Abstract
During spliceosome assembly, the 3′ splice site is recognized by sequential U2AF2 complexes, first with Splicing Factor 1 (SF1) and second by the SF3B1 subunit of the U2 small nuclear ribonuclear protein particle. The U2AF2–SF1 interface is well characterized, comprising a U2AF homology motif (UHM) of U2AF2 bound to a U2AF ligand motif (ULM) of SF1. However, the structure of the U2AF2–SF3B1 interface and its importance for pre-mRNA splicing are unknown. To address this knowledge gap, we determined the crystal structure of the U2AF2 UHM bound to a SF3B1 ULM site at 1.8-Å resolution. We discovered a distinctive trajectory of the SF3B1 ULM across the U2AF2 UHM surface, which differs from prior UHM/ULM structures and is expected to modulate the orientations of the full-length proteins. We established that the binding affinity of the U2AF2 UHM for the cocrystallized SF3B1 ULM rivals that of a nearly full-length U2AF2 protein for an N-terminal SF3B1 region. An additional SF3B6 subunit had no detectable effect on the U2AF2–SF3B1 binding affinities. We further showed that key residues at the U2AF2 UHM–SF3B1 ULM interface contribute to coimmunoprecipitation of the splicing factors. Moreover, disrupting the U2AF2–SF3B1 interface changed splicing of representative human transcripts. From analysis of genome-wide data, we found that many of the splice sites coregulated by U2AF2 and SF3B1 differ from those coregulated by U2AF2 and SF1. Taken together, these findings support distinct structural and functional roles for the U2AF2—SF1 and U2AF2—SF3B1 complexes during the pre-mRNA splicing process.
Keywords: structure–function, RNA splicing, protein–protein interaction, RNA-binding protein, crystal structure, X-ray crystallography, structural biology, U2AF65
Abbreviations: BPS, branch point sequence; DI, detained introns; HEK, human embryonic kidney; IP, immunoprecipitation; Py, polypyrimidine; snRNP, small nuclear ribonucleoprotein; UHM, U2AF homology motif; ULM, U2AF ligand motif; ITC, isothermal titration calorimetry; NCS, noncrystallographic symmetry
The spliceosome assembles on consensus splice site signals of the pre-mRNA in a series of ATP-dependent conformational transitions (reviewed in (1)). In the initial ATP-independent E-complex, the essential pre-mRNA splicing factor U2AF2 recognizes a polypyrimidine (Py) tract preceding the 3′ splice site (2, 3, 4) as a heterodimer with a U2AF1 small subunit, which contacts an AG at the splice site junction (5). First, U2AF2 forms a ternary complex with SF1 (6, 7), which in turn recognizes the branch point consensus sequence (BPS) (8). In the subsequent A-complex, U2AF2 recruits the U2 small nuclear ribonucleoprotein particle (snRNP) of the spliceosome to the 3′ splice site. At this stage, the SF3B1 spliceosome subunit of the U2 snRNP replaces SF1 in the U2AF2 complex (9, 10). Following several ATP-dependent conformational changes among the core snRNP particles and dissociation of U2AF2 (10, 11), the spliceosome ultimately achieves the activated BACT-complex. A final conformational change to the B∗-complex allows the first catalytic reaction of pre-mRNA splicing to generate the branched intron lariat.
This parts list of core spliceosome assemblies has been illuminated by recent cryo-electron microscopy (cryo-EM) structures of B, BACT, C, C∗, and intron-lariat spliceosomes (reviewed in (12)). Nevertheless, cryo-EM approaches have yet to resolve the early stages of 3′ splice site recognition, which is challenging due to a fleeting “dance” of transitions among low-molecular-mass subunits. A cryo-EM structure of a 5′ splice site in the E-like yeast spliceosome assembly revealed weak density that could not be reliably modeled as the U2AF2 and SF1 homologues (Mud2 and BBP) (13). Although SF3B1-containing structures of spliceosomes are available (14, 15, 16, 17, 18, 19), the U2AF2–SF3B1 complex has not been resolved. As such, the field’s structural understanding of U2AF2 and its partners remains limited to piecewise structures of the interacting domains.
A C-terminal “U2AF Homology Motif” (UHM) domain of U2AF2 binds a well-characterized “U2AF Ligand Motif” (ULM) adjoining a coiled-coil region of SF1 (20, 21). Such UHM family members are marked by an RNA recognition motif–like fold with specialized features for recognizing ULM proteins, as opposed to RNA (reviewed in (22)). SF1 appears to specifically bind to U2AF2, whereas an intrinsically unstructured, N-terminal region of SF3B1 contains five ULMs that have been shown to associate with various UHM-containing proteins, including U2AF2 (23, 24, 25) (Fig. 1A). Many of the UHM complexes with SF3B1 ULMs have been structurally characterized, including SPF45, RBM39, and TatSF1 (26, 27, 28). However, the structure of the U2AF2–SF3B1 complex and its relevance for pre-mRNA splicing is unknown.
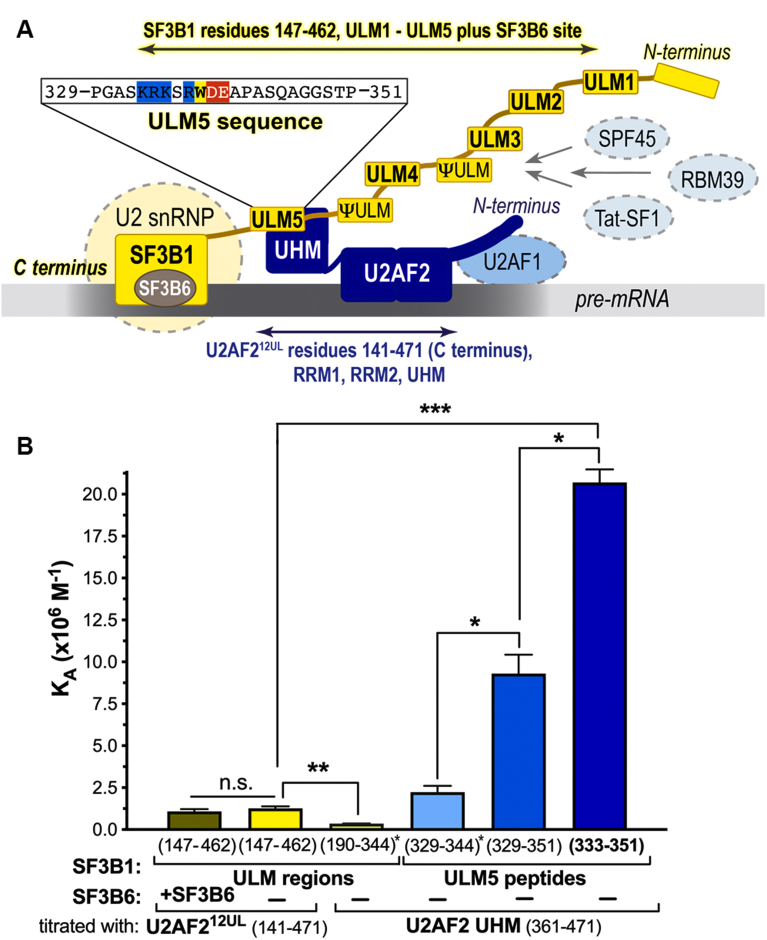
Figure 1.
SF3B1 ULM5 (residues 333–351) is a relevant, high-affinity ligand for the U2AF2 UHM.A, schematic diagram of the SF3B1 (yellow)–U2AF2 (navy) complex at the 3′ splice site (gray). The sequence of the ULM5 region is expanded above, with the ULM consensus marked in color for basic residues (blue), acidic residues (red), or tryptophan (yellow). Boundaries of regions used for isothermal titration calorimetry are indicated by double-headed arrows. B, bar graph of the average apparent binding affinities (KA) and standard deviations of three isothermal titration calorimetry experiments. The residue ranges of the SF3B1 fragment in the sample cell are given on the x-axis and the U2AF2 residues are listed below. SF3B6 (also called p14a) is included in the first experiment of the graph (taupe). Values reproduced from Thickman et al (2006) J. Mol. Biol. v356:664 are marked by asterisks. The thermodynamic values and representative isotherms are given in Table S1 and Fig. S1. Significance was assessed by unpaired t tests with Welch’s correction implemented in GraphPad Prism, here and for all figures: n.s., not significant; ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005.
Here, we determine the U2AF2 UHM–SF3B1 ULM5 crystal structure. The ULM5 ligand conformation diverges from prior UHM–ULM complexes, highlighting the importance of determining new structures in the UHM–ULM family. We find that the SF3B6 subunit has no detectable influence on the U2AF2–SF3B1 binding affinity. We demonstrate the functional importance of the U2AF2 UHM–SF3B1 ULM5 interface for association of these splicing factors and provide evidence for its functional contributions to splicing of pre-mRNA transcripts.
