Highlights
-
•
dsCellNet is a cell–cell interactions inference method for developmental series RNA-seq data.
-
•
dsCellNet respectively predicts interactions according to protein localizations.
-
•
dsCellNet adds dynamic time warping to construct cell–cell communication.
-
•
dsCellNet uses fuzzy clustering to refine key time points and connections.
Keywords: Single-cell RNA sequencing, Cell-cell communications, Developmental series analysis, Computational methods
Abstract
Cell-cell interactions mediated by ligand-receptor complexes are critical to coordinating organismal development and functions. Majority of studies focus on the key time point, the mediator cell types or the critical genes during organismal development or diseases. However, most existing methods are specifically designed for stationary paired samples, there hasn’t been a repository to deal with inferring cell–cell communications in developmental series RNA-seq data, which usually contains multiple developmental stages. Here we present a cell–cell interaction networks inference method and its application for developmental series RNA-seq data (termed dsCellNet) from the developing and aging brain. dsCellNet is implemented as an open-access R package on GitHub. It identifies interactions according to protein localizations and reinforces them via dynamic time warping within each time point and between adjacent time points, respectively. Then, fuzzy clustering of those interactions helps us refine key time points and connections. Compared to other published methods, our methods display high accuracy and high tolerance based on both simulated data and real data. Moreover, our methods can help identify the most active cell type and genes, which may provide a powerful tool to uncover the mechanisms underlying the organismal development and disease.
1. Introduction
Cell-cell communication across multiple cell types is critical to coordinating organismal development, differentiation and physiological function [1]. Diseases or homeostasis disorders occur when cells can’t interact with each other properly. Hence, more and more studies on development or diseases increasingly require consideration of cell–cell interactions in neuroscience field. Cell-cell communication is mediated by ligand-receptor (L-R) complexes, which encompass extracellular or membrane-ligand and receptor interactions. Through communication, receiver cells perceive the signaling then trigger downstream signaling generally altering transcription factor activity and gene expression in receiver cells [2]. Then those receiver cells finally work on the coordination of cellular activities through communication with other cells. Database of protein–protein interactions (PPIs) increasingly relies on techniques including yeast two-hybrid screening, co-immunoprecipitation and so on. These techniques have identified many interactions between proteins that are membrane or secreted/displayed extracellularly to mediate intercellular communication [1], [3]. Mapping these L-R interactions can help us to understand intercellular communication.
The increasing amount of single-cell RNA sequencing (scRNA-seq) further reinforces us to identify and characterize rare cell types within complex tissues which are limited in traditional techniques, and help in understanding their highly specialized functions [4], [5], [6], [7]. scRNA-seq can not only easily discover the hidden cell types and effectively measure their function, but also be used to infer potential interactions between two cell populations by mapping L-R interactions [6], [7]. In contrast to just focusing on the diseases, elucidating dynamic transcriptomic changes of cell compositions and cell interactions during the development of disease can decipher mediator genes in each stage which are inevitable in discovering how molecular processes underlie disease. Recent studies tried to capture continuous molecular processes over time, which are important for understanding disease or tissue development [8], [9], [10], [11]. But a single map of cell–cell interactions can’t provide a global picture of the dynamic interaction changes during development. Overall, with the exponential growth of studies focusing on the development of tissue or disease, the computational methods that decipher the dynamic interaction changes are lacking.
Recent studies used novel methods to infer potential interactions between two cell populations by mapping L-R interactions from scRNA-seq data [1], [12], [13], [14], [15], [16], [17]. There are four main types of methods. The first one infers interactions by a hypothetical test based on random cell-type label permutation. Only the interaction pairs that pass a certain threshold for expression level in respective cell populations are selected for the downstream analysis [12]. The second one identifies connections by calculating the product of receptor and ligand expression in the corresponding cell types and used a one-sided Wilcoxon rank-sum test to estimate the statistical significance of each interaction [13], [14]. The third one constructs interactions between cell populations by identifying significantly co-expressed ligand and receptor genes. The fourth one infers communications based on a tensor decomposition algorithm [15]. Nevertheless, compared with these published methods, there are still many problems to be solved. First, given the different expression profiles of membrane or secreted ligand and membrane receptor interactions, published methods cannot distinguish these two conditions. Second, published methods construct the cell–cell communications based on the expression of L-R genes in a stationary condition, without deciphering the communication changes during the process of development at the temporal scales. Hence, it is necessary to construct a novel computational method using developmental series scRNA-seq data to infer cell–cell interactions, which can not only filter false positive interactions but also identify key time points and mediatory interactions associated with the process of development or aging.
In this study, we developed a new computational method termed dsCellNet. We used known L-R pairs and public protein localization information to generate a multi-species supported membrane or secreted L-R database. Then, we constructed a more robust interaction network, via respectively calculating the significance of membrane or secreted L-R genes. We chose dynamic time warping (DTW) to calculate the correlation of those interactions to highlight the importance of the developmental series information. Finally, fuzzy clustering divided those candidate interactions into several clusters, which help us identify the key time point and the mediator cell types. Taken together, compared to the existing approaches, dsCellNet elucidates the dynamic interactions during the process of development and aging processes, which sheds light on understanding mechanisms underlying brain development and aging disorders.
