Abstract
Background
Since the beginning of the COVID-19 pandemic, researchers have tried to find the reason behind the variety of the symptoms and disease severity among patients. It seems that genetic background may contribute in severity of this infection. The renin-angiotensin system (RAS) is involved in the pathogenesis of COVID-19. An Insertion/Deletion (I/D) polymorphism in the ACE1 gene may explain the genetic risk for disease severity.
Methods
We genotyped 251 COVID-19 patients: 151 patients with mild or asymptomatic disease compared with 100 patients with severe to critical illness (without any comorbidities for the disease severity).
Results
There was a significant association between the ACE1 DD genotype and disease severity (p-value = 1 × 10−2; OR = 2.004, 95%CI = 1.147–3.499) and our results showed that it was inherited under recessive or codominant inheritance patterns. Also, the I allele showed a protective role against the severe form of COVID-19 disease (p-value = 1 × 10−4).
Conclusion
We concluded that ACE1 DD genotype can predict the risk of severe form of COVID-19 infection in the absence of known comorbidities as disease severity risk factors. Further studies with larger sample sizes in other populations are still needed to clarify the role of ACE I/D polymorphism in SARS-CoV-2 infection severity.
Keywords: Covid-19, ACE, Polymorphism, Insertion-deletion
1. Introduction
Following the outbreak of COVID-19 in 2019 in China and the onset of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic, initial observations showed that patients with COVID-19 had symptom diversity (Wu et al., 2020). Even infected patients in a family showed a different phenotypic spectrum of the disease: asymptomatic, mild, severe symptoms (leading to hospitalization) and critical condition (requiring admission to intensive care unit and ventilator respiration as a lethal condition) (Yanes-Lane et al., 2020; Wu and McGoogan, 2020). Although some known risk factors (such as ages above 65 years, chronic respiratory diseases, hypertension, diabetes, etc.) predispose patients to the severe form of COVID-19, the cause of the disease severity is still not fully understood in some patients without these known underlying risk factors. Based on these observations, the researchers were looking for the role of genetic background on the severity of COVID-19 infection. Accordingly, several studies have investigated several candidate genes and single nucleotide polymorphisms (SNPs) that lead to differences in susceptibility to COVID-19 (Schurr, 2020; Kachuri et al., 2020; Smatti et al., 2020). However, the findings in different populations were inconclusive.
Regarding to the entrance mechanism of COVID-19 to the host cells, it seems that the polymorphisms of ACE1 gene may be influential in the disease severity. Angiotensin converting enzymes (ACE1/ACE2) are proteins of renin-angiotensin system (RAS) which play important role in regulation of blood pressure and fluid electrolyte balance (Zhou et al., 2020). It has been shown that the COVID-19 virus enters the human cells through the binding of the viral spike protein to ACE2 as a receptor on cell surface (Zhou et al., 2020). ACE2, is expressed in a variety of human organs and tissues and is a homolog of angiotensin-converting enzyme-1 (ACE1). ACE1 has a variety of biological activities and can counteract the negative role of the renin-angiotensin system in many diseases (Donoghue et al., 2000; Patel et al., 2017; Santos et al., 2018). On the other hand, ACE2 acts as a vasodilator factor and modulates the effects of ACE1. These two enzymes have opposite effects to each other and ACE1/ACE2 balance will play an important role in the severity of coronary disease (Gemmati et al., 2020a). ACE1 encoding gene contains 26 exons and is located on chromosome 17q23.3 and also known as DCP, ACE1, DCP1, CD143 (OMIM number 106180). It encodes a key enzyme in the conversion of angiotensin I to angiotensin II (Tikhomirova et al., 2017). ACE1 insert (I)/deletion (D) polymorphism is related to a 287-bp ALU repeat in intron 16 and based on this I/D polymorphism, there are three genotypes: of heterozygous DI and homozygous II and DD (Rigat et al., 1990a). It has been shown that ACE1 I/D polymorphism importantly contribute in plasma level of ACE protein and DD genotype is related to highest level of ACE protein (Crisan and Carr, 2000). In the development of severe and critical form of COVID-19, various risk factors such as old age, underlying medical conditions, male gender as well as comorbidities such as diabetes, hypertension, heart disease, obesity, chronic lung disease, immunodeficiency and cancers have been identified (Gao et al., 2021). Interestingly, many of these comorbidities are closely associated with ACE I/D polymorphisms and given the role of these polymorphisms in the ACE/ACE2 balance, it can be a genetic risk factor for severe COVID-19 infection (Yamamoto et al., 2021).
Accordingly, we tried to investigate the association between ACE1 I/D polymorphism with susceptibility to severe form of COVID-19 infection in Iranian patients.
