Abstract
Introduction:
The micro-flora of oral cavity is a myriad of micro-organism. Any infection of oral cavity leads to diseased condition which is a transitional transformation of the micro-organism in a specific paradigm depending upon the diseased condition. Periodontitis is one of the predominant chronic diseases which is a multifactorial infection. Porphyromonas gingivalis is a key etiological agent in causing periodontitis. To study the predominance of these bacteria in the diseased condition is important to detect, quantify and to find its efficacy by comparing different methods for identification.
Aim and Objectives:
The aim of the study is to determine the prevalence of P. gingivalis by anerobic culture and by real-time polymerase chain reaction (PCR) from subgingival plaque samples of chronic periodontitis and healthy individual and to compare efficacy of two methods.
Materials and Methods:
A total of 400 subjects were considered, and subgingival plaque was collected using paper points. Individual were equally divided into two groups: chronic periodontitis (200) and healthy individuals (200). Each plaque sample collected was divided into two aliquots of which the first aliquot was subjected for anerobic culture to isolate P. gingivalis. Phenotypical identification was done morphologically and biochemically further quantification of P. gingivalis was done by colony-forming unit. The second aliquot was subjected for DNA extraction and real-time PCR was conducted to detect and quantify P. gingivalis using specific primer.
Results:
Out of 400 samples, 73% showed detection of P. gingivalis by culture method and through reverse transcription-PCR (RT-PCR), the detection was 75%. Individual detection of P. gingivalis by culture in chronic periodontitis was 89.5% and 54.4% in healthy individuals, while detection by RT-PCR was found to be 91.5% in chronic periodontitis and 58% in healthy individuals. However, comparison between two techniques in detection of P. gingivalis was statistically insignificant.
Conclusion:
When we compared RT-PCR with culture RT-PCR showed higher positivity. RT-PCR is more sensitive and requires less time to detect. However, in the present study, culture also showed good positivity, suggesting proper dilution and with extended incubation, the specificity of culture can be improved to a great extent.
Keywords: Culture and reverse transcription-polymerase chain reaction, periodontitis, plaque, Porphyromonas gingivalis, prevalence, sub gingival
INTRODUCTION
Chronic periodontitis is one of the most common diseases to affect the oral cavity of adult human beings. It is a complex, multifactorial, polymicrobial infection characterized by destruction of tooth-supporting tissues.[1] Over the years, substantial data have been accumulated by researchers which implicate only a small proportion of bacteria residing in the subgingival niche in the initiation and progression of periodontal disease.[2] These include Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythia, Treponema species, Prevotella species, Selenomonas, Aggregatibacter actinomycetemcomitans, Filifactor alocis, Synergistetes species to name a few.[3] Among them, P. gingivalis, a Gram-negative anerobic bacillus and a member of the red complex triad, is considered to be a keystone pathogen in the pathogenesis of chronic periodontitis.[4,5]
Extensive researches done in recent years point evidence about an array of virulence factors produced by P. gingivalis which are responsible for tissue damage and complications seen in periodontal disease.[6] In addition, this organism is also associated with several systemic diseases such as atherosclerosis, coronary artery disease, obesity, preterm labor rheumatoid arthritis and cancers.[7,8,9]
Hence, it is of paramount importance to detect and quantify the presence of P. gingivalis in oral and extraoral lesions so that proper preventive and therapeutic measures can be undertaken.
