Abstract
Background:
The apolipoprotein E (APOE) ε4 allele confers higher risk of neurodegeneration and Alzheimer’s disease (AD), but differs by race/ethnicity. We examined this association in American Indians.
Methods:
The Strong Heart Study is a population-based cohort of American Indians who were 64 to 95 years of age in 2010 to 2013. APOE ε4 status, brain imaging, and neuropsychological testing was collected in N = 811 individuals. Summary statistics, graphics, and generalized linear regressions—adjusted for sociodemographics, clinical features, and intracranial volume with bootstrap variance estimator—compared APOE ε4 carriers with non-carriers.
Results:
APOE ε4 carriers comprised 22% of the population (0.7% homozygotes). Participants were mean 73 years, 67% female, and 54% had some college education. The majority were obese (>50%), hypertensive (>80%), and diabetic (>50%). Neither imaging findings nor multidomain cognitive testing showed any substantive differences between APOE ε4 carriers and non-carriers.
Conclusion:
We found no evidence of neurodegenerative risk from APOE ε4 in American Indians. Additional studies are needed to examine potential protective features.
Keywords: Alzheimer’s disease, APOE genotype, cerebral atrophy, cognitive impairment, vascular dementia
1 |. INTRODUCTION
Apolipoprotein E (apoE), the major cholesterol transporter, is polymorphic with three common isoforms encoded by single nucleotide polymorphisms: ε2, ε3, and ε4. The APOE ε4 allele is a strong, well-established, independent, objective risk factor for neurodegeneration, including Alzheimer’s disease1,2 and related disorders (ADRD),3–5 with structural6,7 and neuropsychological8 effects up to 20 years before clinical recognition. However, studies of African American, Hispanic-Latino, and Asian-ancestry populations have suggested differential, lower, or absent associations for the APOE ε4 allele.9–13 Neurodegenerative disease resembling ADRD, endemic to Pacific Islanders, has not been associated with the APOE ε4 allele14,15; and only two small studies have examined APOE ε4 in North American Indigenous peoples.16,17 Our study purports to address these knowledge gaps with an analysis of brain volumes and cognitive testing markers related to ADRD in a large, heterogeneous, population-based study of American Indians.
Systematic review: Risk of Alzheimer’s disease and related disorders (ADRD) associated with the apolipoprotein E (APOE) ε4 allele is well established.
Interpretation: We did not find any evidence of such an association with structural brain volumes, cognitive performance, or function related to AD in a large cohort of American Indians. Overall, our negative findings lend credence to hypotheses of evolutionary pressure on lipid metabolism genes, suggesting that traditional food sources may be critical in sustaining brain health in Indigenous populations. In addition, social, behavioral, cultural, or other features promoting positive aging and resilience may counterbalance endogenous risks. Attributable risk for ADRD in American Indians is still unclear, due in part to inadequate epidemiology and neuropsychology in this underserved population.
Future directions: Future research should carefully estimate prevalence and incidence of dementia and ADRD; evaluate longitudinal changes in imaging and cognition markers in association with the APOE genotype; consider the effect of APOE genotype on endophenotypes of neurodegeneration, such as biomarkers of amyloid beta (Aβ) and tau; and explore sociocultural or behavioral protective features that may help to explain these findings.
The Strong Heart Study (SHS), a longitudinal cohort of American Indian adults residing in tribal communities in three geographic regions, has published prevalence of APOE ε4 allele (≈24%), and ε4/ε4 homozygotes (≈1%),18 with allele frequencies roughly comparable to those in US populations of European descent. In previous reports from the SHS cohort, the APOE ε4allele was associated with higher serum lipid levels, history of cardiovascular disease, and poor glucose control—all important risk factors for vascular brain injury and ADRD.18 Our analysis aims to expand on these previous findings, to explore whether the presence of the APOE ε4allele is associated with cognition and neurodegeneration using data from a follow-up examination within the SHS cohort, including magnetic resonance imaging (MRI), detailed cognitive assessment, and neurological clinical history. Specifically, we will examine whether carriers of the APOE ε4allele have a greater degree of brain or hippocampal atrophy or a greater degree of memory loss, memory-defined cognitive impairment, or multiple-domain cognitive impairment.
2 |. METHODS
Setting.
SHS participants have undergone six waves of data collection, starting with a baseline examination in 1989 to 1991.19 This original cohort of 4549 American Indians, then 45 to 74 years of age, comprised 62% of the total eligible population of tribal members living in 13 communities in the Northern Plains, Southern Plains, and Southwest.19 In 2010 to 2013, a total of 86% of all survivors from that original cohort, by then 64 to 95 years of age, participated in a comprehensive follow-up examination focused on cranial MRI, cognitive and functional assessment, and detailed neurological history, known as the Cardiovascular Disease and its Consequences in American Indians (CDCAI) study.20 A methodological report for the recruitment and examination of these participants, with Consolidated Standards of Reporting Trials (CONSORT) diagram, has been published previously,20 as well as in-depth descriptions of MRI and cognitive findings.21,22 All participants provided written, informed consent; all participating institutional and Indian Health Service review boards, tribal research review boards, and tribal councils reviewed research procedures, as required.
Data Collection.