2. Material and methods
2.1. Ligand-receptor interaction database
To expand upon the default L-R pair list, we incorporated pairs from published methods including: Ramilowski et al., 2015 [1] (2,557 pairs), CellPhoneDB [12], NATMI [13], and SingleCellSignalR [14]. However, users can also provide their own custom L-R pair list for further analysis.
The default L-R database is based on pairs between human/mouse ligands and receptors. To run dsCellNet on other organisms, the homologs interacting pairs can be automatically inferred based on the input gene data using homologene package (version 1.4.68).
To discriminate the secreted and plasma membrane ligand-mediated signaling, we required GO Cellular Compartment annotation “extracellular region” (GO:0005576), “extracellular space” (GO:0005615), “cell surface” (GO:0009986), and “plasma membrane” (GO:0005886) for ligands and receptors [18]. And just keeping those secreted or plasma membrane ligands and plasma membrane receptors used for further analysis.
2.2. dsCellNet implementation
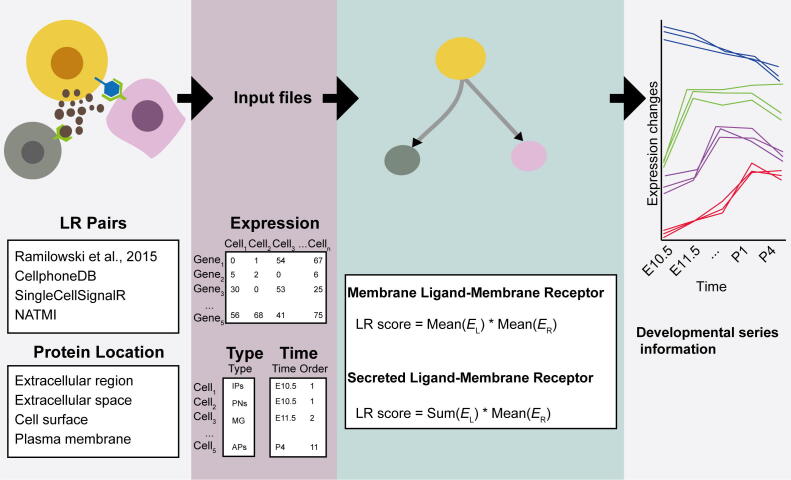
dsCellNet was implemented in R. It requires users to provide three files: a gene expression file with columns corresponding to cells and rows corresponding to genes, a cell type file with the mapping between every single cell and its’ cell type, and a time point file with mapping between every single cell and its’ time point. Input file examples are provided in Fig. 1 and the package documentation. For the gene expression file, users can just submit a raw count matrix but can also submit the preprocessed count matrix. The default data preprocessing includes: cells with less than 5,000 detected reads were excluded; the total number of reads in each single-cell was normalized by the total number of reads and then rescaled by multiplying 1,000,000; genes were retained with detected expression in more than 5 cells; cells with less than 500 detected genes were excluded.
Fig. 1.
dsCellNet workflow. dsCellNet requires three files (a single-cell expression file, a cell type file and a time points file), then respectively infers significant L-R links between cell types based on the subcellular localization of ligands and receptors, finally fuzzing clustering helps users to identify key time point, mediator cell types and key L-R pairs.
To infer cell–cell crosstalk between different cell types, we firstly construct a communication network for each time point. Given the different expression profiles of membrane or secreted ligands and membrane receptor interactions, we construct the communication network by firstly splitting the ligand-receptor (L-R) genes into membrane-ligand, secreted-ligand and membrane-receptor. Let be the weight of membrane-ligands and membrane-receptors mediated interactions between cell type i and cell type j. Let be the expression of L-R gene k in cell type i. Let be the expression of gene k in cell type j. Then the weight is defined as.
| (1) |
The weight of secreted-ligand and membrane-receptor mediated interaction between cell type i and cell type k is defined as.
| (2) |
Finally, we calculated the p value of enrichment of the interactions between two cell types based on random shuffling the cell type labels 100 times. Only the significant interactions (p < 0.05) were kept for constructing communication network. The direction of each interaction is defined as from the cell type expressing the ligand to the cell type expressing the receptor. The width of each edge connecting two cell types represents the number of interactions. The cell–cell communication network was visualized by Cytoscape (version 3.7.1).
Then, to identify cell–cell interactions during development, we calculated the correlation of those significant L-R genes’ developmental series expression using DTW [19]. Let D(X, Y) be the minimum DTW distance between time series X and Y. Let be distance of the optimal alignment between time series X and Y. So the minimum DTW distance is defined as.
| (3) |
Compared with using common methods like Euclidean distance to infer the correlation of developmental series expression between ligand and receptor genes, DTW can be used to dynamically compare them when the time indices between comparison data points do not sync up perfectly which is very useful for scRNA-seq data. As is well known, there is a vast proportion of zeros in scRNA-seq data which may be one of the inherent noises contributed to the lower correlation. So we used tseries package (version 0.10–48) to infer the correlation of developmental series expression between ligand and receptor genes. Only L-R pairs significantly correlated between two developmental series were kept for constructing developmental cell–cell communication networks.
