Abstract
Recent work in this laboratory has shown that the gene coding for acetate kinase (ackA) in Sinorhizobium meliloti is up-regulated in response to phosphate limitation. Characterization of the region surrounding ackA revealed that it is adjacent to pta, which codes for phosphotransacetylase, and that these two genes are part of an operon composed of at least two additional genes in the following order: an open reading frame (orfA), pta, ackA, and the partial sequence of a gene with an inferred peptide that has a high degree of homology to enoyl-ACP reductase (fabI). Experiments combining enzyme assays, a chromosomal lacZ::ackA transcriptional fusion, complementation analysis with cosmid subclones, and the creation of mutations in pta and ackA all indicated that the orfA-pta-ackA-fabI genes are cotranscribed in response to phosphate starvation. Primer extension was used to map the position of the phosphate starvation-inducible transcriptional start sites upstream of orfA. The start sites were found to be preceded by a sequence having similarity to PHO boxes from other phosphate-regulated genes in S. meliloti and to the consensus PHO box in Escherichia coli. Introduction of a phoB mutation in the wild-type strain eliminated elevated levels of acetate kinase and phosphotransacetylase activities in response to phosphate limitation and also eliminated the phosphate stress-induced up-regulation of the ackA::lacZ fusion. Mutations in either ackA alone or both pta and ackA did not affect the nodulation or nitrogen fixation phenotype of S. meliloti.
Through a complex set of biochemical communications, bacteria in the family Rhizobiaceae interact with specific legume hosts to produce root structures known as nodules. Numerous bacterial genes are involved in nodule formation and function. Two of the better-studied examples include the nodulation (nod) genes, which are expressed in response to isoflavones secreted by the plant (reviewed in references 14, 28, and 48), and the dct genes, which are required for uptake of dicarboxylic acids (malate and/or succinate), the primary sources of energy for the bacteria during symbiosis (reviewed in references 13, 33, and 51). In different rhizobia, two-component regulatory systems have been shown to be involved in the positive regulation of both of these activities (20, 21, 40). Two-component regulatory protein systems consist of a sensor histidine kinase which is paired with a cognate-response-regulatory protein. The sensor kinase detects a particular environmental or metabolic signal and transduces the signal to the cell via phosphorylation of the regulator protein (11, 50). When phosphorylated, the regulatory protein is then activated to increase the transcription of target genes under the control of the regulatory pair.
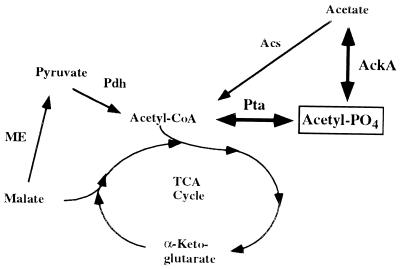
In addition to the sensor kinase, some response-regulatory proteins have been shown to be phosphorylated by other, less specific, secondary mechanisms involving low-molecular-weight phosphorylated compounds such as phosphoramidate, carbamylphosphate, and acetyl phosphate (19, 29–31, 55). Of these, the most extensively studied has been acetyl phosphate. Synthesis of acetyl phosphate is controlled directly by the enzymes acetate kinase and phosphotransacetylase (Fig. 1), which are encoded by the genes ackA and pta, respectively. Both reactions are reversible and, depending on the metabolic activities leading to the synthesis of their substrates, can influence the acetyl phosphate pool size. Factors thus far shown to influence the intracellular concentration of acetyl phosphate include the growth phase, carbon source, and temperature (30, 38, 56). Acetyl coenzyme A (acetyl-CoA) is a substrate for the Pta reaction and is a central metabolic intermediate tied directly to all anabolic processes. Fluctuation of acetyl-CoA levels in response to changing growth conditions could play an important role in determining acetyl phosphate concentrations and, thus, indirectly play a role in affecting gene expression via this less-specific phosphotransferase mechanism. Transcriptional control of ackA and pta would also be expected to be potentially important in this regard; however, comparatively little is known about the regulation of these genes (12).
FIG. 1.
The acetyl phosphate pathway in relation to the tricarboxylic acid cycle, which is a major metabolic pathway in S. meliloti bacteroids. The acetyl phosphate pathway is shown with thick boldfaced arrows. Abbreviations: TCA, tricarboxylic acid; ME, malic enzyme; Acs, acetyl-CoA synthase; AckA, acetate kinase; Pta, phosphotransacetylase; Pdh, pyruvate dehydrogenase.
The potential for acetyl phosphate to act as a signaling metabolite in the Rhizobium-legume symbiosis has been briefly examined for the nod and dct regulatory systems mentioned above. Loh et al. (27) recently reported that Bradyrhizobium japonicum NodW can be phosphorylated by acetyl phosphate in vitro, but Gu et al. (23) were unable to demonstrate this alternative phosphorylation mechanism for Sinorhizobium meliloti DctD. Preston et al. (37) and Smith et al. (46) examined the activities of both Pta and AckA in B. japonicum bacteroids and found that the levels of both enzymes increased in parallel with nodule nitrogen fixation (acetylene reduction) activity. This implied that both enzymes might contribute significantly to bacteroid carbon metabolism and, depending on the relative rates of each enzyme, might allow for the accumulation of acetyl phosphate to levels that could, in turn, influence gene expression.
In this study, we characterized a locus in S. meliloti that contains the genes coding for Pta and AckA. We also created pta and ackA mutants to establish metabolic blockades that would either impede the synthesis of acetyl phosphate or favor its accumulation. We used these mutants to assess the potential importance of acetyl phosphate acting as a signaling metabolite in the S. meliloti-alfalfa symbiosis. These mutations also allowed us to ask whether carbon metabolism via the Pta-AckA pathway is important to alfalfa bacteroids for normal symbiotic function.
MATERIALS AND METHODS
Strains and growth conditions.
The strains of S. meliloti and Escherichia coli used in this study are shown in Table 1. S. meliloti wild-type strain 104A14 (47) was the parent strain from which all mutants were derived. The isolation of the S. meliloti ackA::Tn5B22 mutant strain psi25 was performed as previously described (52), and in this study this strain has been renamed RmMSU4. Transposon Tn5B22 is a derivative of Tn5 (44) and carries a promoterless lacZ gene. When Tn5B22 inserts in the correct orientation relative to a nearby promoter, a lacZ transcription fusion construct that can be used to monitor gene expression is created. The genotypes and construction of other mutants are described below.