There are several laboratory methods that can be used to isolate and identify P. gingivalis from clinical samples. One of the most commonly used methods is cultivation of the organism using a combination of selective and nonselective media and incubation in an anerobic atmosphere.[10] The cultured bacteria can be studied for their physiologic and pathogenic characteristics and for their antimicrobial susceptibility pattern. However, culture takes several days, is labor intensive and quite often it is difficult to separate P. gingivalis from other black-pigmented anerobic bacteria that reside in the oral cavity.[11] The introduction of molecular methods such as polymerase chain reaction (PCR), hybridization, microarray, 16s ribosomal ribonucleic acid (rRNA) sequencing and electrophoresis-based techniques has made detection of oral anerobic bacteria easier and quicker. Among various molecular techniques described, PCR is the most popular because of its ease of performance, high sensitivity and specificity. There are several variations of PCR that is being used with different applications in a clinical microbiology setup. Among them, real-time PCR is most commonly used since it helps to detect even low copy numbers of the organisms and also in quantitation of the number of organisms present in a clinical sample.[12,13,14,15] There are only few studies conducted in the literature comparing culture and PCR in the detection of P. gingivalis, and these studies have been carried out on certain European population,[11,16] but there are no studies that have compared the efficacy of real-time PCR and culture in detection and quantitation of P. gingivalis from subgingival plaque samples among Indian population. It is evident that oral microbe shows variation in the prevalence with respect geographic location,[17] thus making it necessary to check for prevalence among Indian population.
In the light of the background information, the present study was aimed to detect and quantitate P. gingivalis from subgingival plaque sample of healthy subjects and patients with chronic periodontitis using culture and real-time PCR assay.
MATERIALS AND METHODS
The study included a total of 400 subjects of which 200 were apparently healthy individuals (Group I) and 200 were patients with chronic periodontitis (Group II). The participants for the study were selected from patients visiting the outpatient department of our institute. Ethical clearance was obtained from the Institutional Ethical committee. A written informed consent was obtained from each participant before enrolling for the study.
Selection criteria – samples were collected from patients with chronic periodontitis and healthy individuals between the age group of 18 and 60 years belonging to both sexes were enrolled for the study. The inclusion criteria for healthy group were no signs of gingival inflammation, absence of bleeding on probing, probing depth of ≤3 mm, with no clinical attachment loss. The criteria for including chronic periodontitis patients for the study were presence of more than 20 natural teeth in situ, clinical attachment loss ≥5 mm in at least 4 or more teeth, bleeding on probing the presence of gingival inflammation and probing depth of ≥5 mm. The exclusion criteria for both groups included patients with diabetes or any other systemic illness, patients having a habit of tobacco use, patients on any types of medication, pregnant women, lactating mothers, patients who had undergone, periodontal treatment/antimicrobial therapy for a period of 3 months before study and subjects with <20 teeth. Subjects who met inclusion and exclusion criteria were considered for the study. Subgingival plaque sample was collected from each participant after obtaining written informed consent.
Microbiological sampling
After stripping off the supragingival plaque, the subgingival plaque samples were collected for microbiological study using sterile endodontic paper points: at least 4 teeth were sampled, in both healthy and chronic periodontitis. In chronic periodontitis, sample was collected from the deepest pocket site or most diseased site. All paper points from each subject were put in one vial containing reduced transport fluid (RTF) and transferred to the laboratory at the earliest.[18]
Immediately upon receipt in the laboratory, each sample was vortexed for 30 s and was divided into two aliquots; one portion was subjected to DNA extraction and the second one was used for bacterial culture.
Microbial culture
The aliquot to be used for culture was serially diluted in RTF and plated on to Blood Agar and Kanamycin Blood Agar each supplemented with hemin and Vitamin K. The plates were incubated anerobically in an anerobic jar with modified gas pack system for 7 days.[19] At the end of the incubation period, plates were inspected for the presence of small, shiny, circular, black-pigmented and mucoid colonies with or without hemolysis [Figure 1]. The number of colonies was recorded and the morphology was confirmed by Gram staining. Isolated colonies were subcultured on a fresh Blood Agar medium to obtain pure colonies and were subjected to biochemical characterization such as catalase, indole nitrate reductase and sugar fermentation tests. Those colonies which were catalase negative, indole positive, reduced nitrate, did not ferment any carbohydrates and did “not” show any fluorescence under UV light were considered as P. gingivalis [Figure 2]. The number of confirmed colonies on the original plate were multiplied by dilution factor and expressed as colony-forming units (CFUs/ml).
