Abstract
MalF is one of the two integral inner membrane proteins of the maltose-maltodextrin transport system. To identify functional regions in this protein, we characterized a collection of malF mutants obtained by random mutagenesis. We analyzed their growth on maltose and maltodextrins, the steady-state levels and subcellular localization of the mutant proteins, and the subcellular localization of MalK. Only 2 of the 21 MalF mutant proteins allowed growth on maltose and maltodextrins. Most mutations resulting in immunodetectable proteins mapped to hydrophilic domains, indicating that insertions affecting transmembrane segments gave rise to unstable or lethal proteins. All MalF mutant proteins, even those C-terminally truncated or with large N-terminal deletions, were inserted into the cytoplasmic membrane. Having identified mutations leading to reduced steady-state level, to partial mislocation, and/or to misfolding, we were able to assign to some regions of MalF a role in the assembly of the MalFGK2 complex and/or in the transport mechanism.
The maltose-maltodextrin transport system of Escherichia coli constitutes an excellent model to study the folding, assembly, and functioning of an integrated system of proteins involved in the acquisition of molecules by bacteria. This system is, together with the histidine transport system of Salmonella typhimurium, one of the most studied periplasmic-binding-protein-dependent ATP-binding cassette (ABC) transporters, reviewed in reference 1. Maltose and maltodextrins enter the periplasm through LamB, an outer membrane porin. The periplasmic maltose-binding protein (MBP or MalE protein) binds substrates and interacts with MalF and MalG (16), the two integral membrane proteins which constitute, together with two copies of the MalK ATPase, the inner membrane complex MalFGK2. This interaction triggers ATP hydrolysis by MalK, allowing the MalFGK2 complex to translocate the substrate released by MalE into the cytoplasm. The exact mechanism of the transport is not known. Random linker insertion mutagenesis proved to be a powerful tool to identify dispensable or functionally important regions within LamB (2), MalE (9), and MalG (6) proteins. Recently, a transposon-mediated insertion mutagenesis approach was used to further characterize the MalK (18) and MalG (22) proteins. The topology (3, 12, 13, 14), the assembly (29), and the membrane insertion (30) of the MalF protein were studied in some detail. In contrast, little is known about the regions of MalF crucial for transport. In order to identify the MalF regions important for structure and function, we mutagenized the malF gene by random linker insertion. As mutations were isolated independently of an activity test or phenotypic modification, potential bias toward the isolation of any particular class of mutant was minimized, and silent mutations could be obtained.
Random mutagenesis, screening, sequencing, and phenotypes of mutants.
