Abstract
Lon protease of Escherichia coli regulates a diverse set of physiological responses including cell division, capsule production, plasmid stability, and phage replication. Little is known about the mechanism of substrate recognition by Lon. To examine the interaction of Lon with two of its substrates, RcsA and SulA, we generated point mutations in lon which affected its substrate specificity. The most informative lon mutant overproduced capsular polysaccharide (RcsA stabilized) yet was resistant to DNA-damaging agents (SulA degraded). Immunoblots revealed that RcsA protein persisted in this mutant whereas SulA protein was rapidly degraded. The mutant contains a single-base change within lon leading to a single amino acid change of glutamate 240 to lysine. E240 is conserved among all Lon isolates and resides in a charged domain that has a high probability of adopting a coiled-coil conformation. This conformation, implicated in mediating protein-protein interactions, appears to confer substrate discriminator activity on Lon. We propose a model suggesting that this coiled-coil domain represents the discriminator site of Lon.
Energy-dependent Lon (La) protease, first isolated from Escherichia coli (7, 9), has been identified in every organism examined thus far, including gram-positive and gram-negative bacteria, yeast, plants, and humans. Lon consists of four identical subunits (63; reviewed in references 19 and 20) each having an N-terminal highly charged domain (spanning amino acids [aa] 211 to 271), a centrally located ATP binding domain (aa 351 to 421), and a C-terminal proteolytically active domain (1, 8, 16). Mutations in serine 679 suggest that it is the catalytically active residue (2). Mutations in residues H665, H667, and D676, but not D743, also appear to be essential for Lon’s proteolytic activity, yet it is unclear whether these residues belong to a catalytic triad (55). A K362A change in motif A of the ATP-binding domain was found to affect the catalytic efficiency and peptidase activity of Lon (14). To date no physiological function has been defined for the conserved charged domain located at the N terminus.
The characteristic phenotypes of Δlon mutant cells (mucoidy, sensitivity to DNA-damaging agents, and defectiveness in bacteriophage λ and P1 lysogenization and in the degradation of abnormal proteins) (3, 5, 34, 42, 44; reviewed in references 24 and 26) can be directly attributed to the stabilization of Lon substrates with regulatory roles in these pathways. The mucoidy phenotype (overproduction of colanic acid capsular polysaccharide) of Δlon cells is mediated through the stabilization of RcsA (58; reviewed in references 23 and 26). RcsA is a transcriptional activator of the capsular polysaccharide genes (cps): in lon+ cells RcsA is highly unstable and cps transcription is barely detectable, whereas in Δlon cells RcsA stability increases, resulting in a high level of cps transcription (28, 56, 58; reviewed in references 23 and 26). Increasing the amount of RcsA by stabilization of RcsA through the removal of Lon, overexpression of rcsA, or better protection of RcsA by its partner, RcsB, results in expression of the cps genes (27, 56; reviewed in references 23 and 26).
Treatment of cells with DNA-damaging agents (e.g., UV light or methyl methanesulfonate [MMS]) activates expression of SOS genes (reviewed in reference 61) including that encoding SulA, a cell division inhibitor (17, 35–37). UV or MMS treatment of lon+ cells gives rise to short filaments which are eventually resolved into individual cells when normal cell division resumes (25, 35). lon+ cells are resistant to UV or MMS, and SulA is highly unstable in these cells (46; reviewed in reference 26). UV or MMS treatment of Δlon cells gives rise to long, nonseptated filaments. Δlon cells are sensitive to UV or MMS, and SulA is stabilized in these cells (46; reviewed in reference 26). Δlon cells fail to recover after removal of the DNA-damaging agent because the stabilization of SulA renders filamentation irreversible and cell death inevitable. Analogous to the situation for RcsA, increasing the amount of SulA by stabilization of SulA through the removal of Lon, overexpression of sulA, or better protection of SulA by its partner, SulB, results in the formation of lethal filaments (38, 41).
Lon’s function and its substrates vary from organism to organism, and for some Lon enzymes neither function nor substrates have been identified. Very little is known as to how Lon discriminates its substrates from among the hundreds of other nonsubstrate proteins in a cell. In this study we provide genetic and biochemical evidence defining a potential discriminator activity for the conserved charged domain located at the N terminus of Lon.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bacterial strains are described in Table 1. All strains are derivatives of MC4100 unless otherwise indicated. Liquid medium used in this study was LB broth except where noted, and solid media used were LB agar, TB agar, and MacConkey’s lactose agar (Difco). Antibiotics were used at 25 (tetracycline) and 100 (ampicillin) μg/ml. P1vir transductions were performed as described by Silhavy et al. (51).
TABLE 1.
Bacterial strains and plasmid used in this study
| Straina or plasmid | Relevant genotype | Reference(s), source, or construction |
|---|---|---|
| E. coli strains | ||
| CAG12017 | lon+ zba-3054::Tn10 | 52 |
| DDS90 | lon+ rcsA+ rcsA90::lacZ | D. Sledjeski; 47, 54 |
| JM12 | lon E240K (class III) zba-3054::Tn10 | This study |
| JM64 | lon G384D (class II) zba-3054::Tn10 | This study |
| JM98 | lon G374R D483N (class I) zba-3054::Tn10 | This study |
| JT1900 | lon E240K zba-3054::Tn10 cpsB10::lacZ | SG20781 + P1(JM12) |
| JT1916 | lon G384D zba-3054::Tn10 cpsB10::lacZ | SG20781 + P1(JM64) |
| JT1920 | lon G374R D483N zba-3054::Tn10 cpsB10::lacZ | SG20781 + P1(JM98) |
| JT2029 | proCYA221 zaj-403::ΔTn10 rcsA+ rcsA90::lacZ | DDS90 + P1(SG1030) |
| JT2036 | lon E240K zba-3054::Tn10 Δgal-165 | SG21020 + P1(JM12) |
| JT2037 | lon G384D zba-3054::Tn10 Δgal-165 | SG21020 + P1(JM64) |
| JT2038 | lon G374R D483N zba-3054::Tn10 Δgal-165 | SG21020 + P1(JM98) |
| JT2046 | Δlon-510 rcsA+ rcsA90::lacZ | JT2029 + P1(SG4144) |
| JT4000 | Δlon-510 | SG1030 + P1(SG4144) |
| MS100 | lon E240K zba-3054::Tn10 rcsA+ rcsA90::lacZ | DDS90 + P1(JM12) |
| MS101 | lon G384D zba-3054::Tn10 rcsA+ rcsA90::lacZ | DDS90 + P1(JM64) |
| MS102 | lon G374D D483N zba-3054::Tn10 rcsA+ rcsA90::lacZ | DDS90 + P1(JM98) |
| SG1030 | F− Δlac araD proCYA221 zaj-403::ΔTn10 | 60 |
| SG4144 | Δlon-510 | 44 |
| SG20250 | lon+ Δlac | 29 |
| SG20780 | Δlon-510 cpsB10::lacZ | 4 |
| SG20781 | lon+ cpsB10::lacZ | 4 |
| SG21020 | lon+ Δgal-165 | S. Gottesman |
| SG21155 | Δlon Δgal-165 | S. Gottesman |
| Plasmid | ||
| pATC400 | pBR322-rcsA+ | 58 |
All strains except CAG12017 and SG4144 are derived from MC4100 (ΔlacU169 araD flbB rel).