TABLE 1.
Strains of S. meliloti and E. coli and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristica | Reference or source |
|---|---|---|
| S. meliloti strains | ||
| 104A14 | Wild type | 47 |
| RmMSU4 | 104A14 ackA::Tn5B22 | 52 |
| RmMSU5 | 104A14 pta::Ω ackA::Tn5B22 | This study |
| RmMSU6 | 104A14 pta::Ω | This study |
| RmMSU7 | 104A14 phoB::Ω ackA::Tn5B22 | This study |
| E. coli strains | ||
| S17-1 | Pro− Mob+; donor strain in matings | 43 |
| DH5α | Cloning host | 41 |
| Plasmids | ||
| pUC19 | Cloning vector; Ampr | 58 |
| pBluescript KS(+) | Cloning and sequencing vector; Ampr | Stratagene |
| pJQ200KS+ | sacB sacR Gentr SucS | 39 |
| pCPP30 | Broad-host-range plasmid; Gentr | D. Bauer, Cornell University |
| pRL453 | pUC18/19 containing L.EHE1 and C.S3; Ω fragment | 18 |
| p7C9 | Cosmid library clone containing the S. meliloti orfA-pta-ackA-fabI operon | This study |
| pMLS3 | pCPP30 with 2.4-kb XmaI-SacI p7C9 fragment; Gentr | This study |
| pMLS4 | pCPP30 with 3.2-kb BamHI-SacI p7C9 fragment; Gentr | This study |
| pMLS5 | pCPP30 with 4.0-kb BamHI p7C9 fragment; Gentr | This study |
| pMLS6 | pCPP30 with 8.8-kb NotI p7C9 fragment; Gentr | This study |
| pMLS7 | pCPP30 with 5.0-kb EcoRI-SacI p7C9 fragment; Gentr Pta+ | This study |
| pMLS8 | pCPP30 with 9-kb SacI p7C9 fragment; Gentr Pta+ | This study |
| pMLS90 | pJQ200KS+ ligated to linearized pUC19 containing the Ω-interrupted phoB in a 2.3-kb HindIII fragment | 2; this study |
| pMLS101 | pJQ200KS+ containing the linearized 9-kb SacI p7C9 fragment ligated to pBluescript KS(+); pta::Ω | This study |
Ampr, ampicillin resistant; Gentr, gentamicin resistant; SucS, sucrose sensitive.
The basic medium used for culturing all strains was a minimal mannitol ammonium chloride (MMN) medium (47) that was modified to contain no added inorganic phosphate (Pi) salts in the basal medium but was buffered by 10 mM morpholinepropanesulfonic acid (MOPS; pH 7.0) and amended with various amounts of Pi. In some experiments, S. meliloti was cultured on YMB agar (47). E. coli DH5α was used as a host strain for plasmid constructions, and E. coli S17-1 was used for conjugal transfer of plasmids to S. meliloti; both E. coli strains were cultured on Luria-Bertani medium (41). Ampicillin (100 μg · ml−1), gentamicin (25 μg · ml−1 for agar media; 15 μg · ml−1 in broth cultures), streptomycin-spectinomycin (each at 200 μg · ml−1), and/or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 mg · liter−1 for agar media) was included as required.
Plasmids and mutagenesis.
The plasmids used in this study are also shown in Table 1. DNA flanking the transposon cloned from RmMSU4 was used as a probe to identify a hybridizing colony in blots of a S. meliloti cosmid library, the construction of which was previously described (2). The library insert in the hybridizing cosmid p7C9 was subcloned in part or in full into pBluescript KS(+) and pCPP30 for sequencing and complementation experiments, respectively. The S. meliloti phoB gene from strain 104A14 was previously cloned on a 2.3-kb HindIII fragment (2), and for experiments in this study this fragment was subcloned into pUC19 and inactivated by insertion of an Ω cassette (18) into the unique EcoRV site located within the phoB coding region. This entire construct was linearized with XbaI and subcloned into pJQ200SK+ to create pMLS90. A pta mutation was similarly constructed by insertion of the Ω fragment into the pta gene. A 9-kb SacI fragment containing the S. meliloti pta gene was subcloned from p7C9 into pBluescript KS(+) which had been partially digested with KpnI and ligated to a Ω fragment released from pRL453 by KpnI digestion. A clone containing an Ω insertion within pta was identified by restriction mapping, linearized with XbaI, and then ligated to pJQ200SK+ to form pMLS101.
Both pMLS90 and pMLS101 were transformed into E. coli S17-1 for conjugation with S. meliloti. Isolation of phoB::Ω and pta::Ω allelic replacements in transconjugants was performed on MMN agar containing 5 mM Pi and amended with gentamicin, streptomycin, spectinomycin, and 10% (wt/vol) sucrose. Gene replacements were verified by Southern blotting with the wild-type phoB and pta genes as probes (2), by the loss of inducible alkaline phosphatase activity for the phoB mutant (2), and by the lack of Pi starvation-inducible acetate kinase and phosphotransacetylase activities in the different mutants (procedures described below).
Nucleic acid manipulations.
The protocols of Sambrook et al. (41) were used for all manipulations of plasmid and chromosomal DNA. In previous work in which we sequenced the DNA flanking the Tn5B22 insert in RmMSU4, it was determined that the transposon was in the correct orientation for reporting ackA transcription (52). Briefly, total chromosomal DNA was harvested, digested with XmaI or SalI, and then ligated into pBluescript KS(+). The ligated construct was transformed into E. coli DH5α, and plasmids from transformants resistant to ampicillin and gentamicin were analyzed by restriction analysis to verify that each contained a single cloned fragment. Southern blotting was used to verify that the cloned fragment was identical to that in the genome from RmMSU4. Sequencing of the flanking DNA was done by the dideoxynucleotide method, using a kit purchased from United States Biochemical (Cleveland, Ohio). Primers 5′-AACGACGGGATCCATAAT-3′ and 5′-CCATGTTAGGAGGTCACATGGAAGTCAG-3′ were used to initiate sequencing from the lacZ and transposase termini, respectively (44). Large-scale sequencing of segments of p7C9 that contained the complementing pta and ackA genes was accomplished with an ABI 377 DNA sequencer (Perkin-Elmer, Norwalk, Conn.), using synthetic primers complementary to nucleotide sequences determined within the cloned fragments. Sequence homology searches were conducted by using the BLAST network service (3), and sequence alignments were done with the GAP program (15).