To visualize the feature pattern of developmental cell–cell interactions, we performed fuzzy clustering [20], [21] to maximize the ligand or receptor genes’ developmental series expression between cluster distances which can classify those retained genes into different clusters using Mfuzz packages (version 2.50.0). Fuzzy clustering is to compute membership values to minimize the within-cluster distances and maximize the between-cluster distances. Let be the degree to which xj belongs to cluster ci. Let ai is the center of cluster i. Let be the degree of fuzziness. So the fuzzy clustering correlation [22] is defined as.
| (4) |
Besides, in contrast to commonly used hard clustering such as k-means clustering, fuzzy clustering has the advantage of obtaining gradual membership values which allows us to find co-expressed genes and more noise robustness. For this study, we chose eight as the default cluster number. After filtering the membership values less than 0.6 and L-R genes clustered in different clusters, we get the final clusters. Based on the retained genes’ feature pattern, we then obtain the expression trend during development, so that we can find the key time point or genes.
To evaluate the role of each cell type in interaction networks, we used incoming, outgoing and total edge numbers for each cell type per time point. Incoming of one cell type counts the number of L-R pairs sending from this cell type to other cell types. Outgoing of one cell type estimates the number of L-R pairs receiving from other cell types to this cell type. The total edge number of one cell type counts the number of L-R pairs connecting this cell type with other cell types.
2.3. Data preparation
Different single-cell datasets were used to illustrate and benchmark dsCellNet. To demonstrate the efficacy in the inference of cell–cell interactions using developmental series scRNA-seq data, we downloaded scRNA-seq data over the entire period of corticogenesis (Gene Expression Omnibus (GEO): GSE153164 [10]; including embryonic day (E)10.5, E11.5, E12.5, E13.5, E14.5, E17.5, E18.5, P1 and P4). Besides, to illustrate the advantage of dsCellNet in identifying more confident cell–cell interactions, we picked expression data of E15.5 single cells to compare the result of different L-R expression calculations. In order to test the running time, we subsample those cells into 2,000 to 10,000, each with three replicates. Moreover, dsCellNet can also deal with two groups of scRNA-seq data. To illustrate this, we used scRNA-seq data from the human brain of late-stage Alzheimer’s disease and healthy control from GEO: GSE174367 [23].
2.4. Functional enrichment analysis
To analyze the function of L-R genes during development, we carried out the gene ontology (GO) enrichment analysis using clusterProfiler (version 3.10.0).
3. Results
3.1. Overview of dsCellNet workflow
To trace the changes of cell–cell interactions during development, we developed a new computational method termed dsCellNet. Users can provide their data files of gene expression, cell type labels, and time point information to construct developmental series cell–cell networks. Firstly, we collected known L-R pairs and public protein localization information to make a multi-species supported membrane or secreted L-R database. Then, through respectively calculating the significance of membrane or secreted L-R genes, we constructed a more robust interaction network compared to previous methods. Finally, DTW and fuzzy clustering divided those candidate interactions into several clusters (Fig. 1). So, based on dsCellNet method, users can not only infer interactions but can also identify the key time point and the mediator cell types in the progress of divergent biological processes.
3.2. Default ligand-receptor pair list
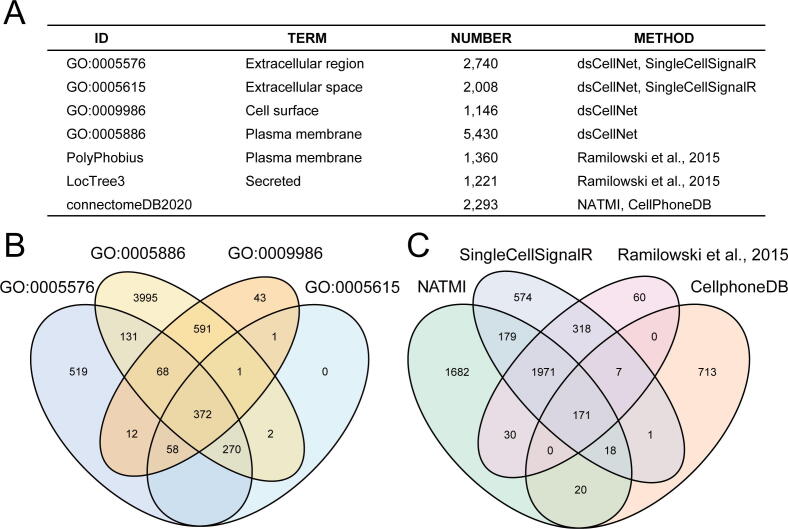
To respectively calculate the secreted or cell membrane ligand-mediated signaling, we collected secreted and localized to the membrane protein (Fig. 2A and 2B). We incorporated pairs from published methods including Ramilowski et al., 2015 (2,557 pairs), CellPhoneDB (1,396 pairs), NATMI (4,071 pairs), and SingleCellSignalR (3,251 pairs) to expand the default L-R pair list (Fig. 2C). In total, 5,744 interactions were kept for the default list of L-R pairs. We annotated genes of the default list as selected subcellular localization (secreted, plasma membrane). Additionally, users can also provide their own custom L-R pair list for further analysis. To facilitate running dsCellNet on other species, the L-R pairs can be inferred based on the default human/mouse homologs using homologene package.