Phenotypic assays.
Mucoidy was assayed on TB agar containing ampicillin, MMS sensitivity on LB agar containing 0.05% MMS, and mitomycin C sensitivity on LB agar containing 0.3 μg of mitomycin C per ml. The UV phenotype was assessed as follows. Overnight cultures were diluted in LB containing ampicillin, grown to an optical density at 600 nm (OD600) of 0.3, pelleted, resuspended in 0.1 volume of 0.01 M MgSO4, and irradiated with UV light (λ = 254) at a dose of 5 mJ/cm2. Cells were diluted, plated on LB agar containing ampicillin, and incubated in the dark.
Mutagenesis.
E. coli CAG12017, which carries a Tn10Tetr 98% linked to the lon+ gene, was mutagenized with nitrosoguanidine (51). P1vir was grown on the mutagenized culture and used to infect a wild-type, nonmucoid lon+ strain (SG20250). Tetr transductants were selected and then screened for acquisition of a mucoid phenotype. These potential lon mutants were moved by P1 into a lon+ cpsB10::lacZ indicator strain (SG20781). Tetr transductants were again selected and screened for lactose utilization as an indication of cps expression. Lac+ transductants, indicating expression of cpsB10::lacZ, were saved.
β-Galactosidase assay.
Three independent candidates of all strains examined were assayed. β-Galactosidase activities were determined by the method of Miller (45). Values reported represent the mean of three assays.
Assessment of filament formation.
Cells were grown to OD600 of 0.2. Nonirradiated cell samples were removed at 0-, 2-, 4-, and 8-h time points, sonicated, pelleted, resuspended in phosphate-buffered saline containing 1% formaldehyde, incubated for 30 min on ice, and then evaluated in light microscopy. Remaining cells in the original 0-h culture were pelleted, resuspended in 0.1 volume of 10 mM MgSO4, and irradiated with UV light (λ = 254) at a dose of 5 mJ/cm2. Irradiated cell samples were removed at 0, 2, 4, and 8 h, fixed in formaldehyde, and evaluated by light microscopy.
RcsA and SulA protein detection.
To examine RcsA, cells were grown to an OD600 of 0.6 and processed as described below. To examine SulA, cells were grown to an OD600 of 0.3, pelleted, resuspended in 0.1 volume of 10 mM MgSO4, and irradiated with UV light (λ = 254) at a dose of 5 mJ/cm2. Cells were diluted and incubated 30 min in foil-lined flasks. Samples were removed, washed twice in 10 mM MgSO4, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (49), and boiled. Protein concentrations were determined by the bicinchoninic acid protein assay method (Pierce). Equal amounts of total cellular protein, resuspended and boiled in SDS-PAGE sample buffer, were fractionated by tricine-SDS-PAGE (50) (16.5% gel for SulA; 14% gel for RcsA). Fractionated samples for SulA analysis were transferred to a polyvinylidene difluoride (PVDF) membrane (Dupont) in 20 mM Tris (pH 8.3)–20% methanol–150 mM glycine. Fractionated samples for RcsA analysis were transferred to a PVDF membrane in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer (pH 11) with 20% methanol (12, 57). After transfer, all PVDF membranes were blocked in Tris-buffered saline (20 mM Tris [pH 7.4], 125 mM NaCl) containing 0.1% Tween 20, incubated with preabsorded antiserum specific to E. coli RcsA or SulA protein, washed in Tris-buffered saline containing 0.1% Tween 20, and incubated with an appropriate dilution of goat anti-rabbit immunoglobulin conjugated to horseradish peroxidase. Immunoreactive protein bands were visualized on autoradiography film (Hyperfilm; Amersham) by using enhanced chemiluminescence (Amersham).
λOts stability assessment.
Stability of the bacteriophage λO temperature-sensitive mutant protein (λOts) was measured as previously reported (11, 30). Briefly, E. coli cells were grown in LB medium, wild-type λ and λOts phages were diluted, and the dilutions were spotted on TB agar plates containing the E. coli cells to be examined in an overlay. Duplicate plates were prepared; one set was incubated at the permissive temperature (32°C), and the other set was incubated at the nonpermissive temperature (39°C). At the nonpermissive temperature in the E. coli lon+ strain, λO is unstable and thus no plaques form. At either the nonpermissive or permissive temperature in the E. coli Δlon strain, λOts is stable and thus plaques form.
DNA sequencing and analysis.
PCR was used to amplify two overlapping fragments of each lon mutant, encompassing the complete open reading frame (ORF) and regulatory region. DNA from this region was amplified directly from the chromosome as previously described (13), the DNA was purified on Qiagen columns, and both strands were sequenced with appropriate primers on an ABI 377 automated sequencer (Center for Gene Research and Biotechnology, Central Services Laboratory, Oregon State University). All oligonucleotide primers used for PCR and DNA sequencing were provided by the Biopolymer Unit of the University of Maryland Medical School, Baltimore County. PCR oligonucleotide primers used were 5′ AGCCTGCCAGCCCTGTTT 3′, 5′ AGTATCTTGCGGTTCAAA 3′, 5′ GGCGTGAAGCACCGTCGTGT 3′, and 5′ GCATAGAACCGATGTAAGTA 3′. Mutations were identified by comparing the newly generated lon mutant sequences to the wild-type lon sequence (GenBank accession no. 146642, 146644, 1773123, and 1786643), using the Genetics Computer Group program Bestfit.