Genotyping procedures,23 MRI,24 and cognitive examinations20 have been described previously in detail. The APOE genotype was determined in a standard fashion18 using blood samples collected at the baseline SHS visit. Full APOE haplotypes were initially defined, but by the time of the neurology examination (ages 64 to 95), the rare haplotypes were so uncommon that the comparison of interest simplified to APOE ε4 carriers and non-carriers. Neuroradiologists blinded to participant data processed 1.5T MRI scans from the neurology visit using FreeSurfer image analysis suite,25 FIRST in FSL 5.0,26 and the ENIGMA1 protocol27 to estimate combined gray and white matter brain volume, right and left hippocampus, and intracranial space. Four standard cognitive tests were administered, including California Verbal Learning Test (CVLT) version II short form, comprising several indices of immediate and delayed verbal learning and memory;28 Weschler Adult Intelligence Scale, 4th Edition (WAIS-IV) digit symbol coding subtest, measuring visuospatial processing speed and working memory29; the Controlled Oral Word Association (COWA) F,A,S test evaluating phonemic verbal fluency and executive functioning30; and the Modified Mini Mental Status Examination (3MSE) offering a global screening tool for general cognitive functioning.31 Of the participants with imaging and cognitive data available for analysis from the CDCAI neurology examination visit (2010 to 2013), N = 811 also have available APOE genotype data.
Age, sex, years of formal education, and anthropometric (body mass index) or clinical co-morbidities were measured by self-reported questionnaire and clinical evaluation. Hypertension was defined as averaged seated systolic blood pressure >140 mm Hg, diastolic blood pressure >90 mm Hg, or use of antihypertensive medications; diabetes mellitus as fasting glucose >126 mg/dL (7 mmol/L) or use of insulin/antidiabetic medications; high serum LDL (low-density lipoprotein) as >140 mg/dL or use of statins; chronic kidney disease (CKD) as estimated glomerular filtration rate <60 mL/min.
Analyses.
Descriptive techniques—count and percent or mean and standard deviation—were used to summarize sociodemographic, clinical, image volumetric, and cognitive test performance features, stratified by APOE ε4 allele carriers (haplotypes 4/2, 4/3, and 4/4) and non-carriers (haplotypes 2/2, 3/2, 3/3). P-values in Table 1 were used to test the likelihood that differences between APOE ε4 carriers and non-carriers in these features would likely exist in the underlying population, under the null hypothesis of no true difference; tests used Student t (continuous) or chi-square (binary, categorical) distributions. Regressions comparing APOE ε4 allele carrier status (independent variable) against standardized, continuous imaging or cognitive features (dependent variables) used generalized linear models with Gaussian (normal) link and a non-parametric bootstrap (50 replication) variance estimator, to reduce model dependence on assumptions of representative selection, homoscedasticity, and normality. Adjustment variables included age, sex, field center, education, body mass index (BMI), diabetes, hypertension, high LDL, and CKD; imaging volumetric features were also adjusted for intracranial volume. Scatterplots with fractional polynomial curves and 95% confidence intervals examined the associations between cognitive performance scores and age, with stratification (coloration) by APOE ε4 allele carrier status. Cognitive impairment was defined using a simplified actuarial method, as deficits (1.5 SD below mean) in either learning (best of trials 3 and 4) or retention (percent retained after learning) on the CVLT tasks; performance on other cognitive tasks, including WAIS-IV digit coding, COWA, and 3MSE tests, were compared using box plots between those with/out impairment those with/out APOE ε4 allele carrier status. All statistical analyses were conducted using Stata version 14 (Stata Corp, LLC. College Station, Texas, USA).
TABLE 1.
Selected participant characteristics of American Indians, 64 to 95 years of age, stratified by apolipoprotein E (APOE) ε4 carrier status
| APOE ε4 status Non-carriers* | Carriers* | |||
|---|---|---|---|---|
| n = 630 (77.7%) | n = 181 (22.3%) | P | ||
| Age (years), mean (SD) | 73.1 (6.0) | 72.7 (5.5) | .34 | |
| Male, n(%) | 204 (32.4%) | 59 (32.6%) | .96 | |
| Field center, n(%) | Northern Plains | 294 (46.7%) | 78 (43.1%) | .52 |
| Southern Plains | 259 (41.1%) | 83 (45.9%) | ||
| Southwest | 77 (12.2%) | 20 (11.0%) | ||
| BMI (kg/m2), mean (SD) | 31.3 (6.4) | 31.8 (7.1) | .39 | |
| Education categories | Up to high school | 126 (20.0%) | 36 (19.9%) | .11 |
| Grad. high school | 170 (27.0%) | 38 (21.0%) | ||
| Any college | 250 (39.7%) | 71 (39.2%) | ||
| College graduate | 84 (13.3%) | 36 (19.9%) | ||
| Hypertension | 510 (81.0%) | 145 (80.1%) | .80 | |
| Diabetes mellitus | 312 (49.5%) | 87 (48.1%) | .73 | |
| High LDL | 245 (38.9%) | 80 (44.2%) | .20 | |
| CKD | 300 (47.6%) | 79 (43.6%) | .35 | |
| Volume: brain (% IC), mean (SD) | 77.4 (4.6) | 77.6 (4.8) | .56 | |
| Volume: hippocampus (% IC), mean (SD) | 0.5 (0.1) | 0.5 (0.1) | .62 | |
| Volume: left hippocampus (% IC), mean (SD) | 0.3 (0.0) | 0.3 (0.0) | .95 | |
| Volume: right hippocampus (% IC), mean (SD) | 0.3 (0.0) | 0.3 (0.0) | .42 | |
| Learning: CVLT best of trials 3 and 4 | 6.9 (1.5) | 6.9 (1.5) | .62 | |
| Learning: CVLT across-trial consistency score | 82.3 (14.3) | 82.7 (14.1) | .74 | |
| Learning & memory: CVLT % retained | −22.7 (27.1) | −21.8 (27.1) | .68 | |
| Memory: CVLT short delay free recall | 5.9 (2.0) | 6.0 (2.1) | .36 | |
| Memory: CVLT long delay free recall | 5.4 (2.3) | 5.5 (2.2) | .63 | |
| Memory: CVLT long delay cued recall | 5.9 (2.0) | 6.1 (2.1) | .26 | |
| Memory: CVLT recognition / discrimination | 2.6 (0.8) | 2.6 (0.8) | .95 | |
| Processing speed: WAIS coding | 44.4 (16.1) | 42.4 (15.0) | .14 | |
| Phonemic fluency, executive function: COWA | 24.1 (11.3) | 25.3 (11.9) | .21 | |
| General cognition: 3MSE | 88.5 (8.8) | 87.9 (11.0) | .48 | |
APOE ε4 non-carriers include haplotypes 3/2 (n = 42, 5.1%) and 3/3 (n = 588, 72.5%) but no 2/2. APOE ε4 carriers include haplotypes 4/2 (n = 5, 0.6%), 4/3 (n = 170, 21.0%), and 4/4 (n = 6, 0.7%).
