Abstract
Nitric oxide (NO) is an ancestral key signalling molecule essential for life and has enormous versatility in biological systems, including cardiovascular homeostasis, neurotransmission and immunity. Although our knowledge of NO synthases (Nos), the enzymes that synthesize NO in vivo, is substantial, the origin of a large and diversified repertoire of nos gene orthologues in fishes with respect to tetrapods remains a puzzle. The recent identification of nos3 in the ray-finned fish spotted gar, which was considered lost in this lineage, changed this perspective. This finding prompted us to explore nos gene evolution, surveying vertebrate species representing key evolutionary nodes. This study provides noteworthy findings: first, nos2 experienced several lineage-specific gene duplications and losses. Second, nos3 was found to be lost independently in two different teleost lineages, Elopomorpha and Clupeocephala. Third, the expression of at least one nos paralogue in the gills of developing shark, bichir, sturgeon, and gar, but not in lamprey, suggests that nos expression in this organ may have arisen in the last common ancestor of gnathostomes. These results provide a framework for continuing research on nos genes’ roles, highlighting subfunctionalization and reciprocal loss of function that occurred in different lineages during vertebrate genome duplications.
Keywords: vertebrate evolution, genome duplication, gene duplication and loss, nos, phylogenomics, synteny
1. Introduction
Historically classified as a pollutant, nitric oxide (NO) was recognized as ‘Molecule of the Year’ in 1992 [1] for its important function as a cellular signalling molecule. NO plays a role in a myriad of physiological processes, including cardiovascular homeostasis [2], neurotransmission [3], immune response [4], and in neurodegenerative diseases [5] and cancer [6].
Nitric oxide synthase (Nos), the enzyme catalysing the biosynthesis of NO in vivo, is ubiquitous among organisms [7]. Three nos gene paralogues have been described in vertebrates: the constitutively expressed nos1 and nos3, and the inducible nos2 [8].
Although the availability of current genomic data covers all major ray-finned fish lineages, the evolutionary history of their nos gene repertoire remains puzzling. Previous studies reported a variable number of nos genes in teleost fishes: nos1 is always present in a single copy and nos2 is lost or in one or two copies, while nos3 has been reported as missing in the genomes of ray-finned fishes. This apparent gene loss contrasts with literature describing a putative Nos3-like protein localized by antibody stains in gills and vascular endothelium of some teleost species [9,10]. The discovery of a nos3 orthologue in the spotted gar Lepisosteus oculatus, a holostean fish (the sister group of teleosts within the ray-finned lineage) [11], and the variable number of teleost nos2 genes prompted us to study in deep the evolution of this important gene family and nos3 expression pattern in fishes representing key nodes in vertebrate evolution. In an attempt to answer these questions, we have studied the Nos family repertoire at unprecedented phylogenetic resolution, investigated conserved syntenies in fish genomes, and studied the expression pattern of all three nos genes during development in multiple species.
2. Results
(a) . Revised evolutionary history of Nos2 and Nos3
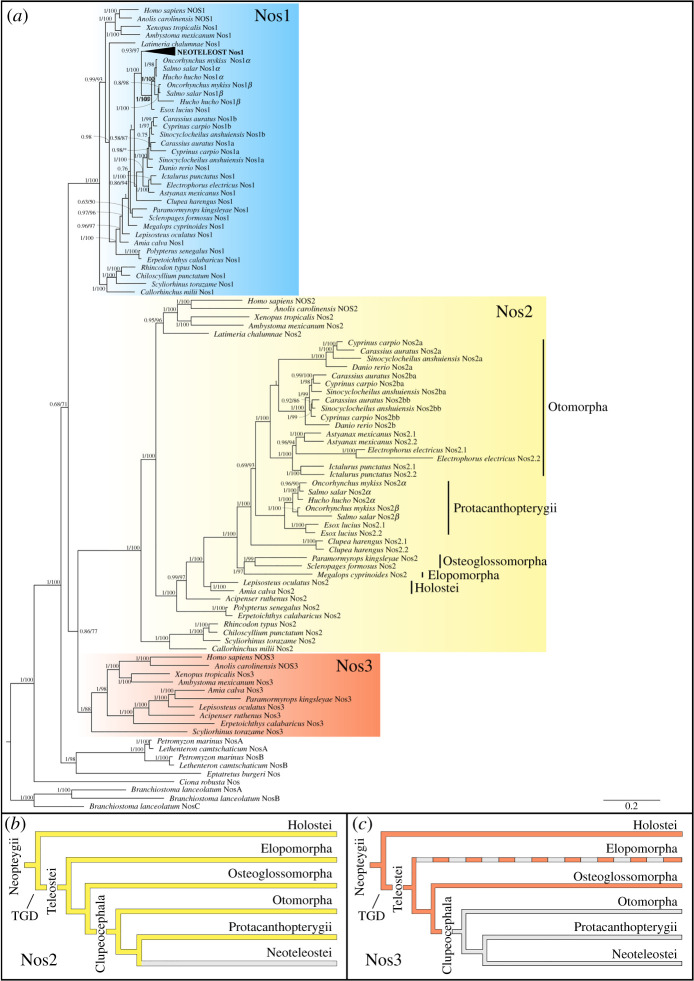
Gaps in our current knowledge of Nos family evolution include the time of origin of the three distinct paralogous nos genes and when some of them were secondarily lost in specific lineages. We reconstructed the Nos phylogeny using 116 protein sequences from 54 species (electronic supplementary material, table S1) providing a broad representation of aquatic vertebrates: cyclostomes (modern jawless fishes), chondrichthyans (cartilaginous fishes), and osteichthyes (bony fishes), including ray- and lobe-finned fishes. Lobe-finned fishes include coelacanths, lungfishes, and tetrapods; ray-finned fishes comprise the non-teleost lineages of polypteriformes (e.g. bichir), acipenseriformes (e.g. sterlet sturgeon), holosteans (lepisosteiformes, e.g. spotted gar, and amiiformes, e.g. bowfin), and the teleosts, subdivided into three major living lineages: elopomorphs (e.g. eels and relatives), osteoglossomorphs (e.g. arowana, mooneyes and the freshwater elephantfish), and clupeocephalans (e.g. zebrafish and medaka) [12] (for clarification see the electronic supplementary material, figure S1).