Fig. 2.
dsCellNet database. (A) Subcellular localization of protein database in published methods. (B) Overlap of protein localization database using in dsCellNet. (C) Overlap of Ligand Receptor database in published methods.
3.3. Impact of protein localizations in cell–cell interactions
To compare the performance of dsCellNet which respectively predicts interactions according to protein localizations with other published methods in inferring cell–cell interactions, we selected the expression data of 11,670 single cells from E15.5 mouse embryonic cerebral cortex to compare the results of cell–cell crosstalk. It seems that inferring significantly enriched L-R interactions using the product of sum expression of L-R pairs captured the least but the highest number of interactions which did not include interactions between the least cell type, such as endothelial cells (ECs), oligodendrocyte precursor cells (OPCs), and other cell types. Inferring significantly enriched L-R interactions based on the product of mean expression of L-R pairs can capture the most but the lowest number of interactions. Significantly enriched L-R interactions predicted by dsCellNet which respectively predicts interactions according to protein localizations captured more general interactions which cover interactions among the least or most cell types (Fig. 3A). The single-cell atlas of E15.5 mouse embryonic cerebral cortex is composed of eight cell types with different proportions, and shows an especially high percentage of projection neurons (PNs) (59.89 %), especially low percentages of vascular and leptomeningeal cells (VLMCs) (0.30 %), microglia (MG) (0.24 %), OPCs (0.02 %) and ECs (0.01 %) (Fig. 3B).
Fig. 3.
Significant cell–cell interactions in Di Bella et al. dataset (GSE153164). (A) L-R pairs were inferred by product of sum ligand expression level and sum receptor expression level (left panel). L-R pairs were inferred by product of sum secreted ligand expression level or mean membrane ligand expression level and mean membrane receptor expression level (middle panel). L-R pairs were inferred by product of mean ligand expression level and mean receptor expression level (right panel). Rows indicate cells expressed ligands, columns indicate cells expressed receptors. Cell-cell interactions are labeled with the number of L-R pairs and also colored from white to orange. (B) Pie chart showing cell type composition in E15.5. Colors indicate different cell types. (C) Overview of the L-R pairs among chosen cell types (PNs, MG and VLMC). Rows represent L-R pairs, columns indicate cells expressed ligands to cells expressed receptors. Right color bars indicate ligand protein localizations.
All of those three methods can identify interactions between PNs and VLMCs, MG and apical progenitors (APs). When we look into the L-R genes in those interactions, the dsCellNet and other methods based on the product of mean expression of L-R pairs have quite similar performance in identification of the membrane-ligand and membrane-receptor interactions, but dsCellNet can capture more interactions between secreted-ligand and membrane-receptor. Methods inferring significantly enriched L-R interactions based on the product of sum expression of L-R pairs tended to identify more interactions in PNs with the highest proportion (Fig. 3C).
3.4. Prediction of cell–cell interactions compared with other approaches
In order to test running time of dsCellNet and other published methods (NATMI, CellPhoneDB, SingleCellSignalR) with default parameters, we randomly subsampled single cells of E15.5 mouse embryonic cerebral cortex into 2,000, 4,000, 6,000, 8,000, 10,000 cells each with three replicates (Supplementary Fig. 1). All methods were conducted on a personal computer equipped with Intel® Xeon® Gold 5218 CPU @ 2.30 GHz, 125G memory and 101G swap memory. We found SingleCellSignalR and CellPhoneDB tend to take a significantly longer time to finish compared with NATMI and dsCellNet dealing with the same single cells (Wilcoxon test, p less than 0.01; Supplementary Fig. 1). SingleCellSignalR takes up to 2 ∼ 3 h to finish for samples with 10,000 single cells. Among the top two performing methods, NATMI takes the least time to finish. But dsCellNet only requires 12 min for 10,000 single cells, this makes it an acceptable choice for constructing developmental series cell–cell interactions (Supplementary Fig. 1). It should be noted that we also used dsCellNet to deal with up to 98,047 single-cells in this study.
We next compared our results with those obtained by other published methods (NATMI, CellPhoneDB, SingleCellSignalR) using expression data of single cells from E15.5 mammalian cerebral cortex (Supplementary Fig. 2). dsCellNet, CellPhoneDB, and NATMI can capture the interactions among eight cell types, while SingleCellSignalR can only capture interactions among six cell types. SingleCellSignalR ignored the interactions between ECs and other cell types which seem to be important in other three methods. Moreover, SingleCellSignalR cannot distinguish the autocrine interactions with the default parameters. The interactions inferred by dsCellNet and NATMI were quite similar. By comparing the performance of dsCellNet and other existing approaches, each approach generates a different view of cell–cell communication networks, users need to consider these differences when interpreting their communication networks. The general characteristics of dsCellNet and other tools are shown in Table 1.
Table 1.