Number and percent, n(%). BMI, body mass index;, kg/m2, kilograms per meter squared; LDL, low-density lipoprotein; CKD, chronic kidney disease; % IC, volumetric measures presented as % intracranial volume; CVLT, California Verbal Learning Test; WAIS, Weschler Adult Intelligence Scale; COWA, Controlled Oral Word Association; 3MSE, Modified Mini Mental Status Examination.
3 |. RESULTS
Among participants of the CDCAI neurology examination (2010 to 2013; N = 811), APOE ε4 non-carriers included haplotypes 3/2 (n = 42, 5.1%) and 3/3 (n = 588, 72.5%) but no 2/2. APOE4 carriers included haplotypes 4/2 (n = 5, 0.6%), 4/3 (n = 170, 21.0%), and 4/4 (n = 6, 0.7%).
As has been detailed in previous reports,20 participants had a mean age 73 and were 67% female (Table 1). Education level was generally high, with 54% having attended at least some college. BMI was also high (mean >30 kg/m2), and many were hypertensive (>80%), diabetic (nearly 50%), dyslipidemic (>39%), or had CKD(>44%). When APOE ε4 carriers were compared with non-carriers, most sociodemographic or clinical features were similar, except for proportion of those with high LDL (44% vs 39%, not significant) and CKD (44% vs 48%, not significant). Performance on cognitive tests was varied: mean WAIS coding task score was 42 to 44 (SD 15 to 16); mean COWA 24 to 25 (SD 11 to 12); mean 3MSE 88 to 89 (SD 9 to 11). CVLT learning tasks were generally successful: mean 7 (SD 1.5) of 9 total, with relatively high consistency across trials; however, retention was lower, with mean 22% to 23% lost (SD 27%), resulting in long delay mean score 1 to 2 points lower (mean 5 to 6, SD 2). Brain, hippocampus, and intracranial (IC) volumes were similar between APOE ε4 carriers and non-carriers, as were all CVLT measures, and the other cognitive test scores (WAIS, COWA, 3MSE).
Generalized linear models (Table 2) of APOE ε4 carrier status with MRI-defined brain volumes, adjusted for sociodemographics and key clinical risk features, demonstrated little evidence of any association. The beta coefficients were close to 0 and the 95% confidence intervals were squarely centered on 0. P-values for volumetric coefficients were robustly null (P > .2); however, P-values for the age coefficients in each model were robustly significant (P < .01). Adjustment for multiple testing was not performed because each of the tests is closely correlated, and part of the same singular, a priori stated hypothesis.
TABLE 2.
Associations* of selected volumetric magnetic resonance imaging (MRI) findings with APOE ε4 carrier status
| N | β (95% CI) | P | P (age) | |
|---|---|---|---|---|
| Brain volume | 758 | 0.02 (−0.05, 0.09) | .594 | <.001 |
| Hippocampus volume | 756 | −0.05 (−0.21, 0.11) | .548 | .003 |
| Left hippocampus volume | 757 | −0.01 (−0.20, 0.18) | .916 | .001 |
| Right hippocampus volume | 757 | −0.08 (−0.21, 0.05) | .217 | .005 |
Generalized linear regression model, Gaussian (normal) link, bootstrap variance-covariance estimator (VCE) with 50 replications; adjusted for intracranial volume, age, sex, field center, education, BMI, diabetes, hypertension, high LDL, and CKD. Standardized volumetric exposure and adjustment variables, for direct comparisons.