All Nos proteins considered in the present study showed conservation of canonical domains organization. Here we confirmed the presence of single Nos1 in all jawed vertebrates examined, except for two gene duplicates in cyprinids (nos1a and nos1b) and salmonids (nos1α and nos1β) (figure 1a blue shading; electronic supplementary material, figure S2-a). Most fish lineages retained Nos2, including chondrichthyans (Callorhinchus milii, Rhincodon typus, Chiloscyllium punctatum, Scyliorhinus torazame), polypteriformes (Polypterus senegalus, Erpetoichthys calabaricus), acipenseriformes (Acipenser ruthenus), holosteans (Amia calva, Lepisosteus oculatus), elopomorphs (Megalops cyprinoides), osteoglossomorphs (Paramormyrops kingsleyae, Scleropages formosus) and coelacanthiformes (Latimeria chalumnae) (figure 1a, yellow shading), although a nos2 gene loss event occurred at the stem of Neoteleostei (figure 1b), since it has not been found in any available genomic or transcriptomic data from this clade. On the other hand, our phylogenetic analysis highlights the occurrence of extra nos2 duplicates in several lineages, for which we adopted a specific nomenclature based on the phylogenetic analysis and synteny conservation: nos2a and nos2b in the zebrafish Danio rerio; nos2a, nos2ba and nos2bb in the goldfish Carassius auratus, the blind golden-line barbel Sinocyclocheilus anshuiensis and the common carp Cyprinus carpio; nos2α and nos2β in salmonids (Salmo salar and Oncorhynchus mykiss); and lastly, nos2.1 and nos2.2 in a characid (the Mexican tetra Astyanax mexicanus), a gymnotid (the electric eel Electrophorus electricus), an ictalurid (the channel catfish Ictalurus punctatus), an esocid (the northern pike Esox lucius), and a clupeid (the Atlantic herring Clupea harengus) (figure 1a, yellow shading).
Figure 1.
Evolution of the Nos gene family. (a) Phylogenetic analysis of Nos proteins in chordates. The tree topology was inferred by Bayesian inference and maximum-likelihood methods, with the exact topology obtained from the former shown here (see the electronic supplementary material, figure S7 for the maximum-likelihood tree). Numbers at nodes represent posterior probability values (left) and maximum-likelihood bootstrap support for 1000 replicates (right). (b,c), Evolutionary scenarios indicating the loss of Nos2 event in Neoteleostei (b) and Nos3 in Clupeocephala (c) as grey lines. Nos3 in Elopomorpha is absent, although parsimony suggests it was present in stem elopomorphs, and it is indicated with a dashed line. TGD stands for teleost-specific genome duplication. (Online version in colour.)
Nos3 deserves special attention since it was previously believed that a loss event predated the lineage of actinopterygians or alternatively that it represents an innovation of tetrapods [7]. Nevertheless, this hypothesis may have been overinterpreted since few ray-finned genome sequences were originally available. The only actinopterygian nos3 reported thus far was in the spotted gar [11]. Here we report the identification of nos3 in genomes of the bichir Po. senegalus, the sterlet sturgeon Ac. ruthenus [13], the bowfin Am. calva [14], and the freshwater elephantfish Pa. kingsleyae [15] (figure 1a, red shading). The absence of nos3 in clupeocephalans indicates a gene loss event at the stem of this group (figure 1c). Furthermore, we did not find nos3 in the tarpon M. cyprinoides, the most complete genome available among Elopomorpha, nor in transcriptomic data of the European eel Anguilla anguilla. On the other hand, we did identify a nos3 orthologue in the cloudy catshark Scy. torazame, suggesting its presence in the ancestor of gnathostomes. Previously, two nos genes had been found in the lamprey, called nosA and nosB [7], with unresolved orthology to gnathostome nos1-nos2-nos3, and derived from a lineage-specific tandem duplication in the lamprey lineage. Based on this finding, we searched for the presence of nos genes in other cyclostomes. We found orthologous genes to Petromyzon marinus nosA and nosB paralogues in the arctic lamprey Lethenteron camtschaticum [16], and a single nos gene in the inshore hagfish Eptatretus burgeri. Our phylogenetic analysis shows that the hagfish Nos remains outside the lamprey NosA-NosB clade, therefore with no clear orthology relationship to any specific gnathostome Nos1, Nos2, Nos3, and suggesting that the duplication giving rise to the lamprey nosA-nosB occurred at least before the last common ancestor of Petromyzontidae.
In order to study the Nos evolution at the protein level and verify if each gene clade is under differential selection pressure, we conducted a branch model (BM) analysis (see the electronic supplementary material). The BM analysis showed significant p-value and ω values less than 1 for all Nos proteins: Nos1 (ω1 = 0.035), Nos2 (ω1 = 0.092) and Nos3 (ω1 = 0.082) (electronic supplementary material, table S2). Therefore, they are under purifying (negative) selection, and in particular, the Nos2 and Nos3 evolution resulted slightly more relaxed with respect to Nos1.
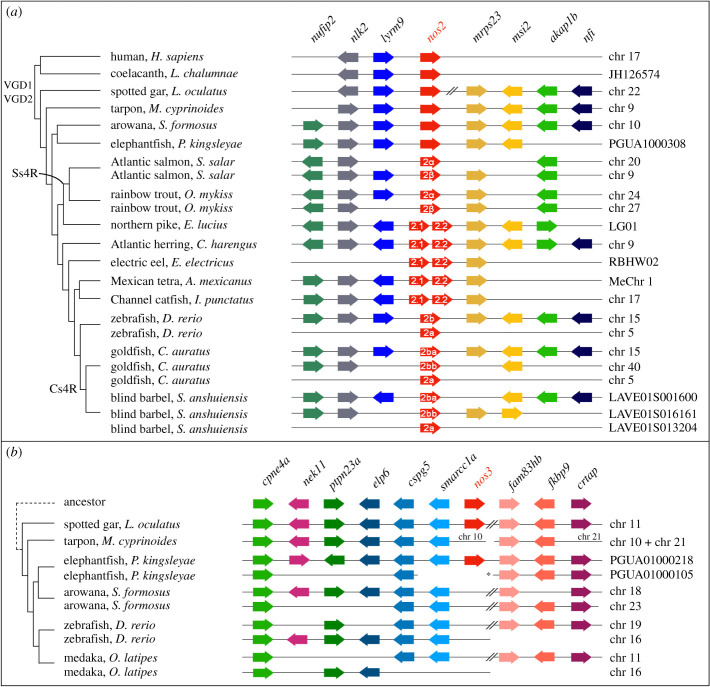
To better understand the gene loss and expansion events highlighted by our phylogenetic analysis, we next analysed the microsynteny (genes linked in proximity) of nos genes in different species. This revealed a complex evolutionary scenario for nos2 compared to nos1 and nos3. Specific nos2 duplications in different lineages are explained by distinct evolutionary events in teleosts. First, the lack of synteny conservation between nos2a and nos2b in cyprinids, and the lack of nos2a in the expected location in non-cyprinid fishes (electronic supplementary material, figure S2-b) indicates that these paralogues originated in a specific gene duplication event in a common ancestor of the lineage, independently from the teleost-specific genome duplication (TGD) (the alternative explanation would require numerous nos2a losses in several fish lineages), in which while nos2b has remained in the ancestral genomic location, nos2a has been translocated to a different position in the genome (figure 2a; electronic supplementary material, figure S2-b). Second, an additional genome duplication event after the TGD specifically occurred independently in several teleost lineages, causing the presence of extra nos2 paralogues. These include some cyprinids, in which the carp-specific genome duplication event (Cs4R) probably occurred before the divergence of Ca. auratus, Si. anshuiensis and Cy. carpio [17], and salmonids (salmonid-specific genome duplication or Ss4R) [18,19], with Sa. salar and O. mykiss in this study. These additional tetraploidization events can explain the origin of the two independent sets of nos2 genes in cyprinid and salmonid species. In the case of cyprinids, both our phylogenetic and synteny analyses clearly show their nos2b orthology, and we denote them as nos2ba and nos2bb (figures 1a and 2a). In the case of salmonids, we name them nos2α and nos2β to distinguish them from the cyprinid nos2a and nos2b paralogues, which have a separate origin (see above; figure 2a). Third, independent tandem gene duplications explain the presence of two nos2 copies, that we named nos2.1 and nos2.2, located next to each other in the same chromosomal fragment in the genomes of the Atlantic herring (Cl. harengus), the Mexican tetra (cavefish, As. mexicanus), the electric eel (El. electricus), the channel catfish (I. punctatus) and the northern pike (Es. lucius) (figure 2a).
