Abstract
Proteins induced by acid or base, during long-term aerobic or anaerobic growth in complex medium, were identified in Escherichia coli. Two-dimensional gel electrophoresis revealed pH-dependent induction of 18 proteins, nine of which were identified by N-terminal sequencing. At pH 9, tryptophan deaminase (TnaA) was induced to a high level, becoming one of the most abundant proteins observed. TnaA may reverse alkalinization by metabolizing amino acids to produce acidic products. Also induced at high pH, but only in anaerobiosis, was glutamate decarboxylase (GadA). The gad system (GadA/GadBC) neutralizes acidity and enhances survival in extreme acid; its induction during anaerobic growth may help protect alkaline-grown cells from the acidification resulting from anaerobic fermentation. To investigate possible responses to internal acidification, cultures were grown in propionate, a membrane-permeant weak acid which acidifies the cytoplasm. YfiD, a homologue of pyruvate formate lyase, was induced to high levels at pH 4.4 and induced twofold more by propionate at pH 6; both of these conditions cause internal acidification. At neutral or alkaline pH, YfiD was virtually absent. YfiD is therefore a strong candidate for response to internal acidification. Acid or propionate also increased the expression of alkyl hydroperoxide reductase (AhpC) but only during aerobic growth. At neutral or high pH, AhpC showed no significant difference between aerobic and anaerobic growth. The increase of AhpC in acid may help protect the cell from the greater concentrations of oxidizing intermediates at low pH. Isocitrate lyase (AceA) was induced by oxygen across the pH range but showed substantially greater induction in acid or in base than at pH 7. Additional responses observed included the induction of MalE at high pH and induction of several enzymes of sugar metabolism at low pH: the phosphotransferase system components ManX and PtsH and the galactitol fermentation enzyme GatY. Overall, our results indicate complex relationships between pH and oxygen and a novel permeant acid-inducible gene, YfiD.
Escherichia coli and related enteric bacteria show a number of genetic responses to change in pH (for reviews, see references 8, 35, and 41). For example, the lysine and arginine decarboxylase systems in E. coli remove acid from the cell and help alkalinize its growth medium. The glutamate decarboxylase system appears particularly important for protection from extreme acid (12, 22, 55). At high pH, the Na+/H+ antiporter helps maintain internal pH and protects cells from excess sodium (11). Many questions remain, however, particularly with regard to the mechanisms of regulation of internal pH, which remains near pH 7.6 over a wide range of pH values of growth (pH 4.5 to 9.0) (see references 43 and 56). Survival in acidic environments contributes to the pathogenesis of E. coli O157:H7 (23) and of Helicobacter pylori (30), and virulence factors such as ToxR in Vibrio cholerae respond to acid (29). Survival in base is significant because enteric bacteria encounter alkaline stresses such as pancreatic secretion, and in complex media, their own metabolism can raise the pH as high as pH 9 (20, 42).
The effects of pH on gene expression are known to intersect with a number of other environmental factors, most notably oxygenation. For example, the acidic induction of amino acid decarboxylases is increased by anaerobiosis (28, 31), whereas the oxygen-induced cytochrome o is repressed by acid (5, 41).
We report the results of a proteomic investigation of the relationships between acid and base stress, and between pH stress and anaerobiosis, in E. coli. Previously, Neidhardt and colleagues have used two-dimensional gel electrophoresis (2-D gels) to elucidate patterns of protein response to many environmental conditions (51), including aerobiosis and anaerobiosis (47, 48) (see reference 24 for a review of oxygen regulation). Nevertheless, relatively few proteomic studies have been done of pH stress (14) or of stress by permeant acids which depress internal pH (19, 38).
In our study, we observed steady-state expression of proteins from cultures grown for several generations in a stress condition. Proteins separated on 2-D gels were stained and isolated for N-terminal sequence analysis (34) and then assigned to known genes and open reading frames within the E. coli genome. Thus, our approach emphasized long-term adaptations to stress, rather than short-term responses to sudden change, such as heat shock or universal stress response. While our particular gel conditions covered only a limited subset of the entire E. coli proteome, they nonetheless revealed unexpected and previously unknown patterns of response.
We addressed the following questions. What proteins are induced at the acidic limit for growth (in our media, pH 4.4 to 4.5)? Are any of them also induced by a permeant acid that depresses internal pH? What proteins are induced at the alkaline limit for growth (pH 9.0 to 9.2)? Does anaerobic or aerobic growth affect pH-dependent proteins? We characterized several intriguing effects of pH and anaerobiosis on expression of proteins which may help the cell resist pH stress.
MATERIALS AND METHODS
Growth conditions.
E. coli K-12 W3110 (48) was grown under various conditions as shown below (Table 1). All media contained LBK broth (10 g of tryptone, 5 g of yeast extract, 6.4 g of KCl) containing a pH-appropriate sulfonate buffer at 100 mM (41, 42). Buffers used included homopiperazine-N,N′-bis-2(ethanesulfonic acid) (HOMOPIPES), 2-(N-morpholino)ethanesulfonic acid (MES), piperazine-N,N′-bis-(2-ethanesulfonic acid) (PIPES), 3-(N-morpholino)propanesulfonic acid (MOPS), 3-[(1,1-dimethyl-2-hydroxyethyl)amino]-2-hydroxypropanesulfonic acid (AMPSO), and 3-[N-tris(hydroxymethyl)methylamino]propanesulfonic acid (TAPS). Media were adjusted for pH with KOH, to avoid high concentrations of sodium ion, which inhibits growth at high pH (15, 17).