2. Materials and methods
2.1. Study population
From September to December 2020, two hundred and fifty-one COVID-19 patients referred to Farmanfarmayan Health center (Tehran, Iran) participated voluntarily in this study. All of the patients had positive COVID-19 RT-PCR test result for their nasopharyngeal swab samples. This study has been approved by the ethics committee of the school of medicine of Tehran University of Medical Sciences (Ethics code: IR.TUMS.MEDICINE.REC.1399.794). This study was also conducted in accordance with the Helsinki Declaration and all experiments and techniques used in this study have been performed in accordance with the relevant regulations. Blood samples were collected from all participants in EDTA-tubes as anticoagulant after signing informed consent form. Patients with asymptomatic or mild form of COVID-19 infection categorized as control group without age limitation (n = 151). Patients with severe to critical symptoms (such as a respiratory rate more than 30 beats per minute, oxygen saturation level (SPO2) less than 90%, lung infiltration more than 50% and organ failure), who also needed to be hospitalized were classified as case group (n = 100). Since our study design was based on the exclusion of known confounding factors for the association of ACE polymorphism with the disease severity (such as ages older than 65 years and underlying diseases predisposing to severe symptoms), all patients with age above 65 years and previous history of comorbidities in electronic health records for diabetes, hypertension, heart failure, stroke, cancer chemotherapy or immunodeficiency, were excluded from case group.
2.2. DNA extraction and ACE genotyping
Total genomic DNA was isolated using standard salting-out method (MWer et al., 1988). The quality and quantity of the purified DNA samples were evaluated by Nano-Drop 2000™ spectrophotometer (Thermo Fisher Scientific, USA). The extracted genomic DNA samples were stored at -20o C until genotyping.
PCR technique was used to investigate the insertion/deletion polymorphism of ACE gene. The PCR rection volume was 10 μl and each reaction consisted of: 5 μl of PCR MasterMix 2× (Yektatajhiz, Iran), 0.4 μl genomic DNA sample, 0.2 μl (10 pmol/μl) of forward primer (5’-CTGGAGACCACTCCCATCCTTTCT-3′), 0.2 μl (10 pmol/μl) of reverse primer (5’-GATGTGGCCATCACATTCGTCAGAT-3′) and 4.2 μl distilled water. For each experiment, a blank was prepared by adding 0.4 μl of water instead of a template DNA. The amplification reactions were performed using ABI Veriti thermal cycler machine (Thermo Fisher Scientific, USA). The PCR program was carried out with initial denaturation step at 94 °C for 5 min, followed by 30 amplification cycles (30 s at 94 °C, 45 s at 59 °C, and 30 s at 72 °C) and a final extension at 72 °C for 7 min. Electrophoresis on 2% agarose gels (SinaClon BioScience, Iran) containing 0.5 mg/L DNA gel stain (SinaClon BioScience, Iran) was used to separate the amplification products and the image of bands visualized under the UV light and captured by gel documentation system (Syngene, UK).
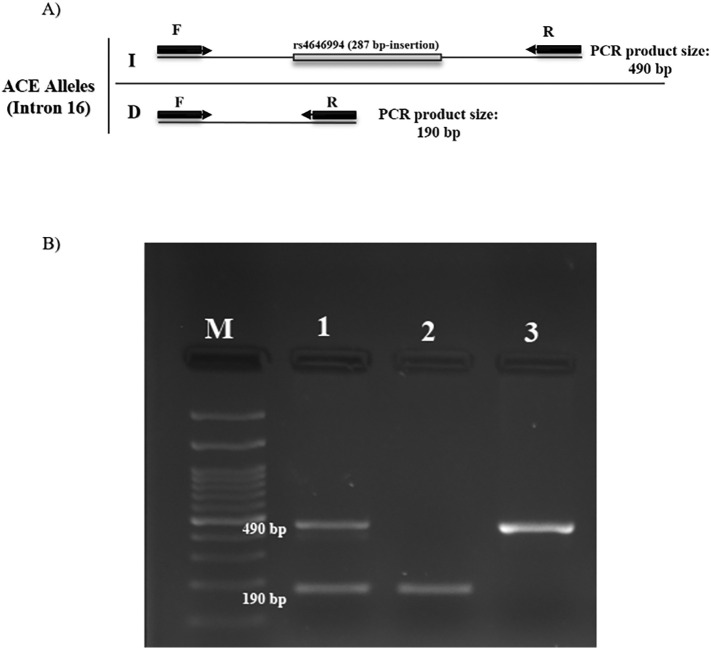
The genotype of each patient was detected based on PCR product band size on agarose gel electrophoresis. Accordingly, if a 287 bp fragment is inserted, the amplified product size is 490 bp, and if the fragment is deletion, the amplified band size is 190 bp. In the presence of both 490 and 190 bp bands, the patient has a heterozygous genotype (DI), and if only the 190 bp or only the 490 bp band is observed, the patient is homozygous for deletion mutation (DD) and insertion mutation (II) in the 287 bp fragment, respectively (Fig. 1 ). PCR-based genotyping was confirmed by sanger sequencing for some DNA samples.