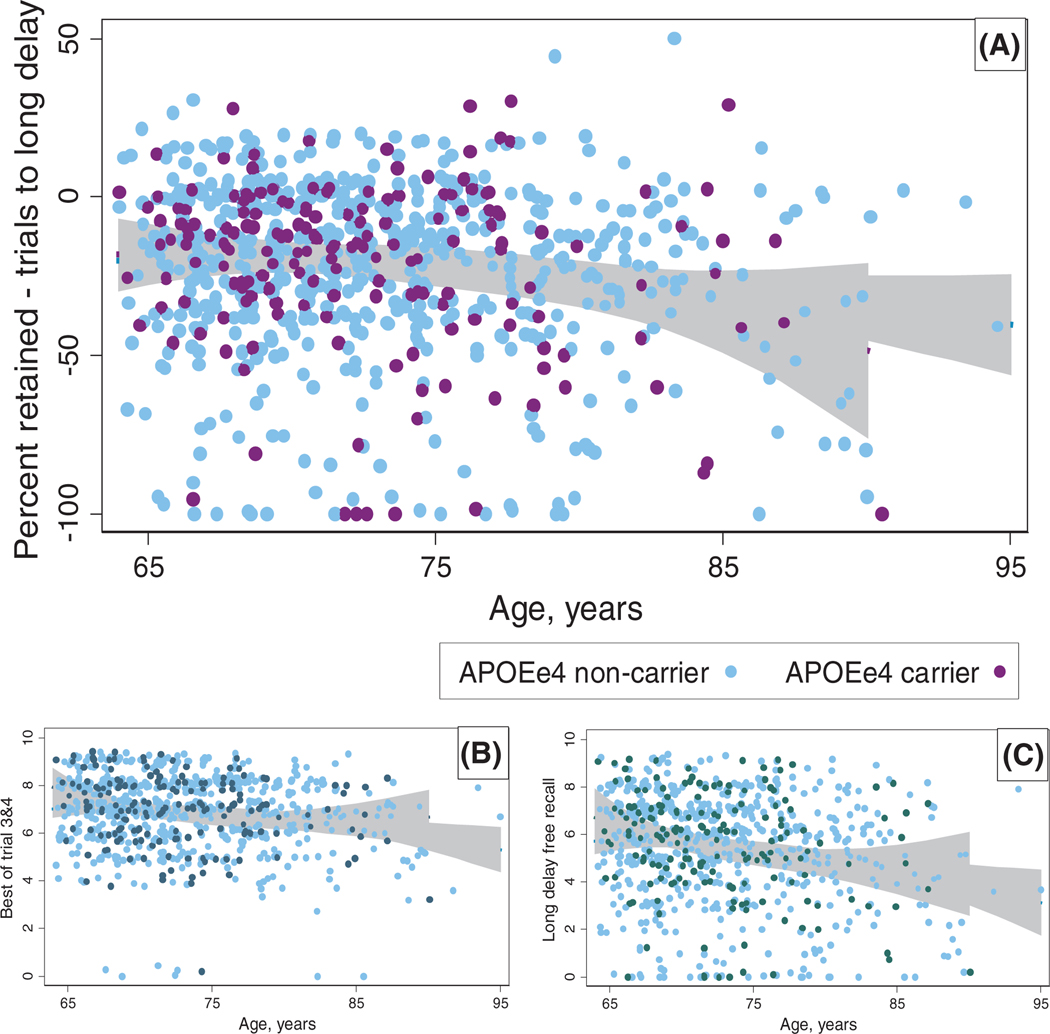
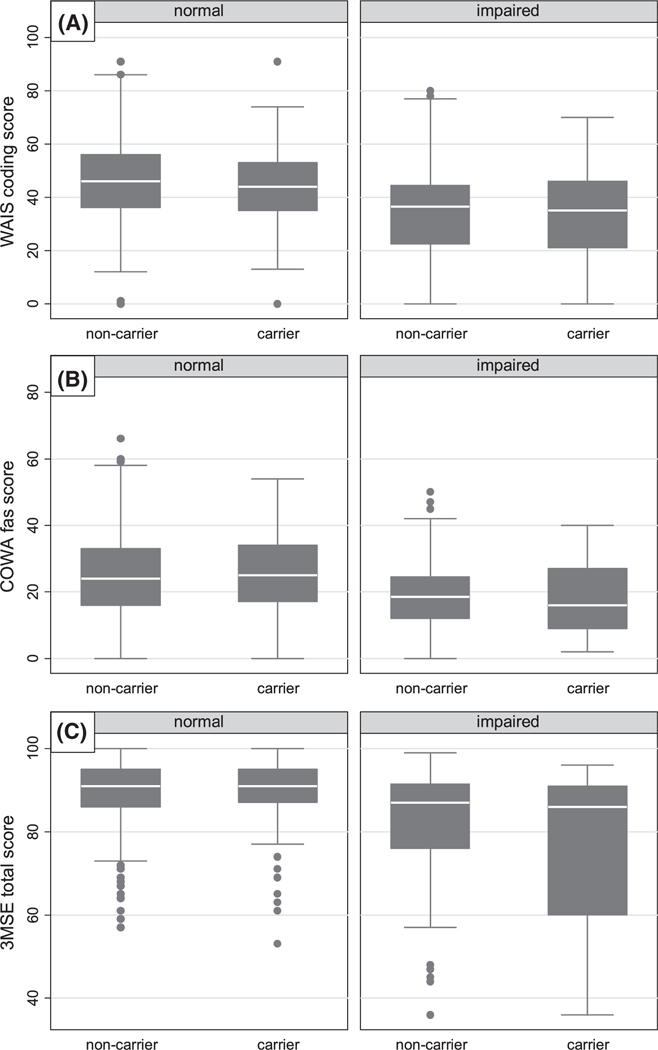
Similarly structured models for multiple measures of cognitive performance on the CVLT test (Tables 3) were also null (P > .3), but with significant positive control coefficients for age (P < .1). Scatterplots of various CVLT performance metrics (Figure 1) over age, stratified by APOE ε4 carrier status, with fractional polynomial fit lines and 95% confidence intervals, showed overlapping data and association for the entire range of age, with no divergence in the oldest groups. Box plots of non-CVLT cognitive tests—WAIS, COWA, and 3MSE, grouped by an actuarial-derived memory impairment categorization based on CVLT performance (Figure 2), and also by APOE ε4 carrier status—showed lower cognitive performance among those with memory deficit, but no differences by APOE ε4 carrier status, both in the impaired and in the non-impaired.
TABLE 3.