TABLE 1.
Growth conditions for culturesa
| Growth condition | Buffer (100 mM) | Oxygen | pHb | Growth time (h) |
|---|---|---|---|---|
| Acid | HOMOPIPES or MES | + | 4.4 | 13–20 |
| − | 4.5 | 40–50 | ||
| Neutral | PIPES or MOPS | + | 7.0 | 1.7 |
| − | 7.0 | 3 | ||
| Base | AMPSO or TAPS | + | 9.2 | 10–35 |
| − | 9.0 | 24–48 | ||
| Propionic acid | MES with 50 mM propionate | + | 6.0 | 53 |
| − | 6.2 | 24 |
All media contained buffered LBK. Cultures were grown to an optical density at 600 nm of 0.35 to 0.50 at 37°C, with or without aeration as indicated.
Initial pH of broth at inoculation. Final pH after growth changed by less than 0.3 U.
Cultures were grown overnight and then diluted 100-fold in fresh medium and grown to an optical density at 600 nm of 0.35 to 0.50 at 37°C, either oxygenated (rotary aeration, in a flask whose capacity was 10 times the culture volume) or without oxygenation (rotated in a closed tube). Growth under stress (extreme pH and/or propionate concentration) was defined as growth requiring more than 10 h to reach mid-log phase. Growth within the range of pH 6 to 7, in the absence of propionic acid, was defined as an unstressed condition.
2-D gel electrophoresis.
The overall procedure for 2-D gels was based on procedures described in references 19 and 51, with the ESA Investigator 2-D electrophoresis system. Thirteen milliliters of each culture was chilled, pelleted, and washed with LBK. Cell pellets were then treated with urea-sodium dodecyl sulfate sample buffer and DNase-RNase, according to ESA procedures. Samples were applied to isoelectric focusing gels containing Ampholine solution pH 3 to 10, generating a functional gradient of approximately pH 4.5 to 6.5. Tube gels were applied to second-dimension electrophoretic gels containing 11.5% Duracryl. For comparative analysis, gels were stained with Coomassie blue. The choice of stain was based on the observation that proteins detected with Coomassie blue are generally present in sufficient quantity to enable determination of N-terminal sequence (52).
For comparison of spot densities between different growth conditions, gels were scanned and digitized. Spot densities were quantified with Paint Shop Pro (Jasc), normalized to the overall density of the gel proteins. For each growth condition, spot densities were determined on three gel images from three independently grown cultures. The mean of the normalized spot densities is reported in Fig. 3. Some proteins appeared in a major spot and a minor spot; in these cases, only the major spot was quantified. The pI and molecular weight scales for the gels are based on comparison with E. coli reference gels in reference 51 and in the SWISS-2DPAGE database of the Swiss Institute of Bioinformatics (50a).
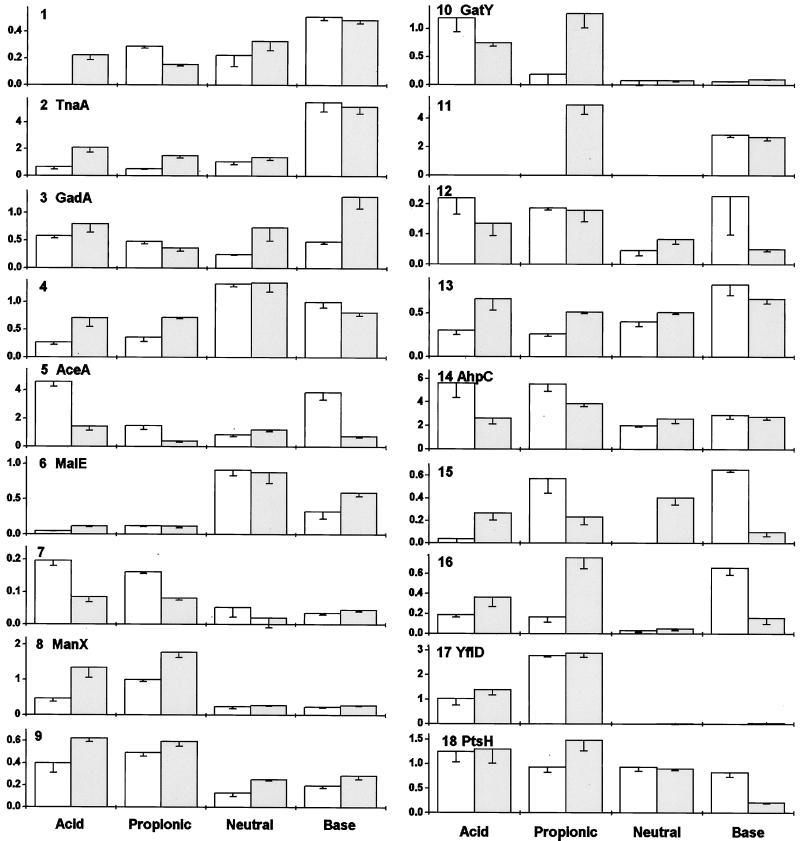
FIG. 3.