Fig. 1.
A) Diagram of PCR for ACE1 genotyping. The amplified band by two forward (F) and reverse (R) primers, if 287 bp fragment inserted, PCR product is 490 bp which represents the I allele, and if the 287 bp fragment is deleted, 190 bp will be obtained, which represents the D allele. B) Electropherogram PCR product. Line M: 100 bp ladder. Line 1 shows DI genotype, line 2 shows DD genotype and line 3 is II genotype.
2.3. Statistical analysis
SPSS software (version 25) was used in statistical analysis. Allele and genotype frequencies between case and control groups were compared for significancy, using χ2 (Fisher's Exact test). Odds ratio (OR) with 95% confidence intervals (CI) were used to describe the strength of association. The genotypes frequencies in study population were evaluated for concordance with Hardy Weinberg equilibrium using SNP Analyzer 2.0 software. Also, association between ACE1 I/D polymorphism and COVID-19 severity were analyzed in recessive, dominant and codominant models by SNP Analyzer 2.0 software. All p-value of less than 0.05 was considered to show statistically significant results.
3. Results
The total number of patients participating in this study was 251. The control group consisted of 151 patients with a mean age of 38.39 ± 11.84 and patients in this group had mild symptoms. The case group consists of 100 patients with severe to critical symptoms with a mean age of 46.35 ± 10.24. The percentage of male patients participated in the control and case groups is 61% and 55%, respectively.
Regarding to the ACE1 gene I/D polymorphism, there were three genotypes (DD, II, and DI) among the study population.
The frequency of genotypes and alleles in the control and case groups was shown in Table 1 . The D allele had more frequency in the case and control groups and in whole study population. Regarding genotypes, DI genotype was more frequent in case and control groups and in all COVID-19 patients.
Table 1.
Genotypes and alleles frequencies of the study population, p-values, odds ratio (OR) and 95% confidence intervals (95% CI).
|
Genotypes na(%) |
Alleles na(%) |
Hardy-Weinberg equilibrium (p-value) |
||||
|---|---|---|---|---|---|---|
| II | DI | DD | I | D | ||
| Control group na=151 |
30 (19.9) |
87 (57.6) |
34 (22.5) |
147 (48.7) |
155 (51.3) |
0.059 |
| Case group na=100 |
18 (18) |
45 (45) |
37 (37) |
81 (40.5) |
119 (59.5) |
0.507 |
| Total study population n = 251 |
48 (19.12) |
132 (52.58) |
71 (28.28) |
228 (45.40) |
274 (54.58) |
0.336 |
| p-value | 6 × 10−1 | 4 × 10−2 | 1 × 10−2 | 1 × 10−4 | 1 × 10−2 | |
| OR (95% CI) | 0.878 (0.459–1.679) |
0.592 (0.356–0.987) |
2.004 (1.147–3.499) |
0.465 (0.342–0.631) |
0.696 (0.526–0.920) |
|
n: sample number.
The statistical analysis of the results showed a significant difference (p-value = 4 × 10−2) between the distribution of ACE genotypes between the case and control groups.
The frequency of II genotypes did not show any significant difference (p-value = 6 × 10−1) between case and control groups. But the frequency of DD genotype in the case group showed a significant increase (p-value = 1 × 10−4) in comparison with the control group. Also, the frequency of DI genotype decreased from 57% in the control group to 45% in the case group (p-value = 4 × 10−2). Also, both I and D alleles showed significant differences between case and control groups.
The frequency distribution of ACE genotypes in case and control groups as well as in the all COVID-19 patient were in accordance with Hardy-Weinberg equilibrium (Table 1). Risk assessment for the ACE gene in recessive, dominant, and co-dominant inheritance models showed that ACE1 I/D polymorphism was associated with the severe form of COVID-19 disease in both recessive (p-value = 0.013) and co-dominant (p-value = 0.041) models (Table 2 ).
Table 2.
