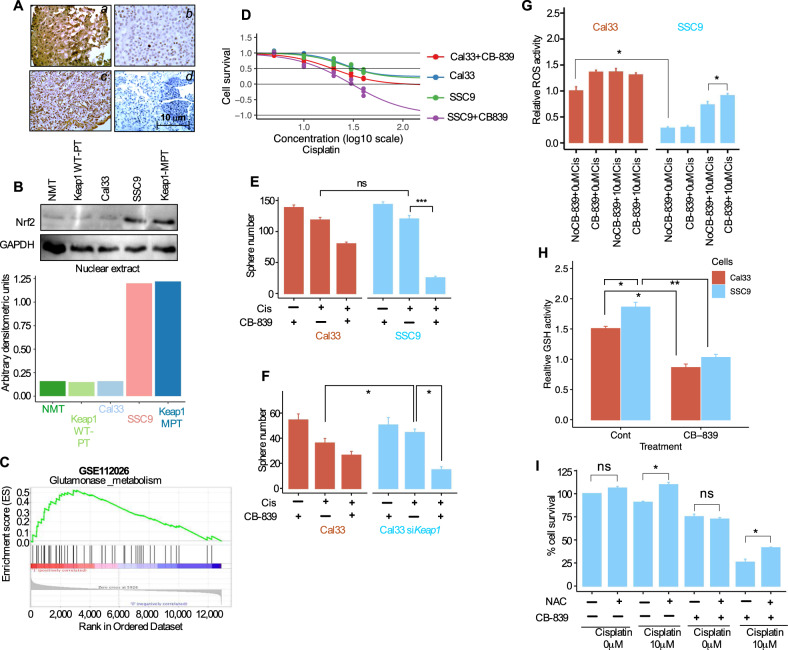
To evaluate the loss of Keap1 and its effects on Nrf2 overexpression and cellular localization in primary HNSCC tumors, we immunoassayed Nrf2 expression using the anti-Nrf2 antibody in HNSCC primary tumor tissues. Strong nuclear and cytoplasmic and nuclear Nrf2 expression was detected in primary tumor tissues harboring Keap1 mutations (Fig. 3A; part a). Five tumor tissues with wild-type Keap1 demonstrated decreased nuclear Nrf2 and weak cytoplasmic Nrf2 expression (Fig. 3A; parts b, c), while the normal tissue did not show Nrf2 expression (Fig. 3A, part d). We also detected nuclear Nrf2 localization in parts of Keap1-wild-type tumor tissues but to a lesser extent than Keap1- mutant tissue (Fig. 3A, part c). Aiming to study the levels of known Nrf2 target genes in tumor and normal samples, we measured the total GSH levels and enzymatic activity of SOD1, NQO1, and GST levels in eleven primary tumors and adjacent normal tissues. Among all tissues, four tumors (4/11; 36%) harbored Keap1 mutations, 2 (2/11; 18%) Nrf2 mutations, and remaining samples (5/11; 45%) from patients with Keap1 wild-type status (Suppl. Fig. 3A). We examined the GSH, SOD1, and NQO1 enzyme activity and GST levels in tumor and normal cells and found that these enzymes were at relatively higher levels in tumor tissues than in their corresponding adjacent normal tissues. Importantly, patients carrying Keap1 and Nrf2 mutations had higher levels of GSH, SOD1, NQO1, and GST compared to wild-type (Suppl. Fig. 3A).
Fig. 3. The biological effect of Keap1 mutations and Nrf2 overexpression in altered HNSCC tumor cells.
A Immunohistochemical assessment of Nrf2 expression in HNSCC tumor tissues. Part ‘a’ shows the strong nuclear and cytoplasmic Nrf2 expression in the Keap1 mutated patient (patient #3). Part ‘b’ and ‘c’ shows comparatively weaker cytoplasmic and nuclear Nrf2 expression in Keap1-wild-type tissue. Part ‘d’ show negative Nrf2 staining in adjacent normal tissue. B Immunoblot analysis of Nrf2 from nuclear protein in patient’s tumor cells, HNSCC cell lines, and non-malignant tissue. (bottom: Quantification of Nrf2 protein band density after normalizing with GAPDH). NMT: Non-malignant tissue; Keap1-WT-PT: Keap1 wild-type patient tumor; Keap1-MPT: Keap1 mutant patient tumor. C Gene set enrichment analysis (GSEA) of previously defined glutamine metabolism signature using RNA-seq data from GSE112026 data set. D Cell survival of Cal33 (Keap1 wild-type) and SSC9 (Keap1 mutant) cells with or without CB-839 (100 nM, 24-hour pre-treatment) and cisplatin (n = 3). Results were normalized with untreated cells. E Relative number of Spheres of Cal33-Keap1 wild-type and Keap1-mutant SSC9 cells with or without CB-839 (100 nM, 24-hour pre-treatment) and cisplatin (n = 3, 10 μM, n = 3, ***P < 0.001). F Relative number of Spheres of Keap1 wild-type Cal33 cells with or without knockdown of Keap1 by Keap1 specific siRNA in the presence or absence of CB-839 (n = 3, 100 nM, 24-hour pre-treatment) and cisplatin (10 μM, n = 3, *P < 0.05). G Intracellular reactive oxygen (ROS) levels measured by DCFDA intensity via FACS in Cal33 (Keap1 wild-type) and SSC9 (Keap1 mutant) cells with or without CB-839 and cisplatin treatment (n = 3, 10 μM, *P < 0.05). Results were normalized with untreated cells. H GHS (Glutathione) activity analysis of Cal33 (Keap1 wild-type) and SSC9 (Keap1 mutant) cells with or without CB-839 (n = 3, 10 μM, *P < 0.05, **P < 0.01). I Cell survival of SSC9 Keap1 mutant cells treated in the absence or presence of cisplatin (10 μM) or CB-839 and with or without NAC (n = 3, P < 0.05).
To determine the nuclear accumulation of Nrf2, we immunostained the Nrf2 protein in Keap1 mutant SSC9 cells. The results showed nuclear accumulation of Nrf2 protein in Keap1- mutated SSC9 cells (Suppl. Fig. 3B). To further examine the nuclear accumulation of Nrf2, we immunoassayed Nrf2 in Keap1 mutant and WT patients’ tumor samples and in two established HNSCC cell lines. Tumor cells with Keap1 mutations (SSC9 and Keap1-mutated patient’s tumor cells) demonstrated increased nuclear localization of Nrf2 in comparison to normal and Keap1 wild-type (Cal33 and Keap1 wild-type patients’ tumor cells) cells (Fig. 3B). Since Nrf2 controls the key components of the glutathione (GSH) and tightly regulates GSH levels by directly controlling glutamate-cysteine ligase complex (GCLC) and GCLM as well as glutathione S-transferase (GST) [28, 29] and thus cells acquire chemotherapeutic resistance, we investigated Nrf2-regulated target genes. Real-time RT-PCR analysis revealed that the majority of the Nrf2-regulated target genes were highly modulated in the cancer cells (Suppl. Fig. 3C). In addition, drug resistance markers MDR1 and ABCG2 were also highly upregulated in the cancer cells but not in the normal cells (Suppl. Fig. 3C). Importantly, as expected, the Keap1 transcripts and proteins were downregulated in Keap1 mutated cells (Suppl. Fig. 3C).