Results
SF3B1 ULM5 is a high-affinity binding site for U2AF2 that is independent of SF3B6
Among the SF3B1 ULMs, we previously showed that the fifth ULM (ULM5) has the highest binding affinity for the U2AF2 UHM (23). The SF3B1 ULM5 is next to the binding site for an SF3B6 subunit (also called p14a). Here, we investigated whether extending the SF3B1 region and adding the SF3B6 subunit would influence its binding affinity for U2AF2. In addition, we extended the boundaries of the U2AF2 construct to include the nearly full-length protein (U2AF212UL), except for an unstructured N-terminal region that binds U2AF1 (Fig. 1A). We characterized the interactions using isothermal titration calorimetry (ITC) and the same conditions as our prior experiments (23) (Table S1 and Fig. S1). The apparent equilibrium binding affinities increased subtly for the lengthened protein constructs (Fig. 1B). Including the SF3B6 subunit had no significant effect on the association of SF3B1 with U2AF212UL, which is consistent with NMR evidence for distinct binding sites of SF3B1 for SF3B6 or the U2AF2 UHM (24).
More notably, the apparent stoichiometry of the lengthened complex decreased to two U2AF212UL bound per SF3B1 ULM-containing region (Table S1), rather than three U2AF2 UHMs (23). Most likely, the smaller size of the U2AF2 UHM than U2AF212UL left room for a third molecule to associate with the SF3B1 ULMs. We note that the ability of excess U2AF212UL to concurrently bind more than one of the SF3B1 ULMs did not necessarily reflect the stoichiometry in the context of the assembling spliceosome, which is thought to contain a single U2AF2 and U2 snRNP per splice site (29).
We further compared the U2AF2 UHM binding affinities for SF3B1 ULM5 peptides with different boundaries. The SF3B1 peptides corresponding to the isolated ULM5 bound to U2AF2 with higher apparent affinity than the SF3B1 region containing all five ULMs, since this average apparent binding affinity included lower-affinity ULMs in addition to ULM5. Including a TP-motif, which is a phosphorylation site in human cells (30, 31), at the C terminus of ULM5 increased its binding affinity for the U2AF2 UHM by 4-fold, whereas extending the N terminus of ULM5 slightly decreased its U2AF2 UHM affinity by 2-fold (Figs. 1, S1 and Table S1). These results defined a high-affinity ULM5 region (residues 333–351) for cocrystallization with the U2AF2 UHM.
The SF3B1 ULM5 binds to the U2AF2 UHM in an atypical trajectory
To view the U2AF2 UHM–SF3B1 ULM5 interactions, we determined the crystal structure at 1.80-Å resolution (Fig. 2A and Table S2). The crystallographic asymmetric unit contained two similar copies of the U2AF2 UHM–SF3B1 ULM5 complex (RMSD 1.4 Å for 113 matching Cα atoms) (Fig. S2, A and B). The electron density maps revealed 12 or 9 ordered residues for the noncrystallographic symmetry (NCS)-related copies of UHM-bound SF3B1 ULM5 (Fig. S2C). The additional ordered residues append a C-terminal α-helical turn (residues 344–346) that makes crystallographic and NCS-related contacts with neighboring UHMs. Otherwise, the two different copies in the crystallographic asymmetric unit of U2AF2 bound to SF3B1 ULM5 shared nearly identical conformations.
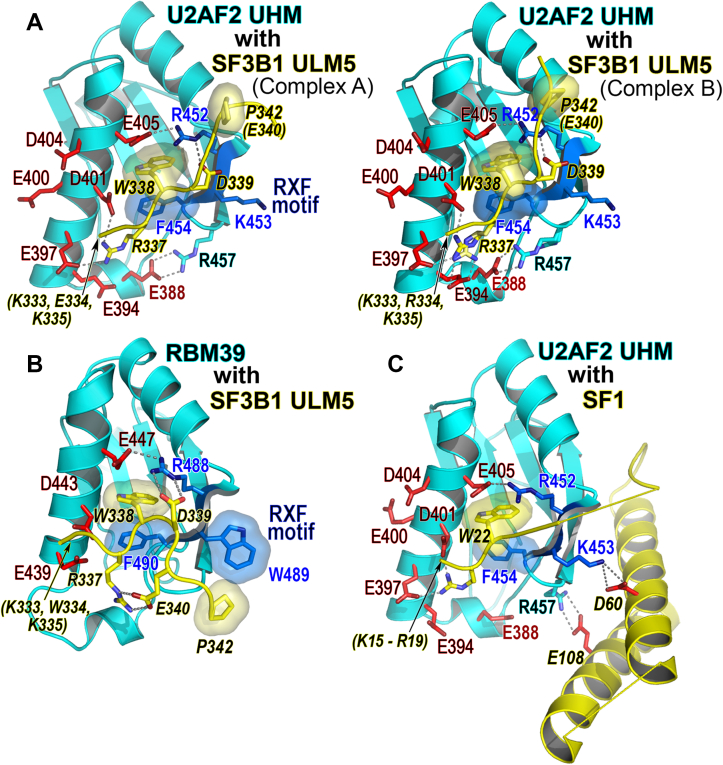
Figure 2.
Structure of the U2AF2 UHM bound to SF3B1 ULM5 and comparison with related complexes.A, views of the two U2AF2 UHM–SF3B1 ULM5 complexes (Complex A and Complex B) in the crystallographic asymmetric unit. The SF3B1 R337 and U2AF2 R457 side chains of complex B have two alternative conformations. B, RBM39 UHM–SF3B1 ULM5 (Protein Data Bank ID 4OZ1) or C, U2AF2 UHM–SF1 (Protein Data Bank ID 4FXW) complex viewed in a similar orientation. The UHM is colored cyan; the ULM ligand is yellow with italicized residue labels; key acidic UHM residues are red; residues in the UHM RXF motif are blue; and disordered residues are labeled in parentheses. The crystallographic data and refinement statistics are given in Table S2. Crystal packing and electron density maps are shown in Fig. S2.
The prominent, SF3B1 ULM5-interacting regions of the U2AF2 UHM were located in a characteristic RXF motif (F454, R452) and acidic α-helix (E405, E397) of the UHM family (22) (Fig. 2A), as compared for the RBM39 (also called CAPERα) UHM bound to SF3B1 ULM5 (27) (Fig. 2B). A central tryptophan (W338) of ULM5 inserted between two α-helices of the UHM. The W338 side chain was sandwiched in a T-type interaction between the F454 aromatic ring and an R452–E405 salt bridge. The R337 side chain further anchored the SF3B1 ULM5 by a distinct salt bridge with the U2AF2 UHM E397. In addition, disordered basic residues at the N terminus of the ULM5 were aligned for electrostatic attraction with the acidic UHM α-helix.
Other than these core features of UHM–ULM family members, both of the SF3B1 ULM5 copies followed unusual linear trajectories, running nearly parallel to the acidic α-helixes of the U2AF2 UHMs (Figs. 2A and S2C). Apart from three additional residues that were resolved at the C terminus of one ULM5 copy (complex B in Figs. 2 and S2), the U2AF2-bound ULMs did not appear to be involved in crystal packing contacts (Fig. S2A). The extended conformation of the SF3B1 ULM5 backbone was similar between the two NCS-related copies of the U2AF2 complex (RMSD 0.1 Å between 6 matching Cα atoms or 0.7 Å between all 55 matching atoms of the core ULM motif, Fig. S2B).
An SF3B1 TP-motif, which can be phosphorylated by cell-division kinases (30, 31), was positioned near an intramolecular R452–E405 salt bridge of the U2AF2 UHM. By contrast, SF3B1 ULMs typically bind other UHMs in a U-shaped conformation with the TP-motif packed against an exposed aromatic side chain from the central “X” position of the UHM RXF motif (e.g., W489 of RBM39 in Fig. 2B). A lysine (K453) at the “X” position of the U2AF2 RXF motif replaces the characteristic aromatic residue of other UHMs, where it may influence the trajectory of the U2AF2-bound ULM.
In the U2AF2 UHM complex with the SF1 factor that precedes SF3B1 during spliceosome assembly, the K453 residue of the U2AF2 RXF motif mediates a specific salt bridge with the SF1 ULM-coiled coil region (20, 21). SF1 lacks a TP-motif in the region following its ULM, which instead forms a disordered loop (Fig. 2C). The angle relative to the U2AF2 UHM α-helix is greater for the SF1 ULM polypeptide than for SF3B1 ULM. The distinct trajectory of the U2AF2-bound SF3B1 ULM5 appears to be anchored by a salt bridge between U2AF2 R452 and SF3B1 D339, which is replaced by an asparagine (N23) in the SF1 counterpart. Despite slightly different orientations, the extended backbone conformations of the U2AF2-bound SF1 and SF3B1 ULMs are similar (RMSD 0.5 Å between five matching Cα atoms of the SF1 ULM and SF3B1 ULM5). Like the U2AF2-bound SF3B1 ULM5 but distinct from other UHM–ULM complexes, the C-terminal residues of the SF1 ULM are located closer to R452 than to the central position of the RXF motif. This comparison suggests that contacts between the TP-motif of SF3B1 and the aromatic residue of other UHM RXF motifs contribute to the U-shaped ULM conformation, whereas the “RKF” of the U2AF2 UHM permits ULM ligands to adopt extended, near-linear conformations.