Figure 1.
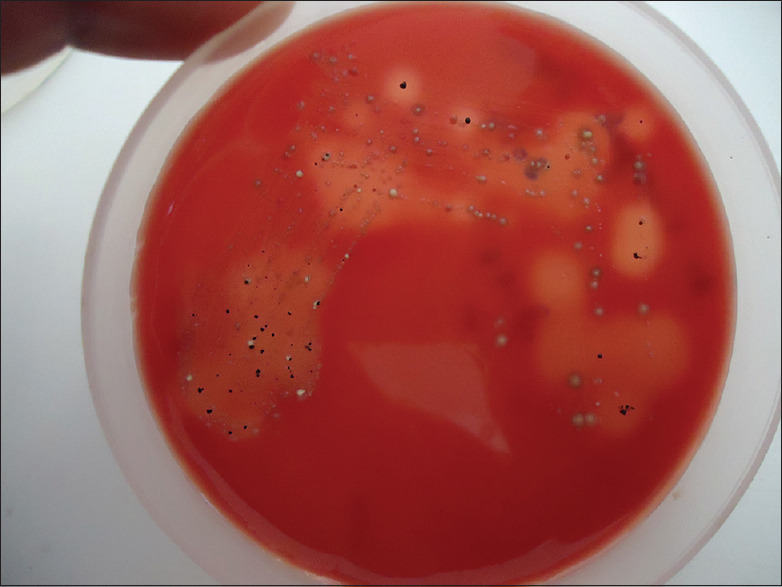
Photograph showing black-pigmented colonies of Porphyromonas gingivalis on blood agar plate
Figure 2.
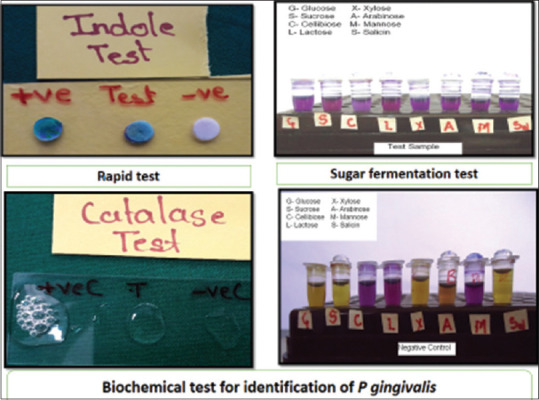
Photograph showing various biochemical reactions for identification of Porphyromonas gingivalis, which shows indole test positive (greenish-black color change) when compared with positive (greenish color) and negative (no color change) control; catalase test did not produce effervescence when compared to positive (effervescence produced) and negative (no effervescence) controls and sugar fermentation test showing no color change
DNA extraction and reverse transcription-polymerase chain reaction procedure
DNA extraction was carried out by “Modified Proteinase K method” as described previously.[20] In brief, the plaque sample was vortexed and washed three times in Tris-ethylenediaminetetraacetic acid (EDTA) buffer (pH 7.5) containing 1M Tris base and 0.5M EDTA. After this, 50 μl of lysis buffer I containing 1M Tris, 0.5M EDTA, Triton X-100 was added followed by addition of 50 μl lysis Buffer II containing 50 mM Tris-HCl (pH 8.0), 50 mM potassium chloride, 50 mM Magnesium chloride, 0.45% Tween-20, 0.45% Nodient P-40. For protein degradation, 10 μl of proteinase-K (10 mg/ml) was added and incubated at 60°C for 2 h and then kept in boiling water bath for 10 min to inactivate the enzyme. The sample was then centrifuged and the supernatant containing DNA was aliquoted in a separate tube and stored at −20°C till further processing.