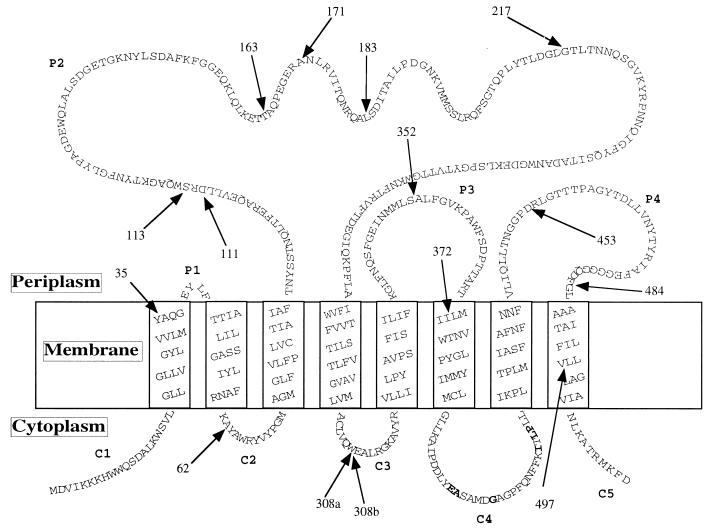
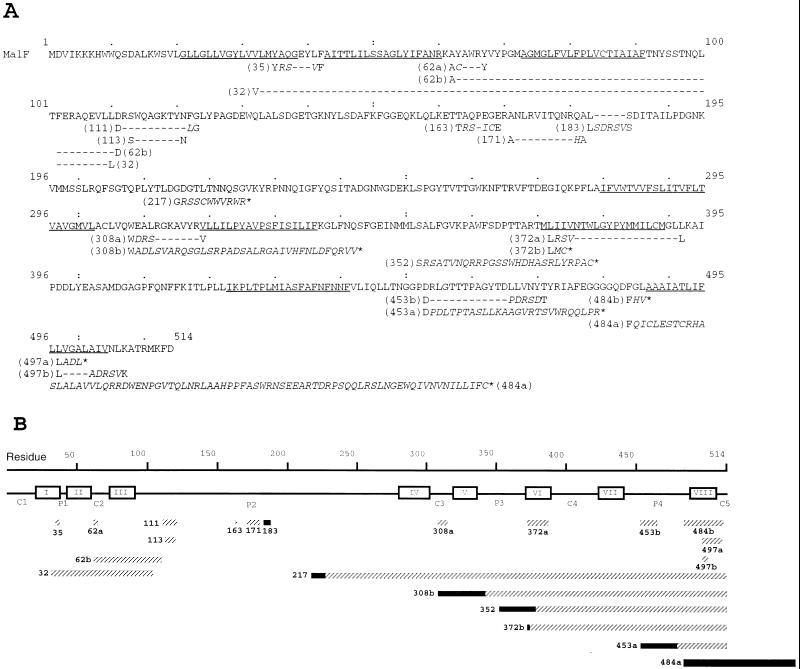
E. coli strains, plasmids, media, and culture conditions were previously described in reference 20. Random linker mutagenesis was performed as described elsewhere (2, 6), except that a double-stranded unphosphorylated BgIII linker, d(CAGATCTG) (Biolabs), was used. Mutated pTAZFQ (malF under the control of the tac promoter) plasmid was transformed into the MalF− strain DHB4. Sixty-two of 533 screened clones had one insertion in or very close to malF. Total cell extracts were prepared according to the method described in reference 20. Thirty-one of these plasmids were found to promote expression of a protein recognized by the MalF antiserum (data not shown). Clones with no immunodetectable MalF could result from the instability of the mutated protein or plasmid, from a large deletion of the protein, or from an early frameshift. We tried to restore the reading frame of the latter nonimmunodetectable mutants. We cut the plasmid DNAs with BgIII and filled in or removed the cohesive ends by using the Klenow fragment of E. coli DNA polymerase I or the mung bean nuclease, respectively. The protein sequence modifications generated by the linker insertions were deduced from DNA sequence analysis. DNA sequencing (25) revealed that several mutants were obtained more than once. They were probably nonindependent clones arising during transformation. Finally, we obtained 21 mutants from 15 independent different random BglII linker insertions in malF. The positions of these primary insertions are shown in Fig. 1, as related to the topological model of MalF predicted by the PHDtoplogy program (24). This model agrees with the experimentally determined topological model (14) and was used to predict the topology of the mutant proteins. Almost all the random insertions resulting in immunodetectable MalF mutant proteins were situated in or near hydrophilic regions (Fig. 1). As insertions occurred at random, this suggests that hydrophilic domains are more prone to modifications than are transmembrane (TM) segments. The sequences of the 21 mutants are shown in Fig. 2A. In Fig. 2B, mutations are represented schematically, and the relative sizes of insertions and deletions are shown. We obtained 12 immunodetectable mutant proteins without large insertions or deletions. These mutations consisted in the insertion of one to five amino acids determined by the linker accompanied by the deletion of 4 to 19 amino acids, except in mutant 183, in which five amino acids were inserted. In five cases (217, 308b, 352, 372b, and 453a), the mutation resulted in a truncated MalF, either by a large deletion or by a shift in the translation reading frame, leading to a premature stop. In one case (mutant 484a), the frameshift created a replacement of the last 30 amino acids by 78 non-MalF amino acids. Two mutants had large in-frame deletions including TM3 (mutant 62b) and TMs 2 and 3 (mutant 32).