Interface mutations reduce U2AF2 UHM–SF3B1 ULM5 binding affinity
We probed key residues at the U2AF2 UHM–SF3B1 ULM interface by ITC of structure-guided mutant proteins (Figs. 3, S1 and Table S1). The amino acid substitutions are unlikely to perturb the overall protein folds since we targeted residues at the UHM surface and the ULM region itself is unstructured (23). First, we tested the relevance of U2AF2 UHM contacts with the C-terminal residues of the SF3B1 ULM5, which were affected by crystal contacts in one of the two copies (Fig. S2A). Since SF3B1 T341 has an intramolecular contact that appears to position M346 of complex A, we investigated a T341A/M346A double mutant (Fig. 3C). We also tested a glycine substitution for P342 that is expected to confer a flexible backbone without a side chain, in contrast with the wildtype proline. The U2AF2 UHM binding affinities of the T341A/M346A and P342G SF3B1 ULM5 mutants were similar to those of the wildtype (WT) counterpart (Fig. 3D), consistent with the variable positions of these residues in the two crystallographically independent copies of the complex.
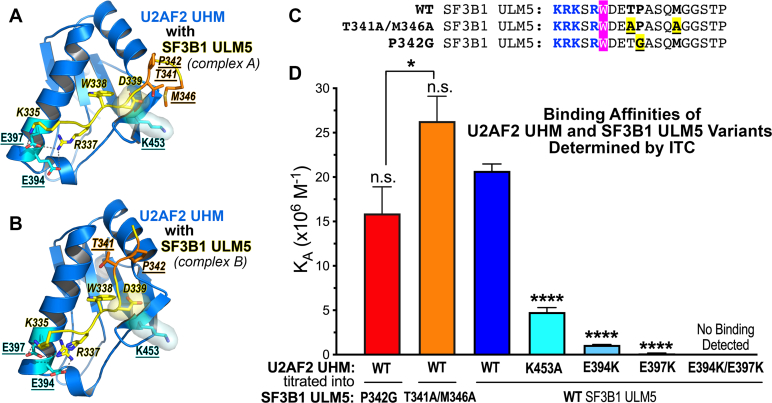
Figure 3.
Contribution of interface residues to the U2AF2 UHM–SF3B1 ULM5 binding affinity.A and B, interactions mediated in the two copies of the U2AF2 UHM (marine)–SF3B1 ULM5 (yellow, italicized labels) complex tested by structure-guided mutagenesis (underlined labels; U2AF2: cyan; SF3B1: orange). C, sequences of the SF3B1 ULM5 peptides (residues 333–351) showing the structure-guided mutants (yellow) tested by isothermal titration calorimetry. The central tryptophan is magenta and the basic residues are blue. D, bar graph of the average apparent binding affinities (KA) and standard deviations of three isothermal titration calorimetry experiments. The thermodynamic values and representative isotherms are given Table S1 and Fig. S1. The significance of the differences between the mutant and WT affinities are given, except where indicated by lines between the P342G and T341A/M346A pair; n.s., not significant, p > 0.05; ∗p < 0.05; ∗∗∗∗p < 0.0005.
Next, we tested a potential role for the singular U2AF2 K453 residue at the central position of the UHM RXF motif. As described above, this U2AF2 lysine differs from a typically aromatic side chain at the corresponding position of other UHMs and appeared to influence the bound ULM conformation (Fig. 2). The distinct trajectory of the U2AF2-bound ULM5 packed SF3B1 D339 against the hydrophobic portion of the U2AF2 K453 side chain (Fig. 3, A and B), which in turn positioned SF3B1 D339 to mediate a salt bridge with U2AF2 R452. Accordingly, replacing K453 with alanine reduced the binding affinity of the U2AF2 UHM–SF3B1 ULM5 complex by 4-fold (Fig. 3D and Table S1). This small, but statistically significant, change is consistent with loss of the observed D339–K453 contact and/or disruption of a weak ionic interaction between the side chains.
Lastly, we examined interactions between the canonical acidic α-helix of the UHM and basic residues of the N-terminal ULM tail (Fig. 3, A and B). An E397K mutation reduced the U2AF2 binding affinity for SF3B1 ULM5 by approximately 100-fold, consistent with disruption of an U2AF2 E397–SF3B1 R337 salt bridge that is present in both copies of the complex in the crystallographic asymmetric unit. A U2AF2 E394K mutation also penalized SF3B1 ULM5 binding, although to a lesser extent (by approximately 20-fold) in agreement with loss of an alternative E394–R337 salt bridge. Substituting lysines for both U2AF2 E394/E397 residues abolished detectable binding to the SF3B1 ULM5 (Fig. 3D and Table S1). The additive effect of the two mutations is expected to decrease the apparent dissociation constant (KD) approximately 100 μM. Although this value would exceed the limit for a reliable isotherm fit, the absence of any detectable heats for SF3B1 binding to the double U2AF2 mutant suggested that the E394/E397 interactions synergize to some extent. The undetectable E394K/E397K U2AF2 binding to SF3B1 ULM5, coupled with our prior observations that the central ULM tryptophan is required for detectable association of the purified U2AF2 UHM with SF3B1 proteins (23, 25), equipped us with structure-guided mutants to investigate the functional importance of the U2AF2–SF3B1 interface.
The UHM–ULM interface contributes to U2AF2–SF3B1 association in human cells
We investigated whether association of the full-length U2AF2 and SF3B1 proteins in coimmunoprecipitations from human cells (human embryonic kidney [HEK] 293T) relied on the UHM–ULM interface (Fig. 4). The N-terminally tagged constructs for U2AF2 (HA-tag, HAU2AF2) and SF3B1 (FLAG-tag, FLAGSF3B1) were transiently coexpressed in HEK 293T cells. The HAU2AF2-associated protein complexes were immunoprecipitated using anti-HA agarose beads. We found that wildtype FLAGSF3B1 efficiently associated with HAU2AF2 (Fig. 4B). The HAU2AF2 E394K/E397K mutation abolished detectable coimmunoprecipitation of FLAGSF3B1, consistent with the ability of this mutation to disrupt U2AF2 UHM binding to the SF3B1 ULM-containing region in ITC experiments.
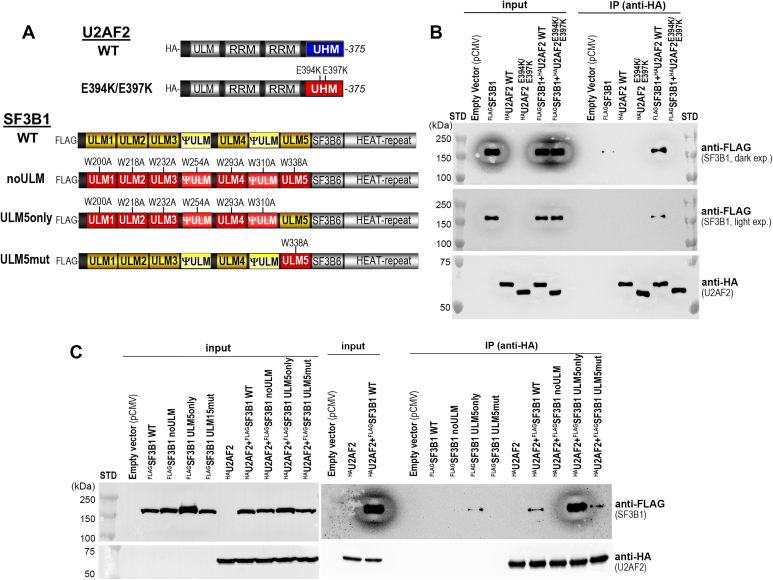
Figure 4.
The U2AF2 UHM contributes to association with SF3B1 in human cell extracts. Coimmunoprecipitation showing the amounts of FLAG-tagged SF3B1 (FLAGSF3B1, 148 kDa) retained by HA-tagged U2AF2 (HAU2AF2, 55 kDa) variants coexpressed in HEK 293T cells. Associations between the wildtype (WT) and structure-guided mutant proteins diagrammed in A were compared, including B, a E394K/E397K U2AF2 UHM-mutant, or C, SF3B1 ULM mutants. “Input” controls correspond to 0.5% and 1% of the total protein amounts used for the coimmunoprecipitations shown in B and C. STD, molecular size markers.