DNA extracted from the standard strain of P. gingivalis ATCC No. 33277 was used in the study to generate standard curve. 16S rRNA species-specific gene of P. gingivalis was amplified by using primers; forward primer 5'-AGG CAG CTT GCC ATA CTG CG-3' and reverse primer 5'-ACT GTT AGC AAC TAC CGA TGT-3'.[18] Real-time PCR was performed in 20 μl total volume with FastStart Universal SYBR Green Master, ×2 concentrated master mix (Roche, Switzerland) that contains 2.5 mM MgCl2, FastStartTaq DNA Polymerase, Reaction Buffer, Nucleotides (dATP, dCTP, dGTP, dUTP) and SYBR Green I dye.
Primers at a concentration of 8 p mole/μl and DNA template of about 100 ng concentration were added to the reaction mixture. The tubes were kept in Realplex master cycler (Eppendorf, Germany) and the thermal cycling conditions included; initial denaturation at 95°C for 5 min followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 30 s. Melting curve analysis was performed to check the specific amplification by the primers. Cycle thresholds (Cts value) for all standard samples were obtained and standard curve was plotted (Ct value against quantity).
Regression line with slope of −3.2 and R2 value close to 1.0 is considered as optimum. Ct for unknown samples was obtained and plotted onto the standard curve and corresponding quantity was obtained [Figure 3].
Figure 3.
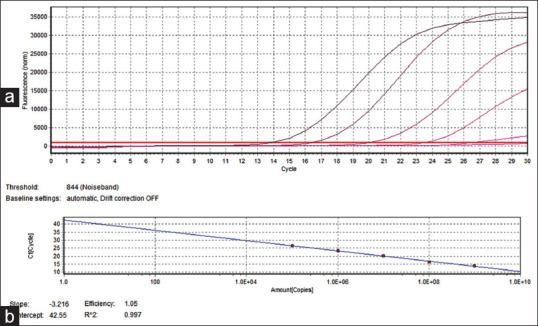
Graph showing identification of Porphyromonas gingivalis through reverse transcription-polymerase chain reaction, (a) amplification of test DNA, (b) standard curves
Statistical analysis
The results obtained were tabulated and statistical analysis was done using GraphPad prism software version 5.1 (GraphPad software Inc., USA).
RESULTS
The study comprised of 200 adult participants in each group belonging to both the sexes. In the healthy group, there were 81 males and 119 females; in diseased group, the numbers of males and females were 92 and 108, respectively. When all samples were analysed we found 72% of samples positive for P.Gingivalis through culture and 75% positivity through RT- PCR [Table 1]. The results obtained were statistically insignificant. On comparing the prevalence of P. gingivalis in healthy and diseased between culture and reverse transcription-PCR (RT-PCR), it was statistically insignificant [Table 2]. It was observed that in both study groups, RT-PCR showed higher percentage compared to culture in detecting P. gingivalis. However, this difference was statistically not significant. However, both methods showed significant difference in the prevalence of P. gingivalis between healthy and diseased groups [Tables 3 and 4].
Table 1.
Prevalence of Porphyromonas gingivalis by culture and reverse transcriptase-polymerase chain reaction
| Methods | Negative | Positive | Total | Fisher’s exact test (P) |
|---|---|---|---|---|
| Culture | 112 (28.0) | 288 (72.0) | 400 (100.0) | 0.7488 (nonsignificant) |
| RT-PCR | 101 (25.0) | 299 (75.0) | 400 (100.0) |
RT-PCR: Reverse transcriptase-polymerase chain reaction
Table 2.