Associations* of selected California Verbal Learning Test short form, 2nd Edition (CVLT) scores, with APOE ε4 carrier status
| N | β (95% CI) | P | P (age) | |
|---|---|---|---|---|
| Best of trials 3 and 4 | 743 | 0.00 (−0.14, 0.15) | .967 | .012 |
| Across-trial consistency | 738 | 0.02 (−0.13, 0.16) | .827 | .085 |
| Percent retained to long delay | 736 | 0.04 (−0.11, 0.20) | .559 | .031 |
| Short free recall | 743 | 0.06 (−0.08, 0.21) | .396 | .001 |
| Long free recall | 743 | 0.03 (−0.14, 0.20) | .740 | .001 |
| Long cued recall | 743 | 0.08 (−0.09, 0.26) | .349 | <.001 |
| Recognition/discrimination | 743 | −0.02 (−0.20, 0.16) | .824 | <.001 |
Generalized linear regression model, Gaussian (normal) link, bootstrap variance-covariance estimator (VCE) with 50 replications; adjusted for intracranial volume, age, sex, field center, education, BMI, diabetes, hypertension, high LDL, and CKD. Standardized cognitive exposure variables, for direct comparisons.
FIGURE 1.
Scatterplots and association curves of age with the California Verbal Learning Test short form, 2nd Edition (CVLT); proportion (percent) retained between learning trials and 10-minute “long” delay recall. Cross-sectional population range of age (64 to 95 years) on the X-axis. Y-axis features include percent retained between learning trials and long delay free recall (A); best performance between trials 3 and 4 (B); and long delay free-recall score (C). Stratification by apolipoprotein E (APOE) ε4 carrier status, with non-carriers in (light) blue and carriers in (dark) purple (or dark navy, dark green). Association lines using fractional polynomial fit (blue, purple) with 95% confidence interval (gray)
FIGURE 2.
Raw cognitive test scores, stratified by cognitive impairment and by apolipoprotein E (APOE) ε4 carrier status. Cognitive test scores include: (A) Weschler Adult Intelligence Scale (WAIS) digit symbol coding for processing speed, (B) Controlled Oral Word Association test (COWA) for phonemic fluency and executive function, and (C) Modified Mini Mental Status Examination (3MSE) for general cognition. Cognitive impairment defined using operationalized (actuarial) method based on California Verbal Learning Test short form, 2nd edition (CVLT) score
4 |. DISCUSSION
This study evaluated associations between the presence of the APOE ε4 allele with both neuroimaging and cognitive markers of vascular brain injury and neurodegeneration in American Indians and found that the presence of the APOE ε4 allele was not associated with sociodemographic, clinical, subclinical, or functional markers of either vascular or degenerative brain disease in this large, heterogeneous cohort of American Indians over age 65. Furthermore, the degree of APOE ε4 allele association was not different between those with or without cognitive or memory impairment, or over the range of age—a departure from prior studies in other populations.9 These findings are consistent with prior small studies in Choctaw16 and Cherokee,17 and with prior studies in this same population-based cohort that APOE genotype does not modify other risk associations.21
Of the neuropsychological domains that we examined, the APOE ε4 allele has been previously associated with decrements in processing speed and memory across the life-course,32,33 independent of lifestyle and other factors,34 in population-based cohorts of primarily non-Hispanic Whites from the United Kingdom and Australia.35,36 Of note, in vascular dementia, APOE ε4 may preferentially impair spatial memory and reaction time, whereas in AD, processing speed and immediate recall tasks may be more strongly affected.37
Despite the negative findings in the current analysis, LDL cholesterol has been previously and significantly associated with APOE ε4 carrier status in the SHS cohort. APOE phenotype influenced LDL, high-density lipoprotein (HDL), and serum apolipoproteins, with differences by sex and menopausal status.18 These findings are consistent with an endogenous role of apoE in lipid metabolism.
The APOE gene is polymorphic with three common isoforms, including ε2, ε3, and ε4–encoded by two single nucleotide polymorphisms. Approximately 25% of the non-Hispanic White and European populations carries at least one APOE ε4 allele.9,32 Indigenous populations living in environments characterized by foraging or limited food availability also present with ε4 as a common allele, including Pygmies (41%), Khoi San (37%), Papuans (37%), Lapps (31%), Malaysian (24%), Australian (26%) aborigines, and some American Indian/Native Americans (28%).38,39 In the SHS baseline cohort of American Indians 45 to 74 years of age, the APOE ε4 allele was characterized as 24%, with ≈1% homozygotes.18 In the CDCAI examination—which included 86% of surviving, eligible participants, who were then 64 to 95 years of age20–these frequencies were 22% and 0.7%, respectively.
Populations with long-established agricultural economies have slowly replaced the ε4 with the ε3 allele as the more common inherited form, suggesting that the high prevalence of type 2 diabetes, obesity, and other conditions related to metabolic disturbance in some populations may be a consequence of genetic variants that have undergone positive selection during periods of erratic food supply.38 This hypothesis posits that carriers of the ε4 allele differentially survive in foraging conditions because they are able to deposit additional fat between famines, bestowing an evolutionary advantage. When food availability increases, excess capacity for adipose tissue deposition becomes a risk, rather than an advantage. Recent studies have further found that ε4 frequency decreases and ε2/ε3 frequencies increase with more southerly latitude,40 suggesting that such natural selection pressures based on environmental temperatures and food availability patterns present since the Paleolithic period, may have resulted in functional differences in lipid metabolism for both Indigenous and non-Indigenous populations.41 Such a finding, if true, could suggest that traditional foods and food sources may be critically important to population health.