RESULTS
Lon-substrate interactions.
In an attempt to define the substrate specificity of Lon, Dervyn et al. in the laboratory of O. Huisman overexpressed sulA in lon+ cells and observed that the cells became mucoid, suggesting that high levels of SulA protected RcsA from Lon-dependent degradation (11). Mutations in SulA which abolished this ability to saturate Lon, evident by the loss of the mucoid phenotype, could be identified; however, the mutant SulA protein was still degraded in a Lon-dependent fashion. These observations led them to hypothesize that Lon protease contains different substrate recognition sites: a high-affinity site for specific substrates such as SulA and RcsA, and a low-affinity site for nonspecific substrates such as abnormal or mutant proteins (11, 22, 26). Presumably, both RcsA and SulA are recognized by the high-affinity site, while the mutant SulA is no longer recognized by this high-affinity site but rather interacts with the low-affinity site. We extended these studies by overexpressing RcsA in lon+ cells and then examining their response to UV light, MMS, and mitomycin C (Table 2). We predicted that if RcsA and SulA had equivalent affinities for Lon, then by overexpressing RcsA to such a level that lon+ cells were mucoid, SulA should be protected from Lon-dependent degradation. This protection would be observed phenotypically as sensitivity to UV, MMS, and mitomycin C. We observed that under conditions of high levels of RcsA, lon+ cells were very mucoid yet still resistant to UV, MMS, and mitomycin C, suggesting that SulA was not protected from Lon-dependent degradation (Table 2, line 2). Failure to express high enough levels of RcsA to protect SulA was ruled out because enough intact and functional RcsA was available to activate cps expression (mucoid phenotype), the level of cps activation (as monitored by a cpsB10::lacZ fusion) was higher than that observed in Δlon cells (data not shown), and RcsA was readily detected by immunoblot analysis (data not shown). Several possibilities exist to explain these results, including that specific substrates, although presumably acting at a high-affinity site, may have different affinities for Lon, thus creating a hierarchical order with respect to substrate selection.
TABLE 2.
High-level expression of RcsA in lon+ cells does not protect SulA
| Relevant genotype (straina) | Phenotype
|
|||
|---|---|---|---|---|
| Mucoidyb | MMSc | UVc | Mitomycin Cc | |
| lon+ (SG20250/pBR322) | − | R | R | R |
| lon+ (SG20250/pATC400) | + | R | R | R |
| Δlon (JT4000/pBR322) | + | S | S | S |
| Δlon (JT4000/pATC400) | + | S | S | S |
All strains are MC4100 derivatives.
Assessed visually. −, nonmucoid; +, mucoid.
Determined as described in Materials and Methods. R, resistant; S, sensitive.
If RcsA and SulA interact at a high-affinity site but their interaction is dictated by a preference order, we predicted that lon mutations which defined the difference in this interaction for these two substrate classes could be isolated. To test this prediction, a strain carrying a Tn10 98% linked to lon+ was mutagenized with nitrosoguanidine. P1vir was grown on the pool mutant cells, and this lysate was used to infect a lon+ strain (SG20250). Tetr transductants were selected and subsequently screened for the Lon− phenotype, mucoidy, indicating accumulating RcsA. Of the 100,000 Tetr transductants screened in this approach, 0.05% were mucoid. The increase in RcsA levels was verified by transducing the lon mutations in to a strain containing cpsB10::lacZ (SG20781). This fusion reports the expression of the cps genes as a function of the available RcsA protein (4). High-level expression of cpsB10::lacZ fusion correlates with high levels of RcsA protein (26, 56). All lon mutants examined showed an increase in cps expression compared to lon+, indicating an increased availability of intact and functional RcsA. The mucoid mutants were screened for sensitivity to the DNA-damaging agents UV light, MMS, and mitomycin C and assayed for filament formation. As shown in Table 3, three distinct classes of lon mutants were identified. Class I mutants were mucoid, had high-level cpsB10::lacZ expression, were completely sensitive to DNA-damaging agents, and had extremely long filaments 8 h after UV exposure. Class II mutants were mucoid, had low-level cpsB10::lacZ expression, were partially sensitive to DNA-damaging agents, and had medium-length filaments 8 h after UV exposure. Class III mutants were mucoid, had medium- to high-level cpsB10::lacZ expression, were completely resistant to DNA-damaging agents, and exhibited no filaments.
TABLE 3.
Phenotypic descriptions of the effects of three classes of lon mutation on the stability of RcsA and SulA protein
| Relevant genotype (strainsa) | RcsA
|
SulA
|
||
|---|---|---|---|---|
| Mucoidyb | cpsB10::lacZ (β-galactosidase activity)c | MMS phenotyped | Filament formation 8 h after UV exposuree | |
| lon+ (SG20250 or SG20781) | − | 5 | R | No filaments (single cells) |
| Δlon (JT4000 or SG20780) | + | 418 | S | Extremely long filaments |
| Class I lon (JM98 or JT1920) | + | 575 | S | Extremely long filaments |
| Class II lon (JM64 or JT1916) | +/− | 117 | S/R | Medium-length filaments |
| Class III lon (JM12 or JT1900) | + | 230 | R | No filaments (single cells) |
All strains are MC4100 derivatives. Strains assayed for β-galactosidase activity contain cpsB10::lacZ.
Assessed visually. −, nonmucoid; +, mucoid.
β-Galactosidase assays were carried out as described by Miller (45). Values represent the average of three independent assays.
Determined on LB agar containing 0.05% MMS. R, resistant; S, sensitive; S/R, intermediate phenotype.
Assayed as described in Materials and Methods.
Previously we reported that rcsA expression was 100-fold higher in Δlon mutant cells than in lon+ cells (13). In these studies, expression of rcsA was measured by using a rcsA::lacZ fusion at the λatt site (54), creating partial diploid strains (rcsA90::lacZ at the λatt site and rcsA at its normal position in the chromosome). The 100-fold increase in rcsA expression in a Δlon strain was due to both an accumulation of functional RcsA protein in the absence of Lon protease and rcsA expression activated by RcsA protein (13). We predicted that if RcsA activated its own expression, then a difference in the level of rcsA activation would be observed between the three lon mutant classes. As shown in Table 4, expression of the rcsA90::lacZ fusion was highest in the class I and class III lon mutants and was approximately 380- to 420-fold higher than in lon+ cells. Expression of the rcsA90::lacZ fusion in the class II lon mutant was somewhat less than in Δlon cells and in the class I and class III lon mutants, yet rcsA expression in the class II lon mutant was still 100-fold higher than in the lon+ strain. These results provide further support that functional and intact RcsA is available to activate rcsA expression in all three classes of lon mutants, with functional RcsA levels highest in the class I and III mutants.
