Abstract
Summary
Ancient DNA datasets are increasingly difficult to visualize for users lacking computational experience. Here, we describe mapDATAge, which aims to provide user-friendly automated modules for the interactive mapping of allele, haplogroup and/or ancestry distributions through space and time. mapDATAge enhances collaborative data sharing while assisting the assessment and reporting of spatiotemporal patterns of genetic changes.
Availability and implementation
mapDATAge is a Shiny R application designed for exploring spatiotemporal patterns in ancient DNA data through a graphical user interface. It is freely available under GNU Public License in Github: https://github.com/xuefenfei712/mapDATAge.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Ancient DNA research focuses on the genetic characterization of archaeological assemblages and sediments within the last 1.5 million-year timescale (van der Valk et al., 2021). With the ever-growing capacity of high throughput sequencing instruments and improved DNA manipulation techniques, it has become increasingly possible to chart patterns of genetic variation through space and time at the scale of uniparental markers, individual Single Nucleotide Polymorphism (SNP) or even the whole genome (Orlando et al., 2021). The temporal stratification of allelic frequencies at individual loci has also provided improved resolution into the detection of selection signatures (Schraiber et al., 2016). Furthermore, spatiotemporal changes in individual ancestry profiles have helped reconstruct the atlas of past population migrations on the planet, mostly in humans (Nielsen et al., 2017), but increasingly across a range of other species, mainly domestic plants (Kistler et al., 2020) and animals (Frantz et al., 2020).
While ancient DNA analysis typically involves the exploration of patterns of genetic variation through space and time, there are currently no user-friendly tools facilitating the underlying visualization steps. mapDATAge provides the first interactive platform to map spatiotemporal patterns in ancient genetic data. It helps users generate hypotheses by identifying regions and/or time periods characterized by important changes in their genetic composition. It also improves the collaborative experience by allowing all stakeholders and project partners to directly interact with the data.
2 Implementation
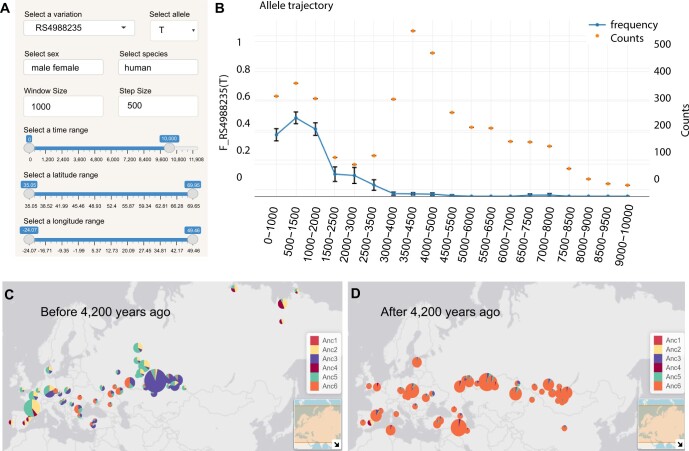
mapDATAge is designed to visualize and explore the presence of geographic and temporal patterns in ancient DNA data. It takes simple tabulated text files as input, providing samples as rows and those data types to be visualized as columns, including age, GPS coordinates, presence/absence of alleles, ancestry components, Principal Component Analysis (PCA) coordinates and more. It displays different modules to interactively: (i) map the spatiotemporal distribution of a given set of samples or alleles (AMAP); (ii) draw temporal trajectories of allele frequencies, estimating mean and confidence intervals assuming binomial sampling for genotype data or iteratively random sampling one read per sample if read counts are provided (TRAJECTORY, Fig. 1A and B); (iii) draw maps of individual ancestry profiles in two user-defined time slices (ANCESTRY, Fig. 1C and D); (iv) PCA and/or Mutidimensional Scaling (MDS) projections (PCA); (v) draw the spatial distribution of alleles at one or multiple loci (MULTISNPS) and (vi) map (sub) haplogroup distributions (HAPLO).
Fig. 1.