Several recent studies have reported that loss of Keap1 alters cellular metabolic requirements and confers sensitivity to glutamine metabolism inhibitors [30–32]. These reports suggest that glutathione (GSH) production is increased by glutamine metabolism. In lung cancer cells, it was shown that glutaminase inhibition can sensitize the radiation-induced treatment resistance in the Keap1 and Nrf2 mutant cells [30]. We, therefore, aimed to determine whether loss of Keap1 preferentially chemosensitizes by targeting glutaminase metabolism. First, we analyzed publicly available RNA-sequence data set GSE112026 and found significant overexpression of genes involved in glutamine metabolism (Fig. 3C). Next, we explored the possibility if targeting glutamine metabolism can chemosensitize Keap1 mutant cells. We used a combination of chemotherapeutic agent cisplatin and a small-molecule glutamine inhibitor CB-839, which is currently under investigation in phase 1 and 2 clinical trials. Our results showed that although treatment of cells with CB-839 alone did not show significant sensitivity to CB-839 in Keap1 mutant SSC9 and Keap1 wild-type Cal33 cells. However, the combination of cisplatin and CB-839 significantly increased the sensitivity to combination treatment and killed a substantial number of cells in Keap1 mutant SSC9 cells (Fig. 3D). In addition, the combination treatment significantly abolished the sphere growth efficiency in Keap1 mutant SSC9 cells suggesting that the combination treatment may exhibit the potential to inhibit the self-renewal capacity of Keap1 mutant cells (Fig. 3E). Furthermore, we noticed substantial inhibition of sphere growth effect in Keap1 wild-type Cal33 cells after silencing by Keap1-siRNA (Fig. 3F). To identify by which mechanisms CB-839 preferentially chemosensitize the cells, we assessed the intracellular ROS levels after treatment with CB-839 and cisplatin. The baseline ROS levels in Keap1 mutant SSC9 cells showed lower than in wild-type cells (Fig. 3G). However, unlike Keap1 wild-type cells, a combination of cisplatin and CB-839 treatment significantly increased the ROS levels in Keap1 mutant SSC9 cells compared to cisplatin alone treatment (Fig. 3G). Additionally, the CB-839 treatment significantly reduced the GSH activity in Keap1 mutant SSC9 cells (Fig. 3H). These results suggest that CB-839 treatment preferentially follows the inhibition of free radical scavenging capacity in Keap1 mutant cells compared with Keap1 wild-type counterpart. We then tested the hypothesis that the addition of exogenous free radical scavenger preferentially rescues the capacity of Keap1 mutant cells from CB-839-mediated chemosensitization. Our results showed that treatment of Keap1 mutant SSC9 cells with a ROS scavenger NAC did not show significant effects on cell survival by NAC alone or either 10 μM cisplatin or CB-839 alone (Fig. 3I). Importantly, treatment of cells by NAC significantly rescued the increased cell death which was caused by the combination treatment of CB-839 and 10 μM of cisplatin (Fig. 3I).
Loss of Keap1 increases the Nrf2 transcriptional activity, cancer stem cells characteristics in HNSCC
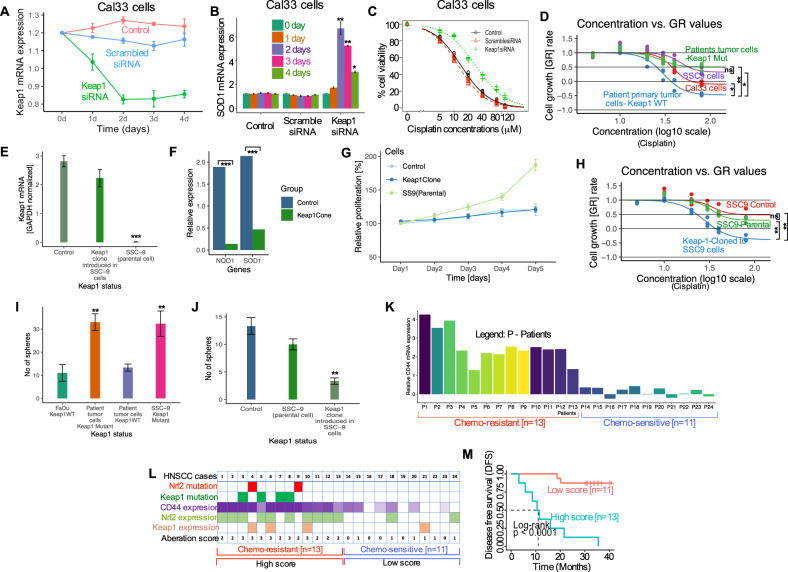
To get more insight into the Keap1 mutations and resulting chemoresistance through Nrf2 activation, we assessed the effects of siRNA knockdown of Keap1 on the sensitivity of Cal33 tumor cells to cisplatin. Keap1 siRNA was transfected into Cal33 cells under the treatment of 10 μM cisplatin. On days 1 and 2, siRNA against Keap1 steadily reduced Keap1 mRNA and maintained at this level for two days and again increased but stayed below baseline levels on days 3 and 4, while untreated control and scrambled-siRNA treated cells retained higher levels or on the baseline level (Fig. 4A). To identify if the knockdown of Keap1 also activates the transcriptional activity of Nrf2 target genes, we assessed SOD1 expression following Keap1 siRNA knockdown. Keap1 knockdown substantially increased the expression of SOD1 transcript by 6.2 and 5.1-fold on days 2 and 3 (Fig. 4B). In contrast, no significant change in SOD1 transcript was observed in control and scrambled siRNA-treated cells (Fig. 4B).
Fig. 4. Loss of Keap1 increases the Nrf2 transcriptional activity, increase cancer stem cell characteristics, and predictor of chemotherapeutic outcome in patients with HNSCC.