Comparison of tools for measuring cell–cell communication.
| Tool | Method | Multiple species | Language | Direction | Network compare | LR database size | Time-series data |
|---|---|---|---|---|---|---|---|
| CellPhoneDB | simulated p value | No | Python | No | No | 1,396 | No |
| SingleCellSignalR | regularized product | Yes | R | Yes | No | 3,251 | No |
| NATMI | cell specificity weight | Yes | Python | Yes | Yes | 4,071 | No |
| dsCellNet | regularized product | Yes | R | Yes | Yes | 5,684 | Yes |
3.5. Application of dsCellNet to developmental series single-cell RNA-seq dataset
The cell type composition during the entire period of corticogenesis trends to have four different stages: E10.5-E12.5; E12.5-E14.5; E14.5-E17.5; E17.5-P4, which is corresponding to the four developmental stages (symmetrically dividing neuroepithelial cells, birth date of the deep layer 6 and 5 excitatory neurons, the birth date of the superficial layer 4 and 2/3 excitatory neurons and gliogenesis) (Fig. 4A). We used expression, cell types and time point information to infer cell–cell interaction network (Supplementary Fig. 3A; Supplementary Table 1). The incoming (Fig. 4B) and outgoing (Fig. 4C) numbers show that the APs are the most-communicating cell type in the early stage (E10.5-E12.5), while IPs are the most-communicating cell type in the middle stage (E12.5-E17.5). ECs, OPCs, astrocytes (ASCs), and PNs interacted with other cell types in the later stages (E18.5-P4).
Fig. 4.
Changing trend of cell–cell interactions in the developing cortex. (A) Flow diagram displaying changing trend of the proportion of each cell types across the entire period of corticogenesis. (B) Heatmap view of the number of L-R pairs directed to each cell type. (C) Heatmap view of the number of L-R pairs released from each cell type. Row indicates each cell type, column indicates each time point. (D) Line chart showing the expression changes of Cluster 1 (upper panel), Cluster 4 (middle panel) and Cluster 5 (lower panel) along the corticogenesis stage. (E) Network-graph view of Cluster 1 (upper left panel), Cluster 4 (middle left panel) and Cluster 5 (lower left panel). Arrows indicate target cell type and thickness is the number of L-R pairs between the two cell types (the number range from 1 to 240, edge range from 0.3 to 6). Stacked histogram showing cell type composition in ligand or receptor of Cluster 1 (upper right panel), Cluster 4 (middle right panel), and Cluster 5 (lower right panel). Representative gene ontology (GO) terms enriched in L-R pairs of Cluster 1 (F), common in Cluster 1 and Cluster 5 (G), and Cluster 5 (H).
The expression changes of L-R pairs were clustered into eight distinct clusters using Mfuzz packages [21] (Supplementary Fig. 3B; Supplementary Table 1). Among those, cluster 1 represents L-R pairs that are highly expressed in E12.5, clusters 4, 5 and 6 represent L-R pairs that are highly expressed in the middle stage, whereas clusters 2, 3, 7 and 8 represent L-R pairs that are highly expressed in the later stage. We selected genes of cluster 1 (E12.5; 564 genes), cluster 4 (E15.5; 407 genes) and cluster 5 (E17.5; 306 genes) which are highly expressed in the early, middle and later stages to identify their interactions, respectively (Fig. 4D). The overall interactions are quantified in Fig. 4E, which shows APs sending most of the signals in cluster 1, intermediate progenitors (IPs) sending most of the signals in cluster 4, no cell types show dominated control in cluster 5. Functional enrichment analysis showed that interactions of cluster 1 significantly enriched for functions related to stem cell proliferation, stem cell division, and stem cell population maintenance (Fig. 4F), indicating those interactions may be involved in regulating the pluripotency of stem cell functions at E12.5. The overlapped functions between cluster 1 and cluster 5 mostly involved in axonogenesis, neuron projection guidance, and calcium ion homeostasis etc. (Fig. 4G). Interactions of cluster 5 significantly enriched in synapse maturation and cell polarity maintenance etc. (Fig. 4H).
3.6. Application of dsCellNet to two single-cell RNA-seq datasets
dsCellNet is not only restricted to construction of developmental series cell–cell interactions, but can also be applied to deal with network comparison between two groups. To illustrate this, we used scRNA-seq data from human brains of six late-stage Alzheimer’s disease patients and six healthy controls to infer cell–cell interaction network (Supplementary Fig. 4A and 4B; Supplementary Table 2). Interestingly, we found that compared with interactions of healthy individuals, same interactions with significantly higher frequency occurrence among the late-stage Alzheimer’s individuals (Supplementary Fig. 4C).