Quantification of protein expression from Fig. 1 and 2. Spot images were measured and normalized as stated in Materials and Methods. The vertical axis represents arbitrary units of expression. Open bars, aerobic growth; gray bars, anaerobic growth. Error bars represent the standard errors of the means.
For protein determination, gels were transferred to Immobilon-P polyvinylidene difluoride membrane (Millipore) with the Investigator Graphite Electroblotter, type II (ESA). The membrane was stained with Coomassie blue; protein spots were excised for sequence determination.
Analysis of N-terminal peptides.
Protein spots cut from the transfer membrane were washed four times in 10% methanol and then dried and frozen. N-terminal sequence analysis was performed by the Biopolymer Facility at the University of California at Davis. Samples were processed with a 470A or 477A ABI sequencer with on-line high-pressure liquid chromatography, capable of picomole analysis. Peptide sequences were matched against all known and putative E. coli proteins in the Swiss-Prot database, with the ExPASy Fasta program. The theoretical molecular weight and pI of each protein were determined with the ExPASy proteomics tool Compute pI/Mw.
RESULTS
Experimental design and controls.
E. coli W3110 was grown in a heavily buffered rich broth, a condition which enables growth to greater extremes of pH than does minimal medium (22). Cultures were grown for several generations under stress conditions designed to force E. coli to grow at the extreme limits of tolerance for pH, aerobically or anaerobically (Table 1). In our media, aerobiosis permitted growth at slightly greater extreme values than did anaerobiosis (pH 4.4 versus pH 4.5; pH 9.2 versus pH 9.0).
A factor which can complicate the study of pH effects is the need for high concentrations of buffers to maintain pH during growth (42). If all the buffers needed are present under all pH conditions, then the concentration of counterions will differ greatly at the two ends of the pH range. By using different buffers at different pH conditions, we minimized the difference in K+ concentration in the different pH media. For each different pH, gels were run with at least two different pH-appropriate buffers (such as PIPES or MOPS at pH 7.0) in order to control for buffer effects; no significant effects of buffers were seen. For the stress conditions containing propionic acid, appropriate controls were run at pH 6 and at pH 6.5. The gel patterns did not differ significantly from those for the pH 7 controls.
We also designed a growth condition to acidify the cell internally while external pH was only mildly acidic (pH 6 to 6.5). This approach used an organic acid which permeates the membrane in the protonated form and then dissociates within the cytoplasm (38). At a mildly acidic pH (pH 6.0 to 6.5), the permeant acid generates a concentration gradient which draws protons into the cell, lowering internal pH. Proteins which showed induction under both extreme pH and permeant-acid conditions could be responding specifically to internal acidification, rather than external acid (41). Previously, we have used benzoic acid for this purpose (19), but in the experiments reported here, we used propionic acid, to avoid induction of proteins specifically regulated by the benzoate anion (37). Appropriate controls were run buffered at pH 6 or 6.5; the gel patterns for these controls did not differ significantly from those for the pH 7 controls.
Overall profile of proteins expressed.
A protein was considered to be pH dependent if gels from three independent cultures produced a spot density significantly different from the spot density on three gels from a different growth condition (Fig. 1 and 2). Proteins which appeared to be induced in 2-D gels were identified by N-terminal sequence analysis. This approach reveals only proteins whose expression remains elevated during long-term growth in a stress condition. It avoids induction of the generalized heat shock and universal stress systems, which occurs during an abrupt shift between growth conditions (19).
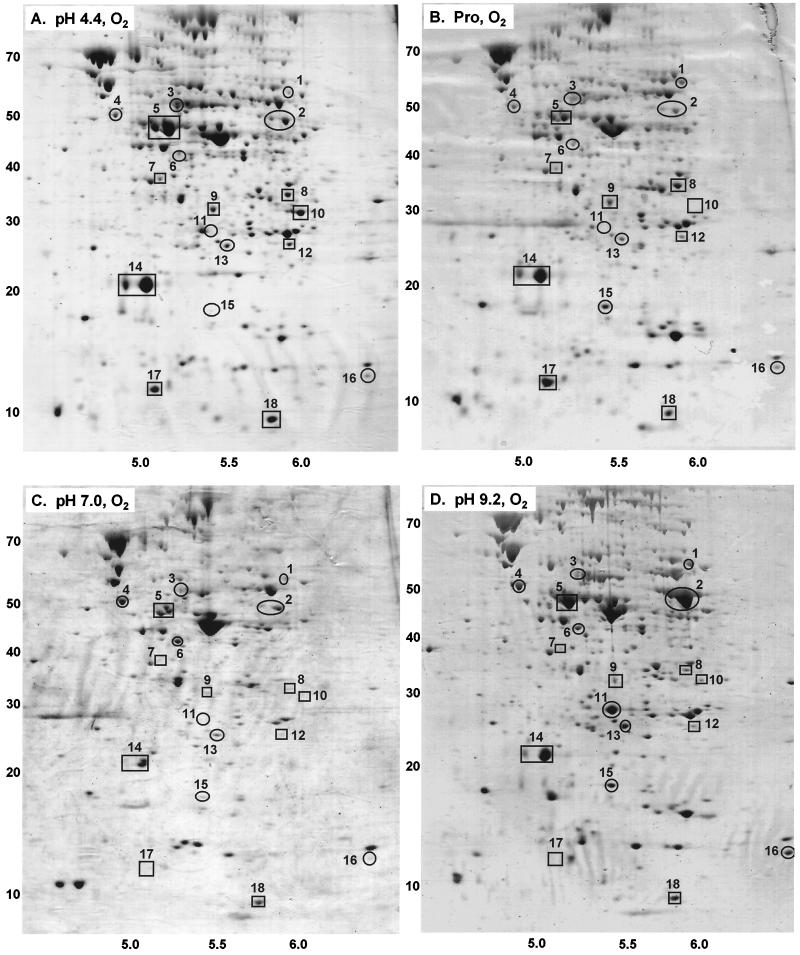
FIG. 1.