A Cal33 cells were transfected with siRNA against Keap1, scrambled, and control for 96 h. Keap1 mRNA was assessed by quantitative RT-PCR. Results expressed as fold-change. B Cal33 cells were transfected as described in A and SOD1 mRNA was assessed by quantitative RT-PCR. C Cells were treated with Keap1 siRNA to knock down the Keap1 gene and assessed the cell viability 72 h after cisplatin treatment in the indicated concentrations in Cal33 cells. Data presented as mean SD of triplicate experiments. D Cell survival at 72 h after cisplatin treatment of indicated HNSCC patient’s primary tumor and HNSCC cell lines (*P < 0.05, **P < 0.01). E qRT-PCR analysis of Keap1 expression in control, Keap1 expressing SSC9 clone and parental SSC9 cells (***P < 0.001). F qRT-PCR analysis of Nrf2 target genes SOD1 and NQO1 in control, Keap1 expressing clone, and parental SSC9 cells (***P < 0.001). G Cell proliferation activity of Keap1 expressing clone, control, and parental SSC9 cells. H Cell survival at 72 h after cisplatin treatment in parental SCC9, mock-transfected and Keap1-expressing clones. I Relative number of tumorspheres generated by the indicated patient’s tumor cells and cell lines. J Relative number of tumorspheres in parental SCC9, mock-transfected, and Keap1-expressing clone (**P < 0.01). K Expression of CD44 in cisplatin-resistant (n = 13) and cisplatin-sensitive (n = 11) HNSCC patients. L Summary of the results for the CD44 expression analysis in the presence of Keap1 or Nrf2 mutations and/or Keap1 or Nrf2 protein expression in each case (n = 24). The number of aberrations in each case was represented as the aberration scores (0, 1, 2, and 3) and all 24 cases were assigned into two groups based on the aberration scores: a “high score group” (n = 13 as aberration score 2 and 3) and low score group (n = 11 as aberration score 1, and 0). M Kaplan–Meier disease-free survival curve for 24 patients was generated according to the aberration score. The high score group was significantly associated with shorter disease-free survival (Log-rank p < 0.0001).
Next, we tested the cisplatin sensitivity in Keap1 siRNA-transfected cells. Untreated control and scrambled-siRNA-treated cells showed sensitivity to cisplatin. Although, Keap1 siRNA treated cells showed resistance at a lower dose but showed sensitivity to cisplatin at higher doses (Fig. 4C), and a higher EC50 value was recorded in Keap1-siRNA (the EC50 to cisplatin was 32.3 μM for Keap1 siRNA-transfected cells and 7.2 μM and 8.7 μM for control and scrambled-siRNA, respectively. (p < 0.05 displays the differences between Keap1 siRNA and control and scrambled groups).
Next, we sought to examine if the alterations of Keap1 show any associations with chemotherapy resistance in HNSCC cells. To establish the functional differences in sensitivity to chemotherapy in Keap1-WT and Keap1-mutant cells, we examined whether loss of Keap1 showed any effect on cell survival. Treatment of cells with cisplatin showed that Keap1-mutant SSC9 and patients’ primary tumor cells had resistance to cisplatin compared to Keap1 wild-type cells (Fig. 4D). To explore whether the restoration of Keap1 in Keap1-mutant cells affects cancer cell growth, we established a clone of SSC9 cells that stably express Keap1 cDNA. The results showed the expression of Keap1 mRNA in control and Keap1 clone cells while the absence of Keap1 mRNA in parental SSC9 cells (Fig. 4E). We further assessed changes in the expression of Nrf2 target genes SOD1 and NQO1 in the Keap1 clone cells. We found that restoration of Keap1 expression eliminated the SOD1 and NQO1 gene expression (Fig. 4F). In addition, we compared the cell proliferation activity in the Keap1-expressing clone. We found that Keap1 clone cells grew comparatively slower than the parental and mock-transfected control cells (Fig. 4G). Next, we examined the cisplatin sensitivity by reintroducing the Keap1 clone in parental SSC9 cells. Keap1-expressing clone demonstrated poorer cell survival after cisplatin treatment for 72 h than parental and control cells (Fig. 4H). Furthermore, loss of Keap1 showed extraordinary self-renewal capacity in Keap1 mutant cells (Fig. 4I). Consistent with baseline differences in cell growth, Keap1 expressing clones showed poorer tumorsphere formation after cisplatin treatment with clear contrast in parental and control cells (Fig. 4J), suggesting additional evidence of therapeutic resistance in HNSCC through self-renewal of the tumor cells.
Next, we examined the level of a well-known cancer stem cell (CSC) marker CD44 mRNA by qRT-PCR in chemoresistant (n = 13) and sensitive (n = 11) HNSCC patients’ samples. Twelve chemoresistant (54.17%) out of 24 patients treated with chemotherapy showed higher expression of CD44 (>50%) compared with the corresponding chemosensitive group (Fig. 4K). In addition, mutation analysis (review Figs. 1 and 2) led us to find Keap1 mutations in four cases (17%) and Nrf2 in two cases (8%). On the basis of a positive aberration score for the mutations of Keap1 and Nrf2 and/or CD44 expression, we assigned 24 cases to two groups: 13 chemoresistant cases with 2–3 aberration scores to a “high group” and 11 chemosensitive cases with 0 and 1 aberration score to a “low score group” (Fig. 4L). Interestingly, the high score group (chemoresistant group) had worse DFS (Fig. 4M; Log-rank P < 0.0001). These results suggest that a fraction of patients treated with chemotherapeutic agents experience resistance to treatment, enhancing the CSC marker CD44 expression in addition to Keap1 mutations and Nrf2 activation in HNSCC and impacting the patients’ overall treatment outcome.
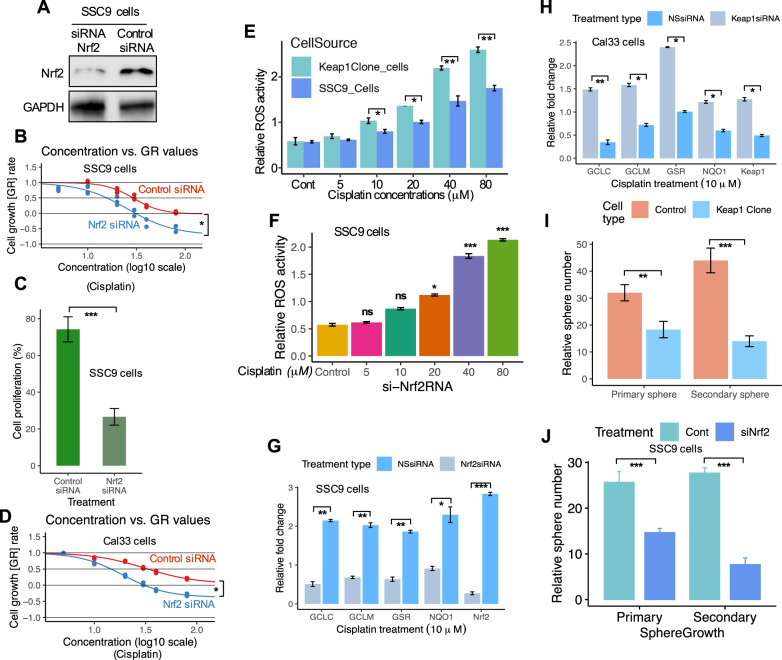
Knockdown of Nrf2 in Keap1 defective cells leads to activation of ROS-mediated stress pathway and enhances the chemosensitivity
Next, we assessed the role of Nrf2 activation and chemoresistance in Keap1-mutant SSC9 cells. First, we silenced Nrf2 expression by siRNA for Nrf2 and assessed chemosensitivity. Silencing Nrf2 by siRNA significantly reduced the endogenous Nrf2 expression and activity in SSC9 cells (Fig. 5A). Cells treated with Nrf2 siRNA showed increased sensitivity to cisplatin treatment in comparison with control siRNA-treated cells (Fig. 5B), concomitant with the decrease in cell proliferation in Keap1-mutant SSC9 cells (Fig. 5C; p < 0.001). In addition, we tested cisplatin sensitivity in Cal33 cells (Keap1 WT) and observed high sensitivity to cisplatin in Nrf2 knockdown cells (Fig. 5D; p < 0.05).