RNA isolation and primer extension.
Total RNA was harvested from cells by the method described by Schmidt-Goff and Federspiel (42). Primer extension and end labeling of the oligonucleotide primer with [32P]ATP (DuPont-NEN; 3,000 Ci mmol−1, 10 mCi ml−1) were performed on 20 μg of strain 104A14 RNA as described by Ausubel et al. (4), using polynucleotide kinase and avian myeloblastosis virus reverse transcriptase from Promega (Madison, Wis.). The primer sequence was 5′-GAGCGGCAAGGCCGCAATCCCAATTAT-3′. Sequencing ladders employing the same primer were generated by the dideoxy termination method, using Sequenase II T7 polymerase (United States Biochemical) and Sequetide (DuPont-NEN). The primer extension product with the corresponding sequencing ladder was separated on 6% polyacrylamide–7 M urea sequencing gels.
Phosphate stress induction experiments.
For phosphate starvation experiments, cells were grown to mid-exponential phase in MMN broth containing 5 mM Pi and antibiotics as required, washed twice with MMN lacking Pi (25°C), resuspended in Pi-free MMN to an A595 of 0.2, and then divided into two cultures. No Pi was added to one of the cultures, while the other culture was amended to 5 mM Pi. Both cultures were incubated in a reciprocal shaking water bath at 30°C. After the desired incubation period, a culture sample was taken for measurement of Pta, AckA, or β-galactosidase activity.
Enzyme assays.
β-Galactosidase activity was measured as previously described (52). For measuring phosphotransacetylase and acetate kinase activities, both Pi-grown and non-Pi-grown cells were washed and resuspended in a buffer that contained 50 mM Tris-HCl (pH 7.5) and 10 mM MgCl2 and then were disrupted by sonication. The sonicate was centrifuged at 10,000 × g for 5 min to pellet debris, and then the resulting supernatant was removed and stored on ice for use in enzyme assays.
Determinations of phosphotransacetylase and acetate kinase activities were based on acetyl phosphate production or consumption and were measured by the hydroxymate assay (26). Phosphotransacetylase activity was assayed by the method described by Stadtman (49). Briefly, the 333-μl assay mixture contained 10 mM Tris-HCl, 6 mM acetyl phosphate, 100 mM potassium chloride, 10 mM cysteine, 1 mM CoA, and 20 to 40 μl of cell extract. After a 10-min incubation at 25°C, the reaction mixture was supplemented with sodium arsenate to 50 mM, incubated for an additional 45 min at 25°C, diluted 1:1 with 2 M neutralized NH2OH-HCl, and incubated for a further 5 min at 25°C. The reaction was stopped by adding trichloroacetic acid to a final concentration of 5%. The final volume was brought to 1.5 ml with 2.5% FeCl2 (dissolved in 2 N HCl) and centrifuged at 13,000 × g for 5 min to clear insoluble material, and then the absorbance was measured at 540 nm against a water blank. Acetate kinase activity was measured as described by Aceti and Ferry (1). In a total volume of 333 μl, the assay mixture contained 145 mM Tris-HCl, 200 mM potassium acetate, 10 mM ATP, and 0.7 M neutralized NH2OH-HCl; cell extract was added to initiate the reaction. After incubation at 25°C (15 to 30 min), trichloroacetic acid was added to a concentration of 5% to stop the reaction. The reaction mixture was then brought to a volume of 1 ml with 2.5% FeCl2 (dissolved in 2 N HCl), incubated at 25°C for 15 min to allow for color development, and centrifuged at 13,000 × g for 5 min to clear insoluble material, and then the A540 was determined. Assays without added acetate and ATP were also conducted to correct for levels of acyl phosphates in cell extracts.
Plant growth and inoculation.
Axenic alfalfa plants were cultured in Magenta growth boxes (Sigma, St. Louis, Mo.) that were described previously (32), using the sand-alumina plant culture technique (22) to control the Pi supply at levels that were Pi limiting and non-Pi limiting for the plants (2).
For each strain, the stability of the transposon and/or Ω cassette during symbiosis was assessed. Nodules were surface sterilized as described previously (2) and then crushed in a 0.85% (wt/vol) saline solution prior to being serially diluted; aliquots were then spread onto YMB agar. Sixty isolated colonies of each mutant were subcultured onto MMN and MMN plus antibiotic to determine whether antibiotic resistance was retained. Southern blot analysis of chromosomal DNA from five nodule isolates for each strain was conducted to verify that the location of the transposon and/or Ω cassette had not changed.
Nucleotide sequence accession number.
The nucleotide sequence of the orfA-pta-ackA-fabI gene sequence has been submitted to GenBank (accession no. AF095903).
RESULTS
Sequence analysis of the S. meliloti ackA locus.
Using transposon Tn5B22 as a promoter probe, we previously identified ackA as one of several S. meliloti genes that are up-regulated in response to Pi stress (52). Identification of the interrupted ackA gene in mutant strain RmMSU4 was accomplished by cloning the transposon and flanking DNA from RmMSU4 (previously designated psi25 [52]), sequencing the transposon-chromosome junctions, and then conducting homology searches of major databases (52). To more extensively characterize this locus, we used a fragment of DNA flanking the transposon to probe an S. meliloti cosmid library (2) and obtained cosmid p7C9, which contained ackA. Restriction mapping, Southern blot analysis, and enzyme assays following Pi starvation treatments confirmed that ackA was preceded by a suitable amount of DNA such that the transcriptional control elements for ackA could be studied (results not shown). A variety of hybridizing DNA fragments were subcloned from p7C9 into pBluescript and either partially or completely sequenced.
The results of this sequencing indicated the presence of three genes in addition to ackA. The first is a 2,199-bp open reading frame (ORF) capable of encoding a protein of 80.5 kDa. It does not have any homologs in the major protein or nucleotide databases and was designated orfA. It is preceded by a putative ribosome binding site and followed by a 182-bp region containing several stem-loops, but no factor-independent terminators were identified by computer analysis. This short segment was followed by a second ORF, 807 nucleotides in length and preceded by a ribosome binding site. This ORF is capable of being translated into a 28.5-kDa protein having an inferred amino acid sequence with 29% identity and 50% similarity to the phosphotransacetylase from Clostridium acetobutylicum (9). Phosphotransacetylases differ in size, and the identified gene is of the smaller variety that includes phosphotransacetylases from C. acetobutylicum (9) and Methanosarcina thermophila (25).