FIG. 1.
Topology model of MalF according to the PHDtopology program from the EMBO server (24). Positions of the different randomly inserted BglII sites are shown by arrows. Numbers indicate the codon numbers preceding the BglII sites. C, cytoplasmic domain; P, periplasmic domain. Boldface letters correspond to the conserved peptidic motif EA----G---------I-LP (7). Two independent insertions, but with different downstream sequences due to a change in the reading frame, were found after codon 308 (308a and 308b).
FIG. 2.
Mutations in protein MalF. The derivatives of malF were sequenced with the Sequenase version 2.0 DNA sequencing kit (U.S. Biochemical and Amersham) with [α-33P]dATP (ICN). Oligonucleotides (Eurogentec) corresponding to 12 segments of the malF gene were used as primers. The approximate location of the insertion was first determined by restriction enzyme analysis, and then the appropriate primer was chosen. In some cases, the sequencing of the antiparallel strand was carried out to resolve ambiguities. (A) Sequences of wild-type MalF and the collection of mutants described in this work. Predicted α-helices are shown underlined in the wild-type sequence. In mutant sequences, amino acids inserted are shown in italics between the positions of original MalF amino acids. Each mutant is named according to the last nonmodified amino acid. Hyphens denote deletions. Stop codons in the frameshift mutations are indicated by asterisks. (B) Mutations in MalF protein, represented schematically in a linear fashion. Predicted TM segments are boxed and numbered in roman numbers. The lengths of rectangles are proportional to the sizes of deletions (solid) or insertions (hatched).
We analyzed the overall activity of MalF mutants by an in vivo genetic complementation assay. The mutant malF genes carried by the plasmid pTAZFQ were expressed in DHB4 (MalF−). Maltose transport phenotypes of mutants were characterized on MacConkey–2% maltose or MacConkey–1% maltodextrin (malto-oligosaccharide mixture from Pfanstiehl Laboratories Inc.) plates. In the absence of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG), the basal expression level of the wild-type malF gene was sufficient to promote a Mal+ Dex+ phenotype. Among the mutant proteins only two (mutants 35 and 163) complemented the malF3 mutation for maltose and maltodextrin fermentation. The phenotypes were significantly improved neither by adding IPTG nor when MalG and/or MalK was co-overexpressed, indicating that none of the proteins was limiting. None of the MalF mutants displayed a MalE-independent phenotype (4). Maltose phenotypes were characterized by measuring the doubling time (DT) of mutants in liquid M63B1-maltose minimal medium (Table 1). Under these conditions, only mutants 35 and 163 displayed DTs similar to that obtained with wild-type MalF. The growth rate of the strains in glycerol-supplemented minimal medium was similar to that of control cells (wild-type malF), indicating that no growth defect was caused by the expression of mutant proteins (data not shown).
TABLE 1.
Summary of characterization of MalF mutant proteins
| Characteristic | Value for wild type | Value for mutant with deletion type:
|
Value for pTAZQ | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Short
|
Large
|
||||||||||||||||||||||
| 35 | 62a | 111 | 113 | 163 | 171 | 183 | 308a | 372a | 453b | 484b | 497a | 497b | 62b | 32 | 217 | 308b | 352 | 372b | 453a | 484a | |||
| DT in M63B1-maltosea | 230 | 226 | 488 | 537 | 598 | 231 | 912 | 555 | 447 | 521 | 465 | NAf | 568 | 937 | NA | NA | NA | NA | NA | NA | NA | NA | 436 |
| Steady-state protein level (% of wild type)b | 100 | <25 | >75 | 25–50 | 50–75 | 50–75 | >75 | >75 | >75 | 25–50 | >75 | 25–50 | 25–50 | >75 | 25–50 | <25 | NA | NA | NA | NA | <25 | 50–75 | |
| Proteolytic productsc | − | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | NDg | + | + | + | + | + | |
| Aggregationd | − | − | − | − | − | − | − | − | − | − | + | − | − | + | − | + | NA | − | − | − | − | − | |
| Cytoplasmic MalK (% total)e | 10 | NA | 23 | 31 | 19 | NA | 33 | 32 | 47 | 98 | 24 | NA | 12 | 36 | NA | NA | NA | NA | NA | NA | NA | NA | |
DT was measured in the ED170 strain transformed with pTAZFGQ and pACYK plasmids, at 0.2% maltose and 10 μM IPTG.