Figure 2.
Conserved microsynteny of nos2 and nos3. (a) The nos2 paralogues derived from different duplication modalities: carp-specific genome duplication (Cs4R) (nos2ba and nos2bb in the goldfish and blind barbel); salmonid-specific genome duplication (Ss4R) (nos2α and nos2β in the Atlantic salmon and rainbow trout); tandem gene duplication occurred independently in five lineages (nos2.1 and nos2.2 in the northern pike, Atlantic herring, electric eel, Mexican tetra and channel catfish). An additional nos2 duplicate (nos2a) is present in cyprinids (zebrafish, goldfish, and blind barbel) (see the electronic supplementary material, figure S2). (b) A conserved synteny map of genomic regions around the nos3 gene locus highlights the loss in Clupeocephala (including zebrafish and medaka), and in Osteoglossomorpha (arowana). Consecutive genes are represented as arrows and are colour coded according to their orthology and ohnology. The direction of arrows indicates gene transcription orientation. // indicates long-distance on the chromosome (>600 kb), * indicates scaffold 72 of the freshwater elephantfish genome [15]. (Online version in colour.)
Bichir, reedfish, sterlet, spotted gar, bowfin and freshwater elephantfish are the only ray-finned fishes that retained a nos3 orthologue. Therefore, we investigated the absence of nos3 in clupeocephalans. First, we looked for the genomic region containing nos3 in fishes that represent outgroups to the clupeocephalans. We found one long scaffold of the Pa. kingsleyae genome (scaffold 217) [15] showing extensive conserved synteny with the nos3-containing segment of the linkage group 11 (LG) in the spotted gar genome (figure 2b). While these appear to correspond to one of the TGD ohnologons (figure 2b), there are two other Pa. kingsleyae scaffold segments (from scaffolds 72 and 104) that together seem to represent the second TGD ohnologon, but lacking the expected nos3 TGD ohnologue (figure 2b). Zebrafish chromosomes 16 and 19 and medaka chromosomes 11 and 16 contain orthologous regions to the two Pa. kingsleyae and Le. oculatus TGD ohnologons, but lack a nos3 gene at the expected locations. The one-to-one relationship between these Pa. kingsleyae scaffolds and zebrafish and medaka chromosomes is challenging to determine (figure 2b). Regardless, the most parsimonious explanation for the nos3 repertoire in ray-finned fishes is that, one of the two nos3 TGD ohnologues was lost in the teleost common ancestor, while the other was retained and later lost in secondary, independent events in the common ancestor of Clupeocephala and, probably, that of Elopomorpha (figures 1c and 2b).
(b) . Expression of nos in vertebrate developing gills
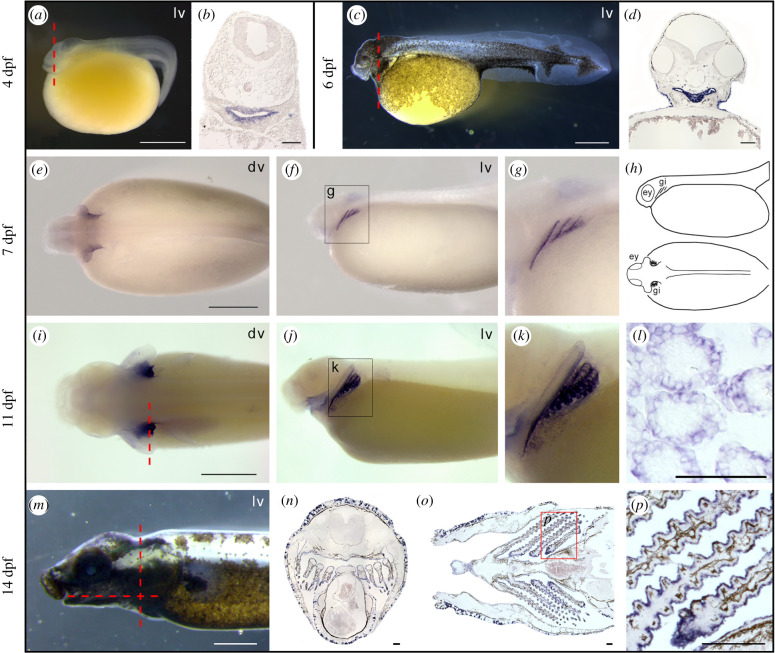
Spotted gar is an important emerging experimental organism representing an evolutionary bridge between teleosts and tetrapods that facilitates cross-species comparisons. The gar genome is slowly evolving compared to that of teleosts and has preserved a more ancient structural organization [20]. Therefore, we examined the expression patterns of nos genes during gar development. As expected, nos1 was expressed in several regions of the developing nervous system (electronic supplementary material, figure S3, and [21]). By contrast, nos2 expression was not detected during the developmental stages covered in the present study, i.e. from 4 to 14 days post fertilization (dpf). Unexpectedly, the expression of nos3 was first detected in embryos in the pharyngeal area at 4 dpf (figure 3a,b) and increased at 6 dpf (figure 3c,d). At 7 dpf, embryos showed clear nos3 expression in developing arches III, IV, and V (figure 3e–g). Later, at 11 dpf, the positive signal is localized in gill filaments (figure 3i–k). Histological sections highlighted the presence of nos3 in the epithelium of branchial lamellae (figure 3l), also confirmed by the signal in gill structures in an advanced stage of maturation in 14 dpf juveniles (figure 3m–p).
Figure 3.