Following pta is an ORF spanning 1,179 bp that encodes an inferred 42.1-kDa peptide with 35% identity and 61% similarity to the acetate kinase from C. acetobutylicum (9). The predicted size, based on translation initiation occurring at the second methionine within the ORF, is supported by protein alignments with other inferred acetate kinases described in the databases. This initiation site would place the translational start site just 6 bp from the termination codon of pta.
Immediately downstream from ackA is an ORF coding for an inferred peptide with 50% identity and 70% similarity (based on partial sequence) to FabI, the enoyl-ACP reductase from E. coli (8). The putative fabI gene is preceded by a strong ribosome binding site in the small, 14-bp intergenic region. The sequenced region of the putative fabI sequence also exhibits similarly high levels of homology to fabI genes of several other organisms, which encode enzymes ranging in length from 262 to 278 amino acids, thus indicating that only approximately 20% of the corresponding S. meliloti fabI gene has been sequenced. The inferred peptide from the putative S. meliloti fabI gene identified here is most closely related to other enoyl-ACP reductases utilizing NADH as a substrate. The close association with ackA suggests the possibility of cotranscription of fabI with upstream genes and is supported by the inability to detect independent transcription in primer extension analyses. Further evidence for cotranscription was supported by Northern analyses which showed the presence of an irresolute 7- to 10-kb transcript unique to phosphate-starved cells (data not shown).
Operon studies: mutational and complementation analysis of gene expression.
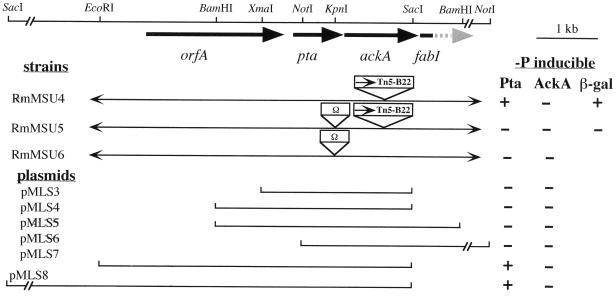
The orfA-pta-ackA-fabI gene arrangement, the locations of Tn5B22 and the Ω cartridge in the mutant strains, and the different plasmid constructs are shown in Fig. 2. Enzyme activity levels observed under Pi-limiting and non-Pi-limiting conditions with different combinations of the mutant strains and plasmid constructs are presented in Table 2. Consistent with the sequence analysis of the Tn5B22 insertion in ackA, S. meliloti RmMSU4 lacked Pi starvation-inducible AckA activity. Pi starvation-inducible AckA activity could be restored to normal levels when this strain contained cosmid p7C9 (results not shown). However, p7C9 DNA fragments contained in pMLS3, pMLS4, pMLS7, or pMLS8 (Fig. 2) did not result in restoration of inducible AckA activity. For pMLS7 and pMLS8, this was apparently due to the absence of the C-terminal 45 amino acids of AckA beyond the SacI site (Fig. 2). When the complete AckA coding region was present in pMLS6 but the upstream sequence was not (Fig. 2), there was a slightly increased basal level of AckA activity that lacked normal inducibility (Table 2) (similar results were obtained for pMLS5 [data not shown]). Strain RmMSU6, containing pta interrupted by the Ω cassette (Fig. 2), showed no Pi stress-inducible Pta activity (Table 2). Furthermore, the mutated pta in strain RmMSU6 also eliminated Pi stress-inducible AckA activity (Fig. 2; Table 2). RmMSU4 containing pMLS7 (data not shown) or pMLS8 (Table 2) displayed very high levels of Pta activity in response to Pi limitation; presumably this was due to a gene dosage effect. A pta::Ω mutation was also introduced into RmMSU4 to create the ackA pta double-mutant strain RmMSU5 (Fig. 2), and as would be predicted, it had no Pi stress-inducible Pta or AckA activity (Table 2). The mutated pta in strain RmMSU5 also eliminated the Pi stress-inducible ackA::lacZ reporter activity (Fig. 2; also see Fig. 5 below). Together, these results indicate that pta and ackA are transcribed as an operon from a promoter located upstream of pta.
FIG. 2.
Operon and complementation analysis of the S. meliloti SacI-NotI fragment containing the orfA-pta-ackA-fabI operon. An abbreviated SacI-NotI fragment is shown at the top, with thick boldfaced arrows delineating the boundaries of each gene shown. The positions of the restriction sites used in subcloning analysis are as shown. Below the fragment, mutant strain numbers with the location of Tn5B22 transposon or the Ω interposon in the pta and ackA genes are shown. At the bottom, plasmid numbers, along with their corresponding subclone inserts, are shown relative to the relevant restriction sites. At the right, the Pi starvation (−P)-inducible activities of the Pta and AckA enzymes (+), or the lack thereof (−), as well as reporter enzyme levels (where relevant) are shown for each construct or strain. The inducibility of all plasmid-borne Pta and AckA activities was assessed in an RmMSU6 mutant background.
TABLE 2.
Acetate kinase and phosphotransacetylase activities in wild-type and mutant S. meliloti strains
| Strain | Genotype | Plasmid (relevant gene) | Phosphotransacetylase activitya
|
Acetate kinase activitya
|
||
|---|---|---|---|---|---|---|
| +Pi | −Pi | +Pi | −Pi | |||
| 104A14 | Wild type | 20 ± 10 | 100 ± 26 | 12 ± 3 | 134 ± 5 | |
| RmMSU4 | ackA::Tn5-B22 | 21 ± 3 | 98 ± 16 | 7 ± 1 | 11 ± 1 | |
| RmMSU5 | pta::Ω ackA::Tn5-B22 | 35 ± 1 | 17 ± 2 | 11 ± 1 | 10 ± 2 | |
| RmMSU6 | pta::Ω | 20 ± 10 | 17 ± 2 | 6 ± 1 | 11 ± 2 | |
| RmMSU7 | phoB::Ω ackA::Tn5-B22 | 21 ± 2 | 28 ± 3 | 9 ± 3 | 13 ± 1 | |
| RmMSU4 | ackA::Tn5-B22 | pMLS8 (pta) | 43 ± 23 | 726 ± 107 | 8 ± 2 | 15 ± 3 |
| RmMSU4 | ackA::Tn5-B22 | pMLS6 (ackA) | 26 ± 7 | 70 ± 18 | 33 ± 1 | 47 ± 9 |
| RmMSU6 | pta::Ω | pMLS8 (pta) | 90 ± 22 | 716 ± 86 | 6 ± 1 | 10 ± 2 |
| RmMSU6 | pta::Ω | pMLS6 (ackA) | 27 ± 6 | 25 ± 3 | 18 ± 2 | 27 ± 5 |
Values are means ± standard errors for data from three to six separate experiments. Activities are expressed in nanomoles of acetyl phosphate consumed (Pta) or generated (AckA) per minute per milligram of protein.