We next examined the effects of mutating the SF3B1 ULM tryptophans to alanine, which is known to prevent detectable U2AF2–SF3B1 binding in ITC experiments (23). The FLAGSF3B1 mutations either affected all ULMs and pseudo-ULMs (“noULM”), left only ULM5 intact (“ULM5only”), or disrupted only ULM5 (“ULM5mut”) (Fig. 4A). As expected, considering ITC results (23), the FLAGSF3B1 noULM variant no longer detectably coimmunoprecipitated with HAU2AF2 (Fig. 4C). Conversely, HAU2AF2 association with FLAGSF3B1 ULM5only appeared to significantly increase in the absence of the other intact ULMs/pseudo-ULMs. Since the ULM5only variant and wildtype SF3B1 ULM regions have similar apparent affinity for the U2AF2 UHM in ITC experiments with recombinant proteins, this enhanced interaction is likely due to disruption of SF3B1 sites for additional regulatory factors present in cells, such as other UHM-containing proteins that could normally occlude U2AF2 binding. Following mutation of only ULM5, the FLAGSF3B1 noULM5 protein continued to coimmunoprecipitate with similar amounts of HAU2AF2 as wildtype FLAGSF3B1, consistent with the ability of other SF3B1 ULMs to bind the U2AF2 UHM (23).
The U2AF2 UHM–SF3B1 ULM interface contributes to splicing of representative transcripts
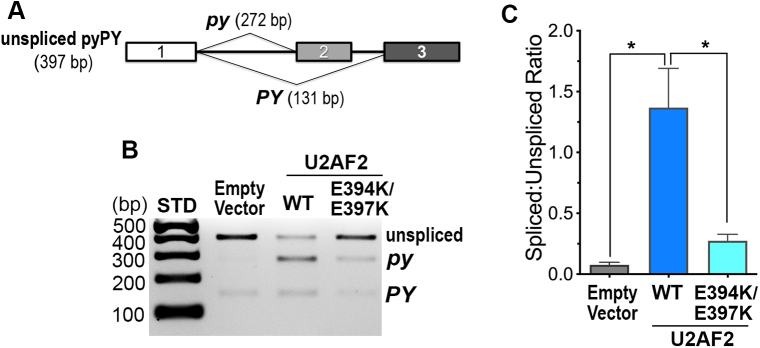
The UHM–ULM-dependent association of U2AF2 with SF3B1 suggested that this interface could contribute to the pre-mRNA splicing functions of these proteins. As a first step toward testing this hypothesis, we made use of a well-characterized, U2AF2-sensitive minigene comprising alternative 3′ splice sites (py and PY) (32, 33) (Fig. 5A), in combination with the E394K/E397K U2AF2 mutation that abolished detectable association with SF3B1. As described previously (33), HEK 293T cells stably expressing the pyPY minigene in our cell culture conditions produced mostly unspliced pyPY transcript as judged by reverse-transcription (RT)-PCR (Fig. 5B). Overexpression of wildtype U2AF2 increased splicing of the py site substantially and the PY splice site moderately as measured by RT-PCR and quantitative real-time (q)RT-PCR (Figs. 5, B and C and S3). By contrast, most of the pyPY transcript remained unspliced following overexpression of the E394K/E397K mutant U2AF2. A small increase in py splicing for E394K/E397K U2AF2 can be explained by mutational disruption of only the U2AF2 UHM, whereas the U2AF2 RNA-binding domain and RS domain for U2 snRNA/pre-mRNA annealing remained functional. Alternatively, U2AF2 regulates levels of other splicing factors (e.g., U2AF1 or SPF45 (32, 34)), which could indirectly increase py splicing. Regardless, the significant effects of the E394K/E397K mutation confirmed that the U2AF2 UHM is important for splicing the pyPY prototype.
Figure 5.
The U2AF2 UHM influences splicing of a prototypical minigene substrate.A, schematic diagram of the pyPY transcript. Alternatively spliced sites generate two differently sized products (py, 272 bp and PY, 131 bp). The unspliced pyPY RT-PCR product is 397 bp. B, representative RT-PCR of total RNA isolated from HEK 293T cells stably expressing the pyPY minigene and transfected with either wildtype (WT) or a double mutant U2AF2 (E394K/E397K). The RT-PCR products were resolved by agarose gel electrophoresis with ethidium bromide staining. C, quantification of pyPY RT-PCR products. The band intensities from three independent replicates were background corrected and averaged. The average ratios of the total spliced product (py + PY) to the unspliced pyPY and standard deviations are plotted for each sample. Primer sequences are listed in Table S3, and immunoblots are shown in Fig. S3. STD, 100 bp ladder molecular size markers. ∗p < 0.05.
In the next step, we asked whether SF3B1 and the SF3B1 ULMs contribute to alternative splicing of endogenous, U2AF2-responsive transcripts. We and others have shown that skipping of THYN1, SAT1, INTS13, and RNF10 exons is sensitive to reduced U2AF2 levels (33, 35, 36). Although we were unable to achieve robust rescue to test the effects of structure-guided U2AF2 variants (33), these precedents offered a means to compare the potential contributions by the SF3B1 ULMs to U2AF-sensitive splicing events.
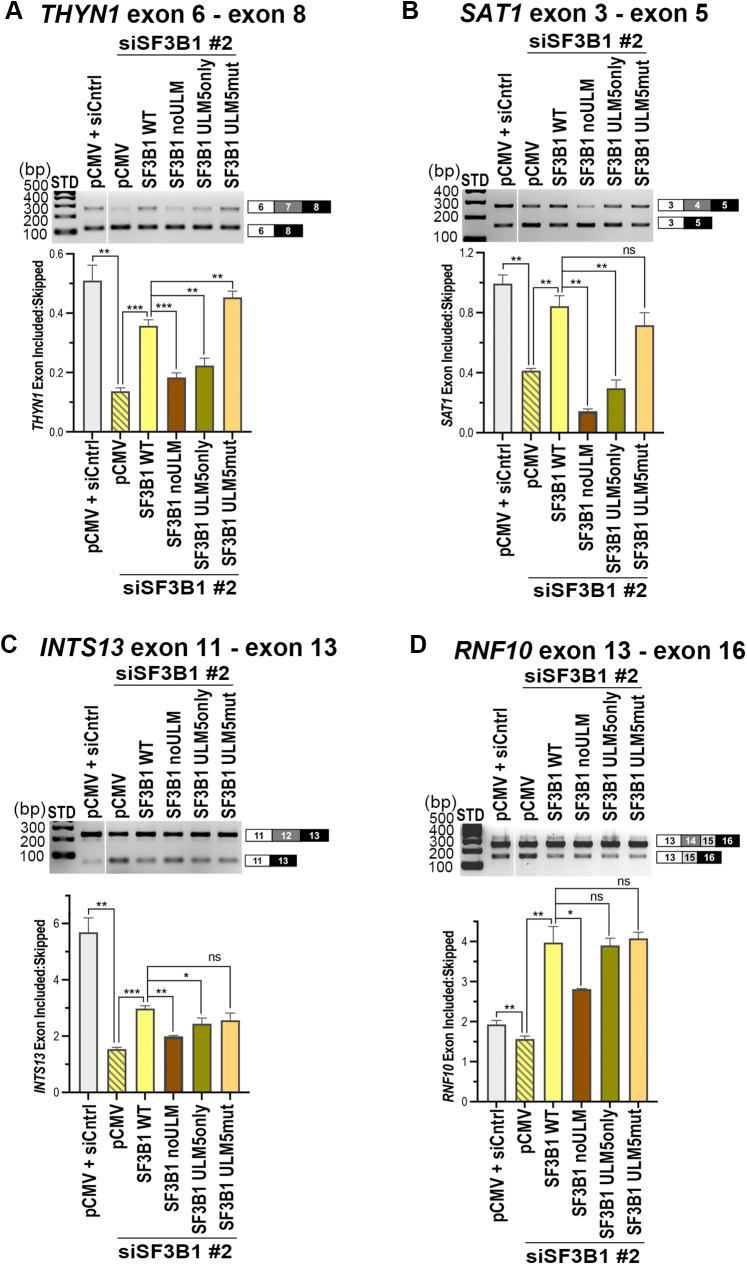
In support of a functional relationship between U2AF2 and SF3B1, the siRNA-mediated knockdown of SF3B1 levels (Fig. S4) increased exon-skipped splicing of representative transcripts in a similar manner as U2AF2 knockdown (Figs. 6 and 7). Reexpression of wildtype SF3B1 had converse effects, either partially rescuing splicing, or for RNF10, increasing the ratio of exon inclusion-to-skipping above native levels (Fig. 6). Disrupting all of the SF3B1 ULMs with tryptophan-to-alanine mutations severely penalized the ability of the SF3B1 noULM variant to enhance exon inclusion, in agreement with negligible binding of this SF3B1 mutant to UHM splicing factors (23, 37). Restoring only ULM5, the preferred binding site of U2AF2, partially restored splicing. SF3B1 ULM5mut, in which all ULMs except ULM5 were preserved, restored splicing to similar levels as WT SF3B1. This result is consistent with the ability of SF3B1 ULM5mut to bind and coimmunoprecipitate with U2AF2, yet leaves open the possibility of some ULM–UHM redundancy (e.g., a potential ability of PUF60 or SPF45 to substitute for U2AF2 (34, 38)). Altogether, these results demonstrate the importance of the U2AF2 UHM and SF3B1 ULMs for pre-mRNA splicing functions in cells.