Spotted gar nos3 localization during development. Expression of nos3 is localized in the pharyngeal area in 4 dpf (a,b) and 6 dpf (c,d) embryos, in pharyngeal arches in 7 dpf larvae (e–g) schematized in (h), in developing gills in 11 dpf late larvae (i–l), and in gill lamellae in 14 dpf juveniles (m–p). Coronal (n) and transversal section (o) planes are indicated with a red dashed line in (m). ey, eye; gi, gill; dv, dorsal view; lv, lateral view. Scale bar is 1 mm in (a,c,e,i,m); 100 µm in (b,d,l,n,o,p). (Online version in colour.)
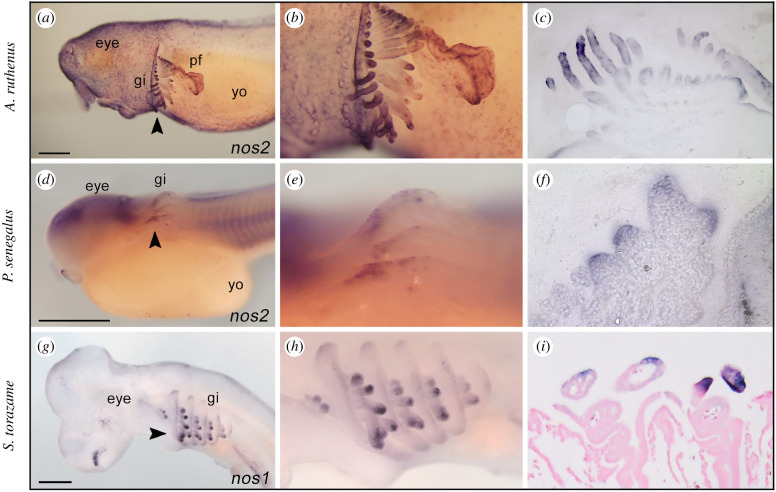
The detection of nos3 transcripts in gills of spotted gar and the established involvement of NO gas in osmoregulatory control and vascular motility in gills of numerous teleosts [22–25] prompted us to investigate whether a similar nos expression pattern occurred in developing gills of other fish species. We investigated nos expression in the sterlet sturgeon and the bichir, members of early branching groups of ray-finned fishes [12]. Moreover, we similarly searched nos expression pattern in the chondrichthyan cloudy catshark to infer the ancestral expression condition among gnathostomes. Unlike gar, we discovered that nos3 was not expressed in gills of other species analysed in this work (electronic supplementary material, figure S3), thus raising questions of whether nos3 expression in gills represents an oddity of holosteans or gars. Surprisingly, we found a different scenario in which other nos genes were expressed in gills of sturgeon, bichir, and shark. In particular, nos2 was expressed in the branchial area of the sterlet sturgeon (figure 4a–c) and bichir embryos (figure 4d–f), while nos1 is expressed in gills of catshark embryos (figure 4g–i).
Figure 4.
Expression of nos genes in developing gills of sturgeon, bichir, and shark embryos. The expression of nos2 in the gills of sterlet sturgeon Acipenser ruthenus (14 mm stage, a–c) and bichir Polypterus senegalus (stage 31, d,e); nos1 in the shark Scyliorhinus torazame (stage 27, g–i). Higher magnification views of the gill structure of (a,d,g) are shown in (b,e,h), respectively. The arrowheads indicate sectioning plane (a,d,g): transversal sections (c,f, 50 µm) and frontal section (I, 10 µm). gi, gill; yo, yolk; pf, pectoral fin. Scale bar in (a,d,g) is 0.5 mm. (Online version in colour.)
Our results show that nos paralogues are expressed in pharyngeal arches and gills in both actinopterygians and chondrichthyans. These findings lead us to question whether nos expression in gills could be a conserved feature also in sarcopterygians, and in particular in amphibians that use gills for gas exchange. Therefore, to investigate the presence of nos transcripts in amphibia, we chose the neotenic axolotl Ambystoma mexicanum because it retains functional external gills throughout life. Gene expression analysis by quantitative polymerase chain reaction (qPCR) revealed that nos1 and nos2 are almost not detectable in adult axolotl gills, while nos3 is highly expressed in gill structures (electronic supplementary material, figure S4). Therefore, we conclude that nos expression in gills is a conserved feature in the neotenic amphibian assayed, previously observed exclusively in fishes.
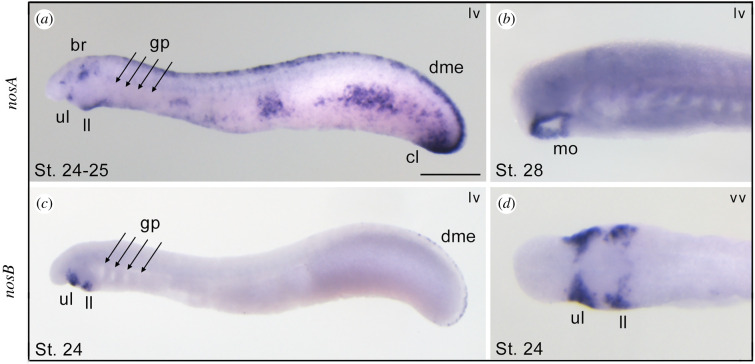
(c) . Expression of nos genes in the lamprey
In cyclostomes (jawless vertebrates, including lampreys and hagfish), cartilaginous and bony gnathostomes (jawed vertebrates), gills are endoderm-derived structures, pointing to a single origin of pharyngeal gills before the divergence of these vertebrate lineages [26,27]. To assess whether nosA and nosB are expressed in gills during embryogenesis, we performed whole-mount in situ hybridization experiments at different embryonic stages. We found that lamprey nosA was expressed in several tissues, including the brain, dorsal midline epidermis, tailbud, mouth and cloaca, but not in gills (figure 5a,b). Conversely, the lamprey nosB paralogue showed restricted expression in the developing mouth, specifically in the cheek process, including upper and lower lip regions (figure 5c,d). These results show that in the arctic lamprey, neither of the two nos paralogues is expressed in immature or mature gills, suggesting a fundamental difference in the role of nos genes in jawless and jawed vertebrates.
Figure 5.
Expression patterns of nosA and nosB in larvae of the arctic lamprey. At stages 24–25 the nosA is expressed in the brain, mouth, upper and lower lip, dorsal midline epidermis, and cloaca (a). At stage 28, nosA expression is restricted to the mouth (b). The nosB is exclusively expressed in the cheek process, consisting of upper and lower lips (c,d), and faint expression in the dorsal midline epidermis (c). br, brain; cl, cloaca; dme, dorsal midline epidermis; gp, gill pouches; mo, mouth; ll, lower lip; ul, upper lip; lv, lateral view; vv, ventral view. Scale bar in (a) is 0.5 mm. (Online version in colour.)