Comparison of prevalence of Porphyromonas gingivalis in chronic periodontitis and healthy by culture and reverse transcriptase-polymerase chain reaction
| Groups | Methods | Negative | Positive | Total | Fisher’s exact test (P) |
|---|---|---|---|---|---|
| Chronic periodontitis | Culture | 21 (10.5) | 179 (89.5) | 200 (100.0) | 0.8056 (nonsignificant) |
| RT-PCR | 17 (8.5) | 183 (91.5) | 200 (100.0) | ||
| Healthy | Culture | 91 (45.5) | 109 (54.5) | 200 (100.0) | 0.7755 (nonsignificant) |
| RT-PCR | 84 (42.0) | 116 (58.0) | 200 (100.0) |
RT-PCR: Reverse transcriptase-polymerase chain reaction
Table 3.
Comparison of prevalence of Porphyromonasgingivalis by culture in chronic periodontitis and healthyindividuals
| Groups | Culture | Total | Fisher’s exact test | |
|---|---|---|---|---|
|
| ||||
| Negative | Positive | |||
| Chronic periodontitis | 21 (10.5) | 179 (89.5) | 200 (100.0) | <0.001 (significant) |
| Healthy | 91 (45.5) | 109 (54.5) | 200 (100.0) | |
| Total | 112 (28.0) | 288 (72.0) | 400 (100.0) | |
Table 4.
Comparison of prevalence of Porphyromonas gingivalis by reverse transcriptase-polymerase chain reaction in chronicperiodontitis and healthy individuals
| Groups | RT-PCR | Total | Fisher’s exact test | |
|---|---|---|---|---|
|
| ||||
| Negative | Positive | |||
| Chronic periodontitis | 17 (8.5) | 183 (91.5) | 200 (100.0) | <0.001 (significant) |
| Healthy | 84 (42) | 116 (53) | 200 (100.0) | |
| Total | 101 (25.25) | 299 (74.75) | 400 (100.0) | |
RT-PCR: Reverse transcriptase-polymerase chain reaction
Further, in culture CFU value for healthy and chronic periodontitis was compared, the difference was statistically significant. Similar finding was observed with RT-PCR where the copy numbers of DNA were higher in chronic periodontitis compared to healthy and were statistically significant [Figure 4].
Figure 4.
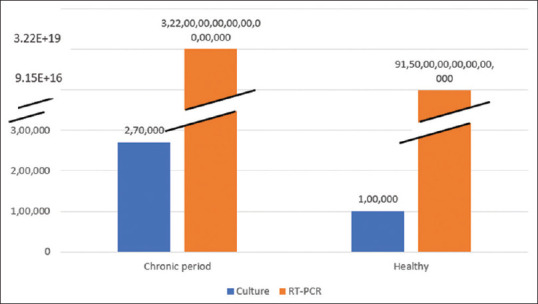
Graph showing for colony-forming unit in culture and copy numbers through reverse transcription-polymerase chain reaction in subgingival plaque of chronic periodontitis and healthy individuals
When culture was considered as gold standard and compare with PCR for sensitivity and specificity, sensitivity was 93.97% and specificity was 93.06% [Table 5].
Table 5.
Correlation between detection of Porphyromonas gingivalis by real-time polymerase chain reaction and anerobic culture in subgingival plaque samples
| Anerobic culture | RT-PCR | Sensitivity (%) | Specificity (%) | ||
|---|---|---|---|---|---|
|
| |||||
| Positive | Negative | Total | |||
| Positive | 281 (97.5) | 7 (2.5) | 288 | 93.97 | 93.06 |
| Negative | 18 (16.10) | 94 (83.90) | 112 | ||
| Total | 299 (74.70) | 101 (25.30) | 400 | ||
RT-PCR: Reverse transcriptase-polymerase chain reaction
DISCUSSION
A number of studies have evaluated the usefulness of detection and quantitation of P. gingivalis and other periodontal pathogens in plaque samples with different techniques.[12,13,15] Several of these assays have limited specificity and sensitivity and are often laborious. Recently, RT-PCR assays have been developed for study of oral bacterial pathogens including P. gingivalis, providing a means of improved detection.[13]
In the present study, we have compared RT-PCR and culture to determine the additional value of PCR in subgingival plaque samples with low numbers of P. gingivalis. The results of both culture and RT-PCR showed that P. gingivalis was predominant in patients with periodontitis in comparison to healthy individuals and this difference was highly significant. Studies from different geographic locations have reported varying prevalence of P. gingivalis in subgingival plaques specimens of healthy and periodontally diseased adult individuals. To a large extent, the results were highly influenced by the technique used for the detection of organism. Investigators have reported 10%–66% prevalence of P. gingivalis in the oral cavity of healthy individuals and 10%–78% in patients with periodontitis by culture.[21,22,23,24,25,26,27] In our study, we observed a positivity rate of 54.5% and 89.5% in healthy and periodontally diseased individuals, respectively. Our findings are on the higher side when compared to results of other authors. We feel that there are two reasons for the detection of P. gingivalis in high numbers in our study. One reason could be serially diluting samples before inoculation so that other rapid-growing bacteria are diluted and the presence of even low number of bacteria could be detected. In addition, we incubated all the culture plates for a period of 7 days, allowing slow-growing strains of P. gingivalis to appear increasing the detection rate.