Figure 6.
The SF3B1 ULMs contribute to accurate splicing of endogenous, U2AF2-responsive gene transcripts. The abilities of indicated SF3B1 ULM variants to rescue splicing of representative U2AF2-regulated transcripts, including A, THYN1; B, SAT1; C, INTS13; and D, RNF10, were compared by RT-PCR. The ethidium bromide–stained agarose gels are shown above. The ratios of exon-included:exon-skipped band intensities for background-corrected and averaged replicates were plotted below. Primer sequences are listed in Table S3, and immunoblots are shown in Fig. S4. STD, 100 bp ladder size standards. n.s., not significant; ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005.
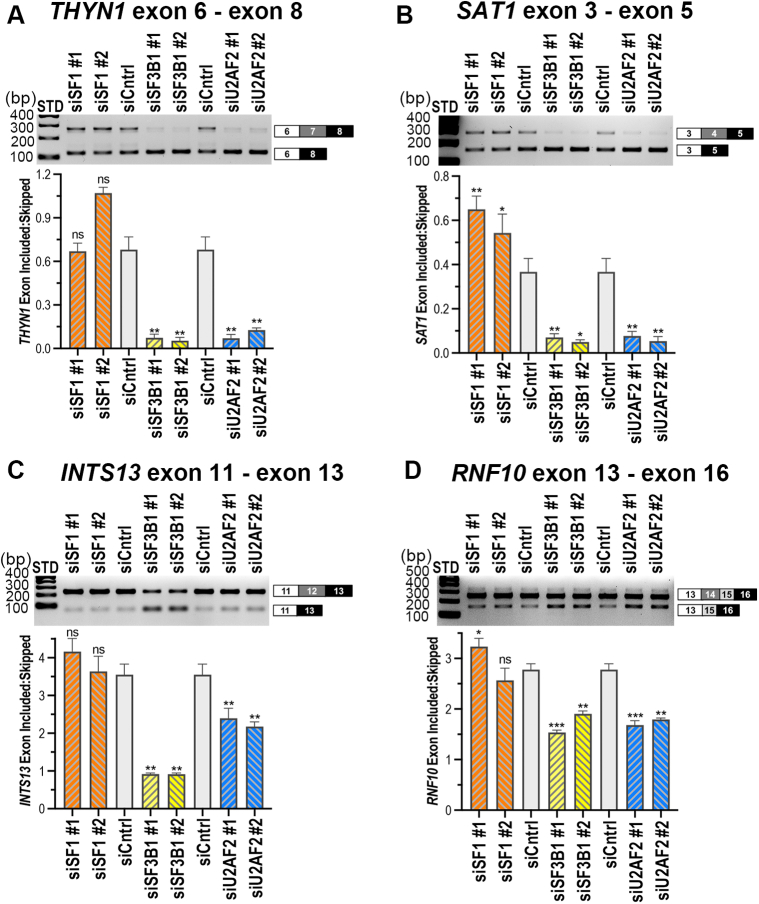
Figure 7.
Comparison of SF3B1 and SF1 modulation of representative U2AF2-responsive gene transcripts. The effects of siRNA-mediated reductions in SF1, SF3B1, or U2AF2 levels on splicing of representative U2AF2-regulated sites, including A, THYN1; B, SAT1; C, INTS13; and D, RNF10, were compared by RT-PCR as described for Figure 6. n.s., not significant; ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005.
SF3B1 and SF1 regulate splicing of U2AF2-sensitive transcripts that are mostly distinct
The U2AF2 UHM interacts with SF1 prior to SF3B1 during spliceosome assembly (6, 7, 9, 10), raising the possibility of some redundancy in the functions of SF1 and SF3B1 for pre-mRNA splice site selection. To distinguish potential overlapping functions of the two U2AF2 partners, we compared alternative splicing of the representative, U2AF2-responsive THYN1, SAT1, INTS13, and RNF10 transcripts following siRNA-mediated reduction of U2AF2, SF1, or SF3B1 (Figs. 7 and S4). In contrast with U2AF2 and SF3B1, SF1 had little or no significant effect on splicing of these transcripts, in agreement with the previously noted, selective requirement of SF1 for splicing of specific human transcripts in ex vivo and in vitro assays of representative pre-mRNA substrates (8, 39, 40).
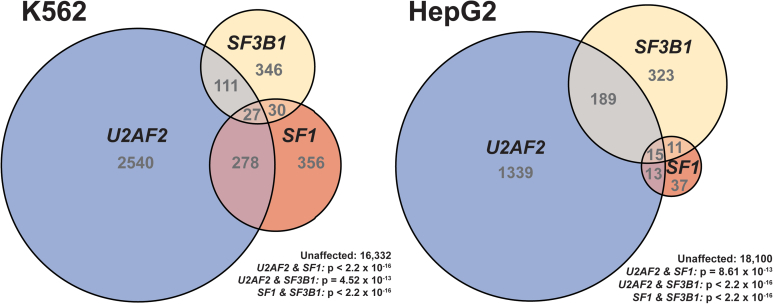
To more comprehensively examine an unbiased set of transcripts, we analyzed RNAseq datasets available from the ENCODE (encyclopedia of DNA elements) project (41, 42, 43) of U2AF2, SF3B1, and SF1 knockdown in K562 erythroid leukemia and HepG2 hepatocellular carcinoma cell lines (Fig. 8 and Table S4). We used STAR Aligner (44) and DESeq (45) with custom Python scripts to identify alternative splicing events as described (46, 47) (Experimental Procedures). We then used DEXseq (48) to quantify differential splicing.
Figure 8.
U2AF2, SF3B1, and SF1 coregulate a subset of transcripts genome-wide. Euler diagrams showing the number of overlapping and unique alternative splicing events that exhibited statistically significant changes upon RNAi knockdown of U2AF2, SF3B1, or SF1 relative to control shRNAs from the ENCODE project. Experiments performed in K562 erythroid leukemia cells are shown in the left diagram, and those in HepG2 hepatocellular carcinoma cells are on the right. The number of events belonging to each region is indicated. All pairwise comparisons showed significantly greater overlap than expected by chance. For each cell line, the total number of unaffected events assayed and the p-values for each overlap (Fisher’s exact test, one sided) are shown.
More than a thousand U2AF2-sensitive splicing events were identified for each cell line. A lesser but still substantial number of differential splicing events were identified for the SF3B1-knockdown samples. In contrast, SF1-associated differential splicing events were less frequent in HepG2, consistent with a conditional, kinetic role for SF1 in metazoan BPS selection (8, 39, 40, 49). A nearly 10-fold increase in the number of SF1-associated splicing changes in K562 cells compared with HepG2 further suggested that SF1-responsive alternative splicing is highly dependent on the cellular context. The modulated transcripts largely differed between SF3B1 and SF1 knockdowns for either cell line. Significant subsets of U2AF2-responsive transcripts also were regulated by either SF3B1 or SF1. With the notable exception of a subset of splicing events in K562 that are responsive to all three knockdowns, including almost 20% of SF3B1-responsive events, the SF3B1/U2AF2 or SF1/U2AF2-responsive subsets showed little overlap. These observations suggested that, for the majority of splicing events, SF3B1 and SF1 contribute differently to cellular pre-mRNA splicing and the splicing functions of U2AF2, such that some introns are more dependent on one factor or the other for efficient splicing.
Discussion
Here, we determined the crystal structure of a cognate U2AF2 UHM–SF3B1 ULM5 complex. We show that this interface is important for U2AF2–SF3B1 association in cell extracts and regulation of pre-mRNA splicing, whereas previous studies have been limited to the interactions of the purified protein domains.
Structurally, the U2AF2-bound SF3B1 ULM5 conformation is unique among UHM-bound SF3B1 ULM structures (Fig. 2). The similar conformations of two NCS-related complexes reinforced that the extended SF3B1 ULM5 conformation was not an artifact of crystal contacts. Instead, the conformation of the U2AF2-bound ULM appeared related to an atypical lysine (K453) at the central residue of the U2AF2 RXF motif, which is occupied by an aromatic residue in nearly all other UHMs (30, 31). Although lysines can be posttranslationally modified, no modifications of U2AF2 K453 have been documented to date. Instead, this K453 side chain appears to serve specific structural roles, for example, mediating a specific salt bridge in the U2AF2 complex with SF1 (Fig. 2C) (20). In the SF3B1 ULM5 complex with the U2AF2 UHM described here, the distinctive U2AF2 K453 packed with the SF3B1 D339 side chain, which in turn positioned D339 for a salt bridge with U2AF2 R452. Accordingly, a K453A mutation decreased the binding affinity of U2AF2 for the SF3B1 ULM5 (Fig. 3). In contrast with the U2AF2 complex, the SF3B1 ULMs bind other UHMs in a curved conformation that stacks a cyclin-dependent kinase–phosphorylation site of SF3B1 against the U2AF2 RXF motif (30, 31). These distinctive interactions raised the possibility that phosphorylation differently regulates SF3B1 association with U2AF2 compared with other UHM-containing partners.