Fig. 5. Knockdown of Nrf2 in Keap1 defective cells leads to activation of ROS-mediated stress pathway and enhances the chemosensitivity.
A Nrf2 expression in SSC9 cells transfected with control or Nrf2-siRNA. GAPDH was shown as a control. B Cell survival at 72 h after cisplatin treatment in control and Nrf2-siRNA-treated SSC9 cells. C Cell proliferation of SSC9 cells after treatment with control and Nrf2-siRNA. D Cell survival at 72 h after cisplatin treatment in control or Nrf2-siRNA-treated Cal33 cells. E Intracellular ROS level measured by DCFDA staining of SSC9 and Keap1-expressing clone cells. F Intracellular ROS level measured by DCFDA staining of SSC9 cells under the treatment of Nrf2-siRNA and cisplatin. G Silencing of Nrf2 in cisplatin-treated SSC9 cells and analysis of Nrf2-dependent genes. H Inhibition of Keap1 expression by Keap1-siRNA in Cal33 cells and analysis of Nrf2-dependent genes. I Analysis of a relative number of spheres generated in primary and secondary sphere cultures in SSC9 and Keap1 clone cells. J Analysis of a relative number of spheres generated in primary and secondary sphere cultures in Nrf2 knockdown SSC9 cells. Each experiment was repeated in triplicates. Data presents as mean ± SEM (*P < 0.05; **P < 0.01, ***P < 0.001).
Cisplatin induces intrinsic apoptosis by producing mitochondrial ROS [33] leading to the induction of apoptosis. Furthermore, various antioxidant enzymes are induced by Nrf2 activation and reduce the intracellular ROS level, therefore, resulting in the cells becoming more resistant to chemotherapies [19]. Moreover, Nrf2 directly affects the homeostasis of ROS by regulating the antioxidant defense system [34]. Nrf2-mediated chemotherapeutic resistance is likely to occur due to the reduction of drug-induced ROS. Considering these, we hypothesized that Nrf2-induced chemotherapy resistance might be partially due to the decrease in drug-induced ROS generation. To address this question, we treated mock and Keap1-expressing SSC9 clones with cisplatin and assessed the mitochondrial ROS production using a fluorescent indicator. At the same time, we analyzed the ROS level in Nrf2-siRNA-treated SSC9 cells. We observed that both the Keap1-expressing clone and Nrf2 knockdown cells showed higher ROS generation under the treatment of cisplatin (Fig. 5E, F), suggesting that increased proliferation (Fig. 5C) was seen in Keap1-mutated cells is largely mediated by Nrf2. To further demonstrate that Nrf2 activation contributes to the increased expression of antioxidants, and xenobiotic metabolism enzymes, we challenged cells with Nrf2 siRNA in SSC9 cells. Transfection of Nrf2siRNA in cells decreased the Nrf2 mRNA by 70–75% with the reduction of Nrf2 target genes (Fig. 5G). Conversely, inhibition of Keap1 expression by siRNA increased the Nrf2 target genes in Cal33 cells (Fig. 5H). In addition, in vitro sphere formation assay revealed that Keap1-expressing cells decreased the growth of spheres by 1.5-fold compared with parental SSC9 cells under the cisplatin treatment condition (Fig. 5I). The growth of the sphere continued to decrease further in the secondary sphere culture approximately by 1.5–2.0-fold (Fig. 5I). In addition, Nrf2 knockdown impaired tumorsphere growth in these cells confirmed that overexpression of Nrf2 contributes to a stem-like phenotype in HNSCC cells (Fig. 5J). These data indicate that loss of Keap1 in HNSCCs leads to increased cell proliferation, and expression of Nrf2, as well as sphere growth efficiency, suggesting that Nrf2 activation and decrease of ROS and chemotherapeutic resistance is the vital mechanistic mediator observed in loss of Keap1.
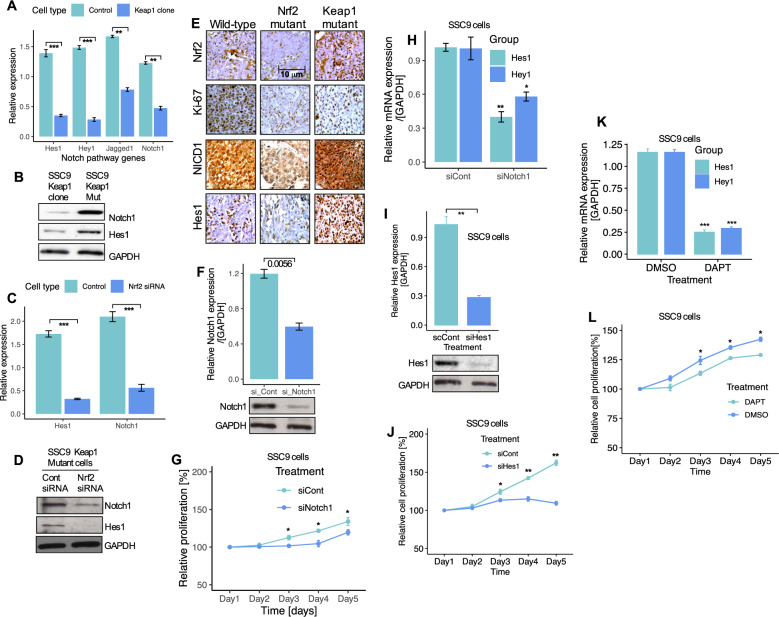
Keap1 mutations and Nrf2 overexpression regulates Notch signaling in HNSCC cells
Our clinical results indicate that Keap1 mutations are strongly associated with chemo-radio resistance. However, patients with Keap1 mutations developed tumor regrowth in the lung. Furthermore, loss of Keap1 and Nrf2 activation has previously been reported to confer resistance to chemotherapy [35, 36]. Recent studies have reported that the Notch target genes show direct downstream transcriptional mediators of Nrf2 signaling [37–40]. Moreover, previous studies have reported that activation of the Notch signaling enhances self-renewal of oral squamous cell carcinoma cancer cells, while its loss impairs the maintenance of self-renewal [41]. Therefore, we hypothesized that the Keap1-Nrf2 pathway likely modulates activation of the Notch pathway. To explore this, we first measured the expression of Notch1 and Notch target genes in Keap1-expressing clone cells. Notch1 and Notch target genes prominently decreased in Keap1-expressing cells (Fig. 6A), while their expression significantly increased in Keap1 mutant cells. Next, we tested if the Nrf2 overexpression and Keap1 mutations may have any effects on Notch activity. The expression levels of Notch1 and Hes1 were decreased in Keap1 expressing cells compared to Keap1 mutant cells (Fig. 6B). On the other hand, Nrf2 knockdown cells expressed significantly lower levels of Notch1 and Hes1 mRNA (Fig. 6C), and protein compared to controls (Fig. 6C, D). These results suggest that Keap1-Nrf2 regulates Notch signaling in HNSCC. We obtained tumor tissues from Keap1 mutant, wild-type, and Nrf2 mutant patients’ samples and immunostained them for the expression of Notch1, Hes1, Ki67, and Nrf2. Immunostaining results confirmed the absence of Nrf2 in Nrf2 mutant tumor tissues and high levels of Keap1 mutant tumors (Fig. 6E). Ki-67, a cell proliferation marker, was highly expressed in Keap1-mutant tumors compared with those in Nrf2 mutant tumors (Fig. 6E). Expression of Notch1 and Hes1 were significantly highly expressed in Keap1 mutant tumors as compared with those of Nrf2-mutant tumors (Fig. 6E). These results indicated the functional role of the Keap1-Nrf2 pathway in regulating the cell proliferation and active involvement of the Notch signaling HNSCC tumors.