Proteins expressed during growth with aeration. Media contained LBK buffered with 100 mM HOMOPIPES, pH 4.4 (A); 100 mM MES–50 mM propionic acid, pH 6 (B); 100 mM MOPS, pH 7.0 (C); or 100 mM AMPSO, pH 9.2 (D). The horizontal axes represent pHs of the isoelectric focusing gradients; the vertical axes represent molecular weights in thousands, based on migration in the sodium dodecyl sulfate electrophoretic gel.
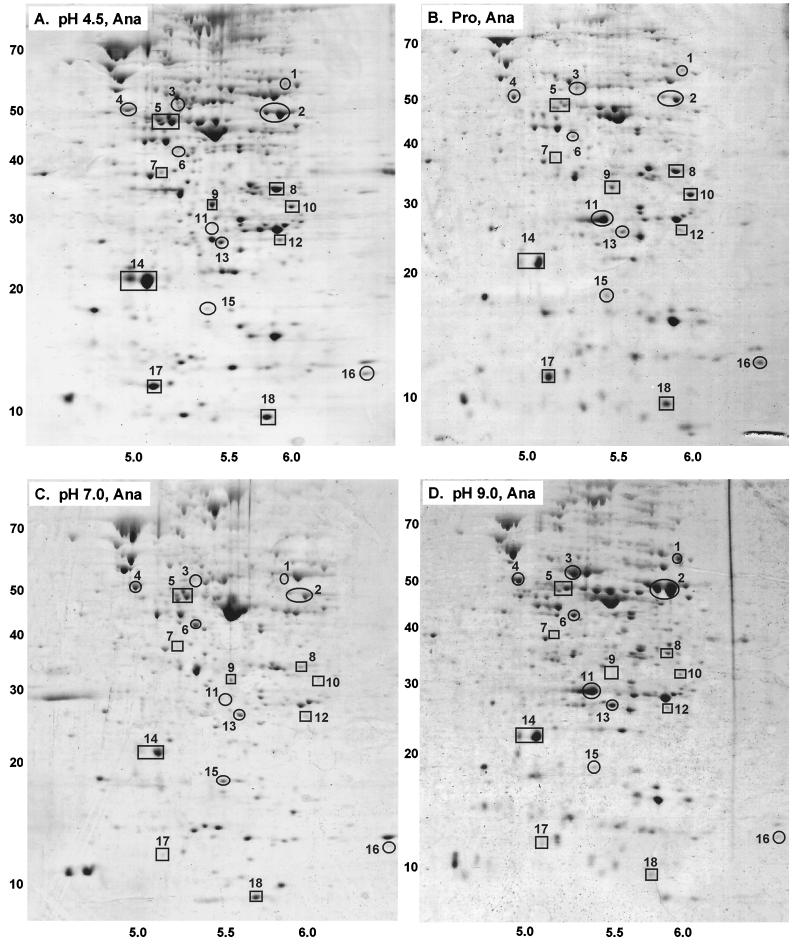
FIG. 2.
Proteins expressed during anaerobic growth. Media contained LBK buffered with 100 mM HOMOPIPES, pH 4.5 (A); 100 mM MES–50 mM propionic acid, pH 6.2 (B); 100 mM MOPS, pH 7.0 (C); or 100 mM AMPSO, pH 9.0 (D). The horizontal axes represent pH values; the vertical axes represent molecular weights in thousands.
In the 2-D gels of cultures grown at neutral pH, an estimated 300 proteins could be detected with Coomassie blue stain (Fig. 1 and 2). Under stressed conditions, somewhat fewer proteins were detected, particularly with propionic acid. Only proteins with the pI range of pH 4.5 to 6.5 would have been detected; this includes about 70% of the proteins in E. coli (51). The electrophoretic conditions and protein loading were optimized for detection and identification of peptides having molecular weights of up to 80,000.
At least 18 proteins showed reproducible differences in expression as a function of pH (Fig. 3). For nine of these, N-terminal analysis provided usable sequence data (Table 2). The computed pI and molecular weights of these proteins corresponded well with estimates based on positions of the spots in the gel images (Fig. 1 and 2).
TABLE 2.
Proteins identified by N-terminal sequence analysis
| Spot no. | N-terminal sequence | Proteina | pIb | Mol wtb |
|---|---|---|---|---|
| 2 | MENFKHLPEPFR | TnaA | 5.88 | 52,773 |
| 3 | QKLLTDFRSE | GadA | 5.22 | 52,685 |
| 5 | MKTRTQQIE | AceA | 5.16 | 47,522 |
| 6 | IEEGKLVIxIN | MalE | 5.22 | 40,707 |
| 8 | IAIVIGT | ManX | 5.74 | 34,916 |
| 10 | MYVVSTKQMLNN | GatY | 5.98 | 31,071 |
| 14 | SLINTKIKPFKN | AhpC | 5.03 | 20,630 |
| 17 | MITGIQITKAAN | YfiD | 5.09 | 14,284 |
| 18 | MFQQEVTITAPN | PtsH | 5.65 | 9,119 |
Proteins identified in the E. coli genome, with the Swiss-Prot database on the ExPASy server, as indicated in Materials and Methods.