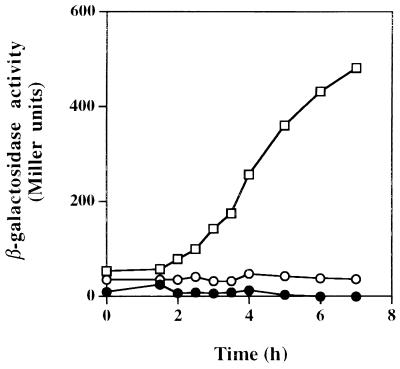
FIG. 5.
Reporter enzyme activity of an ackA::lacZ transcriptional fusion construct in response to phosphate starvation. Cells of each strain were prepared and assayed as described in Materials and Methods. Reporter enzyme levels (in Miller units) for strains RmMSU4 (ackA::Tn5B22) (□), RmMSU5 (pta::Ω ackA::Tn5B22) (●), and RmMSU7 (ackA::Tn5B22 phoB::Ω) (○) are shown. Data points are the averages of data from duplicate cultures from one of three independent experiments documenting these responses. Reporter enzyme levels failed to increase when washed cells of all three strains were resuspended in high-phosphate medium (results not shown to simplify presentation).
To obtain additional information to help identify the region containing the Pi stress-inducible promoter for the putative orfA-pta-ackA-fabI operon, cosmid subclones were analyzed for their ability to complement the loss of inducible Pta activity in RmMSU6. Neither pMLS3 nor pMLS4 (Fig. 2), containing 547 and 1,360 bp of DNA upstream of pta, respectively, was able to complement the Pta− phenotype. However, pMLS7, containing all of pta and orfA and 793 bp of DNA upstream of orfA, was able to complement the Pta− mutant, as did pMLS8, which contains a 9-kb SacI fragment. These results indicated that the promoter is located in the region upstream of orfA.
Promoter analysis and mutational confirmation of PhoB regulation.
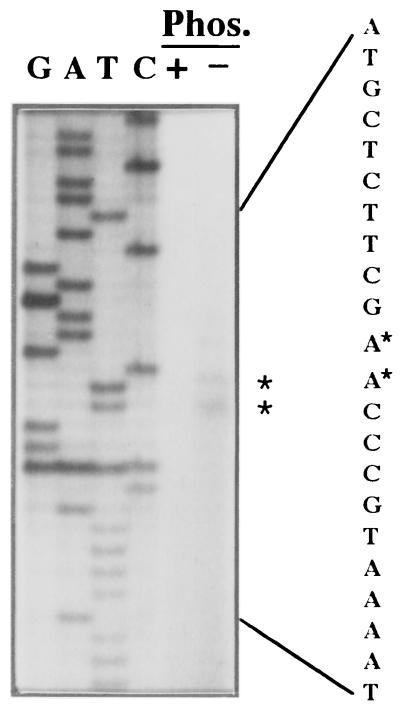
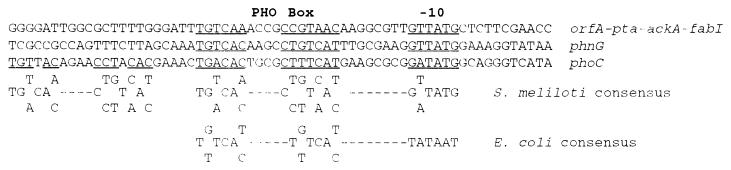
A Pi stress-inducible transcriptional start site was identified by primer extension analysis using a primer complementary to bp 671 to 697. The 73- to 74-nucleotide extension products identified two strong start sites, 171 or 172 bp upstream of the putative translational start predicted for OrfA (Fig. 3). Sequence 5′ of the transcriptional start was found to have significant similarity to PHO box sequences identified for other S. meliloti Pi-sensitive genes (5) (GenBank accession no. M96263). The alignment of this region with other predicted Pi-regulated promoter regions allows the construction of an S. meliloti consensus PHO box (Fig. 4). The resulting consensus sequence shows similarities to that of E. coli (57). In addition to the PHO box, another feature of the promoter region is a less-understood, purine-rich direct repeat with the sequence 5′-AATAAAA-3′. The repeat is separated by 3 bp and is located in the leader sequence beginning 7 bp downstream from the transcriptional start site. An association of this direct repeat with any regulatory function has not yet been determined.
FIG. 3.
Determination of the transcriptional start sites for the S. meliloti orfA-pta-ackA-fabI operon by primer extension. RNA was purified from cells of wild-type strain 104A14 after incubation under high-phosphate culture conditions (Phos. +) or under phosphate starvation conditions (Phos. −). Lanes G, A, T, and C show the sequence ladders generated with the same primer as that used in the extension reaction, with pMLS7 as a template. Relevant complementary DNA sequence is shown on the right, and the positions of the start sites are indicated with asterisks.
FIG. 4.
Suggested structure of a consensus S. meliloti PHO box. The consensus nucleotide sequence is derived from a comparison of single PHO boxes identified in the promoter region of phnG (GenBank accession no. M96263) and the orfA-pta-ackA-fabI operon, as well as the two tandem PHO boxes reported for phoC (5). The E. coli consensus PHO box (57) is shown at the bottom for comparison. Nucleotides matching the S. meliloti consensus sequence for the −10 region and PHO box are underlined.
The sensitivity of this operon to Pi availability and the identification of a putative PHO box suggested that these genes are part of the Pho regulon in this organism and are under the control of the response-regulatory protein PhoB (6). To test this hypothesis, a phoB mutation was introduced into strain RmMSU4 to create strain RmMSU7. Following removal of Pi from the medium, strain RmMSU7 did not show inducible reporter activity during 7 h of Pi starvation (Fig. 5). Inducible β-galactosidase reporter enzyme activity was still not observed following overnight induction (results not shown). The phoB mutation had similar effects on Pta activity (Table 2); Pta activity in Pi-limited RmMSU7 cells did not increase beyond constitutive levels. The lack of reporter enzyme induction in strain RmMSU5 also demonstrated the polar effect of the Ω insert on the expression of the ackA::Tn5B22 transcriptional fusion construct (Fig. 5) and, as with the results of experiments described above, suggested that pta and ackA are cotranscribed and are controlled by the same regulatory element in response to Pi starvation.