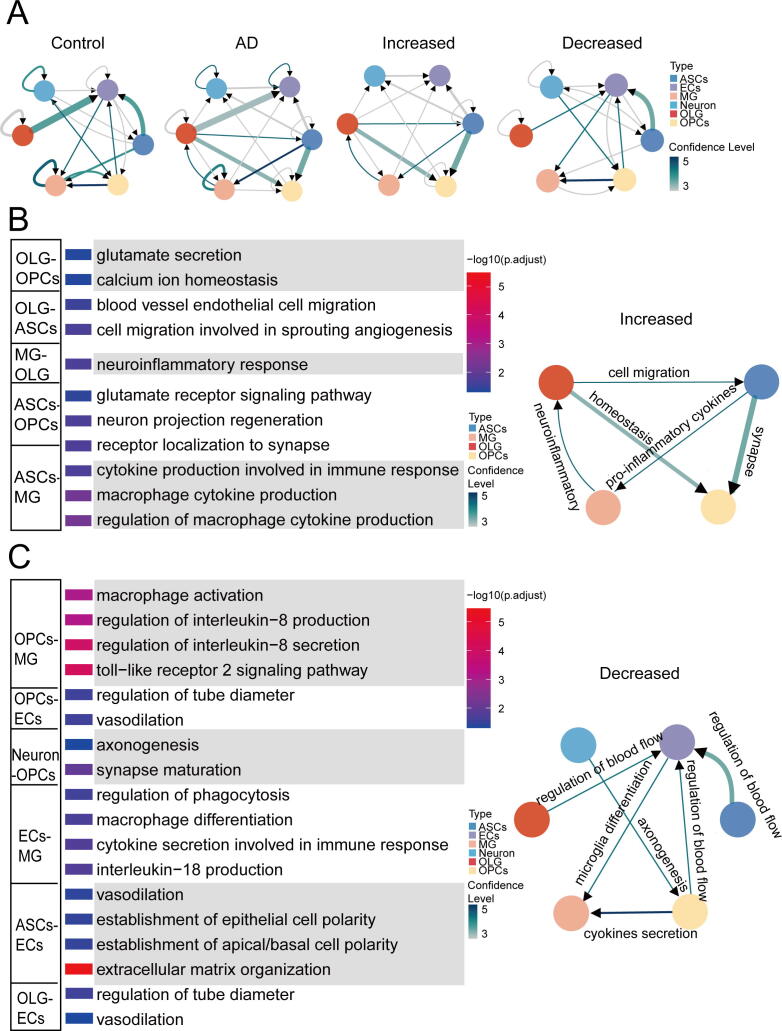
The incoming and outgoing numbers show that each cell type has a relatively similar pattern in sending or receiving signals between healthy controls and Alzheimer’s disease patients (Supplementary Fig. 4D and 4E). Through network extraction, we have identified common interactions in healthy controls, Alzheimer’s disease patients, increased or decreased in Alzheimer’s brains compared to healthy controls (Fig. 5A; Supplementary Table 2). To gain insight into the potential function of those interactions increased in Alzheimer’s brains, the GO enrichment demonstrates MG and ASCs are the primary immune cells that contribute to neuroinflammation which involves cytokine production and glutamate secretion in Alzheimer’s disease (Fig. 5B). Moreover, ASCs induce the function of synapse in OPCs (Fig. 5B). To assess the potential function of interactions decreased in Alzheimer’s brains, we found decreased anti-inflammatory cytokines such as IL8, IL18 are secreted or produced and phagocytosis in MG (Fig. 5C). Consistent with previous studies, functions of ECs are related to the regulation of blood flow in healthy individuals are impaired in AD patients (Fig. 5C). More interestingly, we identified some important receptor genes upregulated such as NRP1, CX3CR1, TLR1 and TLR2 in MG of Alzheimer’s brains, indicating the activation of MG in Alzheimer’s disease.
Fig. 5.
Cell-cell interactions in the human brain with late-stage Alzheimer’s disease and healthy control. (A) Network-graph view of common interactions in healthy control (Control), common interactions in Alzheimer’s disease (AD), increased interactions in Alzheimer’s disease (Increased) and decreased interactions in Alzheimer’s disease (Decreased). Arrows indicate target cell type and thickness is the number of L-R pairs between the two cell types (the number range from 1 to 8, edge range from 0.68 to 4.02). Colors represent confidence level (the number of individuals in which each interaction significantly detected). (B) The enriched gene ontology terms of increased interactions in Alzheimer’s disease (left panel). Summary of functions of cell–cell interactions (at least found in 3 individuals; right panel). (C) The enriched gene ontology terms of decreased interactions in Alzheimer’s disease (left panel). Summary of functions of cell–cell interactions (at least found in 3 individuals; right panel).
4. Discussion
Here, we present dsCellNet, a tool to infer cell–cell interaction networks using developmental series RNA-seq data. First, we demonstrate the workflow of the dsCellNet and compare the effect of dsCellNet which respectively predicts interactions according to protein localizations with other published methods. We found that inferring significantly enriched L-R interactions using the product of sum expression of L-R pairs captured the least but the highest interactions (Fig. 3A). Furthermore, it cannot identify interactions in the least cell type such as ECs and OPCs. Inferring significantly enriched L-R interactions based on the product of mean expression of L-R pairs identified the most but the lowest interactions (Fig. 3A). Second, methods inferring significantly enriched L-R interactions based on the product of mean expression of L-R pairs and dsCellNet didn’t show a bias on the proportion of cell types. So we recommend using dsCellNet or methods inferring significantly enriched L-R interactions based on the product of mean expression of L-R pairs to infer cell–cell communications when dealing with the rare cell proportion.