Values for pI and molecular weight were calculated with the ExPASy proteomics tool Compute pI/Mw.
Alkaline induction of tryptophanase.
Amino acid deaminase activity is known to be increased at high pH, presumably because deaminases release acids which neutralize alkalinity (10). Nevertheless, the genetics of deaminase response to pH have not been characterized.
We found exceptionally high production of tryptophan deaminase (TnaA) at high external pH. TnaA appeared as a double spot (number 2, Fig. 1 and 2). The reason for migration at two positions is unknown; the two forms of the protein may differ with respect to posttranslational processing. The density of the TnaA doublet was equal to or greater than that of any other protein expressed at high pH, either aerobically or anaerobically. The level of expression was 3- to 10-fold greater at high pH than at any other condition tested (Fig. 3); this is probably an underestimate of the true difference, as the image density for the spot was overloaded. Under acidic conditions, anaerobiosis also slightly enhanced expression of TnaA, although never to the levels seen at high pH.
Alkaline induction of GadA during anaerobic growth.
Several amino acid decarboxylase systems are induced by acid and anaerobiosis (2) and have been implicated in acid resistance (22, 55). These systems involve a decarboxylase and a transporter for the substrate and/or product of the decarboxylase (31, 44). The putative transporter GadC for the glutamate system is required for extreme acid resistance of E. coli culture grown to stationary phase in base but is not essential for cultures grown in moderate acid (12). The glutamate system includes two homologous but unlinked glutamate decarboxylases, GadA and GadB (46); one of these, GadB, is cotranscribed with GadC (55).
A protein induced at high pH during anaerobic growth was identified as GadA (number 3, Fig. 3). The degree of induction in comparison with other growth conditions is uncertain because adjacent proteins migrated closely, contributing to the background level. Nevertheless, the N-terminal sequence analysis of GadA (from culture at pH 9, anaerobic) was unambiguous. In the aerated cultures, no induction of GadA was seen at high pH. Expression was repressed at neutral pH, with some increased expression in acid (Fig. 3). Thus, GadA expression showed a complex dependence on pH and oxygenation.
The homologous enzyme GadB has a pI and a molecular weight close to those of GadA. In some gels, a spot adjacent to GadA appeared to be induced similarly; this could be GadB, or it could be a secondary spot for GadA. Other decarboxylases, particularly for lysine (CadA) and arginine (Adi), would be expected to appear induced by acid. These proteins were not clearly separated in our gels, presumably because of their higher molecular weight (above 80,000).
Acid induction of YfiD and AhpC.
A protein induced strongly by acid and by propionate (number 17, Fig. 3) was identified as YfiD. The gene encoding YfiD maps near 58 min; it shows homology to pyruvate formate lyase, including the glycine radical motif (3, 53). In propionate (Fig. 1B and 2B), YfiD was expressed at its highest levels, one of the 10 most highly expressed protein spots observed in the gel. At low pH in the absence of propionate, expression was nearly as high. By contrast, YfiD was nearly undetectable at neutral or high pH. Aeration had no significant effect on expression. These observations make YfiD a strong candidate for internal pH-dependent expression.
Another protein whose steady-state expression was elevated by both acid and propionate was alkyl hydroperoxide reductase (AhpC), a major component of oxidative stress response (50). AhpC appeared as a doublet spot (number 14, Fig. 1 and 2); the density of the smaller component was highly variable. AhpC was expressed to some degree under all growth conditions, but its highest expression was in oxygenated cultures grown in acid or in propionate. The degree of induction under these conditions is probably underestimated, as the protein spots are overloaded. At neutral or high pH, the aerobic and anaerobic cultures showed no difference in AhpC. Anaerobic cultures showed marginal induction by propionate.
Other pH-dependent responses.
Several proteins induced by acid (Fig. 3) function in sugar metabolism (21). These included phosphotransferase system components ManX and PtsH, as well as GatY (galactitol fermentation) (33). ManX was induced either by extreme acid or by propionate, with enhanced levels during anaerobiosis. GatY was also induced by extreme acid or by propionate, but the effect of anaerobiosis appeared more complex. PtsH was expressed at significant levels under all growth conditions but was significantly decreased at high pH.
Acid and propionate repressed expression of the periplasmic maltose binding protein (MalE) (40), a result consistent with the previously reported induction of a malE::lac fusion by base (13). Two unidentified proteins, numbered 1 and 4, were also repressed by growth at low pH or in propionate. Protein number 13 was repressed by acid aerobically but not anaerobically.
A protein induced by either acid or base, compared to pH 7, was isocitrate lyase (AceA). AceA is a glyoxalate shunt enzyme required for growth on acetate or fatty acids and induced by oxygen (6, 16, 25). We found long-term aerobic induction of AceA only in acid or base; at pH 7, oxygenation had no effect. Propionate also had no effect on AceA.