TABLE 4.
Effects of class I, II, and III lon mutations on rcsA expression as measured by rcsA90::lacZ activity
| Relevant genotype (straina) | β-Galactosidase activityb | Fold change from:
|
|
|---|---|---|---|
| lon+ | Δlon | ||
| lon+ (DDS90) | 12 | 0.0045 | |
| Δlon (JT2046) | 2,641 | 220 | |
| Class I lon (MS102) | 5,137 | 428 | 2.0 |
| Class II lon (MS101) | 1,220 | 100 | 0.46 |
| Class III lon (MS100) | 4,658 | 388 | 1.8 |
All strains are MC4100 derivatives carrying a rcsA90::lacZ fusion integrated at the λatt site.
β-Galactosidase assays were carried out as described by Miller (45). Values represent the average of three independent assays.
Results described thus far indicate that phenotypically, class I lon mutants behave most like the previously reported Δlon mutant (44), and class II lon mutants behave phenotypically as if protease activity toward RcsA and SulA has been impaired. The most interesting class, the class III lon mutants, behave phenotypically as if they are Δlon with respect to RcsA (mucoid) yet lon+ with respect to SulA (resistant to DNA-damaging agents). To determine if the phenotypes were a result of decreased levels of Lon protein or truncated Lon, immunoblot analyses of whole-cell extracts of the lon mutants were performed. Intact Lon protein was expressed at normal levels in the three classes of lon mutants (data not shown), indicating that the observed phenotypes were not due to an absent or truncated Lon. A representative from each mutant class was chosen for further analysis.
RcsA and SulA protein levels correspond to the lon mutant phenotypes.
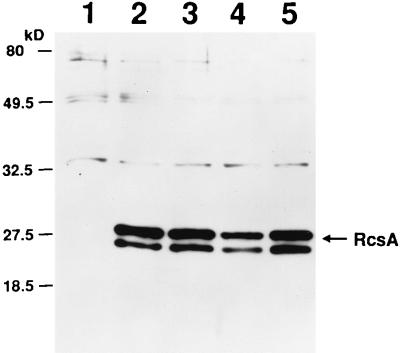
Changes in the level of cps and rcsA expression and in the response to DNA-damaging agents is consistent with changes in RcsA and SulA protein levels. To determine if the protein levels vary as predicted, levels of RcsA (Fig. 1) and SulA (Fig. 2) were evaluated by immunoblotting. RcsA protein was detected in representative extracts from each lon mutant class (Fig. 1, lanes 3 to 5). The level of RcsA in the class I and class III lon mutants (lanes 3 and 5) was comparable to the level detected in the Δlon mutant (lane 2), whereas the level of RcsA in the class II lon mutant was reduced (lane 4). RcsA half-life was measured in the three classes of lon mutants in vivo (data not shown); RcsA half-life in the class I and class III lon mutants was similar to that reported for Δlon cells (greater than 30 min [56]), whereas RcsA half-life in the class II lon mutant appeared to be less than 10 min.
FIG. 1.
Immunodetection of RcsA in lon mutants. Equal amounts of protein from whole-cell extracts were boiled in sample buffer, fractionated on a 14% tricine-SDS-polyacrylamide gel, and analyzed by immunoblotting with preabsorbed polyclonal antiserum specific to RcsA protein. Lanes: 1, SG20781 (lon+); 2, SG20780 (Δlon); 3, JT1900 (class III lon); 4, JT1916 (class II lon); 5, JT1920 (class I lon).
FIG. 2.
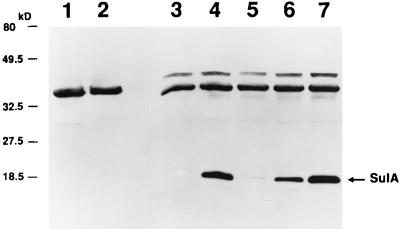
Immunodetection of SulA in lon mutants. Cultures were treated with UV light as described in Materials and Methods. Samples were treated as described for Fig. 1 except that antiserum specific to SulA was used. Lanes: 1, SG20780 (Δlon), no UV; 2, SG20781 (lon+), no UV; 3, SG20781 (lon+), with UV; 4, SG20780 (Δlon), with UV; 5, JT1900 (class III lon), with UV; 6, JT1916 (class II lon), with UV; 7, JT1920 (class I lon), with UV.
In contrast, amounts of detectable SulA varied among the three classes of mutants (Fig. 2). SulA protein is readily detected in the class I lon mutant (Fig. 2, lane 7) at levels comparable to that observed in Δlon cells (lane 4). SulA protein was not detected in the class III lon mutant (lane 5) and was detected at a reduced level in the class II lon mutant (lane 6). Analysis of filament formation (Table 3) corresponds with SulA levels observed by immunoblotting. SulA half-life was measured in the three classes of mutants in vivo (data not shown); SulA half-life in the class I lon mutant was similar to that reported for Δlon cells (20 to 30 min [44, 46, 59]), whereas SulA half-life in the class II lon mutant appeared to be only 10 to 15 min. SulA half-life in the class III lon mutant was similar to that reported for lon+ cells (less than 3 min) (46, 59).
λOts, an abnormal protein, is not affected by the lon mutations.
RcsA and SulA proteins are normal physiological proteins degraded in a Lon-dependent fashion in E. coli and thus are considered specific Lon substrates. Abnormal proteins such as nonsense peptides, protein fragments, missense proteins, and damaged proteins also are substrates for Lon (5, 18, 30, 31, 44). Thus, it seemed reasonable to test the activity of the lon mutants against an abnormal protein. Wild-type λO protein, involved in the replication of phage λ, is not a substrate for Lon, and a λ phage expressing wild-type O protein forms plaques on lon+ (SG21020) and Δlon (SG21155) cells with equal efficiency (1010 plaques/ml). A temperature-sensitive mutation in λOts causes the protein to be abnormal at high temperatures and makes it a substrate for Lon. The degradation of λOts in a Lon-dependent fashion prevents λ phage replication, and thus no plaques form on lon+ (SG21020) cells. In Δlon (SG21155) cells, however, the abnormal λOts protein is stabilized and thus can function, allowing λ phage to replicate and form plaques (efficiency of 1010 plaques/ml). Plaquing efficiencies on the lon mutants JT2036 (class III), JT2037 (class II), and JT2038 (class I) were similar to that seen in the lon+ strain, indicating that λOts protein was still a substrate for these mutant Lons.