To help identify the different L-R genes in those interactions, we just focus on three interactions including PNs-VLMCs, PNs-MG, and PNs-PNs, which include the highest cell proportion and two lower cell proportions [10], [24] at E15.5. dsCellNet and methods based on the product of mean expression of L-R pairs have quite similar performance in membrane-ligand and membrane-receptor, but dsCellNet can infer more interactions in secreted-ligand and membrane-receptor (Fig. 3C). As expected, methods inferring significantly enriched receptor-ligand interactions based on the product of sum expression of L-R pairs have a bias on cell proportion. Hence, dsCellNet can be used to deal with inferring more robust cell–cell interactions.
For the first time, we used developmental series scRNAseq data which contains the entire period of corticogenesis (GSE153164) to discuss the changes of L-R interactions during brain development. Except for cell type composition, the incoming and outgoing edge number of L-R interactions were successfully captured in the early, middle and later stages of corticogenesis, respectively [10]. To identify the L-R pairs in those important stages, L-R pairs highly expressed in early, middle and later stages were obtained through fuzzy clustering. We can capture that APs sending most the signals in cluster 1 which coordinates with mainly APs had interactions with other cell types in early stage through incoming or outgoing signals. L-R genes of cluster 1 significantly enriched in regulating pluripotency of stem cell functions, which coordinates with the results that APs in E12.5 encode cues for neurogenesis [25]. IPs send most of the signals in cluster 4, which is consistent with mainly IPs interacted with other cell types in the middle stage through incoming or outgoing signals. No cell types show dominated control in cluster 5, also corresponding to the results using incoming or outgoing signals. Interactions of cluster 5 significantly enriched in maintenance of cell polarity, which correspond to the neuronal migration at E17.5 [26].
In addition, dsCellNet can also be used to identify changes in cell-to-cell communication by comparing networks using paired samples. Interactions were inferred by using scRNA-seq data from human brains of late-stage Alzheimer’s disease patients and healthy controls. Compared with healthy individuals, same interactions with higher frequency occurrence among late-stage Alzheimer’s disease patients, which indicates Alzheimer’s disease has a general effect on patients. Through network extracting, we found that MG and ASCs are the primary immune cells that contribute to neuroinflammation in Alzheimer’s disease. Anti-inflammatory cytokines such as IL8 and IL18 of Interleukin-1 (IL-1) family are secreted or produced in MG, which could contribute to neuroinflammation [27]. Hence, neuroinflammation may play an important role in the neurodegeneration process. While in healthy individuals, the regulation of cytokine production could prevent excessive neuroinflammation in healthy individuals. The opposite function of MG in late-stage Alzheimer’s disease patients or healthy individuals further suggests and confirms the pivotal roles of MG activation and polarization during the pathology of Alzheimer’s disease. Receptor genes such as NRP1, CX3CR1, and TLR1/2 were found increased in MG of Alzheimer’s brains compared with healthy controls. TLR1 and TLR 2 belong to Toll-like receptor (TLR) family that activates and mediates pro-inflammatory responses in innate immune cells. Nrp1 expresses on ameboid and activated microglia [28], once manipulation of Nrp1 in microglia can alter the immune polarization of microglia and the adaptive immune response [29]. Cx3cl1-Cx3cr1 interactions temper microglial response polarized to inflammatory stimuli [30]. We actually identified the interaction between MG and Neurons in each AD patient (Supplementary Fig. 4B). But when we look into the same interactions in control or AD patients, we can’t found the MG–Neurons interactions (Fig. 5A). This maybe owing to the limitation that the size of default L-R genes, users can support their L-R genes as supplementary. This also might the different ligand and receptor genes mediated interaction between them, which implies the immensely complex mechanisms among AD patients.
In sum, dsCellNet is a new computational tool to facilitate the analysis of cell-to-cell communication networks based on developmental series or paired scRNAseq data. Note that dsCellNet is not restricted to developmental series or paired scRNAseq data analysis, but can also apply to developmental series bulk RNA-seq data or proteomic data. The advantage of dsCellNet using single-cell data for building these networks is that inferring more robust interactions in secreted-ligand and membrane-receptor and rare cell proportion. Thus the cell–cell connection network for developmental series expression data will help users to highlight the key time points and the mediator cell types that are driving these interactions.
CRediT authorship contribution statement
Zhihong Song: Conceptualization, Methodology, Formal analysis, Data curation, Writing – original draft. Ting Wang: Methodology, Software, Validation. Yan Wu: Resources, Visualization. Ming Fan: Visualization, Supervision. Haitao Wu: Conceptualization, Visualization, Supervision, Project administration, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank all members of the Wu laboratory for discussion. This work was supported by the National Key Research and Development Program of China (Grants 2021ZD0202500 and 2021YFA1101801), the National Natural Science Foundation of China (32171148 and 31770929), and the Beijing Municipal Science and Technology Commission (Z181100001518001 and Z161100000216154) of China.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.07.047.
Contributor Information
Zhihong Song, Email: songzhihongcooll@163.com.