With RT-PCR, the detection rates of P. gingivalis were 58% in healthy individuals and 91.5% in periodontally diseased subjects. Other authors have reported a prevalence rate of 9%–60% in healthy adults and 11%–93.3% in patients with periodontitis.[25,26,27,28,29,30] Such a wide variation in the results could be due to selection and number of study subjects, personal habits and most importantly, selection of primers that target different areas of 16S rRNA gene in various studies.
When the results of culture and RT-PCR were compared in both the study groups, it was evident that RT-PCR was superior to culture in its ability to detect the presence of P. gingivalis. However, this difference in detection rate was not statistically significant. Several other investigators have also compared the efficacy of these two techniques and have reported contrasting results. Some studies have shown RT-PCR to be much superior to culture in detecting P. gingivalis by culture,[27,30,31] whereas few authors have come out with reports that are similar to our findings.[21] We also performed quantitation of P. gingivalis in both study groups by culture and RT-PCR. It could be seen that the CFU levels were significantly higher in periodontitis patients than in healthy individuals. Similar findings were seen with RT-PCR wherein there was a significant difference in the copy numbers of P. gingivalis between the two groups. These findings are in accordance with the reports from several other authors.[21,22,23] When sensitivity and specificity were checked keeping culture as gold standard, we obtained sensitivity 93.97% and specificity 93.06%. Similar correlation was made by Boutaga et al. and they obtained sensitivity of 100% and specificity of 94%.[11] Our findings showed that larger number of positive samples detected by PCR compared to the number detected by culture is due to the detection limit of culture.
CONCLUSION
Our study clearly shows that numerically and quantitatively P. gingivalis is present in much higher proportion in periodontally diseased than in healthy individuals. The findings also reveal that by proper dilution and extended incubation, the specificity of culture can be improved to a great extent. However, RT-PCR is technically simple, more sensitive and has the ability to detect specific organism in a few hours' time.
Financial support and sponsorship
Nil.
Conflict of interest
There are no conflicts of interest.
Acknowledgment
The authors would like to thank the Principal, Maratha Mandal's NGH Institute of Dental Sciences and Research Center, Belagavi, for encouraging to conduct the study.