DHB4 cells carrying the pTAZFQ mutant plasmid were grown in M63B1 minimal medium containing ampicillin, glucose, and 0.004% of a mixture of nucleotides and amino acids lacking methionine and cysteine (Sigma). At A600 = 0.5, cells were induced for 30 min with 1 mM IPTG. Cultures (1 ml) were normalized to the same A600 and labelled with 40 μCi of Tran35S-label (ICN) for 30 min at 30°C. Particulate fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% acrylamide, 18-cm-long gels, Tris-Tricine buffer) and subjected to autoradiography. Mutant proteins were clearly resolved, and the radioactivity in bands corresponding to each full-length mutant protein was quantified with a PhosphorImager (Molecular Dynamics) and normalized for the number of methionines and cysteines.
See Fig. 3.
See Fig. 3. Mutant proteins partially extracted with Triton X-100 formed aggregates (+); the rest (−) were completely extracted with Triton X-100.
See Fig. 4.
NA, not analyzed.
ND, not detected.
Steady-state levels, subcellular localization, and stability of MalF mutant proteins.
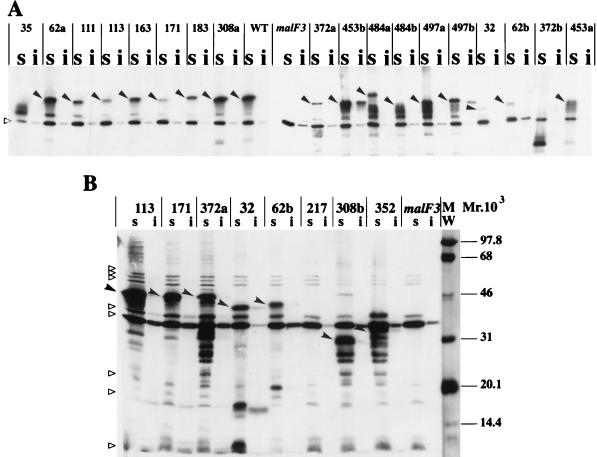
We examined the steady-state levels of the MalF mutants (Table 1). All mutant proteins accumulated at lower steady-state levels than that of the wild-type protein, indicating that mutations led to reduced protein production or to unstable proteins. Mutant 35 was functional in spite of its low protein level. This result suggests that the loss of activity of MalF− mutants was not due merely to a low steady-state level of protein in the cells. The ability of the nonionic detergent Triton X-100 to solubilize the MalF proteins was examined. This detergent specifically solubilizes cytoplasmic membrane proteins (8) and has been used to show the membrane localization of the MalFGK2 complex (20). All mutants except two (453b and 497b) were completely extracted with Triton X-100 (Fig. 3) and therefore localized exclusively in the cytoplasmic membrane like wild-type MalF. Mutants 453b and 497b were only partially extracted with Triton X-100 (Fig. 3A). Hence, a fraction of each of these mutant proteins consisted of aggregated rather than membrane-associated proteins. The insertion of MalF into the membrane was not prevented by the deletion of any of the TMs. These results agree with previously reported findings showing that individual membrane-spanning sequences act as autonomous insertion domains (14, 27) and with earlier studies on polytopic cytoplasmic membrane proteins (6, 19, 22). Immunoblots of mutants with short deletions displayed a single band recognized by the antibody (Fig. 3A, lanes 2 to 8). By contrast, mutants affected in TMs (mutants 35, 372a, 484b, 497a, and 497b), with large in-frame deletions (mutants 32 and 62b), or with frameshifts (mutants 308b, 352, 372b, 453a, and 484a) showed several bands of higher electrophoretic mobility, which most likely correspond to proteolytic degradation products of MalF mutant proteins (Fig. 3).