DISCUSSION
Our 2-D gels revealed several long-term responses to pH and oxygenation. Previously, Hickey and Hirshfield observed a number of proteins induced in supplemented minimal medium by a shift from pH 7 to pH 5 (14); about half of these persisted during growth at pH 5. Proteins identified include dihydrolipoamide acetyltransferase, RNA polymerase B, and DNA polymerase I. The reason for their induction at lower pH is not clear. In cultures shocked by addition of the permeant acid benzoate (19), 33 proteins are induced, of which 14 are identified, all but two of them heat shock or general stress proteins. In our present study, cultures were grown for several generations in buffered complex medium, at the extreme low and high limits of pH.
Most studies of stress response aim to isolate a single factor, such as temperature or oxygen. Yet evidence shows that in complex natural environments, such as intracellular growth, the actual induction patterns look nothing like the results from isolated stress conditions (1). While single-stress studies are important, it is also useful to observe combinations of stress conditions, such as the interaction between pH and aerobiosis. The value of the proteomic approach is that it reveals genes of potential interest that might otherwise have gone unstudied. These observations can then be extended by detailing the regulation of individual genes.
Alkaline induction of TnaA.
Tryptophanase is best known for its role in degrading amino acids for catabolism (26); this would account for its presence at significant levels across the pH range in LBK, a medium rich in amino acids. TnaA expression is induced by tryptophan, via transcription antitermination (49), and requires catabolite activator protein (7). The role of pH has not previously been recognized.
Why is this particular deaminase elevated to one of the most abundant proteins in the cell? TnaA deaminates not only tryptophan, but also serine and cysteine, producing pyruvic acid (26, 32), which can be further degraded anaerobically to acetic and formic acids. Thus, induction of TnaA could offer a particularly effective means of neutralizing excess alkali. The stress conditions were designed to force E. coli to grow at the extreme limits of tolerance for pH. In order to test the relationship between pH and anaerobiosis, each growth condition was selected for stress under aerobic or anaerobic conditions. In our media, aerobiosis permitted growth at slightly greater extreme values than did anaerobiosis (pH 4.4 versus pH 4.5; pH 9.2 versus pH 9.0).
The alkaline induction of TnaA could have broader significance for other stress studies. Growth in unbuffered Luria broth is known to alkalinize the culture (42), but only recently has the role of alkalinization been recognized in the study of stationary-phase response. The SurA stationary-phase survival factor, a periplasmic foldase, is actually required for survival at high pH (20). Similarly, TnaA might contribute to stationary-phase survival by retarding alkalinization. It also might act as a virulence factor for pathogens infecting alkaline regions of the body, such as the pancreatic duct.
Anaerobic alkaline induction of GadA.
The amino acid decarboxylases are expected to be induced in acid, in order to produce a basic amine which alkalinizes the cell. Growth in base strongly represses the lysine and arginine decarboxylases (28, 31, 42). Thus, it was surprising to find the highest levels of GadA during anaerobic growth in base.
The glutamate system in particular contributes to survival of E. coli at the most extreme low pH (22, 44, 55). The relationship of GadA to GadBC, however, is not understood. We indeed found some induction of GadA at pH 4.4, compared to pH 7.0 (Fig. 1 and 2), but the protein was expressed most highly at high pH, under anaerobiosis (Fig. 3). Previously, we showed that GadC is required for cells grown in base to survive sudden exposure to extreme acid (12). It may be that the glutamate decarboxylase system is turned on at high pH during anaerobiosis, when the culture is likely to acidify by fermentative metabolism. Alternatively, GadA responds specifically to anaerobiosis at high pH.
It should be noted that glutamate has other crucial functions in the cell, particularly related to osmotic shock. Following an increase in osmolarity, synthesis of glutamate is required for maintenance of electrical potential (27) and for recovery of cytoplasmic pH in the presence of acetate at external pH 6 (36). Thus, under some acidic conditions the cell may need to limit the breakdown of glutamate.
Acid induction of YfiD and of AhpC.
The effects of permeant weak acids are interesting because of their ability to depress internal pH and because fermentative metabolism naturally produces permeant acids at concentrations high enough to retard growth. In interpreting the responses to propionate, however, the possibility of several other kinds of effects must be considered, such as lowering of proton potential, as well as effects of the organic anion (38).
YfiD showed strong induction by low pH or by pyruvate, with complete repression at or above pH 7. Growth below pH 5 causes some depression of internal pH, comparable to that caused by a permeant acid (44). Thus, YfiD is an even better candidate for response to internal pH than are known permeant acid-inducible proteins such as InaA and MarA, which do not respond to external acidification (37, 42).
Genetically, YfiD shares a key glycine radical motif with pyruvate formate lyase (PflB and PflD) (53). Pyruvate formate lyase cleaves pyruvate to formate and acetyl coenzyme A without reduction of NAD+, a reaction important for anaerobic fermentation (18); it is induced by anaerobiosis and pyruvate (39, 48). YfiD, however, showed no difference in expression between aerobic and anaerobic growth. Instead, YfiD might be an enzyme which directs metabolism into a pathway critical under conditions of depressed internal pH.
The induction of AhpC by acid adds to a growing number of connections between acid and oxidative stress, perhaps related to the increased production of oxygen radicals at low pH. In Salmonella typhimurium, AhpC participates in protection from peroxides and from reactive nitrogen intermediates; its expression is induced by oxidative stress (4, 50). AhpC is also induced by benzoate shock (19). Our finding of the long-term elevation of AhpC by external or internal acid could be related to the increased production of reactive intermediates at low pH. The increased level of AhpC at low pH could explain the observation that acid shock proteins protect S. typhimurium from oxidative stress (9).