Two examples of data visualization with mapDATAge. (A) Configuration options; (B) Temporal trajectory for the T allele at the rs4988235 locus in Europeans (data from the Allen Ancient DNA Resources, https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data). Colored dots represent the number of individuals considered in each time bin. (C) and (D) Horse ancestry profiles prior to and following 4200 years ago (data from Librado et al., 2021). Six genetic ancestry components were considered
Users can select the geographic and/or temporal range of interest, color palette options and the list of annotations displayed on each individual location. The final ONECLICK module allows users to automatically generate figures in html format, applying a preselected range of temporal and spatial parameters. This can prove useful to contrast data from different loci and/or species.
3 Application
To demonstrate the versatility of mapDATAge, we prepared three example files providing geolocated and time-stamped ancient DNA data. The first tabulates the frequency of T allele at rs4988235, responsible for lactose tolerance, in 2120 ancient and modern Europeans, together with sex and mitochondrial haplotype information. The second provides the ancestry profiles and PCA components of 271 horses from Librado et al. (2021). The last dataset tabulates allele counts for 427 previously published ancient horses (Fages et al., 2019; Librado et al., 2015, 2017, 2021), at nine loci, causative for locomotory, stature and coat-coloration phenotypes. Figure 1A shows menus allowing users to select specific visualization parameters, such as the time and geographic range, etc. Figure 1B illustrates the AMAP panel, which shows the previously reported rise of the rs4988235 T frequency within the last ∼3000 years in Europe (Segurel et al., 2020). Figure 1C and D were generated using the ANCESTRY module to illustrate the massive change in the horse genomic makeup that followed the expansion of the DOM2 bloodline approximately ∼4200 years ago (Librado et al., 2021). Installation instructions, guidance for formatting input files and further illustrations of additional features are provided as Supplementary Information.
4 Conclusion
mapDATAge facilitates the interactive visualization of ancient DNA data through space and time. It provides a user-friendly platform for the discovery of spatiotemporal shifts in the genetic composition of populations of interest, which can serve as the basis for generating new hypotheses. It also enhances the collaborative experience by allowing all stakeholders, including those lacking genetic and/or bioinformatic expertise, to actively explore data content.
Supplementary Material
Acknowledgements
We thank Dr Pablo Librado for fruitful comments on earlier versions of the manuscript and for formatting the horse ancestry data.
Funding
This work was supported by the Marie Curie Intra-European Fellowship programme [grant number 101027750]; and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme [grant agreement 681605].
Conflict of interest: none declared.
Contributor Information
Xuexue Liu, Centre for Anthropobiology and Genomics of Toulouse, CNRS UMR 5288, Université de Toulouse, Université Paul Sabatier, Toulouse 31000, France.
Ludovic Orlando, Centre for Anthropobiology and Genomics of Toulouse, CNRS UMR 5288, Université de Toulouse, Université Paul Sabatier, Toulouse 31000, France.
References
- Fages A. et al. (2019) Tracking five millennia of horse management with extensive ancient genome time series. Cell, 177, 1419–1435.e1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz L.A.F. et al. (2020) Animal domestication in the era of ancient genomics. Nat. Rev. Genet., 21, 449–460. [DOI] [PubMed] [Google Scholar]
- Kistler L. et al. (2020) Ancient plant genomics in archaeology, herbaria, and the environment. Annu. Rev. Plant Biol., 71, 605–629. [DOI] [PubMed] [Google Scholar]
- Librado P. et al. (2015) Tracking the origins of Yakutian horses and the genetic basis for their fast adaptation to subarctic environments. Proc. Natl. Acad. Sci. USA, 112, E6889–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. et al. (2017) Ancient genomic changes associated with domestication of the horse. Science, 356, 442–445. [DOI] [PubMed] [Google Scholar]
- Librado P. et al. (2021) The origins and spread of domestic horses from the Western Eurasian steppes. Nature, 598, 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. et al. (2017) Tracing the peopling of the world through genomics. Nature, 541, 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando L. et al. (2021) Ancient DNA analysis. Nat. Rev. Methods, 1, 14. [Google Scholar]
- Schraiber J.G. et al. (2016) Bayesian inference of natural selection from allele frequency time series. Genetics, 203, 493–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurel L. et al. (2020) Why and when was lactase persistence selected for? Insights from Central Asian herders and ancient DNA. PLoS Biol., 18, e3000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Valk T. et al. (2021) Million-year-old DNA sheds light on the genomic history of mammoths. Nature, 591, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.