3. Discussion
Actinopterygians experienced one of the largest radiations in the animal kingdom and their history represents a valuable resource for the formulation of hypotheses regarding the evolution of vertebrate gene families. In this work, we employed data from recent genome projects to clarify and update the evolution of the Nos family across vertebrates. Our phylogenetic analysis confirmed that Nos1 is ubiquitously present as single copy gene across the gnathostome lineage. The only two events of duplication for nos1 were observed in cyprinids and salmonids, as a consequence of their specific Cs4R and Ss4R tetraploidizations, respectively. Furthermore, our phylogenetic data, complemented with syntenic analyses, highlighted for the first time, to our knowledge, a highly complex scenario of Nos2 evolution, for which we suggest a dedicated nomenclature that attempts to incorporate evolutionary origins into gene names. Previous analyses showed the presence of two nos2 genes (nos2a and nos2b) in zebrafish and goldfish [28,29], probably originated from an event of gene duplication that occurred specifically at the stem of the group, and not related to the classic TGD [30,31]. This result is supported by synteny analysis since the chromosomal position of nos2a and nos2b genes is not conserved, as it would be expected if they were retained after whole-genome duplication. Here we show the presence of a nos2a paralogue also in other two cyprinids, Cy. carpio and Si. anshuiensis (figures 1a and 2a). On the other hand, the cyprinid nos2b paralogue independently duplicated in carps after the Cs4R [17], as the conserved synteny suggests (figure 2a). In salmonids, synteny analysis also indicates that the two Nos2 paralogues originated secondarily after the Ss4R (figure 2a) [18,19]. Here, we call these genes nos2ba and nos2bb in carps to emphasize and clarify their relationships to zebrafish genes, and nos2α and nos2β in salmonids to indicate their distinct evolutionary origin. Additionally, the present work shows that nos2 has undergone several independent lineage-specific tandem gene duplication events (nos2.1 and nos2.2) (figure 2a). The search of nos2 in available fish genomes, covering all main groups, failed to find it in any Neoteleostei, and for this reason, we hypothesized a nos2 gene loss event occurred at the stem of Neoteleostei (figures 1 and 6). Importantly, NO produced upon stimulation of the inducible nos2 is considered one of the most versatile players of the immune system [4]. For this reason, it would be important in the future to investigate the impact of Nos2 loss on the immune response in Neoteleostei and if any compensatory mechanisms occurred through the activation of other nos paralogues, as well as to understand if nos2 duplicates underwent neofunctionalization or subfunctionalization, thus providing new functional features to the organism.
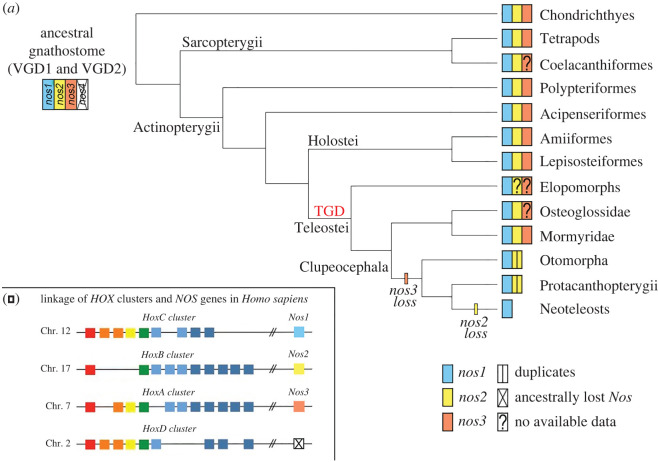
Figure 6.
Nos evolution in light of recent gene findings in vertebrates. The proposed evolution of nos genes in gnathostomes (a) supposes an ancestral loss of a predicted fourth nos gene, based on the linkage of human Nos and Hox clusters (b). Loss of nos3 occurred in stem Clupeocephala and loss of nos2 in stem Neoteleostei (a). Species-specific nos2 duplications occurred in some Otomorpha, including Cyprinidae and Characidae families. (Online version in colour.)
Concerning nos3, our understanding of its evolutionary history had a twist with the finding of a nos3 orthologue in the spotted gar genome [11], proving that the previously postulated actinopterygian-specific loss of nos3 was an incorrect inference. Fostered by this discovery, we specifically searched for the presence of nos3 orthologues in a wide range of fish species to infer the ancestral condition. We identified a nos3 gene in bowfin, thus confirming the presence of nos3 in the other reference genus of the holostean clade, in addition to gar (figure 6). Furthermore, the presence of nos3 in genomes of bichir and sterlet sturgeon, which diverged prior to the teleostean and holostean split, confirmed the hypothesis that nos3 was already present in the common ancestor of extant osteichthyes, rather than an innovation of tetrapods [7] or neopterygians (holosteans plus teleosts) [11] (figure 6). We did not find nos3 gene in the tarpon M. cyprinoides genome (figure 2b), and to date, the limited genomic and transcriptomic data of eels, congers, and morays cannot endorse the presence of a nos3 in Elopomorpha. Therefore, more genome sequences are necessary to confirm its absence in this key group. We also did not find nos3 in any Clupeocephala (non-elopomorph and non-osteoglossomorph teleosts) suggesting that a loss event took place in the common ancestor of clupeocephalans. Notably, we found a nos3 gene in the osteoglossomorph elephantfish Pa. kingsleyae, and it allowed us to confirm that the loss of nos3 did not occur in the last common teleost ancestor, as previously thought [11]. These findings suggest instead the following evolutionary scenario for the nos3 gene: first, since we only find a maximum of one nos3 gene in those cases where it is present, we assume that one of the two TGD ohnologues was immediately lost after the TGD, and the other one was retained. This nos3 gene was then lost in the ancestors of elopomorphs–although further research is needed to confirm this– and clupeocephalans independently in separate events (figure 6).
The discovery of nos3 in sharks (Scy. torazame in this study) suggests that the origin of nos3 predates the divergence of gnathostomes and that three distinct nos paralogues were already present in the last common ancestor of gnathostomes (figure 6), probably originating after the two rounds of whole-genome duplication that took place during early vertebrate evolution (vertebrate gene duplications (VGD) VGD1 and VGD2, 2R hypothesis) [7,32,33]. The origin of nos genes is, in fact, supported by the linkage to the evolutionarily conserved Hox gene clusters and several other syntenic genes (figure 6b; electronic supplementary material, figure S5). Under this scenario, then a fourth nos gene (putative nos4) should have existed but was apparently lost early in the gnathostome evolution (figure 6a).
The apparent lack of nos3 in some vertebrate lineages, such as in coelacanth La. chalumnae (an extant basally diverging sarcopterygian), in arowana Scl. formosus (an osteoglossomorph), and in elopomorph fishes, remains to be clarified in the future.
The protein evolution analysis highlighted that the three Nos clades show negative selection pressure at different rates, being Nos1 under stronger negative selection, in respect to Nos2 and Nos3 that resulted under more relaxed negative selection based on significant ω values. These results are in agreement with the high degree of conservation of nucleotidic and amino acidic sequences during Nos family evolutionary history in vertebrates.