Symbiotic phenotype of AckA− and Pta− mutants.
There were no apparent effects of either the single ackA mutation or the pta ackA double mutation on symbiosis. Dry-matter production of plants inoculated with either mutant was not significantly different from that of plants inoculated with the wild-type strain. When averaged across plants inoculated with the wild-type strain, RmMSU4, and RmMSU5, the mean shoot dry weights ± the standard deviations were 11.3 ± 0.8 and 15.2 ± 0.4 mg · plant−1 for Pi-limited and non-Pi-limited plants, respectively. Uninoculated control plants averaged 5.5 ± 0.2 and 5.7 ± 0.3 mg · plant−1, respectively. The numbers of nodules per plant averaged 5.1 ± 0.3 and 7.9 ± 0.2 for Pi-limited and non-Pi-limited plants, respectively, and the nodule fresh weight per plant for Pi-limited plants was 4.8 ± 0.3 mg · plant−1, compared to 7.5 ± 0.3 mg · plant−1 for plants cultured with high Pi levels. As determined by analysis of variance, all differences in plant dry matter and nodulation between Pi-treated and non-Pi-treated inoculated plants were statistically significant.
DISCUSSION
In this study, the sequence, organization, and regulation of S. meliloti genes coding for phosphotransacetylase and acetate kinase activity were examined. These genes were determined to be located within an operon that includes an unidentified ORF and, based on partial sequence analysis, likely also contains fabI. The order of these genes in the operon was determined to be orfA-pta-ackA-fabI. A close physical association of pta with ackA and their cotranscription have also been observed in C. acetobutylicum (9), E. coli (24), and M. thermophila (45). Inclusion of the unknown orfA in the operon is unique to S. meliloti, and the future identification of OrfA’s function may offer an important clue for determining the role of this operon in the physiology of the cell in general and in response to Pi limitation in particular.
This operon is sensitive to Pi levels in the medium, being up-regulated in response to Pi limitation. The location of the Pi-sensitive transcriptional start site upstream of orfA was confirmed by combining complementation experiments with mutation and primer extension analyses. Two strong adjacent transcriptional start sites were identified, and sequence analysis of the −35 region revealed the presence of a putative PHO box. In other bacteria, the PHO box serves as a target for the phosphorylated response-regulatory protein PhoB (see reference 57 for a review). When a phoB mutation was introduced into the various strains, increased levels of Pta and AckA enzyme activities and ackA::lacZ reporter activity in response to phosphate limitation were abolished. Taken together, the combined evidence strongly argues that S. meliloti genes coding for enzymes directly involved in the synthesis of acetyl phosphate are in an operon controlled by the Pho regulon in response to Pi availability.
Selective mutation of pta and ackA provided an opportunity to assess whether these particular enzymes are important for S. meliloti during symbiosis. Up-regulation of these genes could potentially influence S. meliloti symbiotic function at two different levels. First, the acetyl phosphate concentration could influence alternative signaling routes and thus affect gene expression during symbiosis. There is now overwhelming evidence showing that dicarboxylic acids are the primary energy source for bacteroids (13, 33, 51), with a large proportion of the malate and succinate being routed to acetyl-CoA via a malic enzyme (Fig. 1) (16, 17) for metabolism through the tricarboxylic acid cycle (13, 33, 51). The acetyl-CoA pool size could therefore be substantial, with the flow of carbon through other pathways, such as the Pta-AckA pathway, being possible. Since S. meliloti bacteroid isolation protocols require a significant amount of time and incubation conditions can have significant effects on intracellular concentrations of acetyl-CoA (53), an important direct precursor of acetyl phosphate, reliable measurement of metabolites such as acetyl phosphate in bacteroids can be problematic. However, we reasoned that by creating metabolic blocks at Pta and/or AckA it should be possible to assess whether acetyl phosphate is potentially an important alternative signaling metabolite during symbiosis. If appreciable acetyl-CoA was channeled through this pathway in S. meliloti bacteroids, then inactivation of Pta would diminish acetyl phosphate synthesis, while a block at AckA should provide for an opportunity for acetyl phosphate pools to accumulate. If these mutations had either effect on acetyl phosphate pool size in bacteroids, there appeared to be no consequence for symbiosis, since neither the ackA mutant nor the pta ackA double mutant was different from the wild-type strain with respect to symbiotic phenotype.
A second level at which mutational analysis of pta and ackA could aid in our understanding of S. meliloti bacteroid physiology concerns the relative importance of acetate as an energy source for bacteroids or as a contributor to the acetyl-CoA pool. B. japonicum bacteroid AckA and Pta levels are correlated with nitrogenase activity during soybean nodule ontogeny (37, 46), and B. japonicum bacteroids are capable of using acetate as an energy source to support nitrogen fixation in vitro (36). Together, these reports suggest these enzymes may be important to carbon metabolism in soybean nodule bacteroids. By contrast, Miller et al. (34) reported that while isolated alfalfa nodule bacteroids could use succinate, fumarate, malate, or oxaloacetate to support nitrogenase activity, acetate was ineffective in this regard. This is not inconsistent with the symbiotic phenotype observed for the mutants studied in our experiments, for which the Nod+ Fix+ symbiotic phenotype of the S. meliloti ackA and pta-ackA mutants suggests that acetate metabolism is not a critical aspect of alfalfa bacteroid physiology. However, because of potential metabolic overlap between the Pta-AckA pathway and that of acetyl-CoA synthase (Acs) (Fig. 1), an ackA-acs double mutant would be required to completely assess this issue.