Modern population groups—including those with American Indian, African, Asian, or Hispanic-Latino heritage—may be difficult to categorize and define in the context of pre–Industrial Era population migrations and diaspora. Nevertheless, many studies have identified inconsistent, weak, absent, or otherwise differential associations for the APOE ε4 allele in non-White populations.9–13,42–45 Although some studies may be limited by selection bias, small sample size, and unvalidated cognitive measures, ADRD risk from APOE may be mediated or modified by race-confounded features such as socioeconomic status, diabetes,46 depression,47 and cardiovascular disease.48 Indeed, when large, sensitive, robust analyses are applied to account for socioeconomic differences, racial-risk estimates for APOE genotype appear similar across populations, with socioeconomic accounting for much of the risk difference.49,50 Despite methodological clarifications, inference on genetic studies with racial stratification should be structured with care, to avoid errors of interpretation. Ancestry is only one facet in racial/ethnic population identity, and racial status should not be considered as a surrogate marker for ancestry, and such ancestry should not be considered as homogeneous.
This study found that the APOE ε4 allele does not appear to be linked to neurogenerative disease or cognition in American Indians. Despite strong evidence of risk from the APOE ε4 allele in the majority population and conflicting reports of risk in African Americans, Hispanic/Latinos, and Asians, possibly due to race-linked residual confounding and bias, our population-based cohort study was powered and structured to detect an effect, if one exists. Effects for other participant features were successfully identified, including age, serving as positive statistical controls. Furthermore, a sensitivity power analysis—given the observed study sample (n1 = 630; n2 = 181)—found that a two-tailed t-test with 0.05 alpha can detect a small to medium effect size (Cohen d = 0.27 to 0.30) with 90% to 95% power and a small effect size (d < 0.25) with 80% to 85% power.
Furthermore, research in non-Hispanic Whites has shown that, due to selective survival, the detectable differential effect of APOE genotype on neurodegeneration peaks at around age 70 for ε4/ε4 homozygotes and at around age 75 for APOE ε4 carriers/heterozygotes, compared with non-carriers,7 with detectability reduced to null by age 80. Because of the selection protocol inherent in the SHS and CDCAI cohorts,20 selective survival could increase Type 2 error with bias toward the null, if APOE genotype is associated with the likelihood of participation. Such error is critical to interpretation of these study results because the most likely finding is that the APOE ε4 allele status increases the risk of neurodegeneration and dementia in American Indians in a similar manner and magnitude as that observed in non-Hispanic White or European ancestry populations. However, a few findings argue against a Type 2 error solely explaining our null findings. First, the prevalence of the APOE ε4 allele was similar among SHS participants who did and did not survive to participate in the CDCAI neurology examination, suggesting that selective survival is not likely to have been affected by APOE ε4 allele status in this study. Second, the mean age range in this study was close to the age of peak discoverability in non-Hispanic Whites from prior studies.7 Third, previous studies in the SHS and CDCAI cohort have detected little evidence of selective survival for vascular and degenerative brain outcomes. Similarly, although AD pathogenetic severity could influence detectability of associations between APOE ε4 genetic exposures and markers of AD pathogenesis, the absence of validated cognitive standards by which to stage AD22,51 precludes population stratification until such standards can be established.
Research in context:
Overall, these negative findings lend credence to hypotheses of evolutionary pressure on lipid metabolism genes, suggesting that traditional food sources may be critical in sustaining cognitive function and avoiding neurodegeneration Indigenous populations. Alternatively, positive aging features may counterbalance risk features, contributing to resilient aging. Attributable risk for dementia and ADRD outcomes in American Indians is still unclear, due in part to inadequate information on the epidemiology of these conditions; therefore, future research should carefully estimate the prevalence and incidence of dementia and ADRD; evaluate longitudinal changes in imaging and cognition markers in association with APOE genotype; consider the effect of the APOE genotype on endophenotypes of neurodegeneration, such as biomarkers of Aβ and tau; and explore the cultural, environmental, or genetic protective features that may help to explain these findings.
ACKNOWLEDGEMENTS
The authors wish to thank all Strong Heart Study staff, participants, and communities. The opinions expressed in this paper are solely the responsibility of the author(s) and do not necessarily reflect the official views of the Indian Health Service or the National Institutes of Health (NIH). The original Strong Heart Study has been funded in whole or in part with federal funds from the NIH under contract numbers 75N92019D00027, 75N92019D00028, 75N92019D00029, and 75N92019D00030; cooperative agreements U01HL41642, U01HL41652, U01HL41654, U01HL65520, and U01HL65521; and research grants R01HL109315, R01HL109301, R01HL109284, R01HL109282, and R01HL109319. Funding for the ancillary neurology examinations and these analyses included research grants R01HL093086 (Buchwald), P50AG005136 (Grabowski), and K01AG057821 (Suchy-Dicey). ASD reports funding for this research from NIH research grants; funding for other research from NIH grants and University funds; and funding for meeting attendance from NIH. BH reports funding for consulting unrelated to this research; and funding for meeting attendance from NIH. WL reports funding for other research from NIH research grants. ER reports funding for this research from NIH grants and the Arizona Alzheimer’s Consortium; funding for other research from NIH, Lilly/Avid, Genentech/Roche, and foundation/philanthropy grants; and leadership roles with ALZ-Pathway and Flinn Foundation. DB reports funding for this and other research from NIH research grants; funding for consulting unrelated to this research; and funding for meeting attendance from NIH grants.