Fig. 6. Nrf2 regulates Notch signaling in HNSCC cells.
A Expression of Notch1 and Notch target genes mRNA in control and Keap1-expressing SSC9 clone cells. B Expression of Notch1 and Hes1 proteins in Keap1-mutant and Keap1-expressing SSC9 cells. C Notch1 and Hes1 mRNA and, D protein expression after Nrf2 knockdown in SSC9 cells. E Immunohistochemistry staining and expression of Nrf2, Ki67, Notch1, and Hes1 in HNSCC clinical samples from wild-type, Nrf2, and Keap1 mutant patients tumor tissues. F Notch1 expression in non-targeting control and Notch1 siRNA-treated SSC9 cells G Cell proliferation of SSC9 cells after knockdown of Notch1 by siRNA. H Relative mRNA expression of Hes1 and Hey1 after Notch1 knockdown in SSC9 cells. I Hes1 mRNA expression and, J Cell proliferation after knockdown of Hes1 siRNA in SSC9 cells. K Effects of Notch inhibitor DAPT and, L Assessment of cell growth after treating the cells with Notch inhibitor DAPT for 5-days. The mRNA expression levels were calculated and normalized relative to GAPDH. All experiments were run in triplicate and compared with the control group. Data presents as mean ± SEM (**P < 0.01, ***P < 0.001).
As shown in Figs. 5J and 6D, knockdown of Nrf2 significantly impaired tumorspheres and downregulation of Notch1 and Hes1. Therefore, we tested the possibility of the Notch pathway as a target for directed therapy by exploring its functional consequences in Nrf2-activated HNSCC cells with Keap1 mutations. First, we inhibited Notch1 activity by siRNA in SSC9 cells. Following transfection of cells with Notch1 siRNA, cells showed a significant decrease in cell proliferation (Fig. 6F, G), coupled with a significant reduction in two Notch pathway target genes, Hes1 and Hey1 (Fig. 6H). We then assessed whether inhibition of Hes1 also shows any impact on cell growth. Congruent with the results obtained for Notch1 inhibition in Fig. 6G, inhibition of Hes1 significantly decreased cell proliferation (Fig. 6I, J). Similarly, treatment of cells with a Notch pathway inhibitor DAPT significantly inhibited cell growth (Fig. 6K, L).
Keap1 mutation is a strong predictor of chemotherapeutic outcomes in patients with advanced HNSCC
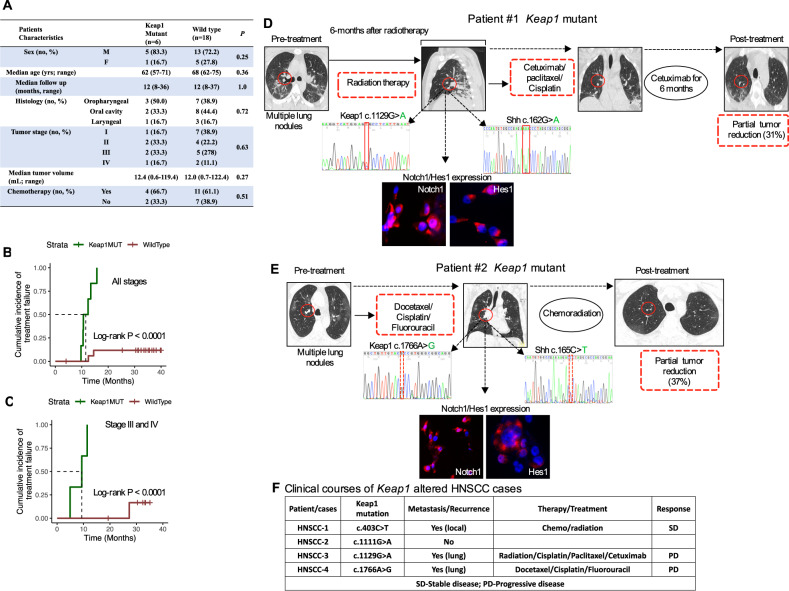
Given the fact that Keap1 mutations and Nrf2 overexpression lead to chemotherapeutic resistance, we, therefore, hypothesized that Keap1 mutations might lead to an increased rate of local recurrence in advanced HNSCC patients treated with chemotherapeutic agents. As described in Figs. 1 and 2, patients with Keap1 mutations were predicted to be deleterious and had a cumulative incidence of local treatment failure at ten months which was 80% in patients whose tumors carried Keap1 mutations as compared with <12% in patients with wild-type tumors (Log-rank p < 0.0001), while analyzing the patients with higher stages, particularly in patients with stage III–IV had higher rates of treatment failure in patients who harbored Keap1 mutations (Log-rank p < 0.0001) (Fig. 7A–C).
Fig. 7. Combination therapy with cetuximab, paclitaxel, and cisplatin led to a partial response in a patient with Keap1 mutant advanced-stage metastatic HNSCC.
A Clinical characteristic of head and neck cancer patient cohort treated with chemotherapy and analyzed for Keap1 and Nrf2 mutation by Sanger sequencing. B, C Association Keap1 mutations and local treatment failure in patients with HNSCC treated with chemotherapy. B Patient cohort and, C Stage III–IV patients who were treated with chemotherapy. D, E Tumor progression in an index patient with lung metastasis was associated with the identification of Keap1 and Shh mutations and tumors from index patients with Keap1 mutant strongly expressed Notch1 and Hes1. In both cases, patients with Keap1 mutations achieved a partial response to 31% and 37% reduction, respectively, in the metastatic lung region upon treatment with two lines of chemoradiation/cetuximab (patient case #1) and three cycles of TPE (docetaxel, cisplatin, and fluorouracil (TPF) followed by chemoradiation with cisplatin treatment (patient case #2). F Clinical courses/outcomes of Keap1 mutant HNSCC patient treated with chemoradiation therapy. SD stable disease, PD progressive disease.