It is important to note that the pta mutants constructed in these experiments still retained some Pta activity, as demonstrated by the extract-dependent disappearance of acetyl phosphate. Likewise, based on the acetate- and ATP-dependent production of acetyl phosphate, AckA activity was judged to still be present in all of the mutants. While the levels of both apparent enzyme activities in these mutants were reduced by 80 to 90% (Table 2), the presence of acetyl phosphate-synthesizing and acetyl phosphate-consuming enzyme activities at least suggests that S. meliloti has other Pta and AckA enzymes and that these particular enzymes could be actively expressed during symbiosis. Functional reiteration is not an uncommon feature in bacteria in general, and in the context of the S. meliloti-alfalfa symbiosis, perhaps the best example is the occurrence of two distinct S. meliloti malic enzymes; the NAD-dependent malic enzyme is required for symbiosis, whereas the NADP-dependent malic enzyme is not (16, 17). Under the high-stringency conditions employed in the Southern blot analyses of the various mutants in this study, potential additional pta or ackA genes were not apparent (results not shown).
It is also important to note that the lack of a symbiotic phenotype associated with the mutants of these specific Pi stress-inducible enzymes is consistent with our earlier observations regarding the expression of Pi-sensitive genes in S. meliloti bacteroids. We have previously found that PhoB is not required for an effective symbiosis (2). Also, we have shown that several other genes subject to PhoB control in response to Pi limitation are also not required for symbiotic function and do not appear to be up-regulated in bacteroids, regardless of the Pi nutrition status of the host plant (52). Taken together, our data seem to suggest that S. meliloti bacteroids are not limited for Pi, that alfalfa nodule bacteroids are buffered from Pi deprivation when the host plant experiences Pi limitation, and that expression of genes under the control of the regulatory protein PhoB is not necessary for a normal symbiotic relationship with alfalfa. However, there are inconsistencies to this pattern. The Pi stress-inducible high-affinity Pi transport system encoded by phoCDET in S. meliloti has been reported to be required for normal symbiotic function (5, 10). The phoCDET operon is under the control of PhoB (6, 7), which also negatively regulates the recently identified S. meliloti pit low-affinity Pi transport system (6, 7). In cultured cells of S. meliloti, the Pit transporter is expressed under high-phosphate conditions and repressed under low-phosphate conditions (6, 7). In E. coli, most mutations that inhibit Pi transport through the high-affinity Pi transport system also result in constitutive expression of the Pho regulon (57). Mutations in S. meliloti phoC (the promoter-proximal gene in the phoCDET operon) have the same effect (6) and also result in the repression of the low-affinity Pit system (54). Spontaneous mutations that disconnect PhoB control of pit in the phoC mutant restore normal symbiotic function (10, 35). Therefore, it is possible that the symbiotic phenotype resulting from mutations in S. meliloti phoCDET is actually not due to the lack of Pi uptake through the Pi stress-inducible high-affinity Pi transport system system per se but rather is due to mutations that suppress the normal symbiotic expression of Pit and that coincidentally eliminate the other primary mechanism of Pi uptake. Additional studies involving manipulation of the control of Pho regulon expression and Pi transport may provide important information to help resolve this issue.
ACKNOWLEDGMENT
This material is based on work supported by the National Science Foundation under grant no. IBN-9420798.
REFERENCES
- 1.Aceti D J, Ferry J G. Purification and characterization of acetate kinase from acetate-grown Methanosarcina thermophila. J Biol Chem. 1988;263:15444–15448. [PubMed] [Google Scholar]
- 2.Al-Niemi T S, Summers M L, Elkins J G, Kahn M L, McDermott T R. Regulation of the phosphate stress response in Rhizobium meliloti by PhoB. Appl Environ Microbiol. 1997;63:4978–4981. doi: 10.1128/aem.63.12.4978-4981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D L. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1993. [Google Scholar]
- 5.Bardin S, Dan S, Osteras M, Finan T M. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J Bacteriol. 1996;178:4540–4547. doi: 10.1128/jb.178.15.4540-4547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardin S D, Finan T M. Regulation of phosphate assimilation in Rhizobium (Sinorhizobium) meliloti. Genetics. 1998;148:1689–1700. doi: 10.1093/genetics/148.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardin S D, Voegele R T, Finan T M. Phosphate assimilation in Rhizobium (Sinorhizobium) meliloti: identification of a pit-like gene. J Bacteriol. 1998;180:4219–4226. doi: 10.1128/jb.180.16.4219-4226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergler H, Wallner P, Ebeling A, Leitinger B, Fuchsbichler S, Aschauer H, Kollenz G, Hogenauer G, Turnowsky F. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J Biol Chem. 1989;269:5493–5496. [PubMed] [Google Scholar]
- 9.Boynton Z L, Bennett G N, Rudolph F B. Cloning, sequencing, and expression of genes encoding phosphotransacetylase and acetate kinase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1996;62:2758–2766. doi: 10.1128/aem.62.8.2758-2766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles T C, Newcomb W, Finan T M. ndvF, a novel locus located on megaplasmid pRmeSU47b (pEXO) of Rhizobium meliloti, is required for normal nodule development. J Bacteriol. 1991;173:3981–3992. doi: 10.1128/jb.173.13.3981-3992.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charles T C, Jin S, Nester E W. Two-component sensory transduction systems in phytobacteria. Annu Rev Phytopathol. 1992;30:463–484. doi: 10.1146/annurev.py.30.090192.002335. [DOI] [PubMed] [Google Scholar]
- 12.Clark D P, Cronan J E., Jr . Two-carbon compounds and fatty acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 343–357. [Google Scholar]
- 13.Day D A, Copeland L. Carbon metabolism and compartmentation in nitrogen-fixing legume nodules. Plant Physiol Biochem. 1991;29:185–201. [Google Scholar]
- 14.Denarie J, Debelle F, Prome J-C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli V, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driscoll B T, Finan T M. NAD+-dependent malic enzyme of Rhizobium meliloti is required for symbiotic nitrogen fixation. Mol Microbiol. 1993;7:865–873. doi: 10.1111/j.1365-2958.1993.tb01177.x. [DOI] [PubMed] [Google Scholar]