FIG. 3.
Subcellular localization of MalF mutant proteins. Cells were induced at an A600 of 0.5 with 1 mM IPTG. Cell fractionation was performed according to the method described in reference 20, with the following modifications. The protease inhibitor PEFABLOC (Interchim) was added at an 0.5 mM final concentration to all buffers. Membrane extracts were recovered after 1 h of centrifugation at 20,000 × g. To solubilize cytoplasmic membrane proteins, Triton X-100 was used as described in reference 15. Equivalent volumes of Triton-soluble (s) and Triton-insoluble (i) fractions were mixed with an equal volume of double-strength sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, boiled for 5 min in a water bath, loaded onto sodium dodecyl sulfate–12% (wt/vol) polyacrylamide gel electrophoresis gels, and subjected to immunoblotting. Nitrocellulose membranes were probed with the specific antibody and with a horseradish peroxidase anti-rabbit immunoglobulin conjugate (Bio-Rad). Immune complexes were revealed by ECL Western blotting detection reagents (Amersham). Relative intensities were quantified by scanning the ECL films with an Image Master VDS apparatus (Pharmacia Biotech). The solid arrowheads indicate the full-length MalF mutant proteins. The empty triangles indicate cross-reacting proteins that are visible in all lanes. The amount of protein loaded in immunoblot B was threefold higher than that loaded in immunoblot A. WT, wild type. MW, molecular weight markers, in thousands.
Subcellular localization of MalK.
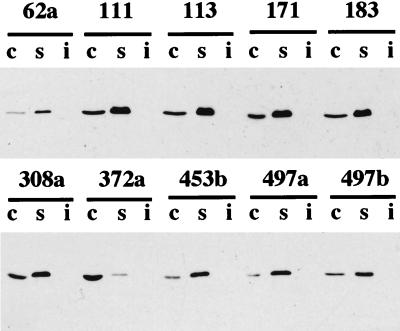
Both in MalF and MalG, the conserved region EA----G---------I-LP is very probably important for the interactions between the hydrophobic membrane proteins and the MalK ATPase (20). Mutants affected in this region were not able to stably maintain MalK in the membrane. We analyzed the cellular localization of MalK in cells containing MalF mutant proteins unable to grow on maltose and maltodextrin, in order to examine if regions other than the conserved region were important for the interaction with MalK. Strain ED169 was transformed with recombinant or wild-type pTAZFGQ and pACYK. Plasmid genes were induced with 10 μM IPTG. In MalF+ MalG+ cells (ED169 transformed with nonmutated pTAZFGQ and pACYK), we found about 90% MalK in the particulate fraction, from which it was completely extracted with Triton X-100. In MalF− MalG− cells (ED169 transformed with pTAZQ and pACYK), we found about 100% MalK in the cytoplasmic fraction. The membrane association of MalK was altered in mutant 308a (47% of MalK in the cytoplasm) and more drastically in mutant 372a (98% of MalK in the cytoplasm) (Fig. 4; Table 1). Mutant 497a behaved like wild-type MalF, and in the other mutants tested, the amount of cytoplasmic MalK ranged from 19 to 36%.
FIG. 4.
Subcellular localization of MalK. MalK subcellular localization experiments were performed as described elsewhere (20), except that strain ED169 instead of strain ED170 was used and extracts were centrifuged at 20,000 × g instead of 200,000 × g. Under these conditions, all MalK protein was solubilized by Triton X-100, and thus, no additional fractionation of the Triton-insoluble fraction with urea was made. Triton-soluble (s), cytoplasmic (c), and Triton-insoluble (i) fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to immunoblotting. The volume loaded for the Triton-soluble and Triton-insoluble fractions was double that for the cytoplasmic fractions.