Combination therapy with cetuximab, paclitaxel, and cisplatin led to a partial response in a patient with Keap1 mutant advanced-stage metastatic HNSCC
The Keap1-Nrf2 pathway has been shown to cancer cell survival and mutations in Keap1 or Nrf2 are clinically relevant predictive biomarkers of chemo-radio resistance [30]. The standard chemotherapy for advanced-stage HNSCC patients are cisplatin, 5-fluorouracil, and docetaxel/paclitaxel, and show improved progression-free and overall survival [42]. Furthermore, the most common sites of distant metastases were reported to be the lung (70%) followed, by the liver (42%) and bones (15%) [43, 44]. However, chemotherapy resistance results in poor treatment outcomes, and the reasons for chemotherapy resistance are diverse and multifaceted. To identify the Keap1 mutations and associated chemo-radio resistance, we present two case reports that recently underwent a combination of chemotherapy and related resistance to therapy. Patient #1 is a 59-year-old male visiting the clinic with advanced metastatic squamous cell carcinoma of the laryngeal. The patient received initial radiotherapy followed by two lines of chemotherapy (cetuximab/paclitaxel/cisplatin [CPP]). The patient had rapid disease progression and underwent biopsy and genotyping of lung metastasis that revealed Keap1 mutations (Fig. 7D). Following CPP, the patient was then treated with cetuximab weekly for six months and achieved a partial response (PR) but had rapid disease progression and eventually succumbed as a consequence of the disease. Patient #2 is a 64-year-old male diagnosed with metastatic oral cavity cancer and initially treated with laryngeal-preservation surgery, followed by three cycles of docetaxel, cisplatin, and continuous infusion of fluorouracil followed by a chemoradiation with cisplatin. The patient achieved a PR. Approximately after 9 months, recurrence was observed, and CPP was initiated. Following CPP, the patients had rapid disease progression and underwent biopsy and genotyping of lung metastasis and found Keap1 mutations (Fig. 7E). Further disease progression of oral cavity cancer was observed shortly after the completion of cycle 3. In both cases, sequencing of cells from lung metastatic site showed Keap1 and Shh mutations and strong expression of Notch1 and Hes1 confirmed the activation of the Notch pathway. Thus, among recurrent and metastatic HNSCC patients, Keap1 mutations appear to be the most significant cause of clinical chemo-radio resistance (Fig. 7F) and are coupled with the activation of the Notch signaling pathway.
Discussion
Loss of Keap1 enhances the nuclear accumulation of Nrf2 followed by the elevated expression of anti-oxidative, anti-xenobiotic stress enzymes and drug efflux pumps [45]. In the present study, we sought to elucidate the alterations of the Keap1-Nrf2 pathway and identify the mechanisms of chemotherapeutic resistance in HNSCC. We identified mutations that occur more frequently in the oral cavity in HNSCC, associated with the changes in amino acids in 4 (17%) tumors out of 24 HNSCC patient tumors sequenced, which is much higher than the TCGA data set. The possible putative reason for the higher incidence of Keap1 mutations in our case is unknown however, we speculate that demographic and genetic makeup may play roles in the higher incidence of Keap1 mutations in HNSCC patients. In this study, we found that all Keap1 mutated tumors exhibited nuclear accumulation of Nrf2 as assessed by immunohistochemical analysis. Keap1 mutations result in the accumulation and activation of Nrf2 and may partly confer the resistance to chemotherapeutic treatment in HNSCC patients and reduce drug-induced ROS production. Importantly, all these mutations involved functionally relevant domains of Keap1 protein, including BTB (c.403 C > T and c.1129 G > A), IVR domain (c.1111 G > A), and KR3 region (c.1766A > G) of Keap1, which are responsible for the ubiquitination and binding of Nrf2 [46, 47]. The activation of the Nrf2 pathway has been proposed to be the leading cause of chemoresistance in several cancers [48, 49]. To identify the mechanisms associated with Nrf2 pathway activation in HNSCC, we sequenced the Keap1 and Nrf2 genes and identified mutations in Nrf2 immunopositive tumors. Interestingly, the Nrf2 mutations involved the Neh2 domain where Keap1 binds. Importantly, we did not detect any mutations in the matched normal tissues, confirming that the mutation is somatic in origin. Furthermore, the overall frequency of mutations (17%; 4/24) for the Keap1 gene in HNSCC tissues suggests that Keap1 mutations are likely a frequent genetic alteration in HNSCC at least in our case which is much higher than that of data reported in TCGA. In addition, we assessed whether the Nrf2 activation pathway is engaged in mediating chemotherapeutic resistance in HNSCC. We used human HNSCC primary tumor cells and HNSCC cell lines to evaluate the functional relationship between Nrf2 activation and chemotherapeutic resistance. Knockdown of Keap1 by siRNA in HNSCC cells demonstrated enhanced Nrf2 pathway activity, which led to enhanced transcriptional activity thereby rendering HNSCC cells resistant to chemotherapy. We have achieved a comparable result in HNSCC tumor and normal tissue, where the loss of functional Keap1 gene and subsequently increased staining intensity of Nrf2 corroborate the above findings. In concordance with the above findings, as expected, GST, NQO1, and SOD1 enzyme levels and GSH levels were highly significantly elevated in the tumor tissues compared with matched normal individuals. This suggests that upregulation of these Nrf2-dependent genes likely contributed to the resistance to chemotherapy treatment and cell survival. This result is in agreement with previously published results, where high antioxidant capacity increases cell survival and proliferation and protects against oxidants, radiation therapy, and chemotherapies [10].
Loss of Keap1 and Nrf2 overexpression induces many stress resistance genes and can restore cancer cell proliferation. It has previously shown that constitutive activation of Nrf2 contributes to tumorigenesis, ROS detoxification, and modulation of redox state and also contributes to resistance to many anticancer drugs [35, 50]. Loss of Keap1 has been identified as a possible mechanism of chemoradiation resistance in many cancers [51]. In the lung cancer cells, Keap1 mutations showed more resistance to etoposide and carboplatin than to Keap1 wild-type cells [45]. Thus, overexpression of Nrf2 due to Keap1 loss may confer resistance to cisplatin, a widely used chemotherapy regimen for head and neck cancer, by regulating ROS and cancer stem cell pathways. In concordance with these previous results, our results demonstrated that downregulation of the Nrf2 expression in Keap1 mutated cancer cells or introduction of Keap1 cDNA in Keap1 mutant cells significantly enhances the sensitivity to cisplatin. In analyzing the clinical cases, all four Keap1 mutated HSNCC patients had a history of recurrence within a year all cases were treated with 2 and 3 lines of chemotherapy and exhibited poor clinical response, suggesting therapeutic resistance due to Keap1 mutations as well as the potential existence of CSC populations in these tumors.