Sequence analysis reveals changes at highly conserved residues.
Sequencing the full lon ORF and its regulatory region from the three classes of lon mutants revealed unique changes in each class. In the class I lon mutant, there were two nucleotide changes: a G-to-A transition resulting in a glycine-to-arginine change at position 374 (G374R), and a G-to-A transition resulting in an aspartate-to-asparagine change at position 483 (D483N). In the class II lon mutant there was a single-nucleotide G-to-A transition, resulting in a glycine-to-aspartate change at position 384 (G384D). In the class III lon mutant there was a single-nucleotide G-to-A transition, resulting in a glutamate-to-lysine change at position 240 (E240K).
Amino acid substitutions G374 and G384 are located in the region between the A and B motifs comprising the ATP binding domain (Fig. 3). Both amino acid substitutions represent a change of a small aliphatic residue to a charged residue. Comparison of Lon sequences from other organisms revealed that glycine 374 is conserved in Lon sequences from gram-positive and gram-negative bacteria and Arabidopsis. In Saccharomyces cerevisiae and Homo sapiens, an asparagine residue is found at this position. A G384D substitution (class II) resulted in reduced catalytic behavior toward both RcsA and SulA. The G384 residue is conserved in all known Lon sequences; it is positioned 15 residues downstream from the last residue comprising motif A of the ATP binding domain. The sequence surrounding the G384 residue (boldface) of Lon is highly conserved among bacterial species: X(S/A)GGVRDE (where X = M, I, or L). Lon sequences from Arabidopsis, S. cerevisiae, and H. sapiens also have similar residues in this region, with residues G383, G384, and D387 being identical.
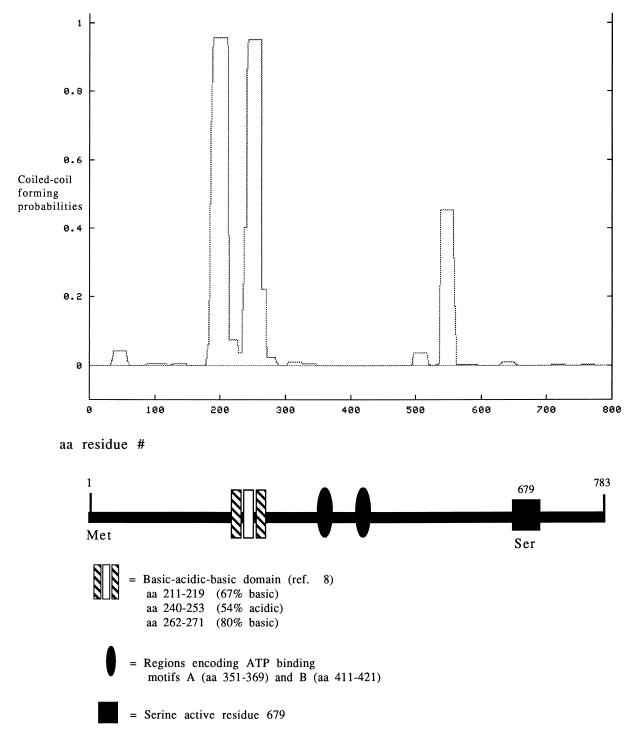
FIG. 3.
Comparison of the predicted coiled-coil structure (generated with COILS version 2.2 [39, 40, 48]) to the predicted domain structure of E. coli Lon protease.
The class I lon mutant, which has no proteolytic activity toward RcsA and SulA, has an additional amino acid substitution, D483N, which represents a change of a charged residue to a noncharged residue. The D483 residue is conserved in all Lon sequences. As in the case of the G384 residue, the residues surrounding the D483 residue (boldface) are highly conserved in bacterial species, with a P(A/G)PLXDRME(V/I) consensus (where X = L, P, Q, or R) evident. Arabidopsis, S. cerevisiae, and H. sapiens Lon sequences also contain similar residues in this region, with residues P478, P480, L481, D483, R484, M485, and E486 being identical.
The Lon sequence of the class III mutant revealed an amino acid substitution (E240K) in the proposed acidic region of the conserved charged domain located at the N terminus which substitutes an acidic residue with a basic residue. Glutamate 240 is conserved among all Lon isolates. The residues surrounding the E240 residue (boldface) are highly conserved in bacterial species, with a (K/Q)AIQKELG(D/E) consensus evident. Arabidopsis, S. cerevisiae, and H. sapiens Lon sequences contained similar residues in this region, with residues I237, E240, L241, and G242 being identical.
Coiled-coil analysis.
Coiled-coil regions are frequently solvent-exposed regions believed to be involved in protein-protein interactions (39, 40, 48). The COILS program (39, 40, 48), which is specific for solvent-exposed, left-handed coiled coils, was used to analyze the primary amino acid sequence of Lon and to subsequently make a prediction as to the likelihood of a particular domain adopting a coiled-coil conformation. Analysis of the E. coli Lon sequence identified a region spanning residues 185 to 228 and 237 to 280 at the N terminus (Fig. 3) as a region with a high probability (ca. 95%) of adopting coiled-coil structures. In this region, approximately 50% of the residues are charged, with an equal distribution of acidic and basic residues. The residues comprising the A and B motifs of the ATP binding domain and the residues surrounding the proposed catalytic active residue (S679) had little probability of forming coiled coils (Fig. 3). Interestingly, the region spanning residues 538 to 558 (approximately 120 residues upstream of the active site S679 residue) is predicted to adopt a coiled-coil structure but at a much lower probability than the N-terminal region. Approximately 48% of residues 538 through 558 are charged, with two-thirds of the charged residues basic. Because the prediction of coiled-coil regions is biased toward hydrophilic, highly charged sequences, the analysis was performed with a weighted and an unweighted matrix, which did not reveal any differences between the two types of analysis. A similar analysis was performed on all known Lon sequences, revealing very similar coiled-coil profiles for all Lon isolates.