Other pH-dependent responses.
The elevation of AceA during aerobic growth in acid or base, but not at pH 7, was intriguing. AceA is subject to complex regulatory mechanisms in the control of respiratory metabolism, which generally alkalinizes complex media (6, 16). On the other hand, a possible basis for the response to acid is the buildup of acetate, an inducer of AceA (25). At low external pH, acetate as a permeant acid is driven into the cell down the pH gradient and will thus reach far greater cytoplasmic concentrations in acidic media.
The acidic induction of enzymes of sugar metabolism suggests that growth in acid may somehow favor these pathways. The reason for such a preference is unclear.
Overall, our results have shown that the patterns of acid and base induction intersect with aerobiosis and anaerobiosis in unpredicted ways. The advantage of the 2-D gel proteomic technique is that it reveals responses that might not otherwise have been sought. For the future, we plan to dissect the molecular basis of these pH-dependent responses.
ACKNOWLEDGMENTS
This work was supported by grant MCB 960963 from the National Science Foundation and by a faculty development grant from Kenyon College.
We are most grateful to Ruth VanBogelen at Parke-Davis for training in 2-D gel analysis.
REFERENCES
- 1.Abshire K Z, Neidhardt F C. Analysis of proteins synthesized by Salmonella typhimurium during growth within a host macrophage. J Bacteriol. 1993;175:3734–3743. doi: 10.1128/jb.175.12.3734-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auger E A, Redding K E, Plumb T, Childs L C, Meng S-Y, Bennett G N. Construction of lac fusions to the inducible arginine and lysine decarboxylase genes of Escherichia coli K-12. Mol Microbiol. 1989;3:609–620. doi: 10.1111/j.1365-2958.1989.tb00208.x. [DOI] [PubMed] [Google Scholar]
- 3.Borodovsky M, Rudd K E, Koonin E V. Intrinsic and extrinsic approaches for detecting genes in a bacterial genome. Nucleic Acids Res. 1994;22:4756–4767. doi: 10.1093/nar/22.22.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L, Xie Q W, Nathan C. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol Cell. 1998;1:795–805. doi: 10.1016/s1097-2765(00)80079-9. [DOI] [PubMed] [Google Scholar]
- 5.Cotter P A, Chepuri V, Gennis R B, Gunsalus R P. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J Bacteriol. 1990;172:6333–6338. doi: 10.1128/jb.172.11.6333-6338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cronan J. Tricarboxylic acid cycle and glyoxalate bypass. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 206–216. [Google Scholar]
- 7.Deeley M C, Yanofsky C. Transcription initiation at the tryptophanase promoter of Escherichia coli. J Bacteriol. 1982;151:942–951. doi: 10.1128/jb.151.2.942-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster J W. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit Rev Microbiol. 1995;21:215–237. doi: 10.3109/10408419509113541. [DOI] [PubMed] [Google Scholar]
- 9.Foster, J. W. Personal communication.
- 10.Gale E F, Epps H M R. The effect of the pH of the medium during growth on the enzymic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem J. 1942;36:600–619. doi: 10.1042/bj0360600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerchman Y, Olami Y, Rimon A, Taglicht D, Schuldiner S, Padan E. Histidine-226 is part of the pH sensor of NhaA, a Na+/H+ antiporter in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1212–1216. doi: 10.1073/pnas.90.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hersh B M, Farooq F T, Barstad D N, Blankenhorn D, Slonczewski J L. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyde M, Coll J L, Portalier R. Identification of Escherichia coli genes whose expression increases as a function of external pH. Mol Gen Genet. 1991;229:197–205. doi: 10.1007/BF00272156. [DOI] [PubMed] [Google Scholar]
- 14.Hickey E W, Hirshfield I N. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl Environ Microbiol. 1990;56:1038–1045. doi: 10.1128/aem.56.4.1038-1045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa T, Hama H, Tsuda M, Tsuchiya T. Isolation and properties of a mutant of Escherichia coli possessing defective Na+/H+ antiporter. J Biol Chem. 1987;262:7443–7446. [PubMed] [Google Scholar]
- 16.Iuchi S, Lin E C C. ArcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karpel R, Alon T, Glaser G, Schuldiner S, Padan E. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J Biol Chem. 1991;266:21753–21759. [PubMed] [Google Scholar]
- 18.Knappe J, Blaschkowski H P, Grobner P, Schmitt T. Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur J Biochem. 1974;50:253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 19.Lambert L A, Abshire K, Blankenhorn D, Slonczewski J L. Proteins induced in Escherichia coli by benzoic acid. J Bacteriol. 1997;179:7595–7599. doi: 10.1128/jb.179.23.7595-7599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazar S W, Almirón M, Tormo A, Kolter R. Role of the Escherichia coli SurA protein in stationary-phase survival. J Bacteriol. 1998;180:5704–5711. doi: 10.1128/jb.180.21.5704-5711.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 307–357. [Google Scholar]
- 22.Lin J, Lee I S, Frey J, Slonczewski J L, Foster J W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J, Smith M P, Chapin K C, Baik H S, Bennett G N, Foster J W. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch A S, Lin E C C. Responses to molecular oxygen. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1526–1538. [Google Scholar]