Haitao Wu, Email: wuht@bmi.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
No new data were generated for this study. All data analyzed within this paper are publicly available.
Code availability
dsCellNet is implemented as an open-access R package accessible to entry level users on Github (https://github.com/songzh523/dsCellNet).
References
- 1.Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., et al. A draft network of ligand–receptor-mediated multicellular signalling in human. Nat Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Shlomo I., Hsu S.Y., Rauch R., Kowalski H.W., Hsueh A.J.W. Signaling receptome: a genomic and evolutionary perspective of plasma membrane receptors involved in signal transduction. Sci STKE. 2003;2003 doi: 10.1126/stke.2003.187.re9. re9–re9. [DOI] [PubMed] [Google Scholar]
- 4.Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–271.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845.e20. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O’Brien S.A., et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459.e29. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 8.Zhong S., Zhang S., Fan X., Wu Q., Yan L., Dong J., et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–528. doi: 10.1038/nature25980. [DOI] [PubMed] [Google Scholar]
- 9.Yao J., Cui Q., Fan W., Ma Y., Chen Y., Liu T., et al. Single-cell transcriptomic analysis in a mouse model deciphers cell transition states in the multistep development of esophageal cancer. Nat Commun. 2020;11:3715. doi: 10.1038/s41467-020-17492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Bella D.J., Habibi E., Stickels R.R., Scalia G., Brown J., Yadollahpour P., et al. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature. 2021;595:554–559. doi: 10.1038/s41586-021-03670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen M., Giladi A., Gorki A.-D., Solodkin D.G., Zada M., Hladik A., et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. 2018;175:1031–1044.e18. doi: 10.1016/j.cell.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. Cell PhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 13.Hou R., Denisenko E., Ong H.T., Ramilowski J.A., Forrest A.R.R. Predicting cell-to-cell communication networks using NATMI. Nat Commun. 2020;11:5011. doi: 10.1038/s41467-020-18873-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabello-Aguilar S., Alame M., Kon-Sun-Tack F., Fau C., Lacroix M., Colinge J. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucl Acids Res. 2020;48 doi: 10.1093/nar/gkaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuyuzaki K., Ishii M., Nikaido I. Uncovering hypergraphs of cell-cell interaction from single cell RNA-sequencing data. BioRxiv. 2019;566182 doi: 10.1101/566182. [DOI] [Google Scholar]
- 16.Armingol E., Officer A., Harismendy O., Lewis N.E. Deciphering cell–cell interactions and communication from gene expression. Nat Rev Genet. 2021;22:71–88. doi: 10.1038/s41576-020-00292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.-H., et al. Inference and analysis of cell-cell communication using Cell Chat. Nat Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayamizu T.F., Baldock R.A., Ringwald M. Mouse anatomy ontologies: enhancements and tools for exploring and integrating biomedical data. Mamm Genome. 2015;26:422–430. doi: 10.1007/s00335-015-9584-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giorgino T. Computing and Visualizing Dynamic Time Warping Alignments in R: The dtw Package 2009. doi: 10.18637/JSS.V031.I07.
- 20.Futschik M., Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005 doi: 10.1142/S0219720005001375. [DOI] [PubMed] [Google Scholar]
- 21.Kumar L., Futschik E., Mfuzz M. A software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang M.-S. A survey of fuzzy clustering. Math Comput Modell. 1993;18:1–16. doi: 10.1016/0895-7177(93)90202-A. [DOI] [Google Scholar]
- 23.Morabito S., Miyoshi E., Michael N., Shahin S., Martini A.C., Head E., et al. Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat Genet. 2021;53:1143–1155. doi: 10.1038/s41588-021-00894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016;167:566–580.e19. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan X., Kang B., Qi C., Lin W., Wang J., Zhang X. Progenitor cell diversity in the developing mouse neocortex. Proc Natl Acad Sci. 2021;118 doi: 10.1073/pnas.2018866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molnár Z., Luhmann H.J., Kanold P.O. Transient cortical circuits match spontaneous and sensory driven activity during development. Science. 2020;370 doi: 10.1126/science.abb2153. eabb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arend W.P., Palmer G., Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 28.Sherafat A., Pfeiffer F., Reiss A.M., Wood W.M., Nishiyama A. Microglial neuropilin-1 promotes oligodendrocyte expansion during development and remyelination by trans-activating platelet-derived growth factor receptor. Nat Commun. 2021;12:2265. doi: 10.1038/s41467-021-22532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyauchi J.T., Caponegro M.D., Chen D., Choi M.K., Li M., Tsirka S.E. Deletion of neuropilin 1 from microglia or bone marrow-derived macrophages slows glioma progression. Cancer Res. 2018;78:685–694. doi: 10.1158/0008-5472.CAN-17-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickman S., Izzy S., Sen P., Morsett L., Khoury J.E. Microglia in neurodegeneration. Nat Neurosci. 2018;21:1359–1369. doi: 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated for this study. All data analyzed within this paper are publicly available.
Code availability
dsCellNet is implemented as an open-access R package accessible to entry level users on Github (https://github.com/songzh523/dsCellNet).