DISCUSSION
Goldberg and Waxman (21, 62–64) proposed a model for Lon proteolysis which accounted for the energy requirements and the processive nature of this enzyme (reviewed in references 19, 26, and 43). In this model, ATP hydrolysis manipulates the conformation of Lon, controlling the accessibility of Lon’s active site. In an expansion of this model (26), a substrate capture function that has two binding sites, one for nonspecific substrates (initiator site) and one for specific substrates (discriminator site), was proposed. If a putative substrate had a low affinity, it would bind the initiator site and be partially degraded; if it had no affinity, it would be released from the initiator site without being cleaved. On the other hand, the discriminator site would bind specific substrates (those with very high affinity) and hold them long enough to activate ATP hydrolysis, resulting in a conformational change and accessibility of Lon’s active site. The substrate would be retained under these conditions and subsequently cleaved.
If Lon protease has two binding sites, a discriminator site for specific substrates, such as RcsA and SulA, and an initiator site for nonspecific substrates, such as abnormal proteins, then where do these sites reside? Dervyn et al. in the laboratory of O. Huisman demonstrated that saturating Lon with SulA protects RcsA from degradation (11), suggesting that these two substrates compete for the same binding site. In contrast, we observed that overexpressing RcsA did not appear to protect SulA from degradation, possibly indicating a hierarchy among these substrates. We propose that if RcsA and SulA interact with different affinities at the same site, then this site would be identifiable by mutations which discriminated between these two substrates. In this study, we provide evidence supporting the hypothesis that discriminator activity can be assigned to a domain located at the N terminus. An E240K substitution in this domain abolished Lon’s activity on RcsA but had no affect on Lon’s activity on SulA. This E240 residue resides in a domain that has a high probability of adopting a coiled-coil conformation conducive to protein-protein interactions, and this conformation and its location are highly conserved among all Lons reported to date. Furthermore, this domain is a strong candidate for protein-protein interactions by virtue of its highly charged, thus potentially sticky, “velcro” behavior. Clearly, these studies provide direct support for the interaction of RcsA with Lon at the velcro domain. Observations made in overexpression studies with both RcsA and SulA provide indirect support for interaction of SulA with this domain. Direct evidence supporting SulA’s interaction with Lon’s velcro domain will come from mutations in this region that impede Lon-SulA interaction. Further studies are under way to identify this class of mutation.
Our study also uncovered other novel mutations, for example, G384D (class II), which resides between motifs A and B of the ATP binding domain and which affects the catalytic behavior of Lon toward both RcsA and SulA but not λOts. In support of this, RcsA and SulA levels of detection and half-life were between the values obtained for lon+ and Δlon cells. Furthermore, rcsA::lacZ expression was not as high in this mutant as in the other lon mutants, indicating that reduced amounts of functional RcsA were available to activate the expression of rcsA. These results suggest that Lon with a G384D substitution can still affect the stability of RcsA and SulA but not as efficiently as wild-type Lon activity. The combination of G374R and D483N substitutions (class I) completely abolishes proteolytic activity toward RcsA and SulA but, like the G384 substitution, does not affect degradation of λOts. Possibly, proteolytic activity toward certain nonspecific abnormal proteins is retained with the G384 or G374R and D483N substitutions, yet Lon-dependent degradation of specific substrates such as RcsA and SulA is abolished due to an inability of Lon to hydrolyze ATP or to assemble into a functional multimeric enzyme, suggesting that catalytic-proteolytic efficiency rather than discriminator activity has been impaired.
If discriminator activity resides in the N-terminal velcro domain of Lon, then where does the initiator site, or the site for nonspecific substrate binding, reside? Functional analysis of the abnormal protein λOts demonstrated that an E240K, a G384D, or a D483N in combination with a G374R substitution in Lon had no effect on λOts stability. These results suggest that Lon’s interaction with λOts protein was at a site distinct from that of specific substrates and that reducing the catalytic behavior toward specific substrates had no apparent affect on λOts stability. While these studies were not designed to identify the binding site for abnormal proteins, they have provided a testable hypothesis for where this domain may reside, namely, the highly conserved residues 538 to 558 predicted to adopt a coiled-coil structure, to which no function has been assigned.
Goldberg and coworkers (8, 10) proposed that the basic regions (aa 211 to 219 and aa 262 to 271) of the velcro domain might be involved in Lon’s ability to bind DNA (6, 65) whereas the acidic region (aa 240 to 253) of this domain might be involved in its activation by polybasic peptides (8). Sequence specific binding of E. coli Lon protease to the peri-Ets site of the human immunodeficiency virus type 2 enhancer was recently reported (15). This information has led several investigators to hypothesize that DNA binding of Lon protease might be involved in its substrate recognition. The velcro domain of Lon does not display characteristics common to DNA binding proteins. Whether DNA binding is specific or nonspecific and whether DNA binding is involved in substrate recognition remain to be determined.
Interestingly, the E240K substitution does not significantly alter the predicted coiled-coil conformation of the velcro domain. However, this substitution was sufficient to abolish Lon’s ability to recognize and degrade RcsA, without affecting the Lon-SulA interaction. Numerous possibilities exist to account for this observation. The velcro domain, a region which spans approximately 100 residues, may have multiple binding sites for different specific substrates, and only the site which interacts with RcsA was affected by the E240K substitution. Alternatively, both substrates may bind at the same site, and thus a more drastic substitution at position 240 is needed to impede SulA binding. In support of this, overexpression studies suggest that RcsA and SulA bind at the same site, yet there may be a hierarchy defining these interactions. Furthermore, coiled-coil analysis of RcsA and SulA revealed a striking difference between these two substrates. SulA has a high probability (60%) of adopting a coiled-coil at the C terminus (residues 124 to 137), and residues in this region are predominately acidic (85%). In contrast, RcsA has a low (10%) probability of adopting a coiled-coil structure (C-terminally located, residues 172 to 188), and the residues in this region are predominately basic. Are the residues comprising the predicted coiled-coils of SulA and RcsA involved in Lon-substrate interactions? If protease-substrate discriminator activity can be assigned to residues involved in coiled-coil conformations, then it remains to be determined if substrate affinity is a reflection of the overall charge of the coiled-coil region of these proteins.