Potential functional domains of the MalF protein.
In several cases, the insertion of the linker introduced a change in specific domains of MalF. We will discuss these changes, with a focus on short deletions, in order to assign tentatively a function to these domains.
The first periplasmic loop (P1) was changed in mutation 35, together with the last amino acids of TM1. This mutation did not affect the utilization of maltose and maltodextrins and mapped in a region of MalF that had been previously defined as highly permissive (11). The mutation introduced at this point seemed to affect the stability of the protein. The amount of nondegraded mutant protein inserted in the membrane, even under noninducing conditions, would be enough to account for the observed Mal+ Dex+ phenotype. This region would not be crucial for the assembly of the MalFGK2 complex or for the constitution of the maltose translocation pathway but would contribute to the stability of the protein.
The large periplasmic domain P2 is characteristic of MalF proteins in enterobacteria. MalF proteins in other bacteria lack this domain and have in some cases six TMs rather than eight (17). Although its sequence in enterobacterial MalF is less conserved than that of other parts of the protein (5, 26), our results strongly suggest that P2 is essential for function. Mutations in P2 had little influence on the steady-state level of MalF, and they did not cause the appearance of proteolytic products. For the five mutations affecting P2, only mutant 163 was competent for maltose-maltodextrin transport. The other mutations affected MalK localization. Mourez et al. (20) showed that both MalF and MalG are needed to stably maintain MalK in the membrane and that residues in C4 of MalF are essential for the binding of MalK to MalFG. The dislocation of MalK provoked by the above mutations could be explained by assuming that P2 participates in anchoring MalK into the membrane. According to the proposed model of the eukaryotic ABC transporter P-glycoprotein determined by electron microscopy and image analysis (23), it is possible that extracytoplasmic domains of ABC transporters are in direct contact with cytoplasmic domains. Alternatively, the effect might be indirect, the mutation affecting either the conformation of C4 or the interaction of MalF with MalG. The latter hypothesis is consistent with the finding that the conformation or the accessibility of MalF, and most notably of the P2 domain, is modified depending on the presence of MalG (21, 28).
Loop P4 was partly deleted in mutant 453b. The mutation led to a high steady-state level of protein, which was defective in transport and partially mislocated since a small fraction of the protein was not extracted from membranes with Triton X-100. The almost correct subcellular localization of MalK in the mutant suggests that the interaction of MalF with MalG is not dramatically affected. Hence, the mutation probably affected transport either by the modification of a substrate binding site or by hampering an interaction with MalE. Interestingly, mutations in the last periplasmic loop of MalG have been proposed to affect the recognition of substrates (6) or the interaction with MalE (22). If these interpretations are correct, the last periplasmic loops of MalF and MalG might cooperate in such functions.
Cytoplasmic loop C2 was altered in mutant 62a. Although nonfunctional, this mutant behaved like wild-type MalF with respect to all other criteria examined. This mutation would affect a transport-specific function.
C3 was partially deleted in mutant 308a. The mutant protein was well expressed and correctly localized into the membrane, but 47% of MalK was found in the cytoplasm. Like loop P2, C3 might be directly or indirectly implicated in the attachment of MalK to the membrane.
Mutants in TMs displayed in general a low steady-state level of protein. This was probably due to proteolytic degradation. The deletion of TM6 (mutant 372a) led to the complete dislocation of MalK. This could be due to the fact that C4 would probably be translocated to the periplasm in this mutant. By contrast, the deletion of C5 and of the C-terminus of TM8 (mutant 497a) did not affect the localization of MalK. Therefore, the C-terminal part of MalF would not be essential for the interaction with MalK or MalG. The properties of mutant 497a are similar to those of mutant 453b (P4), suggesting that residues involved in the transport mechanism or in the translocation pathway of substrates are present in P4, in TM8, and in C5. This conclusion is in agreement with the results of reference 10, which reported that several amino acid substitutions in TM8 led to Mal+ Dex− phenotypes.