The importance of NO in the ontogeny and function of vertebrate gills has already been documented in the context of physio-pharmacological studies, primarily using inhibitors of Nos activity. In gills, NO acts as a paracrine and endocrine vasoactive modulator and, therefore, plays a crucial role in the distribution of oxygenated blood [34]. Moreover, NO has an osmoregulatory function controlling the movement of ions across the gill epithelium [24,35–37], and represents an important molecular component of the immune system employed by macrophages to attack and destroy pathogens [38]. Nevertheless, documentation of Nos enzymatic activity in fish gills has relied exclusively upon techniques unable to discriminate among individual Nos proteins, such as NADPH-diaphorase activity and immunolocalization with heterologous mammalian antibodies [34,36,37,39]. Therefore, the detected enzymatic activity has for a long time been indicated generically as ‘Nos-like’. Here, using a specific messenger RNA transcript detection methodology, we showed, for the first time, to our knowledge, that indeed nos genes are expressed in gills during development in various vertebrates. Surprisingly different Nos paralogues are expressed in gills in different animals tested: nos1 in shark, nos2 in bichir and sterlet sturgeon, and nos3 in spotted gar. The most parsimonious hypothesis to explain this result is that the ancestral nos gene had a number of roles in gills, immune system, brain, and other organs that was controlled by separate regulatory elements and, owing to subfunctionalization after the vertebrate 2R (according to the duplication-degeneration-complementation model) [40], these physiological roles partitioned to different nos ohnologues as lineages diverged and reciprocal loss of the gill expression function occurred in a lineage-specific way. Further support for this hypothesis comes from the identification of nos1-positive cells in gill of zebrafish at 5 dpf, in addition to brain, eye, periderm and NaK ionocytes, according to the recently released developmental single-cell transcriptome atlas [41] (electronic supplementary material, figure S6).
Additionally, to corroborate the involvement of NO in normal gill physiology, we searched for nos expression in gills of a paedomorphic amphibian, the Mexican axolotl, which maintains gill structures in adulthood. Taking into account the different evolutionary and developmental origin of internal and external gills [42], the conservation of nos3 expression in gills indicated that the NO signalling system could be fundamental for the physiology and development of this structure in the axolotl, and perhaps generally in pre-metamorphic amphibians. Therefore, our data highlighted that the expression of at least one nos gene has a functional role in gnathostome gills.
Recently, a single origin of pharyngeal gills predating the divergence of cyclostomes and gnathostomes was suggested [26]. Therefore, we investigated whether either of the two arctic lamprey nos paralogues is expressed in developing gills, but found them expressed mainly in the nervous system, mouth and pharynx, similar to the expression pattern previously reported in the cephalochordate amphioxus [43,44]. This led us to speculate that either the expression of nos genes in gills was acquired in gnathostomes after the divergence from cyclostomes, or alternatively, gill expression was a feature of their last common ancestor but lost in the lineage of cyclostomes.
In conclusion, our findings pave the way for future studies that aim to investigate the ontogenetic role of NO in gill development of aquatic vertebrates. It would be interesting to understand more about species-specific regulatory mechanisms that drive different nos genes expression patterns in gills in different species.
4. Methods
(a) . Phylogenetic analysis
Nos sequences used for evolutionary analyses were retrieved from NCBI, Ensembl, Skatebase and DDBJ databases (electronic supplementary material, table S1). We used proteins from Homo sapiens, Anolis carolinensis and Xenopus tropicalis as internal references, and two non-vertebrate chordates as outgroups: the cephalochordate Branchiostoma lanceolatum NosA, NosB and NosC, and the tunicate Ciona robusta Nos.
For phylogenetic analysis, Nos amino acid sequences were aligned using the MUSCLE algorithm [45] as implemented in MEGAX (v. 10.2.4) [46]. The alignment was trimmed by trimAl v. 1.2rev59 [47] and then formatted into a nexus file using readAl (bundled with the trimAl package) (electronic supplementary material, File S1). The Bayesian inference tree was constructed using MrBayes v. 3.2.6 [48], under the assumption of an LG + I + G evolutionary model. Two independent MrBayes runs of 2 000 000 generations were performed, with four chains each and a temperature parameter value of 0.05. The tree was considered to have reached convergence when the standard deviation stabilized under a value of less than 0.01. A burn-in of 25% of the trees was performed to generate the consensus tree (1 500 000 post-burnt-in trees). The maximum-likelihood phylogenetic tree was inferred on the same multi-sequence alignment (electronic supplementary material, file S1) using IQ-Tree v. 2.1.3 [49] with 1000 replicates, using automatic selection of best-fit model with ModelFinder [50] and branch support assessed with the ultrafast bootstrap approximation [51] (electronic supplementary material, figure S7).
(b) . Synteny
With the aim of finding synteny blocks flanking the nos2 and nos3 orthologues, we employed the Synteny Database [52,53]. Additional information was retrieved in NCBI, Ensemble (v. 102) and Genomicus (v. 100.01) [52].
(c) . Gene expression analysis by in situ hybridization
Whole-mount in situ hybridization experiments were performed for all nos paralogues following species-specific protocols previously described: spotted gar [54], bichir and sturgeon [55], lamprey [56] and shark [57]. Embryo and tissue collection, and protocol modifications to the in situ hybridization are reported in the electronic supplementary material, Extended methods.
Supplementary Material
Acknowledgements
We thank Fumiaki Sugahara for the interpretation of arctic lamprey results, Anna Pospisilova for technical assistance with bichir and sturgeon in situ hybridizations, and Martin Psenicka, Roman Franek, Michaela Fucikova, Marek Rodina, David Gela, Martin Kahanec for sterlet sturgeon spawns. A special thanks to Robert Cerny for the establishment of the African bichir colony at the Charles University in Prague. The sturgeon work was supported by the Ministry of Education, Youth and Sports of the Czech Republic, project Cenakva (LM2018099) and project Biodiversity (CZ.02.1.01/0.0/0.0/16_025/007370). We also thank Jordi Paps and Giacinto De Vivo for their help on phylogenetic inferences, and to Silvia Perea and Iker Irisarri for their help in the selection analyses.
Data accessibility
Accession numbers of protein sequences used in the phylogenetic analysis are available in the electronic supplementary material, table S1. Primer sequences used for the synthesis of in situ hybridization riboprobes and in qRT-PCR experiments are given in the electronic supplementary material, table S3. Electronic supplementary material is available online [58].