Funding information
NIH, Grant/Award Numbers: 75N92019D00027, 75N92019D00028, 75N92019D00029, 75N92019D00030, U01HL41642, U01HL41652, U01HL41654, U01HL65520, U01HL65521, R01HL109315, R01HL109301, R01HL109284, R01HL109282, R01HL109319, R01HL093086, P50AG005136, K01AG057821
REFERENCES
- 1.Cuyvers E, Sleegers K. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 2016;15:857–868. [DOI] [PubMed] [Google Scholar]
- 2.Shen L, Jia J. An Overview of Genome-Wide Association Studies in Alzheimer’s Disease. Neurosci Bull. 2016;32:183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahoney-Sanchez L, Belaidi AA, Bush AI, Ayton S. The Complex Role of Apolipoprotein E in Alzheimer’s Disease: an Overview and Update. J Mol Neurosci. 2016;60:325–335. [DOI] [PubMed] [Google Scholar]
- 4.Yu JT, Tan L, Hardy J. Apolipoprotein E in Alzheimer’s disease: an update. Annu Rev Neurosci. 2014;37:79–100. [DOI] [PubMed] [Google Scholar]
- 5.Reiman EM, Arboleda-Velasquez JF, Quiroz YT, Huentelman MJ, Beach TG, Caselli RJ, et al. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun. 2020;11:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, et al. Preclinical evidence of Alzheimer’s disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–758. [DOI] [PubMed] [Google Scholar]
- 7.Ghisays V, Goradia DD, Protas H, et al. Brain imaging measurements of fibrillar amyloid-beta burden, paired helical filament tau burden, and atrophy in cognitively unimpaired persons with two, one, and no copies of the APOE epsilon4 allele. Alzheimers Dement. 2020;16:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caselli RJ, Langlais BT, Dueck AC, Chen Y, Su Y, Locke DEC, et al. Neuropsychological decline up to 20 years before incident mild cognitive impairment. Alzheimers Dement. 2020;16:512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 10.Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279:751–755. [DOI] [PubMed] [Google Scholar]
- 11.Kwon OD, Khaleeq A, Chan W, Pavlik VN, Doody RS. Apolipoprotein E polymorphism and age at onset of Alzheimer’s disease in a quadriethnic sample. Dement Geriatr Cogn Disord. 2010;30:486–491. [DOI] [PubMed] [Google Scholar]
- 12.Morris JC, Schindler SE, McCue LM, Moulder KL, Benzinger TLS, Cruchaga C, et al. Assessment of Racial Disparities in Biomarkers for Alzheimer Disease. JAMA Neurol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajabli F, Feliciano BE, Celis K, Hamilton-Nelson KL, Whitehead PL, Adams LD, et al. Ancestral origin of ApoE epsilon4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet. 2018;14:e1007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris HR, Al-Sarraj S, Schwab C, Gwinn-Hardy K, Perez-Tur J, Wood NW, et al. A clinical and pathological study of motor neurone disease on Guam. Brain. 2001;124:2215–2222. [DOI] [PubMed] [Google Scholar]
- 15.Galasko D, Salmon D, Gamst A, Olichney J, Thal LJ, Silbert L, et al. Prevalence of dementia in Chamorros on Guam: relationship to age, gender, education, and APOE. Neurology. 2007;68:1772–1781. [DOI] [PubMed] [Google Scholar]
- 16.Henderson JN, Crook R, Crook J, Hardy J, Onstead L, Carson-Henderson L, et al. Apolipoprotein E4 and tau allele frequencies among Choctaw Indians. Neurosci Lett. 2002;324:77–79. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg RN, Richter RW, Risser RC, Taubman K, Prado-Farmer I, Ebalo E, et al. Genetic factors for the development of Alzheimer disease in the Cherokee Indian. Arch Neurol. 1996;53:997–1000. [DOI] [PubMed] [Google Scholar]
- 18.Kataoka S, Robbins DC, Cowan LD, Go O, Yeh JL, Devereux RB, et al. Apolipoprotein E polymorphism in American Indians and its relation to plasma lipoproteins and diabetes. The Strong Heart Study. Arterioscler Thromb Vasc Biol. 1996;16:918–925. [DOI] [PubMed] [Google Scholar]
- 19.Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, et al. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–1155. [DOI] [PubMed] [Google Scholar]
- 20.Suchy-Dicey AM, Shibata D, Best LG, Verney SP, Longstreth WT, Lee ET, et al. Cranial Magnetic Resonance Imaging in Elderly American Indians: design, Methods, and Implementation of the Cerebrovascular Disease and Its Consequences in American Indians Study. Neuroepidemiology. 2016;47:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cholerton B, Omidpanah A, Madhyastha TM, Grabowski TJ, Suchy-Dicey AM, Shibata DK, et al. Total Brain and Hippocampal Volumes and Cognition in Older American Indians: the Strong Heart Study. Alzheimer Dis Assoc Disord. 2017;31:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suchy-Dicey A, Shibata D, Cholerton B, Nelson L, Calhoun D, Ali T, et al. Cognitive Correlates of MRI-defined Cerebral Vascular Injury and Atrophy in Elderly American Indians: the Strong Heart Study. J Int Neuropsychol Soc. 2020;26:263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.North KE, Goring HH, Cole SA, Diego VP, Almasy L, Laston S, et al. Linkage analysis of LDL cholesterol in American Indian populations: the Strong Heart Family Study. J Lipid Res. 2006;47:59–66. [DOI] [PubMed] [Google Scholar]
- 24.Suchy-Dicey AM, Shibata DK, Madhyastha TM, Grabowski TJ, Longstreth WT Jr, Buchwald DS. Findings of Vascular Brain Injury and Structural Loss from Cranial Magnetic Resonance Imaging in Elderly American Indians: the Strong Heart Study. Neuroepidemiology. 2017;48:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage. 1999;9:179–194. [DOI] [PubMed] [Google Scholar]