REFERENCES
- 1.Hajishenegallis G, Lammont RJ. Breaking bad: Manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 2014;44:328–38. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.How KY, Song KP, Chan KG. Porphyromonas gingivalis: An overview of periodontopathic pathogen below the gum line. Front Microbiol. 2016;7:53. doi: 10.3389/fmicb.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora N, Mishra A, Chugh S. Microbial role in periodontitis: Have we reached the top. Some unsung bacteria other than red complex? J Indian Soc Periodontol. 2014;18:9–13. doi: 10.4103/0972-124X.128192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in sub gingival plaque. J. Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 5.Olsen I, Lambris JD, Hajishongalis G. Porphyromonas gingivalis disturbs host commensal homeostasis by changing complement function. J Oral Microbiol. 2017;9:1340085. doi: 10.1080/20002297.2017.1340085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajishengalis G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genco RJ, Van Dyke TE. Prevention: Reducing the risk of CVD in patients with periodontitis. Nat Rev Cardiol. 2010;7:479–80. doi: 10.1038/nrcardio.2010.120. [DOI] [PubMed] [Google Scholar]
- 8.Lungberg K, Wegner N, Yucer-Lindberg T, Venables PJ. Periodontitis in RA: The citrullinatedenolase connection. Nat Rev Rheumatol. 2010;6:727–30. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- 9.Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10:e1003933. doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes BP, Jacinto RC, Pinheiro ET, Sousa EL, Zaia AA, Ferraz CC, et al. Porphyromonas gingivalis, Porphyromonas endodontalis, Prevotella intermedia and Prevotella nigrescens in endodontic lesions detected by culture and by PCR. Oral Microbiol Immunol. 2005;20:211–5. doi: 10.1111/j.1399-302X.2005.00214.x. [DOI] [PubMed] [Google Scholar]
- 11.Boutaga K, van Winkelhoff AJ, Vandenbrouke-Grauls CM, Savelkoul HM. Comparison of real time PCR and culture for detection of Porphyromonas gingivalis in subgingival plaque samples. J Clin Microbiol. 2003;41:4950–4. doi: 10.1128/JCM.41.11.4950-4954.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gohler A, Hetzer A, Holtfreter B, Giesel MH, Schmidt OC, Steinmetz I, et al. Quantitative molecular detection of putative periodontal pathogens in clinically healthy and periodontally diseased subjects. PLoS One. 2014;9:e99244. doi: 10.1371/journal.pone.0099244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asikainen S, Karched M. Molecular techniques in oral microbial taxonomy, identification and typing. In: Rogers A, editor. Molecular Oral Microbiology. UK: Caister Academic Press; 2008. pp. 1–27. [Google Scholar]
- 14.D'Ercole S, Catamo G, Piccolomini R. Diagnosis in periodontology: A further aid through microbiological tests. Crit Rev Microbiol. 2008;34:33–41. doi: 10.1080/10408410701693317. [DOI] [PubMed] [Google Scholar]
- 15.Sanz M, Lau L, Herrera D, Morillo JM, Silva A. Methods of detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in periodontal microbiology, with special emphasis on advanced molecular techniques: A review. J Clin Periodontol. 2004;31:1034–47. doi: 10.1111/j.1600-051X.2004.00609.x. [DOI] [PubMed] [Google Scholar]
- 16.Lyons SR, Griffen AL, Leys EJ. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000;38:2362–5. doi: 10.1128/jcm.38.6.2362-2365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contreras A, Moreno SM, Jaramillo A, Pelaez M, Duque A, Botero JE, et al. Periodontal microbiology in Latin America. Periodontol 2000. 2015;67:58–86. doi: 10.1111/prd.12074. [DOI] [PubMed] [Google Scholar]