In summary, the incapacity of some MalF mutant proteins to participate in maltose transport could be due at least partially to a defective insertion into the membrane, increased rates of proteolysis, or partial aggregation in the cytoplasm. By contrast, mutations in C2, P4, TM8, and C5 probably affected the transport mechanism while mutations in C3 and P2 probably affected the assembly of the MalFGK2 complex. Regions potentially assigned to specific functions will be further dissected by site-specific mutagenesis in order to refine our model.
Acknowledgments
M.I.T. was supported by a European Commission Research Training Grant (Biotechnology Programme, contract number BIO4-CT96-5068).
We are grateful to Alain Charbit, Pierre Martineau, and Jean-Michel Betton for their helpful technical advice and to Jesus A. G. Ochoa de Alda and Wolfgang Köster for careful reading of the manuscript and for helpful suggestions. We thank Beth Traxler and Erwin Schneider for the gift of MalF and MalK antibodies. We thank Muguette Jéhanno for the help in MalK localization.
REFERENCES
- 1.Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulain J C, Charbit A, Hofnung M. Mutagenesis by random linker insertion into the lamB gene of E. coli K12. Mol Gen Genet. 1986;205:339–348. doi: 10.1007/BF00430448. [DOI] [PubMed] [Google Scholar]
- 3.Boyd D, Beckwith J. Positively charged amino acid residues can act as topogenic determinants in membrane proteins. Proc Natl Acad Sci USA. 1989;86:9446–9450. doi: 10.1073/pnas.86.23.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covitz K M Y, Panagiotidis C H, Hor L I, Reyes M, Treptow N A, Shuman H A. Mutations that alter the transmembrane signalling pathway in an ATP binding cassette (ABC) transporter. EMBO J. 1994;13:1752–1759. doi: 10.1002/j.1460-2075.1994.tb06439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahl M K, Francoz E, Saurin W, Boos W, Manson M D, Hofnung M. Comparison of sequences from the malB regions of Salmonella typhimurium and Enterobacter aerogenes with Escherichia coli K12: a potential new regulatory site in the interoperonic region. Mol Gen Genet. 1989;218:199–207. doi: 10.1007/BF00331269. [DOI] [PubMed] [Google Scholar]
- 6.Dassa E. Sequence-function relationships in MalG, an inner membrane protein from the maltose transport system in Escherichia coli. Mol Microbiol. 1993;7:39–47. doi: 10.1111/j.1365-2958.1993.tb01095.x. [DOI] [PubMed] [Google Scholar]
- 7.Dassa E, Hofnung M. Sequence of malG gene in E. coli K12: homologies between integral membrane components from binding protein-dependent transport systems. EMBO J. 1985;4:2287–2293. doi: 10.1002/j.1460-2075.1985.tb03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diedrich D L, Summers A O, Schnaitman C A. Outer membrane proteins of Escherichia coli. V. Evidence that protein 1 and bacteriophage-directed protein 2 are different polypeptides. J Bacteriol. 1977;131:598–607. doi: 10.1128/jb.131.2.598-607.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duplay P, Szmelcman S, Bedouelle H, Hofnung M. Silent and functional changes in the periplasmic maltose-binding protein of Escherichia coli K12. I. Transport of maltose. J Mol Biol. 1987;194:663–673. doi: 10.1016/0022-2836(87)90243-9. [DOI] [PubMed] [Google Scholar]
- 10.Ehrle R, Pick C, Ulrich R, Hofmann E, Ehrmann M. Characterization of transmembrane domains 6, 7, and 8 of MalF by mutational analysis. J Bacteriol. 1996;178:2255–2262. doi: 10.1128/jb.178.8.2255-2262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrmann M, Beckwith J. Proper insertion of a complex membrane protein in the absence of its amino-terminal export signal. J Biol Chem. 1991;266:16530–16533. [PubMed] [Google Scholar]