Our observation from this study suggests a pivotal role for Keap1 mutations during HNSCC oncogenesis due to the deletion of Keap1 led to the increased self-renewal activity of cancer cells and subsequent therapeutic resistance. Importantly the impact of Keap1 loss leads to increased self-renewal activity and therapeutic resistance compared to Keap1 wild-type cells or Keap1 reintroduced clones which define the greater clinical relevance. Previous reports have demonstrated the molecular mechanisms of cisplatin resistance in cancer cells and cisplatin treatment increased mitochondrial ROS generation [52] and triggered the apoptosis process [53, 54]. In our analysis, when we restored Keap1 or silenced Nrf2 observed increased mitochondrial ROS generation and limited the cell growth in cells treated with cisplatin. This suggests that constitutive Nrf2 activation due to Keap1 loss influences oxidative stress and reduces the cellular damage induced by the increase of ROS, therefore, triggering the resistance of chemotherapy, particularly for cisplatin. Han and colleagues [41] recently reported the interaction of Nrf2 and Notch signaling in OCSC (oral squamous cell carcinoma) cells. Activation of Nrf2 in Keap1 deleted cells resulted in hyperproliferation of squamous epithelial cells and activation of Notch signaling [41]. Congruent with this finding, our results report a role in chemoresistance on HNSCC cells which is mediated through increased Notch signaling and increased Notch pathway components upon Nrf2 activation in Keap1 mutated cells, suggesting that Notch also plays a role in mediating the effects of Nrf2 activation in HNSCC cells.
Reviewing all our results, it appears that once HNSCC cells acquire a mutation in Keap1, activation of Nrf2 and the Notch signaling pathway promotes cellular metabolic reprogramming that sustains cellular proliferation. This reprogramming feature leads to the clonal expansion of mutant cells thus acquiring self-renewal capacity and triggering resistance pathways and outpacing the Keap1 WT cells and potentially acquiring additional genetic changes and subsequently leading to therapeutic failures. All these features explain the therapeutic failure and adverse and poor survival outcomes conferred by Keap1 mutations in rapidly progressing HNSCC cells. In a recent study, it was reported the Keap1-Nrf2 double mutations in lung cancer leads to poor outcome and severe therapeutic resistance in radiation therapy [38]. In our study, we found two patients with Nrf2 mutation and none of the patients had concurrent Keap1-Nrf2 mutations. This may be due to the small number of patients assessed in this study and we cannot rule out that Keap1-Nrf2 dual mutations may have somatic or epigenetic consequences and may play a devastating role in therapeutic failures and poor outcomes, disease recurrence in Keap1-Nrf2-mutant tumors. Thus, future studies should include large numbers of patients’ samples to examine the effects of Keap1 mutation as well as to explore the role of Keap1-Nrf2-dual mutation in chemoresistance.
In conclusion, although Keap1–Nrf2 alterations are known to play roles in chemotherapeutic resistance particularly cisplatin resistance in HNSCC, surprisingly, the mutation status is not widely used to make a treatment decision in head and cancer. It would be interesting to investigate whether loss of Keap1 and overexpression of Nrf2 status in tumor samples are clinically relevant mechanisms of chemotherapeutic resistance in HNSCC and develop alternative therapy to counter Nrf2 activation. This may improve personalized therapy in a subset of HNSCC patients who are prone to therapeutic resistance.
Supplementary information
Reporting summer/Reproducibility checklist
Acknowledgements
The authors would like to sincerely acknowledge all the healthcare personnel and patients involved in this study. We deeply acknowledge the assistance of Dr. A.K. Murugan in designing primers for sequence analysis. The authors acknowledge that they received no funding in support of this research.
Author contributions
SSI has contributed to the conception and design and overall supervision of the study, SSI, ASMN, KQ, and BK contributed to data analysis. SSI, ASMN contributed to writing the manuscript. SSI, ASMN, HY, WAF, AA, and BK contributed to reviewing the manuscript. SSI, ASMN, KQ, and BK have contributed to literature searches, data collection, data interpretation, and clinical data acquisition. All authors reviewed the manuscript and have given final approval to the submitted version.
Data availability
The data that support our results in this follow-up study are available from the corresponding authors upon reasonable request with prior permission from Parkview hospital, Chittagong, Bangladesh.
Ethical approval
The patient and patient specimen collection was conducted according to the protocols approved by the ethics committee of the Chittagong Medical College Hospital, Bangladesh, Bangladesh Medical Research Council (BMRC), and the King Faisal Specialist Hospital and Research Centre (KFSH&RC), and informed consent was obtained from all patients before the specimens and information collection.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Professor Rami Aqeilan
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-022-05126-8.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chow LQM. Head and neck cancer. N. Engl J Med. 2020;382:60–72. doi: 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- 3.López-Verdín S, Lavalle-Carrasco J, Carreón-Burciaga RG, Serafín-Higuera N, Molina-Frechero N, González-González R, et al. Molecular markers of anticancer drug resistance in head and neck squamous cell carcinoma: a literature review. Cancers. 2018;10:376. doi: 10.3390/cancers10100376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drinberg V, Bitcover R, Rajchenbach W, Peer D. Modulating cancer multidrug resistance by sertraline in combination with a nanomedicine. Cancer Lett. 2014;354:290–8. doi: 10.1016/j.canlet.2014.08.026. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taguchi K, Yamamoto M. The KEAP1–NRF2 system in cancer. Front Oncol. 2017;7:85. doi: 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Wijst MGP, Brown R, Rots MG. Nrf2, the master redox switch: the Achilles’ heel of ovarian cancer? Biochim Biophys Acta. 2014;1846:494–509. doi: 10.1016/j.bbcan.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Torrente L, Maan G, Rezig AO, Quinn J, Jackson A, Grilli A, et al. High NRF2 levels correlate with poor prognosis in colorectal cancer patients and with sensitivity to the kinase inhibitor AT9283 in vitro. Biomolecules. 2020;10:1–16. doi: 10.3390/biom10101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu T, Harder BG, Wong PK, Lang JE, Zhang DD. Oxidative stress, mammospheres and Nrf2 – new implication for breast cancer therapy? Mol Carcinog. 2015;54:1494. doi: 10.1002/mc.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price KAR, Cohen EE. Current treatment options for metastatic head and neck cancer. Curr Treat Options Oncol. 2012;13:35–46. doi: 10.1007/s11864-011-0176-y. [DOI] [PubMed] [Google Scholar]