We propose that the simplest way Lon recognizes and prioritizes its interactions with specific physiological substrates would be through the discriminator activity of the velcro domain. The charge interactions occurring here would define the substrate’s affinity for Lon and thus dictate a hierarchy for substrates as they were being positioned for cleavage at the proteolytic active site located at the C terminus. Proteolysis of nonspecific substrates, such as abnormal proteins, would not require the discriminator activity of the velcro domain. Correspondingly, specific inhibitors, such as T4 PinA (32, 33, 53), may interact at the velcro domain, thus preventing the capture of specific substrates. Many more intriguing questions as to how Lon selects its substrates from among other nonsubstrate proteins in the cell remain unanswered. Answering these questions may provide a means by which to identify new biological pathways controlled by Lon-dependent degradation in organisms other than E. coli.
ACKNOWLEDGMENTS
W.E., M.M.S., K.P.D., and J.M.S. contributed equally to this work.
W.E. and K.P.D. were partially supported by a predoctoral fellowship from the N. L. Tartar Foundation, and K.P.D. was supported by a predoctoral fellowship from the Eckleman Foundation. We are grateful to N. Ambulos of the Biopolymer Unit of University of Maryland Medical School, Baltimore County, for the oligonucleotides used in this study. This work was supported by grants from the Medical Research Foundation of Oregon, the Dr. Harry B. and Ralph H. Levey Philanthropic Fund, and the National Science Foundation to J.E.T.
REFERENCES
- 1.Amerik A, Antonov V K, Ostroumova N I, Rotanova T V, Chistiakova L G. Cloning, structure and expression of the full-size lon gene in Escherichia coli coding for ATP-dependent La-proteinase. Bioorg Khim. 1990;16:869–880. [PubMed] [Google Scholar]
- 2.Amerik A Y, Antonov V K, Gorbalenya A E, Kotova S A, Rotanova T V, Shimbarevich E V. Site-directed mutagenesis of La protease. A catalytically active serine residue. FEBS Lett. 1991;287:211–214. doi: 10.1016/0014-5793(91)80053-6. [DOI] [PubMed] [Google Scholar]
- 3.Apte B N, Rhodes H, Zipser D. Mutation blocking the specific degradation of reinitiation polypeptides in E. coli. Nature. 1975;257:329–331. doi: 10.1038/257329a0. [DOI] [PubMed] [Google Scholar]
- 4.Brill J A, Quinlan-Walshe C, Gottesman S. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1988;170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukhari A I, Zipser D. Mutants of Escherichia coli with a defect in the degradation of nonsense fragments. Nature. 1973;243:238–241. doi: 10.1038/newbio243238a0. [DOI] [PubMed] [Google Scholar]
- 6.Charette M F, Henderson G W, Doane L L, Markovitz A. DNA-stimulated ATPase activity on the lon (CapR) protein. J Bacteriol. 1984;158:195–201. doi: 10.1128/jb.158.1.195-201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charette M F, Henderson G W, Markovitz A. ATP hydrolysis-dependent protease activity of the lon (capR) protein of Escherichia coli K-12. Proc Natl Acad Sci USA. 1981;78:4728–4732. doi: 10.1073/pnas.78.8.4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin D T, Goff S A, Webster T, Smith T, Goldberg A L. Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La. J Biol Chem. 1988;263:11718–11728. [PubMed] [Google Scholar]
- 9.Chung C H, Goldberg A L. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci USA. 1981;78:4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung C H, Goldberg A L. DNA stimulates ATP-dependent proteolysis and protein-dependent ATPase activity of protease La from Escherichia coli. Proc Natl Acad Sci USA. 1982;79:795–799. doi: 10.1073/pnas.79.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dervyn E, Canceill D, Huisman O. Saturation and specificity of the Lon protease of Escherichia coli. J Bacteriol. 1990;172:7098–7103. doi: 10.1128/jb.172.12.7098-7103.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dierksen K P, Trempy J E. Identification of a second RcsA protein, a positive regulator of colanic acid capsular polysaccharide genes, in Escherichia coli. J Bacteriol. 1996;178:5053–5056. doi: 10.1128/jb.178.16.5053-5056.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebel W, Trempy J E. Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions to activate its own expression. J Bacteriol. 1999;181:577–584. doi: 10.1128/jb.181.2.577-584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer H, Glockshuber R. A point mutation within the ATP-binding site inactivates both catalytic functions of the ATP-dependent protease La (Lon) from Escherichia coli. FEBS Lett. 1994;356:101–103. doi: 10.1016/0014-5793(94)01244-x. [DOI] [PubMed] [Google Scholar]
- 15.Fu G K, Smith M J, Markovitz D M. Bacterial protease Lon is a site-specific DNA-binding protein. J Biol Chem. 1997;272:534–538. [PubMed] [Google Scholar]
- 16.Gayda R C, Stephens P E, Hewick R, Schoemaker J M, Dreyer W J, Markovitz A. Regulatory region of the heat shock-inducible capR (lon) gene: DNA and protein sequences. J Bacteriol. 1985;162:271–275. doi: 10.1128/jb.162.1.271-275.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George J, Castellazzi M, Buttin G. Prophage induction and cell division in E. coli. III. Mutations in sfiA and sfiB restore division in tif and lon strains and permit the mutator properties of tif. Mol Gen Genet. 1975;140:309–332. [PubMed] [Google Scholar]
- 18.Goff S A, Goldberg A L. An increased content of protease La, the lon gene product, increases protein degradation and blocks growth in Escherichia coli. J Biol Chem. 1987;262:4508–4515. [PubMed] [Google Scholar]
- 19.Goldberg A L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992;203:9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg A L, Sreedhara Swamy K H, Chung C H, Larimore F S. Proteases of Escherichia coli. Methods Enzymol. 1983;80:680–702. doi: 10.1016/s0076-6879(81)80052-3. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg A L, Waxman L. The role of ATP hydrolysis in the breakdown of proteins and peptides by protease La from Escherichia coli. J Biol Chem. 1985;260:12029–12034. [PubMed] [Google Scholar]
- 22.Gottesman S. Genetics of proteolysis in Escherichia coli. Annu Rev Genet. 1989;23:163–198. doi: 10.1146/annurev.ge.23.120189.001115. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman S. Regulation of capsule synthesis: modification of the two-component paradigm by an accessory unstable regulator. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 253–262. [Google Scholar]