- 12.Ehrmann M, Boyd D, Beckwith J. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc Natl Acad Sci USA. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Froshauer S, Beckwith J. Nucleotide sequence of the gene for MalF protein, an inner membrane component of the maltose transport system. J Biol Chem. 1984;259:10896–10903. [PubMed] [Google Scholar]
- 14.Froshauer S, Green G N, Boyd D, McGovern K, Beckwith J. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of E. coli. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 15.Hjelmeland L M. Solubilization of native membrane proteins. Methods Enzymol. 1990;182:253–264. doi: 10.1016/0076-6879(90)82021-s. [DOI] [PubMed] [Google Scholar]
- 16.Hor L I, Shuman H A. Genetic analysis of periplasmic binding protein dependent transport in Escherichia coli—each lobe of maltose-binding protein interacts with a different subunit of the MalFGK(2) membrane transport complex. J Mol Biol. 1993;233:659–670. doi: 10.1006/jmbi.1993.1543. [DOI] [PubMed] [Google Scholar]
- 17.Horlacher R, Xavier K B, Santos H, DiRuggiero J, Kossmann M, Boos W. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1998;180:680–689. doi: 10.1128/jb.180.3.680-689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lippincott J, Traxler B. MalFGK complex assembly and transport and regulatory characteristics of MalK insertion mutants. J Bacteriol. 1997;179:1337–1343. doi: 10.1128/jb.179.4.1337-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manoil C, Traxler B. Membrane protein assembly: genetic, evolutionary and medical perspectives. Annu Rev Genet. 1995;29:131–150. doi: 10.1146/annurev.ge.29.120195.001023. [DOI] [PubMed] [Google Scholar]
- 20.Mourez M, Hofnung M, Dassa E. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 1997;16:2066–2077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mourez M, Skouloubris S, Betton J M, Dassa E. Heat shock induction by a misassembled cytoplasmic membrane protein complex in Escherichia coli. Mol Microbiol. 1997;26:821–831. doi: 10.1046/j.1365-2958.1997.6271992.x. [DOI] [PubMed] [Google Scholar]
- 22.Nelson B D, Traxler B. Exploring the role of integral membrane proteins in ATP-binding cassette transporters: analysis of a collection of MalG insertion mutants. J Bacteriol. 1998;180:2507–2514. doi: 10.1128/jb.180.9.2507-2514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg M F, Callaghan R, Ford R C, Higgins C F. Structure of the multidrug resistance P-glycoprotein to 2.5 nm resolution determined by electron microscopy and image analysis. J Biol Chem. 1997;272:10685–10694. doi: 10.1074/jbc.272.16.10685. [DOI] [PubMed] [Google Scholar]
- 24.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;7:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider E, Francoz E, Dassa E. Completion of the nucleotide sequence of the maltose B region in Salmonella typhimurium—the high conservation of the malM gene suggests a selected physiological role for its product. Biochim Biophys Acta. 1992;1129:223–227. doi: 10.1016/0167-4781(92)90492-i. [DOI] [PubMed] [Google Scholar]
- 27.Stochaj U, Fritz H J, Heibach C, Markgraf M, von Schaewen A, Sonnewald U, Ehring R. Truncated forms of Escherichia coli lactose permease: models for study of biosynthesis and membrane insertion. J Bacteriol. 1988;170:2639–2645. doi: 10.1128/jb.170.6.2639-2645.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traxler B, Beckwith J. Assembly of a hetero-oligomeric membrane protein complex. Proc Natl Acad Sci USA. 1992;89:10852–10856. doi: 10.1073/pnas.89.22.10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Traxler B, Lee C, Boyd D, Beckwith J. The dynamics of assembly of a cytoplasmic membrane protein in Escherichia coli. J Biol Chem. 1992;267:5339–5345. [PubMed] [Google Scholar]
- 30.Traxler B, Murphy C. Insertion of the polytopic membrane protein MalF is dependent on the bacterial secretion machinery. J Biol Chem. 1996;271:12394–12400. doi: 10.1074/jbc.271.21.12394. [DOI] [PubMed] [Google Scholar]