- 12.Bauml JM, Vinnakota R, Park Y-HA, Bates SE, Fojo T, Aggarwal C, et al. Cisplatin every 3 weeks versus weekly with definitive concurrent radiotherapy for squamous cell carcinoma of the head and neck. J Natl Cancer Inst. 2019;111:490. doi: 10.1093/jnci/djy133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D, Wang C-Y. Targeting cancer stem cells in squamous cell carcinoma. Precis Clin Med. 2019;2:152. doi: 10.1093/pcmedi/pbz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noman ASM, Parag RR, Rashid MI, Islam S, Rahman MZ, Chowdhury AA, et al. Chemotherapeutic resistance of head and neck squamous cell carcinoma is mediated by EpCAM induction driven by IL-6/p62 associated Nrf2-antioxidant pathway activation. Cell Death Dis. 2020;11:1–15. doi: 10.1038/s41419-020-02907-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noman ASM, Parag RR, Rashid MI, Rahman MZ, Chowdhury AA, Sultana A et al. Widespread expression of Sonic hedgehog (Shh) and Nrf2 in patients treated with cisplatin predicts outcome in resected tumors and are potential therapeutic targets for HPV-negative head and neck cancer. Ther Adv Med Oncol 2020;12:1758835920911229. [DOI] [PMC free article] [PubMed]
- 16.Sparaneo A, Fabrizio FP, Muscarella LA. Nrf2 and notch signaling in lung cancer: near the crossroad. Oxid Med Cell Longev. 2016;2016. [DOI] [PMC free article] [PubMed]
- 17.Chen X, Zhao W, Chen S, Yu D. Mutation profiles of oral squamous cell carcinoma cells. Adv Oral Maxillofac Surg. 2021;2:100026. doi: 10.1016/j.adoms.2021.100026. [DOI] [Google Scholar]
- 18.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, et al. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget. 2014;5:8906–23. doi: 10.18632/oncotarget.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–68. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K et al. Molecular cloning and functional characterization of a new Cap‘n’ collar family transcription factor Nrf3. Biol Chem. 1999;274:6443–52. [DOI] [PubMed]
- 21.Hartikainen JM, Tengström M, Kosma VM, Kinnula VL, Mannermaa A, Soini Y. Genetic polymorphisms and protein expression of NRF2 and sulfiredoxin predict survival outcomes in breast cancer. Cancer Res. 2012;72:5537–46. doi: 10.1158/0008-5472.CAN-12-1474. [DOI] [PubMed] [Google Scholar]
- 22.Martin D, Degese MS, Vitale-Cross L, Iglesias-Bartolome R, Valera JLC, Wang Z et al. Assembly and activation of the Hippo signalome by FAT1 tumor suppressor. Nat Commun. 2018;9:2372. [DOI] [PMC free article] [PubMed]
- 23.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagnostics. 2015;17:251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88:101. doi: 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:1–22. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian Y, Wu K, Liu Q, Han N, Zhang L, Chu Q et al. Modification of platinum sensitivity by KEAP1/NRF2 signals in non-small cell lung cancer. J Hematol Oncol. 2016;9:83. [DOI] [PMC free article] [PubMed]
- 28.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–47. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 29.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed]
- 30.Binkley MS, Jeon YJ, Nesselbush M, Moding EJ, Nabet BY, Almanza D, et al. KEAP1/NFE2L2 mutations predict lung cancer radiation resistance that can be targeted by glutaminase inhibition. Cancer Discov. 2020;10:1826. doi: 10.1158/2159-8290.CD-20-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res. 2019;79:3251. doi: 10.1158/0008-5472.CAN-18-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, Leboeuf SE, et al. Keap1 loss promotes Kras-driven lung cancer andresults in a dependence on glutaminolysis. Nat Med. 2017;23:1362. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleih M, Böpple K, Dong M, Gaißler A, Heine S, Olayioye MA, et al. Direct impact of cisplatin on mitochondria induces ROS production that dictates cell fate of ovarian cancer cells. Cell Death Dis. 2019;10:1–12. doi: 10.1038/s41419-019-2081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Q. Role of Nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–26. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife. 2017;6:e18970. [DOI] [PMC free article] [PubMed]
- 36.Sanghvi VR, Mohan P, Singh K, Cao L, Berishaj M, Wolfe AL, et al. NRF2 activation confers resistance to eIF4A inhibitors in cancer therapy. Cancers (Basel) 2021;13:1–13. doi: 10.3390/cancers13040639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul M, Bisht B, Darmawan D, Chiou R, Ha V, Wallace W, et al. Dynamic changes in intracellular ROS levels regulate airway basal stem cell homeostasis through Nrf2-dependent Notch signaling. Cell Stem Cell. 2014;15:199. doi: 10.1016/j.stem.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiotherapy response prediction. Cancer Discov. 2017;7:86. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak M-K, et al. Regulation of Notch1 signaling by Nrf2: implications for tissue regeneration. Sci Signal. 2010;3:ra52. doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan H, Paiboonrungruan C, Zhang X, Prigge JR, Schmidt EE, Sun Z, et al. Nrf2 regulates cellular behaviors and Notch signaling in oral squamous cell carcinoma cells. Biochem Biophys Res Commun. 2017;493:833. doi: 10.1016/j.bbrc.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vermorken JB, Remenar E, van Herpen C, Gorlia T, Mesia R, Degardin M, et al. Cisplatin, fluorouracil, and docetaxel in unresectable head and neck cancer. N Engl J Med. 2007;357:1695–704. doi: 10.1056/NEJMoa071028. [DOI] [PubMed] [Google Scholar]
- 43.Zbären P, Lehmann W. Frequency and sites of distant metastases in head and neck squamous cell carcinoma: an analysis of 101 cases at autopsy. Arch Otolaryngol Neck Surg. 1987;113:762–4. doi: 10.1001/archotol.1987.01860070076020. [DOI] [PubMed] [Google Scholar]
- 44.Garavello W, Ciardo A, Spreafico R, Gaini RM. Risk factors for distant metastases in head and neck squamous cell carcinoma. Arch Otolaryngol Neck Surg. 2006;132:762–6. doi: 10.1001/archotol.132.7.762. [DOI] [PubMed] [Google Scholar]
- 45.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–9. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 46.Magesh S, Chen Y, Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med Res Rev. 2012;32:687. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canning P, Cooper CDO, Krojer T, Murray JW, Pike ACW, Chaikuad A, et al. Structural basis for Cul3 protein assembly with the BTB-kelch family of E3 ubiquitin ligases. J Biol Chem. 2013;288:7803. doi: 10.1074/jbc.M112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91:386–96. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 49.Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–32. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 50.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez VD, Vucic EA, Thu KL, Pikor LA, Lam S, Lam WL. Disruption of KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex components by multiple genetic mechanisms: association with poor prognosis in head and neck cancer. Head Neck. 2015;37:727–34. doi: 10.1002/hed.23663. [DOI] [PubMed] [Google Scholar]
- 52.So H, Kim H, Lee J-H, Park C, Kim Y, Kim E, et al. Cisplatin cytotoxicity of auditory cells requires secretions of proinflammatory cytokines via activation of ERK and NF-κB. JARO J Assoc Res Otolaryngol. 2007;8:338. doi: 10.1007/s10162-007-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Ježek P, Hlavatá L. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int J Biochem Cell Biol. 2005;37:2478–503. doi: 10.1016/j.biocel.2005.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reporting summer/Reproducibility checklist
Data Availability Statement
The data that support our results in this follow-up study are available from the corresponding authors upon reasonable request with prior permission from Parkview hospital, Chittagong, Bangladesh.