- 18.Loesche WJ, Hockett RN, Syed SA. The predominant cultivable flora of tooth surface plaque removed from institutionalized subjects. Arch Oral Biol. 1972;17:1311–25. doi: 10.1016/0003-9969(72)90164-1. [DOI] [PubMed] [Google Scholar]
- 19.Vaidyalingam PK, Lakshminarayana CS, Doraswami MA. Internal gas generator – A system suitable for creating anaerobic atmosphere. Ind J Surg. 1981;43:154–9. [Google Scholar]
- 20.Evan PV, van Belkum A, Hayes JP. The different types and varieties of nucleic acid target molecules. In: Principles and Technical Aspects of PCR Amplification, Dordrecht. Netherlands: Springer; 2008. p. 34. [Google Scholar]
- 21.Lau L, Sanz M, Herrera D, Morillo JM, Martín C, Silva A. Quantitative real-time polymerase chain reaction versus culture: A comparison between two methods for the detection and quantification of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in subgingival plaque samples. J Clin Periodontol. 2004;31:1061–9. doi: 10.1111/j.1600-051X.2004.00616.x. [DOI] [PubMed] [Google Scholar]
- 22.Kulkarni M, Bhat KG, Thomos BS, Bhat SG, Kulkarni RD. Identification of Multiple strains of Porphyromonas gingivalis using heteroduplex polymerase chain reaction in varying severity of chronic periodontitis. Indian j Med Microbiol. 2019;36:81–6. doi: 10.4103/ijmm.IJMM_17_434. [DOI] [PubMed] [Google Scholar]
- 23.Avila-Campos MJ, Velásquez-Melendez G. Prevalence of putative periodontopathogens from periodontal patients and healthy subjects in São Paulo, Sp, Brazil. Rev Inst Med Trop Sao Paulo. 2002;44:1–5. doi: 10.1590/s0036-46652002000100001. [DOI] [PubMed] [Google Scholar]
- 24.D'Ercole S, Catamo G, Tripodi D, Piccolomini R. Comparison of culture method and multiplex PCR for the detection of periodontopathogenic bacteria in biofilm associated with severe forms of periodontitis. New Microbiol. 2008;31:383–91. [PubMed] [Google Scholar]
- 25.Savitt ED, Vaccaro S. Comparison of cultural methods and DNA probe analysis for detection of Actinobacillus actinomycetemcomitans, Bacteriodesgingivalis and Bacteriodesintermedia in subgingival plaque sample. J Periodontal. 1988;59:431–8. doi: 10.1902/jop.1988.59.7.431. [DOI] [PubMed] [Google Scholar]
- 26.Slot J, Hafstom C, Rosling B, Dahlen G. Detection of Actinobacillus actinomycetemcomitans and Bacteriodesgingivalis in subgingival smear by the indirect fluorescent antibody technique. J Periodonol Res. 1985;20:613–20. doi: 10.1111/j.1600-0765.1985.tb00845.x. [DOI] [PubMed] [Google Scholar]
- 27.Nonnenmacher C, Dalpke A, Mutters R, Heeg K. Quantitative detection of periodontopathogens by real-time PCR. J Microbiol Methods. 2004;59:117–25. doi: 10.1016/j.mimet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Ploeg Jan R, Giertsen E, Ludin B, Morgeli C, Zinkernagel A, Gmur R. Quantitative detection of Porphyromonas gingivalis fim – A genotypes in dental plaque. FEMS Microbiol Lett. 2004;232:31–7. doi: 10.1016/S0378-1097(04)00064-3. [DOI] [PubMed] [Google Scholar]
- 29.Teixeira SR, Mattarazo F, Feres M, Figueiredo LC, de Faveri M, Simionato MR, et al. Quantification of Porphyromonas gingivalis and fim – A genotypes in smoker chronic periodontitis. J Clin Peridontol. 2009;36:482–7. doi: 10.1111/j.1600-051X.2009.01411.x. [DOI] [PubMed] [Google Scholar]
- 30.Kotsilkov K, Popova C, Boyanova L, Setchanova L, Mitov I. Comparison of culture method and real-time PCR for detection of putative periodontopathogenic bacteria in deep periodontal pockets. Biotechnol Biotechnol Equip. 2015;29:996–1002. [Google Scholar]
- 31.Jervoe-Storm PM, Koltzacher M, Falk W, Dorfier A, Jepsen S. Comparison of culture and real-time PCR for detection and quantification of five putative periodontopathogenic bacteria in subgingival plaque sample. J Clin Periodontol. 2005;32:778–83. doi: 10.1111/j.1600-051X.2005.00740.x. [DOI] [PubMed] [Google Scholar]