Authors' contributions
G.A.: conceptualization, data curation, formal analysis, investigation, methodology, writing—original draft, writing—review and editing; I.S.: investigation, methodology; J.P.-A.: conceptualization, data curation, formal analysis, investigation, methodology, validation, writing—original draft, writing—review and editing; D.O.: data curation, formal analysis, writing—review and editing; I.B.: writing—review and editing; R.V.: data curation, investigation, writing—review and editing; J.S.: data curation, investigation, writing—review and editing; V.S.: conceptualization, data curation, investigation, methodology, writing—review and editing; A.F.: data curation, formal analysis, investigation, writing—review and editing; Q.F.: data curation, formal analysis, investigation, writing—review and editing; S.K.: supervision; J.H.P.: conceptualization, data curation, investigation, methodology, supervision, writing—review and editing; S.D.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, supervision, validation, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
The authors declare no competing interests.
Funding
G.A. was supported by the Research grant POR Campania FSE 2014/2020 (IT) and by the EMBO Short Term Fellowship (no. 6936) to visit the Postlethwait laboratory in Oregon (USA) and for the field trip in Louisiana (USA). J.S. is supported by a Marie Skłodowska-Curie grant agreement no. 897949. V.S. is supported by the Charles University Research Centre program no. 204069 and grant no. SVV260571/2020. R.V. and the Ambystoma Genetic Stock Center are supported by US NIH (P40OD019794). J.H.P. and I.B. are supported by the R01 OD011116 grant from the US NIH. I.B. is supported by the US NSF EDGE grant no. 2029216. S.D. is supported by the NOEVO grant from the SZN.
References
- 1.Koshland D. 1992. The molecule of the year. Science 258, 1861. ( 10.1126/science.1470903) [DOI] [PubMed] [Google Scholar]
- 2.Strijdom H, Chamane N, Lochner A. 2009. Nitric oxide in the cardiovascular system: a simple molecule with complex actions. Cardiovasc. J. Afr. 20, 303-310. [PMC free article] [PubMed] [Google Scholar]
- 3.Esplugues JV. 2002. NO as a signalling molecule in the nervous system. Br. J. Pharmacol. 135, 1079-1095. ( 10.1038/sj.bjp.0704569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogdan C. 2001. Nitric oxide and the immune response. Nat. Immunol. 2, 907-916. ( 10.1038/ni1001-907) [DOI] [PubMed] [Google Scholar]
- 5.Knott AB, Bossy-Wetzel E. 2009. Nitric oxide in health and disease of the nervous system. Antioxid Redox Signal. 11, 541-553. ( 10.1089/ars.2008.2234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamm A, Przychodzen P, Kuban-Jankowska A, Jacewicz D, Dabrowska AM, Nussberger S, Wozniak M, Gorska-Ponikowska M. 2019. Nitric oxide and its derivatives in the cancer battlefield. Nitric Oxide 93, 102-114. ( 10.1016/j.niox.2019.09.005) [DOI] [PubMed] [Google Scholar]
- 7.Andreakis N, D'Aniello S, Albalat R, Patti FP, Garcia-Fernandez J, Procaccini G, Sordino P, Palumbo A. 2011. Evolution of the nitric oxide synthase family in metazoans. Mol. Biol. Evol. 28, 163-179. ( 10.1093/molbev/msq179) [DOI] [PubMed] [Google Scholar]
- 8.Förstermann U, Sessa WC. 2012. Nitric oxide synthases: regulation and function. Eur. Heart J. 33, 829-837. ( 10.1093/eurheartj/ehr304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tota B, Amelio D, Pellegrino D, Ip YK, Cerra MC. 2005. NO modulation of myocardial performance in fish hearts. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 142, 164-177. ( 10.1016/j.cbpb.2005.04.019) [DOI] [PubMed] [Google Scholar]
- 10.Agnisola C, Pellegrino D. 2007. Role of nitric oxide in vascular regulation in fish. Adv. Exp. Biol. 293-310. ( 10.1016/S1872-2423(07)01013-7) [DOI] [Google Scholar]
- 11.Donald JA, Forgan LG, Cameron MS. 2015. The evolution of nitric oxide signalling in vertebrate blood vessels. J. Comp. Physiol. B 185, 153-171. ( 10.1007/s00360-014-0877-1) [DOI] [PubMed] [Google Scholar]
- 12.Hughes LC, et al. 2018. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl Acad. Sci. USA 115, 6249-6254. ( 10.1073/pnas.1719358115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du K, et al. 2020. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 4, 841-852. ( 10.1038/s41559-020-1166-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson A, et al. 2021. The genome of the bowfin (Amia calva) illuminates the developmental evolution of ray-finned fishes. Nat. Genet. 53, 1373-1384. ( 10.1038/s41588-021-00914-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallant JR, Losilla M, Tomlinson C, Warren WC. 2017. The genome and adult somatic transcriptome of the mormyrid electric fish Paramormyrops kingsleyae. Genome Biol. Evol. 9, 3525-3530. ( 10.1093/gbe/evx265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta TK, et al. 2013. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc. Natl Acad. Sci. USA 110, 16 044-16 049. ( 10.1073/pnas.1315760110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu P, et al. 2019. The allotetraploid origin and asymmetrical genome evolution of the common carp Cyprinus carpio. Nat. Commun. 10, 4625. ( 10.1038/s41467-019-12644-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berthelot C, et al. 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5, 3657. ( 10.1038/ncomms4657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lien S, et al. 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533, 200-205. ( 10.1038/nature17164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braasch I, et al. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 48, 427-437. ( 10.1038/ng.3526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Annona G, Ferran JL, De Luca P, Conte I, Postlethwait JH, D'Aniello S. 2022. Expression pattern of nos1 in the developing nervous system of ray-finned fish. Genes 13, 918. ( 10.3390/genes13050918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbins IL, Olsson C, Holmgren S. 1995. Distribution of neurons reactive for NADPH-diaphorase in the branchial nerves of a teleost fish, Gadus morhua. Neurosci. Lett. 193, 113-116. ( 10.1016/0304-3940(95)11680-U) [DOI] [PubMed] [Google Scholar]
- 23.Mauceri A, Fasulo S, Ainis L, Licata A, Rita lauriano E, Martfnez A, Mayer B, Zaccone G. 1999. Neuronal nitric oxide synthase (nNOS) expression in the epithelial neuroendocrine cell system and nerve fibers in the gill of the catfish, Heteropneustes fossilis. Acta Histochem. 101, 437-448. ( 10.1016/S0065-1281(99)80044-0) [DOI] [PubMed] [Google Scholar]
- 24.Evans DH. 2002. Cell signaling and ion transport across the fish gill epithelium. J. Exp. Zool. 293, 336-347. ( 10.1002/jez.10128) [DOI] [PubMed] [Google Scholar]