- 24.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 25.Gottesman S, Halpern E, Trisler P. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J Bacteriol. 1981;148:265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottesman S, Maurizi M R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesman S, Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol Microbiol. 1991;5:1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 28.Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol. 1985;162:1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol. 1985;162:1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottesman S, Zipser D. Deg phenotype of Escherichia coli lon mutants. J Bacteriol. 1978;133:844–851. doi: 10.1128/jb.133.2.844-851.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossman A D, Burgess R R, Walter W, Gross C A. Mutations in the lon gene of E. coli K-12 phenotypically suppress a mutation in the sigma subunit of RNA polymerase. Cell. 1983;32:151–159. doi: 10.1016/0092-8674(83)90505-6. [DOI] [PubMed] [Google Scholar]
- 32.Hilliard J J, Maurizi M R, Simon L D. Isolation and characterization of the phage T4 PinA protein, an inhibitor of the ATP-dependent Lon protease of Escherichia coli. J Biol Chem. 1998;273:518–523. doi: 10.1074/jbc.273.1.518. [DOI] [PubMed] [Google Scholar]
- 33.Hilliard J J, Simon L D, Van Melderen L, Maurizi M R. PinA inhibits ATP hydrolysis and energy-dependent protein degradation by Lon protease. J Biol Chem. 1998;273:524–527. doi: 10.1074/jbc.273.1.524. [DOI] [PubMed] [Google Scholar]
- 34.Howard-Flanders P, Simson E, Theriot L. A locus that controls filament formation and sensitivity to radiation in Escherichia coli K12. Genetics. 1964;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huisman O, D’Ari R, George J. Further characterization of sfiA and sfiB mutation in Escherichia coli. J Bacteriol. 1980;144:185–191. doi: 10.1128/jb.144.1.185-191.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huisman O, D’Ari R, Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci USA. 1984;81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huisman O, D’Ari R. An inducible DNA-replication-cell division coupling mechanism in E. coli. Nature. 1981;290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- 38.Jones C, Holland I B. Role of the SulB (FtsZ) protein in division inhibition during the SOS response in Escherichia coli: FtsZ stabilizes the inhibitor SulA in maxicells. Proc Natl Acad Sci USA. 1985;82:6045–6049. doi: 10.1073/pnas.82.18.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 40.Lupas A, Dyke M V, Stock J. Predicting coiled-coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 41.Lutkenhaus J, Sanjanwala B, Lowe M. Overproduction of FtsZ suppresses sensitivity of lon mutants to division inhibition. J Bacteriol. 1986;166:756–762. doi: 10.1128/jb.166.3.756-762.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markovitz A. Regulatory mechanisms for the synthesis of capsular polysaccharide in mucoid mutants of Escherichia coli K-12. Proc Natl Acad Sci USA. 1964;51:239–246. doi: 10.1073/pnas.51.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurizi M R. Proteases and protein degradation in Escherichia coli. Experientia. 1992;48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- 44.Maurizi M R, Trisler P, Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J Bacteriol. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 46.Mizusawa S, Gottesman S. Protein degradation in Escherichia coli: the lon gene controls the stability of SulA protein. Proc Natl Acad Sci USA. 1983;80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Painbeni E, Mouray E, Gottesman S, Rouviere-Yaniv J. An imbalance of HU synthesis induces mucoidy in Escherichia coli. J Mol Biol. 1993;234:1021–1037. doi: 10.1006/jmbi.1993.1656. [DOI] [PubMed] [Google Scholar]
- 48.Parry D A D. Coiled-coils in alpha-helix-containing proteins: analysis of the residue types within the heptad repeat and the use of these data in the prediction of coiled-coils in other proteins. Biosci Rep. 1982;2:1017–1024. doi: 10.1007/BF01122170. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 51.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 52.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skorupski K, Tomaschewski J, Ruger W, Simon L D. A bacteriophage T4 gene which functions to inhibit Escherichia coli Lon protease. J Bacteriol. 1988;170:3016–3024. doi: 10.1128/jb.170.7.3016-3024.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starkova N N, Koroleva E P, Runsh L D, Ginodman L M, Rotanova T V. Mutations in the proteolytic domain of Escherichia coli protease Lon impair the ATPase activity of the enzyme. FEBS Lett. 1998;422:218–220. doi: 10.1016/s0014-5793(98)00012-x. [DOI] [PubMed] [Google Scholar]
- 56.Stout V, Torres-Cabassa A, Maurizi M R, Gutnick D, Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991;173:1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szewczyk B, Kozloff L M. A method for the efficient blotting of strongly basic proteins from sodium dodecyl sulfate-polyacrylamide gels to nitrocellulose. Anal Biochem. 1985;150:403–407. doi: 10.1016/0003-2697(85)90528-7. [DOI] [PubMed] [Google Scholar]
- 58.Torres-Cabassa A S, Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trempy J E, Gottesman S. Alp, a suppressor of lon protease mutants in Escherichia coli. J Bacteriol. 1989;171:3348–3353. doi: 10.1128/jb.171.6.3348-3353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trisler P, Gottesman S. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J Bacteriol. 1984;160:184–191. doi: 10.1128/jb.160.1.184-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular Biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1346–1357. [Google Scholar]
- 62.Waxman L, Goldberg A L. Protease La from Escherichia coli hydrolyzes ATP and proteins in a linked fashion. Proc Natl Acad Sci USA. 1982;79:4883–4887. doi: 10.1073/pnas.79.16.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waxman L, Goldberg A L. Protease La, the lon gene product, cleaves specific fluorogenic peptides in an ATP-dependent reaction. J Biol Chem. 1985;260:12022–12028. [PubMed] [Google Scholar]
- 64.Waxman L, Goldberg A L. Selectivity of intracellular proteolysis: protein substrates activate the ATP-dependent protease (La) Science. 1986;232:500–503. doi: 10.1126/science.2938257. [DOI] [PubMed] [Google Scholar]
- 65.Zehnbauer B A, Foley E C, Henderson G W, Markovitz A. Identification and purification of the lon+ (capR+) gene product, a DNA-binding protein. Proc Natl Acad Sci USA. 1981;78:2043–2047. doi: 10.1073/pnas.78.4.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]