- 17.Driscoll B T, Finan T M. NADP+-dependent malic enzyme of Rhizobium meliloti. J Bacteriol. 1996;178:2224–2231. doi: 10.1128/jb.178.8.2224-2231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elhi J, Wolk C P. A versatile class of positive selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 19.Feng J, Atkinson M R, McCleary W, Stock J B, Wanner B L, Ninfa A J. Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J Bacteriol. 1992;174:6061–6070. doi: 10.1128/jb.174.19.6061-6070.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giblin L, Boesten B, Turk S, Hooykaas P, Ogara F. Signal transduction in the Rhizobium meliloti dicarboxylic acid transport system. FEMS Microbiol Lett. 1995;126:25–30. doi: 10.1111/j.1574-6968.1995.tb07385.x. [DOI] [PubMed] [Google Scholar]
- 21.Gottfert M, Grob P, Hennecke H. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Brahyrhizobium japonicum. Proc Natl Acad Sci USA. 1990;87:2680–2684. doi: 10.1073/pnas.87.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gourley C J P, Allen D L, Russelle M P, Bloom P R. Evaluation and improvements of a sand-alumina culture technique to screen plants for low phosphorus tolerance. Soil Sci Soc Am J. 1993;57:103–110. [Google Scholar]
- 23.Gu B, Lee J, Hoover T, Scholl D, Nixon T. Rhizobium meliloti DctD, a ς54-dependent transcriptional activator, may be negatively controlled by a subdomain in the C-terminal end of its two component receiver module. Mol Microbiol. 1994;13:51–66. doi: 10.1111/j.1365-2958.1994.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 24.Kakuda H, Hosono K, Shiroishi K, Ichihara S. Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J Biochem. 1994;116:916–922. doi: 10.1093/oxfordjournals.jbchem.a124616. [DOI] [PubMed] [Google Scholar]
- 25.Latimer M T, Ferry J G. Cloning, sequence analysis, and hyperexpression of the genes encoding phosphotransacetylase and acetate kinase from Methanosarcina thermophila. J Bacteriol. 1993;175:6822–6829. doi: 10.1128/jb.175.21.6822-6829.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipmann F, Tuttle L C. A specific micromethod for the determination of acyl phosphates. J Biol Chem. 1945;159:21–28. [Google Scholar]
- 27.Loh J, Garcia M, Stacey G. NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J Bacteriol. 1997;179:3013–3020. doi: 10.1128/jb.179.9.3013-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long S R. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukat G S, McCleary W R, Stock A M, Stock J B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 31.McCleary W R, Stock J B, Ninfa A J. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol. 1993;175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDermott T R, Kahn M L. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J Bacteriol. 1992;174:4790–4797. doi: 10.1128/jb.174.14.4790-4797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDermott T R, Griffith S M, Vance C P, Graham P H. Carbon metabolism in Bradyrhizobium japonicum bacteroids. FEMS Microbiol Rev. 1989;63:327–340. [Google Scholar]
- 34.Miller R W, McRae D G, Al Jobore A, Berndt W B. Respiration supported nitrogenase activity of isolated Rhizobium meliloti bacteroids. J Cell Biochem. 1988;38:35–49. doi: 10.1002/jcb.240380105. [DOI] [PubMed] [Google Scholar]
- 35.Oresnik I J, Charles T C, Finan T M. Second site mutations specifically suppress the Fix− phenotype of Rhizobium meliloti ndvF mutations on alfalfa: identification of a conditional ndvF-dependent mucoid colony phenotype. Genetics. 1994;136:1233–1243. doi: 10.1093/genetics/136.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson J B, LaRue T A. Utilization of aldehydes and alcohols by soybean bacteroids. Plant Physiol. 1981;68:489–493. doi: 10.1104/pp.68.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preston G G, Zeiher C, Wall J D, Emerich D W. Acetate-activating enzymes of Bradyrhizobium japonicum bacteroids. Appl Environ Microbiol. 1989;55:165–170. doi: 10.1128/aem.55.1.165-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruss B, Wolfe A J. Regulation of acetyl phosphate synthesis and degradation and the control of flagellar expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 39.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 40.Ronson C W, Astwood P M, Nixon B T, Ausubel F M. Deduced products of C-4 dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 1987;15:7921–7934. doi: 10.1093/nar/15.19.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Frisch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Schmidt-Goff C M, Federspiel N A. In vivo and in vitro footprinting of a light-regulated promoter in the cyanobacterium Fremyella diplosiphon. J Bacteriol. 1993;175:1806–1813. doi: 10.1128/jb.175.6.1806-1813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. BioTechnology. 1983;1:784–791. [Google Scholar]
- 44.Simon R, Quandt J, Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 45.Singh-Wissmann K, Ferry J G. Transcriptional regulation of the phosphotransacetylase-encoding and acetate kinase-encoding genes (pta and ack) from Methanosarcina thermophila. J Bacteriol. 1995;177:1699–1702. doi: 10.1128/jb.177.7.1699-1702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith M T, Preston G G, Emerich D W. Development of acetate and pyruvate metabolic enzyme activities in soybean nodules. Symbiosis. 1994;17:33–42. [Google Scholar]
- 47.Somerville J E, Kahn M L. Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J Bacteriol. 1983;156:168–176. doi: 10.1128/jb.156.1.168-176.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stacey G. Bradyrhizobium japonicum nodulation genetics. FEMS Microbiol Lett. 1995;127:1–9. doi: 10.1111/j.1574-6968.1995.tb07441.x. [DOI] [PubMed] [Google Scholar]
- 49.Stadtman E R. Phosphotransacetylase from Clostridium kluyveri. Methods Enzymol. 1955;1:596–599. [Google Scholar]
- 50.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Streeter J G. Review: recent developments in carbon transport and metabolism in symbiotic systems. Symbiosis. 1995;19:175–196. [Google Scholar]
- 52.Summers M L, Elkins J G, Elliot B A, McDermott T R. Expression and regulation of phosphate stress inducible genes in Sinorhizobium meliloti. Mol Plant-Microbe Interact. 1998;11:1094–1101. doi: 10.1094/MPMI.1998.11.11.1094. [DOI] [PubMed] [Google Scholar]
- 53.Takamura Y, Nomura G. Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon source and energy metabolism of Escherichia coli K12. J Gen Microbiol. 1988;134:2249–2253. doi: 10.1099/00221287-134-8-2249. [DOI] [PubMed] [Google Scholar]
- 54.Voegele R T, Bardin S, Finan T M. Characterization of the Rhizobium (Sinorhizobium) meliloti high- and low-affinity phosphate uptake systems. J Bacteriol. 1997;179:7226–7232. doi: 10.1128/jb.179.23.7226-7232.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wanner B L. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174:2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wanner B L. Multiple controls of the Escherichia coli Pho regulon by the Pi sensor PhoR, the catabolite regulatory sensor CreC, and acetyl phosphate. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms: cellular and molecular biology. Washington, D.C: ASM Press; 1994. pp. 13–21. [Google Scholar]
- 57.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1357–1381. [Google Scholar]
- 58.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]