- 25.Maloy S R, Nunn W D. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J Bacteriol. 1982;149:173–180. doi: 10.1128/jb.149.1.173-180.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFall E, Newman E B. Amino acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 358–379. [Google Scholar]
- 27.McLaggan D, Naprstek J, Buurman E T, Epstein W. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J Biol Chem. 1994;269:1911–1917. [PubMed] [Google Scholar]
- 28.Meng S-Y, Bennett G N. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol. 1992;174:2659–2669. doi: 10.1128/jb.174.8.2659-2669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 30.Mobley H L T. Helicobacter pylori factors associated with disease development. Gastroenterology. 1997;113:S21–S28. doi: 10.1016/s0016-5085(97)80006-6. [DOI] [PubMed] [Google Scholar]
- 31.Neely M N, Dell C L, Olson E R. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J Bacteriol. 1994;176:3278–3285. doi: 10.1128/jb.176.11.3278-3285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton W A, Snell E E. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc Natl Acad Sci USA. 1964;51:382–389. doi: 10.1073/pnas.51.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nobelmann B, Lengeler J W. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J Bacteriol. 1996;178:6790–6795. doi: 10.1128/jb.178.23.6790-6795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyström T, Neidhardt F C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol Microbiol. 1992;6:3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x. [DOI] [PubMed] [Google Scholar]
- 35.Olson E R. Microreview: influence of pH on bacterial gene expression. Mol Microbiol. 1993;8:5–14. doi: 10.1111/j.1365-2958.1993.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 36.Roe A J, McLaggan D, Davidson I, O’Byrne C, Booth I R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosner J L, Slonczewski J L. Dual regulation of inaA by the multiple antibiotic resistance (Mar) and superoxide (SoxRS) stress response systems of Escherichia coli. J Bacteriol. 1994;176:6262–6269. doi: 10.1128/jb.176.20.6262-6269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salmond C V, Kroll R G, Booth I R. The effect of food preservatives on pH homeostasis in Escherichia coli. J Gen Microbiol. 1984;130:2845–2850. doi: 10.1099/00221287-130-11-2845. [DOI] [PubMed] [Google Scholar]
- 39.Sawers G, Böck A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J Bacteriol. 1988;170:5330–5336. doi: 10.1128/jb.170.11.5330-5336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuman H A. Active transport of maltose in Escherichia coli K12. Role of the periplasmic maltose-binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J Biol Chem. 1982;257:5455–5461. [PubMed] [Google Scholar]
- 41.Slonczewski J L, Foster J W. pH-regulated genes and survival at extreme pH. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1539–1549. [Google Scholar]
- 42.Slonczewski J L, Gonzalez T N, Bartholomew F M, Holt N J. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J Bacteriol. 1987;169:3001–3006. doi: 10.1128/jb.169.7.3001-3006.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slonczewski J L, Rosen B P, Alger J R, Macnab R M. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc Natl Acad Sci USA. 1981;78:6271–6275. doi: 10.1073/pnas.78.10.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Small P L, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Small P L C, Waterman S R. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 1998;6:214–216. doi: 10.1016/s0966-842x(98)01285-2. [DOI] [PubMed] [Google Scholar]
- 46.Smith D K, Kassam T, Singh B, Elliott J F. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J Bacteriol. 1992;174:5820–5826. doi: 10.1128/jb.174.18.5820-5826.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith M W, Neidhardt F C. Proteins induced by aerobiosis in Escherichia coli. J Bacteriol. 1983;154:344–350. doi: 10.1128/jb.154.1.344-350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith M W, Neidhardt F C. Proteins induced by anaerobiosis in Escherichia coli. J Bacteriol. 1983;154:336–343. doi: 10.1128/jb.154.1.336-343.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166:217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storz G, Jacobson F S, Tartaglia L A, Morgan R W, Silveira L A, Ames B N. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Swiss Institute of Bioinformatics. 12 February 1999, revision date. SWISS-2DPAGE database. [Online.] ExPASy server. http://expasy.hcuge.ch/. [26 February 1999, last date accessed.]
- 51.VanBogelen R A, Abshire K Z, Pertsemlidis A, Clark R L, Neidhardt F C. Gene-protein database of Escherichia coli K-12, edition 6. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2067–2117. [Google Scholar]
- 52.VanBogelen R A, Olson E R. Application of two-dimensional protein gels in biotechnology. Biotechnol Annu Rev. 1995;1:69–103. doi: 10.1016/s1387-2656(08)70048-6. [DOI] [PubMed] [Google Scholar]
- 53.Wagner A F V, Frey M, Neugebarger F A, Schäfer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wanner B L, McSharry R. Phosphate-controlled gene expression in Escherichia coli K-12 using Mudl-directed lacZ fusions. J Mol Biol. 1982;158:347–363. doi: 10.1016/0022-2836(82)90202-9. [DOI] [PubMed] [Google Scholar]
- 55.Waterman S R, Small P L. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21:925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 56.Zilberstein D, Agmon V, Schuldiner S, Padan E. Escherichia coli intracellular pH, membrane potential, and cell growth. J Bacteriol. 1984;158:246–252. doi: 10.1128/jb.158.1.246-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]