We established that an intact UHM is necessary for U2AF2 to detectably associate with SF3B1 in human cell extracts (Fig. 4). This result is a refreshing confirmation that the long-known interaction between the purified U2AF2 UHM and the SF3B1 ULMs (23, 24, 25) is relevant for association of the full-length factors in cells. The high affinity of the purified U2AF2 UHM for a minimal SF3B1 ULM5 (Table S1) further reinforced the case for a cognate interaction between these regions. Nevertheless, the binding affinity of U2AF2 for SF3B1 is moderate compared with SF1 (50). Other factors are likely to enhance and regulate U2AF2 specificity for SF3B1 versus SF1 in the context of the full-length, spliceosome-associated proteins, including pre-mRNA interactions by the U2 snRNA and SF3B1 HEAT repeats, dynamic RNA unwindases, and kinases/phosphatases.
Our results show that the U2AF2 UHM is important for splicing of a minigene prototype and that the SF3B1 ULMs contribute to representative alternative pre-mRNA splicing events (Figs. 5 and 6). In principle, the consequences of disrupting the U2AF2 UHM and SF3B1 ULMs in these systems could result from interactions with other partners, e.g., SF1 for U2AF2 or other UHM-containing splicing factors such as SPF45 or Tat-SF1 for SF3B1 (22, 34). However, we note that roles for SF1, SPF45 or Tat-SF1 in pre-mRNA splicing are conditional and rare (8, 34, 40, 51). Indeed, reduced SF1 levels had little effect on splicing of the U2AF2-responsive splice sites examined here (Fig. 7). Moreover, the graded restoration of splicing following ablation of all ULMs, only ULM5, or all but ULM5 (Fig. 6) agreed with the preference of U2AF2 to bind SF3B1 ULM5 while retaining the capacity to bind other ULMs ((23) and Fig. 1). Therefore, we believe that the impact of the U2AF2 UHM and SF3B1 ULMs on pre-mRNA splicing observed here is likely to arise in part or full from disrupting the U2AF2–SF3B1 complex.
On a transcriptome-wide scale, we documented numerous U2AF2-responsive splicing events, in agreement with a central role for U2AF2 in identifying the major class of 3′ splice sites (52), as well as previous findings of ubiquitous U2AF2 CLIP-seq sites (53, 54). The lower, but still significant, number of SF3B1-responsive splicing events was consistent with decreased availability of a spliceosome subunit that targets the intronic BPS (55, 56), which could generally penalize splicing of sensitive introns or select an alternative BPS without detectably switching the splice site (e.g., (57)). We documented a similar number of SF1-responsive as SF3B1-responsive splicing events in K562 cells but very few SF1-responsive splicing events in HepG2 cells. This finding reinforces conditional and cell type–dependent roles for SF1 in cellular pre-mRNA splicing (8, 39, 40). A substantial subset of splice sites that are responsive to both U2AF2 and SF3B1 knockdown, which are largely separate from the sites sensitive to both U2AF2 and SF1 depletion, underscores a distinct, functional relationship between U2AF2 and SF3B1.
In summary, we have demonstrated that the UHM–ULM interface is important for U2AF2–SF3B1 association and provided evidence for its functional contribution to pre-mRNA splicing. The singular conformation of the U2AF2-bound SF3B1 ULM5 diversifies known modes of UHM–ULM interaction and suggests that phosphorylation of the SF3B1 TP motifs may regulate U2AF2 differently compared with other UHM-containing partners. Altogether, these results lay a groundwork for future expansions in our understanding of the structural and functional distinctions among UHM-containing proteins and their dynamic associations with the SF3B1 ULMs.
Experimental procedures
DNA constructs
All protein and peptide sequences correspond to the human homologues. The U2AF2 UHM construct was described previously (20, 23). The U2AF212UL construct includes residues 141 to the C terminus (residue 471) of NCBI RefSeq NP_009210. The SFB1147-462 construct includes residues 147 to 462 of NCBI RefSeq NP_036565. The full-length SF3B6 construct matches to NCBI RefSeq NP_057131. For transfections, the plasmids encoding full-length U2AF2 and SF3B1 were described previously (28, 33, 58). Structure-guided mutations were introduced by Genscript.
Preparation of purified proteins
All proteins were expressed using the pGEX-6p vector, purified by glutathione affinity, proteolytic cleavage to remove the GST-tag, ion exchange, and a final size exclusion chromatography step in 50 mM NaCl, 25 mM Hepes pH 7.4, 0.2 mM TCEP through a Superdex-75 column (Cytiva). Synthetic peptides were purchased with >98% purity (Biomatik Corp).
Isothermal titration calorimetry
A MicroCal VP-ITC (Malvern Panalytical) was used to inject 28 aliquots of 10 μl each at a rate of 2 s μl−1 separated by a 4-min relaxation time into the sample cell. The experiments were run at 30 °C, 15 μcal s−1 reference power, and with constant stirring at 307 rpm. Concentrations were typically 5 μM SF3B1 in the sample cell and 50 to 100 μM U2AF2 in the syringe. Most isotherms were corrected for the heats of dilution by subtracting the last three data points of the saturated region, then fit using Origin v7.0 (Malvern). To account for the very low SF3B1 ULM5 binding affinities, the concentrations used for the U2AF2 mutants were approximately 20 μM SF3B1 ULM5 in the sample cell/200 μM U2AF2 UHM in the syringe, and the isotherms of the E397K variant were corrected by subtracting the average heat from a titration of the E397K U2AF2 UHM into buffer. The ITC results are detailed in Table S1 and isotherms are shown in Fig. S1.
Crystallization and structure determination
The U2AF2 UHM–SF3B1 ULM5 complex was crystallized by the hanging drop vapor diffusion method at 4 °C from 20.5 mg/ml U2AF2 UHM in the presence of a 1.5-fold molar excess of SF3b155 ULM5 peptide. The reservoir solution contained 0.1 M sodium citrate tribasic dihydrate pH 5.6, 15% v/v 2-propanol, 20% v/v PEG 4000. For cryoprotection, crystals were sequentially transferred to reservoir solution supplemented with 10% v/v glycerol, then flash cooled in liquid nitrogen. Crystallographic data sets at 100 K were collected by remote data collection at the Stanford Synchrotron Radiation Light (SSRL) source Beamline 12-2 (59). The data were processed using the SSRL AUTOXDS script (A. Gonzalez and Y. Tsai), which implements XDS (60) and CCP4 packages (61). The structure was determined by molecular replacement using Phaser (62) with the U2AF2 UHM from the SF1 complex (Protein Data Bank ID: 4FXW) as the search model (20). A top solution with LLG of 1253 and a top TFZ-score of 26.7 showed clear electron density for the SF3B1 peptide bound to both copies of the U2AF2 UHM in the crystallographic asymmetric unit (feature-enhanced electron density maps (63) are shown in Fig. S2). The structure was refined in Phenix.refine (64) and manually adjusted in Coot (65). The crystallographic data collection and refinement statistics are reported in Table S2.
Cell culture and transfections
Human embryonic kidney epithelial cells (HEK 293T, ATCC CRL-3216) were maintained at 37 °C in a humidified atmosphere containing 5% CO2 as described (33). Cells were transfected in six-well plates at 50 to 60% confluency with the indicated siRNAs and/or DNA plasmids, using jetPRIME (Polyplus-transfection SA) as instructed by the manufacturer. For experiments with the pyPY minigene, a stable 293T cell line expressing the pyPY transcript was transfected with plasmids expressing U2AF2 variants, then cells were harvested 24 h after transfection as described (33). For knockdown experiments, cells were transfected with 25 nM of Stealth siRNAs (Thermo Fisher Scientific), targeting either U2AF2 (catalog nos. HSS117616, HSS117617), SF1 (catalog nos. HSS187735, HSS144483), SF3B1 (catalog nos. HSS146413, HSS146415), or a “Lo GC” control (catalog no. 12935200) and harvested 2 days after transfection. For “rescue” experiments with the SF3B1 variants, the samples were harvested 2 days after cotransfection of the siRNAs and plasmid DNAs. Immunoblots of protein expression levels are provided in Figs. S3 and S4.
Coimmunoprecipitation
HEK 293T cells were transfected in 10-cm plates with combinations of wildtype or mutated plasmids encoding HA-tagged U2AF2, FLAG-tagged SF3B1, or empty vector control (pCMV5-XL6). Since the ULM mutations appeared to alter FLAGSF3B1 expression, the amounts of transfected SF3B1 constructs and empty control vector were adjusted to achieve similar levels of FLAGSF3B1 variants among the coimmunoprecipitation inputs, while maintaining equivalent amounts of total transfected DNA. After 24 h, cells were harvested and lysed in immunoprecipitation (IP) buffer (50 mM Tris pH 8.0, 75 mM NaCl, 5% v/v glycerol, 10 mM CaCl2, cOmplete EDTA-free protease inhibitor [Sigma-Aldrich], β-glycerophosphate, 0.5 mM DTT) plus 0.5% v/v Triton X-100. Resuspended cells were sheared then centrifuged to remove debris. Equal amounts of total protein (DC Protein Assay, Bio-Rad) were used for the immunoprecipitation reactions. First, a fraction of each sample was set aside as an input control. Then, 1 mg of each of the remaining lysates was diluted 4-fold with IP buffer plus 0.1% v/v Triton X-100 and incubated for 2 h at 4 °C with protein G-Sepharose (GE Healthcare catalog no. 17061801), prebound to HA-specific antibody (rabbit anti-HA from Sigma-Aldrich, catalog no. H6908) (3.5 μg antibody per reaction). Beads were collected by centrifugation and washed 6 times with IP buffer before analysis by SDS-PAGE.