- 26.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56:907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Naturegenetics. 2012;44:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delis DC, Kramer JH, Kaplan E, Ober BA, California Verbal Learning Test (CVLT-II). 2 ed. US: the Psychological Corporation; 2000. [Google Scholar]
- 29.Wechsler D. Wechsler Adult Intelligence Scale. 4th ed. 2008. [Google Scholar]
- 30.Benton AL, Hansher K. Multilingual aphasia examination. 2nd ed. AJA Associates; 1976. [Google Scholar]
- 31.Teng EL, Chang Chui H. The Modified Mini-Mental (3MS) Examination. Journal Clinical Psychiatry. 1987;48:314–318. [PubMed] [Google Scholar]
- 32.Marioni RE, Campbell A, Scotland G, Hayward C, Porteous DJ, Deary IJ. Differential effects of the APOE e4 allele on different domains of cognitive ability across the life-course. Eur J Hum Genet. 2016;24:919–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weissberger GH, Nation DA, Nguyen CP, Bondi MW, Han SD. Meta-analysis of cognitive ability differences by apolipoprotein e genotype in young humans. Neurosci Biobehav Rev. 2018;94:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyall DM, Celis-Morales C, Lyall LM, Graham C, Graham N, Mackay DF, et al. Assessing for interaction between APOE epsilon4, sex, and lifestyle on cognitive abilities. Neurology. 2019;92:e2691–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batterham PJ, Bunce D, Cherbuin N, Christensen H. Apolipoprotein E epsilon4 and later-life decline in cognitive function and grip strength. Am J Geriatr Psychiatry. 2013;21:1010–1019. [DOI] [PubMed] [Google Scholar]
- 36.Ready RE, Baran B, Chaudhry M, Schatz K, Gordon J, Spencer RM. Apolipoprotein E-e4, processing speed, and white matter volume in a genetically enriched sample of midlife adults. Am J Alzheimers Dis Other Demen. 2011;26:463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuinness B, Carson R, Barrett SL, Craig D, Passmore AP. Apolipoprotein epsilon4 and neuropsychological performance in Alzheimer’s disease and vascular dementia. Neurosci Lett. 2010;483: 62–66. [DOI] [PubMed] [Google Scholar]
- 38.Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele?. Ann Hum Genet. 1999;63:301–310. [DOI] [PubMed] [Google Scholar]
- 39.Shaw JT, Tate J, Kesting JB, Marczak M, Berkholz JR, Lovelock PK, et al. Apolipoprotein E polymorphism in indigenous Australians: allelic frequencies and relationship with dyslipidaemia. Med J Aust. 1999;170:161–164. [DOI] [PubMed] [Google Scholar]
- 40.Demarchi DA, Salzano FM, Altuna ME, Fiegenbaum M, Hill K, Hurtado AM, et al. APOE polymorphism distribution among Native Americans and related populations. Ann Hum Biol. 2005;32:351–365. [DOI] [PubMed] [Google Scholar]
- 41.Eisenberg DT, Kuzawa CW, Hayes MG. Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. Am J Phys Anthropol. 2010;143:100–111. [DOI] [PubMed] [Google Scholar]
- 42.Logue MW, Schu M, Vardarajan BN, Buros J, Green RC, Go RC, et al. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol. 2011;68:1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajan KB, Barnes LL, Wilson RS, Weuve J, McAninch EA, Evans DA. Apolipoprotein E Genotypes, Age, Race, and Cognitive Decline in a Population Sample. J Am Geriatr Soc. 2019;67:734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knopman DS, Mosley TH, Catellier DJ, Coker LH. Atherosclerosis Risk in Communities Study Brain MRIS. Fourteen-year longitudinal study of vascular risk factors, APOE genotype, and cognition: the ARIC MRI Study. Alzheimers Dement. 2009;5:207–214. [DOI] [PubMed] [Google Scholar]
- 45.Duara R, Loewenstein DA, Lizarraga G, Adjouadi M, Barker WW, Greig-Custo MT, et al. Effect of age, ethnicity, sex, cognitive status and APOE genotype on amyloid load and the threshold for amyloid positivity. Neuroimage Clin. 2019;22:101800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sapkota B, Subramanian A, Priamvada G, Finely H, Blackett PR, Aston CE, et al. Association of APOE polymorphisms with diabetes and cardiometabolic risk factors and the role of APOE genotypes in response to anti-diabetic therapy: results from the AIDHS/SDS on a South Asian population. J Diabetes Complications. 2015;29:1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irie F, Masaki KH, Petrovitch H, Abbott RD, Ross GW, Taaffe DR, et al. Apolipoprotein E epsilon4 allele genotype and the effect of depressive symptoms on the risk of dementia in men: the Honolulu-Asia Aging Study. Arch Gen Psychiatry. 2008;65:906–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haan MN, Mayeda ER. Apolipoprotein E Genotype and Cardiovascular Diseases in the Elderly. Curr Cardiovasc Risk Rep. 2010;4:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawyer K, Sachs-Ericsson N, Preacher KJ, Blazer DG. Racial differences in the influence of the APOE epsilon 4 allele on cognitive decline in a sample of community-dwelling older adults. Gerontology. 2009;55:32–40. [DOI] [PubMed] [Google Scholar]
- 50.Weuve J, Barnes LL, Mendes de Leon CF, Rajan KB, Beck T, Aggarwal NT, et al. Cognitive Aging in Black and White Americans: cognition, Cognitive Decline, and Incidence of Alzheimer Disease Dementia. Epidemiology. 2018;29:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verney S, Suchy-Dicey A, Cholerton B, Calhoun D, Nelson L, Montine T, et al. The associations among sociocultural factors and neuropsychological functionings in older American Indians: the Strong Heart Study. Neuropsychology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]