- 25.Pellegrino D, Sprovieri E, Mazza R, Randall D, Tota B. 2002. Nitric oxide-cGMP-mediated vasoconstriction and effects of acetylcholine in the branchial circulation of the eel. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 132, 447-457. ( 10.1016/S1095-6433(02)00082-X) [DOI] [PubMed] [Google Scholar]
- 26.Gillis JA, Tidswell ORA. 2017. The origin of vertebrate gills. Curr. Biol. 27, 729-732. ( 10.1016/j.cub.2017.01.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warga RM, Nüsslein-Volhard C. 1999. Origin and development of the zebrafish endoderm. Development 126, 827-838. ( 10.1242/dev.126.4.827) [DOI] [PubMed] [Google Scholar]
- 28.Poon K-L, Richardson M, Korzh V. 2008. Expression of zebrafish nos2b surrounds oral cavity. Dev. Dyn. 237, 1662-1667. ( 10.1002/dvdy.21566) [DOI] [PubMed] [Google Scholar]
- 29.Lepiller S, Franche N, Solary E, Chluba J, Laurens V. 2009. Comparative analysis of zebrafish nos2a and nos2b genes. Gene 445, 58-65. ( 10.1016/j.gene.2009.05.016) [DOI] [PubMed] [Google Scholar]
- 30.Postlethwait J, Amores A, Force A, Yan YL. 1998. The zebrafish genome. Methods Cell Biol. 60, 149-163. ( 10.1016/S0091-679X(08)61898-1) [DOI] [PubMed] [Google Scholar]
- 31.Amores A. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282, 1711-1714. ( 10.1126/science.282.5394.1711) [DOI] [PubMed] [Google Scholar]
- 32.Dehal P, Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3, e314. ( 10.1371/journal.pbio.0030314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakatani Y, Shingate P, Ravi V, Pillai NE, Prasad A, McLysaght A, Venkatesh B. 2021. Reconstruction of proto-vertebrate, proto-cyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nat. Commun. 12, 1-4. ( 10.1038/s41467-020-20314-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tota B, Amelio D, Cerra MC, Garofalo F. 2018. The morphological and functional significance of the NOS/NO system in the respiratory, osmoregulatory, and contractile organs of the African lungfish. Acta Histochem. 120, 654-666. ( 10.1016/j.acthis.2018.08.011) [DOI] [PubMed] [Google Scholar]
- 35.Tipsmark CK. 2003. Regulation of Na+/K+-ATPase activity by nitric oxide in the kidney and gill of the brown trout (Salmo trutta). J. Exp. Biol. 206, 1503-1510. ( 10.1242/jeb.00284) [DOI] [PubMed] [Google Scholar]
- 36.Ebbesson LOE. 2005. Nitric oxide synthase in the gill of Atlantic salmon: colocalization with and inhibition of Na+,K+-ATPase. J. Exp. Biol. 208, 1011-1017. ( 10.1242/jeb.01488) [DOI] [PubMed] [Google Scholar]
- 37.Hyndman KA, Choe KP, Havird JC, Rose RE, Piermarini PM, Evans DH. 2006. Neuronal nitric oxide synthase in the gill of the killifish, Fundulus heteroclitus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 144, 510-519. ( 10.1016/j.cbpb.2006.05.002) [DOI] [PubMed] [Google Scholar]
- 38.Campos-Perez JJ, Ward M, Grabowski PS, Ellis AE, Secombes CJ. 2000. The gills are an important site of iNOS expression in rainbow trout Oncorhynchus mykiss after challenge with the Gram-positive pathogen Renibacterium salmoninarum. Immunology 99, 153-161. ( 10.1046/j.1365-2567.2000.00914.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mistri A, Kumari U, Mittal S, Mittal AK. 2018. Immunohistochemical localization of nitric oxide synthase (NOS) isoforms in epidermis and gill epithelium of an angler catfish, Chaca chaca (Siluriformes, Chacidae). Tissue Cell 55, 25-30. ( 10.1016/j.tice.2018.09.008) [DOI] [PubMed] [Google Scholar]
- 40.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531-1545. ( 10.1093/genetics/151.4.1531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farnsworth DR, Saunders LM, Miller AC. 2020. A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 459, 100-108. ( 10.1016/j.ydbio.2019.11.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stundl J, Pospisilova A, Jandzik D, Fabian P, Dobiasova B, Minarik M, Metscher BD, Soukup V, Cerny R. 2019. Bichir external gills arise via heterochronic shift that accelerates hyoid arch development. Elife 8, e43531. ( 10.7554/eLife.43531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Annona G, Caccavale F, Pascual-Anaya J, Kuratani S, De Luca P, Palumbo A, D'Aniello S. 2017. Nitric oxide regulates mouth development in amphioxus. Sci. Rep. 7, 8432. ( 10.1038/s41598-017-08157-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caccavale F, Annona G, Subirana L, Escriva H, Bertrand S, D'Aniello S. 2021. Crosstalk between nitric oxide and retinoic acid pathways is essential for amphioxus pharynx development. Elife 10, e58295. ( 10.7554/eLife.58295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792-1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547-1549. ( 10.1093/molbev/msy096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972-1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ronquist F, et al. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539-542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268-274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587-589. ( 10.1038/nmeth.4285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518-522. ( 10.1093/molbev/msx281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen NTT, Vincens P, Roest Crollius H, Louis A. 2018. Genomicus 2018: karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 46, D816-D822. ( 10.1093/nar/gkx1003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catchen JM, Conery JS, Postlethwait JH. 2009. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 19, 1497-1505. ( 10.1101/gr.090480.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jowett T, Yan Y-L. 1996. Double fluorescent in situ hybridization to zebrafish embryos. Trends Genet. 12, 387-389. ( 10.1016/S0168-9525(96)90091-8) [DOI] [PubMed] [Google Scholar]
- 55.Minarik M, et al. 2017. Pre-oral gut contributes to facial structures in non-teleost fishes. Nature 547, 209-212. ( 10.1038/nature23008) [DOI] [PubMed] [Google Scholar]
- 56.Sugahara F, Murakami Y, Kuratani S. 2015. Gene expression analysis of lamprey embryos. In situ hybridization methods, pp. 263-278. New York, NY: Humana Press. [Google Scholar]
- 57.Adachi N, Takechi M, Hirai T, Kuratani S. 2012. Development of the head and trunk mesoderm in the dogfish, Scyliorhinus torazame: II. Comparison of gene expression between the head mesoderm and somites with reference to the origin of the vertebrate head. Evol. Dev. 14, 257-276. ( 10.1111/j.1525-142X.2012.00543.x) [DOI] [PubMed] [Google Scholar]
- 58.Annona G, et al. 2020. Evolution of the nitric oxide synthase family in vertebrates and novel insights in gill development. Figshare. ( 10.6084/m9.figshare.c.6136963) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers of protein sequences used in the phylogenetic analysis are available in the electronic supplementary material, table S1. Primer sequences used for the synthesis of in situ hybridization riboprobes and in qRT-PCR experiments are given in the electronic supplementary material, table S3. Electronic supplementary material is available online [58].