Immunoblotting
For immunoblots to assess total protein levels (Figs. S3 and S4), harvested cells were lysed in 50 mM Tris pH 8.0, 10 mM EDTA, 1% w/v SDS, 1 mM DTT, phosphatase inhibitors, and protease inhibitors. Total protein concentrations were measured (DC Protein Assay, Bio-Rad), and equal amounts of protein were loaded per lane of SDS-PAGE. Separated proteins were transferred to polyvinylidene difluoride membranes and immunoblotted with antibodies specific for SF3B1 (Abcam, catalog no. ab170854), SF1 (Bethyl Laboratories, catalog no. A303-213A), U2AF2 (Sigma-Aldrich, catalog no. U4758), FLAG (Sigma-Aldrich, catalog no. F1804), HA (Sigma-Aldrich, catalog no. H6908), or GAPDH (Cell Signaling, catalog no. 14C10), all diluted 1:1000 v/v with 5% w/v dry milk in TBS-T. Secondary antibodies included anti-rabbit IgG horseradish peroxidase (Invitrogen, catalog no. 31460) or anti-mouse horseradish peroxidase (Invitrogen, catalog no. 31340). The chemiluminescence signal from Clarity Western ECL substrate (Bio-Rad, catalog no. 170–5061) was detected on a Chemidoc Touch Imaging System (Bio-Rad).
RT-PCR of minigene and endogenous gene transcripts
The protocols used for RT-PCR were described previously (33). Briefly, total RNA was isolated from harvested cells and DNase I treated using the RNeasy kit (Qiagen). The cDNAs were synthesized using random primers and Moloney murine leukemia virus RT (Invitrogen). The RT-PCR products were separated on a 2% w/v agarose-TBE gel, stained with ethidium bromide, and visualized using a Gel Doc XR+ gel documentation system (Bio-Rad). The band intensities of three technical replicates were quantified and background corrected using ImageJ (66) and are representative of multiple biological replicates. The primer sequences are listed in Table S3.
Bioinformatics and statistical analysis
RNAseq read alignment
RNAseq analysis was performed as described (46, 47). FASTQ files were downloaded from the ENCODE project site (https://www.encodeproject.org/; sample accession numbers are listed in Table S4). These included eight control replicates treated with the control shRNA and two biological replicates for each knockdown (SF3B1, U2AF2, SF1) from both K562 erythroid leukemia and HepG2 hepatocellular carcinoma cell lines. Reads were mapped to the Hg38 human reference genome using STAR Aligner (44). Mapping statistics are listed in Table S4.
Identification of alternative splicing events
Splice junctions determined by STAR mapping were combined for all samples and collapsed into a nonredundant set of introns. Alternative and constitutive intron classifications were performed using custom Python scripts and are agnostic with regard to existing annotations other than known gene boundaries (46, 47). The workflow takes a set of intron coordinates, assigns them to a gene, and divides them into subgroups based on overlapping coordinates. If no overlapping introns exist for a given intron, it is assigned to the constitutive class. The subgroups containing overlapping introns are assigned a splicing classification if the start and end coordinates of all of the constituent introns fall into a pattern representing a known splice type (cassette, mutually exclusive, alternative 5′ splice site, alternative 3′ splice site). For identification of detained introns (DI), STAR-mapped reads were filtered to remove intron-spanning reads and reads aligning to annotated exons or repeat RNAs. To remove polyadenylation sites within introns that might contribute to false-positive DI identification, the genome-wide coordinates of known polyadenylation sites were extracted from the GENCODE annotation (67), and introns containing these sites were not considered in assignment of DI status. Finally, alternative splicing annotations from the steps above were used to further filter out any introns that might contain exons or other introns to produce a nonoverlapping set of introns spanning each gene locus. Mapped and filtered reads were assigned to introns using Bedtools (68). For each gene, the sum of normalized intronic read counts was used to allocate reads to individual introns under a null model based on their length and mappability. A variance stabilizing transform based on the square root of intron effective length adjusted by RNAseq read length (√(Ld) where L=mappability adjusted intron length, d = RNAseq read length) was then used to weight individual introns. The sum of normalized intronic reads per gene in each RNAseq replicate was then partitioned and allocated to each intron proportional to its weight. This results in an in silico null model replicate corresponding to each RNAseq replicate. Differential analysis using DESeq (45) was then used to determine introns enriched in read coverage (in the RNAseq replicates) compared with the in silico null model replicates using an false discovery rate–adjusted p value threshold of 0.01 and fold change threshold of 2.
Differential splicing analysis
Annotated alternative and constitutive exons were used as an input to generate an “exon part” gtf that was compatible with DEXSeq, using the script dexseq_prepare_annotation.py (48). Reads were counted from mapped bam files using the counting script dexseq_count.py to generate count tables for each exon part. Differential expression of the alternative splicing events and DI was then determined using standard DEXSeq analysis with a padj. <0.05 as the cutoff for significant changes, comparing the eight control replicates against each of the two replicate sets for each knockdown. Overlapping events that were differentially spliced in each of the knockdowns were then counted, and the proportional Euler diagrams were produced in R using the package eulerr (https://CRAN.R-project.org/package=eulerr). Statistical significance of overlaps was determined using one-sided Fisher’s exact test calculated in R version 3.6.3 (69).
Data availability
Atomic coordinates and structure factors of U2AF2 UHM bound to SF3B1 ULM5 (accession code 7SN6) have been deposited at the Protein Data Bank (http://wwpdb.org).
Supporting information
This article contains supporting information.This article includes Tables S1–S4 and Figs. S1–S4.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Maria Carmo-Fonseca (U. Lisbon, Portugal) for providing the pyPY and wildtype HA-U2AF2 plasmids, Esther Obeng (St Jude) and Benjamin Ebert (Dana-Farber) for the SF3B1 template plasmid, and Steven Horner and Justin Leach for contributions to ITC. Data from the ENCODE consortium were courtesy of Brenton R. Graveley (U. Conn. Health). The crystallographic data were collected at SSRL, which is supported by the US DOE (Contract No. DE-AC02–76SF00515) and NIH (P41 GM103393).
Author contributions
C. L. K. conceptualization; S. L., J. L. J., M. J. P., P. L. B., C. L. K. methodology; S. L., J. L. J., M. J. P. validation; P. L. B. formal analysis; J. W. G., V. N. B., N. J., X. H., E. G., M. J. P., P. L. B. investigation; J. L. J. data curation; J. W. G., S. L., P. L. B., C. L. K. writing – original draft; J. W. G., X. H., S. L., J. L. J., M. J. P., P. L. B., C. L. K. writing – review & editing; J. W. G., S. L., P. L. B., C. L. K. visualization; P. L. B., C. L. K. funding acquisition.
Funding and additional information
This study was supported by National Institutes of Health (NIH) grants R01 GM070503 to C. L. K., R01 GM141544 to P. L. B., and T32 GM135134 supported J. W. G. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biography

Justin W. Galardi is a fourth-year PhD candidate in the laboratory of Prof. Clara Kielkopf in the Biochemistry & Molecular Biology program of the University of Rochester School of Medicine and Dentistry. His current work reveals an unusual interaction mode between splicing factors that are critical for the early stages of spliceosome assembly. As a next step, Justin is excited to investigate how this “healthy” interaction is dysregulated in cancers and viral infections.
Edited by Karin Musier-Forsyth
Footnotes
Present address for Sarah Loerch: Dept. Chem. & Biochem., UC Santa Cruz, Santa Cruz, CA 95064, USA.
Supporting information
References
- 1.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamore P.D., Green M.R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl. Acad. Sci. U. S. A. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal A.A., Salsi E., Chatrikhi R., Henderson S., Jenkins J.L., Green M.R., et al. An extended U2AF65-RNA-binding domain recognizes the 3' splice site signal. Nat. Commun. 2016;7 doi: 10.1038/ncomms10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackereth C.D., Madl T., Bonnal S., Simon B., Zanier K., Gasch A., et al. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 5.Wu S., Romfo C.M., Nilsen T.W., Green M.R. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 6.Rain J.C., Rafi Z., Rhani Z., Legrain P., Kramer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berglund J.A., Abovich N., Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanackovic G., Kramer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol. Biol. Cell. 2005;16:1366–1377. doi: 10.1091/mbc.E04-11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gozani O., Potashkin J., Reed R. A potential role for U2AF-SAP155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agafonov D.E., Deckert J., Wolf E., Odenwalder P., Bessonov S., Will C.L., et al. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell Biol. 2011;31:2667–2682. doi: 10.1128/MCB.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett M., Michaud S., Kingston J., Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017;18:655–670. doi: 10.1038/nrm.2017.86. [DOI] [PubMed] [Google Scholar]
- 13.Li X., Liu S., Zhang L., Issaian A., Hill R.C., Espinosa S., et al. A unified mechanism for intron and exon definition and back-splicing. Nature. 2019;573:375–380. doi: 10.1038/s41586-019-1523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan C., Wan R., Bai R., Huang G., Shi Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science. 2016;353:904–911. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 15.Plaschka C., Lin P.C., Nagai K. Structure of a pre-catalytic spliceosome. Nature. 2017;546:617–621. doi: 10.1038/nature22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertram K., Agafonov D.E., Dybkov O., Haselbach D., Leelaram M.N., Will C.L., et al. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell. 2017;170:701–713.e711. doi: 10.1016/j.cell.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., Yan C., Zhan X., Li L., Lei J., Shi Y. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018;28:307–322. doi: 10.1038/cr.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haselbach D., Komarov I., Agafonov D.E., Hartmuth K., Graf B., Dybkov O., et al. Structure and conformational dynamics of the human spliceosomal B(act) complex. Cell. 2018;172:454–464 e411. doi: 10.1016/j.cell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Bai R., Wan R., Yan C., Lei J., Shi Y. Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation. Science. 2018;360:1423–1429. doi: 10.1126/science.aau0325. [DOI] [PubMed] [Google Scholar]
- 20.Wang W., Maucuer A., Gupta A., Manceau V., Thickman K.R., Bauer W.J., et al. Structure of phosphorylated SF1 bound to U2AF65 in an essential splicing factor complex. Structure. 2013;21:197–208. doi: 10.1016/j.str.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Madl T., Bagdiul I., Kern T., Kang H.S., Zou P., et al. Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition. Nucl. Acids Res. 2013;41:1343–1354. doi: 10.1093/nar/gks1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loerch S., Kielkopf C.L. Unmasking the U2AF homology motif family: A bona fide protein-protein interaction motif in disguise. RNA. 2016;22:1795–1807. doi: 10.1261/rna.057950.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thickman K.R., Swenson M.C., Kabogo J.M., Gryczynski Z., Kielkopf C.L. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J. Mol. Biol. 2006;356:664–683. doi: 10.1016/j.jmb.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spadaccini R., Reidt U., Dybkov O., Will C., Frank R., Stier G., et al. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cass D.M., Berglund J.A. The SF3b155 N-terminal domain is a scaffold important for splicing. Biochemistry. 2006;45:10092–10101. doi: 10.1021/bi060429o. [DOI] [PubMed] [Google Scholar]
- 26.Corsini L., Bonnal S., Basquin J., Hothorn M., Scheffzek K., Valcarcel J., et al. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 27.Loerch S., Maucuer A., Manceau V., Green M.R., Kielkopf C.L. Cancer-relevant splicing factor CAPERα engages the essential splicing factor SF3b155 in a specific ternary complex. J. Biol. Chem. 2014;289:17325–17337. doi: 10.1074/jbc.M114.558825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loerch S., Leach J.R., Horner S.W., Maji D., Jenkins J.L., Pulvino M.J., et al. The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface. J. Biol. Chem. 2019;294:2892–2902. doi: 10.1074/jbc.RA118.006764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Weinmeister R., Kralovicova J., Eperon L.P., Vorechovsky I., Hudson A.J., et al. Stoichiometries of U2AF35, U2AF65 and U2 snRNP reveal new early spliceosome assembly pathways. Nucl. Acids Res. 2017;45:2051–2067. doi: 10.1093/nar/gkw860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beausoleil S.A., Jedrychowski M., Schwartz D., Elias J.E., Villen J., Li J., et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rimel J.K., Poss Z.C., Erickson B., Maas Z.L., Ebmeier C.C., Johnson J.L., et al. Selective inhibition of CDK7 reveals high-confidence targets and new models for TFIIH function in transcription. Genes Dev. 2020;34:1452–1473. doi: 10.1101/gad.341545.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pacheco T.R., Coelho M.B., Desterro J.M., Mollet I., Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3' splice site. Mol. Cell Biol. 2006;26:8183–8190. doi: 10.1128/MCB.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maji D., Glasser E., Henderson S., Galardi J., Pulvino M.J., Jenkins J.L., et al. Representative cancer-associated U2AF2 mutations alter RNA interactions and splicing. J. Biol. Chem. 2020;295:17148–17157. doi: 10.1074/jbc.RA120.015339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukumura K., Yoshimoto R., Sperotto L., Kang H.S., Hirose T., Inoue K., et al. SPF45/RBM17-dependent, but not U2AF-dependent, splicing in a distinct subset of human short introns. Nat. Commun. 2021;12:4910. doi: 10.1038/s41467-021-24879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu X.D., Ares M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatrikhi R., Feeney C.F., Pulvino M.J., Alachouzos G., MacRae A.J., Falls Z., et al. A synthetic small molecule stalls pre-mRNA splicing by promoting an early-stage U2AF2-RNA complex. Cell Chem Biol. 2021;28:1145–1157.e1146. doi: 10.1016/j.chembiol.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kielkopf C.L., Rodionova N.A., Green M.R., Burley S.K. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 38.Hastings M.L., Allemand E., Duelli D.M., Myers M.P., Krainer A.R. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS One. 2007;2:e538. doi: 10.1371/journal.pone.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kralovicova J., Houngninou-Molango S., Kramer A., Vorechovsky I. Branch site haplotypes that control alternative splicing. Hum. Mol. Genet. 2004;13:3189–3202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- 40.Corioni M., Antih N., Tanackovic G., Zavolan M., Kramer A. Analysis of in situ pre-mRNA targets of human splicing factor SF1 reveals a function in alternative splicing. Nucl. Acids Res. 2011;39:1868–1879. doi: 10.1093/nar/gkq1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Nostrand E.L., Freese P., Pratt G.A., Wang X., Wei X., Xiao R., et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583:711–719. doi: 10.1038/s41586-020-2077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucl. Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boutz P.L., Bhutkar A., Sharp P.A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun C.J., Stanciu M., Boutz P.L., Patterson J.C., Calligaris D., Higuchi F., et al. Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell. 2017;32:411–426.e411. doi: 10.1016/j.ccell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S., Reyes A., Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guth S., Valcarcel J. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 2000;275:38059–38066. doi: 10.1074/jbc.M001483200. [DOI] [PubMed] [Google Scholar]
- 50.Chatrikhi R., Wang W., Gupta A., Loerch S., Maucuer A., Kielkopf C.L. SF1 phosphorylation enhances specific binding to U2AF65 and reduces binding to 3’-splice-site RNA. Biophys. J. 2016;111:2570–2586. doi: 10.1016/j.bpj.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller H.B., Robinson T.J., Gordan R., Hartemink A.J., Garcia-Blanco M.A. Identification of Tat-SF1 cellular targets by exon array analysis reveals dual roles in transcription and splicing. RNA. 2011;17:665–674. doi: 10.1261/rna.2462011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruskin B., Zamore P.D., Green M.R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 53.Shao C., Yang B., Wu T., Huang J., Tang P., Zhou Y., et al. Mechanisms for U2AF to define 3' splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 2014;21:997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zarnack K., Konig J., Tajnik M., Martincorena I., Eustermann S., Stevant I., et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gozani O., Feld R., Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 56.Das B.K., Xia L., Palandjian L., Gozani O., Chyung Y., Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol. Cell Biol. 1999;19:6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darman R.B., Seiler M., Agrawal A.A., Lim K.H., Peng S., Aird D., et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3' splice site selection through use of a different branch point. Cell Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 58.Obeng E.A., Chappell R.J., Seiler M., Chen M.C., Campagna D.R., Schmidt P.J., et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30:404–417. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soltis S.M., Cohen A.E., Deacon A., Eriksson T., Gonzalez A., McPhillips S., et al. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr. D Biol. Crystallogr. 2008;64:1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bunkoczi G., Echols N., McCoy A.J., Oeffner R.D., Adams P.D., Read R.J. Phaser.MRage: automated molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2013;69:2276–2286. doi: 10.1107/S0907444913022750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afonine P.V., Moriarty N.W., Mustyakimov M., Sobolev O.V., Terwilliger T.C., Turk D., et al. FEM: feature-enhanced map. Acta Crystallogr. D Biol. Crystallogr. 2015;D71:646–666. doi: 10.1107/S1399004714028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 66.Abramoff M.D., Magalhaes P.J., Ram S.J. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 67.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors of U2AF2 UHM bound to SF3B1 ULM5 (accession code 7SN6) have been deposited at the Protein Data Bank (http://wwpdb.org).