Analysis of ACE I/D genotypes association with COVID-19 disease severity under three different inheritance models. p-values, odds ratio (OR) and 95% confidence intervals (95% CI).
| Inheritance models | Recessive (DD vs. DI + II) |
Co-dominant (DI vs. II + DD) |
Dominant (DD + DI vs. II) |
|---|---|---|---|
| p-value | 0.013 | 0.041 | 0.713 |
|
OR (95% CI) |
2.021 (1.157–3.529) |
1.814 (0.859–3.829) |
1.129 (0.591–2.16) |
4. Discussion
Global experience about COVID-19 infection showed that it cannot be a simple respiratory viral infection and there is a complex pathophysiology behind its severe presentations. Several investigations have shown that disturbance of the renin-angiotensin (RAS)system balance is involved in the clinical complications of COVID-19 patients (Wiese et al., 2020; Gemmati et al., 2020b). Abnormal changes in the ACE1/ACE2 balance, involves in the of several medical problems such as heart failure, thrombotic disorders, nephropathy and severe acute respiratory distress (Haznedaroglu, 2020; Xiao et al., 2020). The I/D polymorphism in ACE1 gene can influence on the ACE1 level in both blood circulation and tissue. It has been shown that the D allele in this polymorphism, is associated with higher ACE1 activity and it is estimated that about 60% of the ACE1 levels be determined by the ACE1 I/D polymorphism (Tiret et al., 1992). Indeed, previous investigations have been shown that individuals with DD genotype have double levels of ACE1 activity in comparison with II genotype (Rigat et al., 1990b).
According to the contribution of ACE1 in RAS system and pathophysiology of COVID-19 complications in one hand and the influence of ACE1 gene I/D polymorphism in ACE1 level in another hand, in the present study, we tried to determine the role of ACE I/D polymorphisms in the severity of symptoms in COVID-19 patients. The results of our study showed that the risk of developing severe COVID-19 disease in patients with D allele (p-value = 1 × 10−2) and DD genotype (p-value = 1 × 10−2) was significantly higher than two other genotypes (DI, II). Also, the risk of severe form of COVID-19, was increased 2-folds in individuals with DD genotype (OR = 2.004, 95%CI = 1.147–3.499) and our results showed that ACE1 D/I variant was inherited under recessive or co-dominant inheritance patterns. Our findings in Iranian patients are in concordance with the results of other studies in Indian and Spanish populations (Verma et al., 2021; Gómez et al., 2020). In another investigation on Italian patients, in spite of limited sample size (26 ICU-admitted COVID-19 patients), the results were consistent with our findings (Annunziata et al., 2020). Conversely, an investigation about ACE1 D/I association with COVID-19 risk on Czechs (as a west Slavic ethnic group in central Europe), showed un concordant results with our findings (Hubacek et al., 2021). This study reported that the frequency of II genotype was significantly higher in symptomatic COVID-19 patients. It is considerable that the participants in this study consisted of asymptomatic and symptomatic (without hospitalization) COVID-19 patients. Although the reason of this discrepancy is unclear now, but it seems that lack of severe to critical form of COVID-19 in the study population along with ethnicity background can be influential.
Comparison of ACE1 genotypes frequencies data in different ethnicities with reported COVID-19 cases and mortality rate, showed that the frequency of ACE1 II genotype was negatively correlated with the number of SARS-CoV-2 infected cases and number of deaths due to SARS-CoV-2 infection (Yamamoto et al., 2020). Our findings showed that the allele frequency of I allele was significantly higher in control group and it can be a protective allele, but the II genotype had not association with asymptomatic or mild form of COVID-19 infection in our study population.
5. Conclusion
The results of our study showed that ACE1 DD genotype is associated with the severity of COVID-19 in Iranian patients and it can double the risk of severe form of the COVID-19 disease. Also, our findings showed that the II genotype did not play a significant role against the severe form of COVID-19 disease. Thus, determining ACE genotype can help in prediction of COVID-19 infection progression to a severe form as a background genetic risk factor. Identifying the role of genetic factors on susceptibility to severe COVID-19 disease can play an important role in the better management of patients and personalized medicine as well as the development of better treatment strategies for these high-risk groups. The advantage of our study was the exclusion of patients with known risk factors for severe form of COVID-19 infection from case group to better evaluation of the influence of genetic background on disease presentation. Further studies with larger sample sizes in other populations are needed to confirm the role of ACE I/D polymorphism in SARS-CoV-2 infection as well as to identify other genetic risk-factors.
Author contributions
J.S.L. conducted the experiments and wrote the manuscript. and J.S.R contributed in laboratory works. M.S.Y. performed statistical analysis and data interpretation. P.I. designed the study, supervised the project and revised the manuscript. All of the authors reviewed and approved the final manuscript.
Declaration of Competing Interest
No potential conflict of interest was reported by the authors.
Acknowledgements
This study was a part of a M.Sc. thesis supported by Tehran University of Medical Sciences (Tehran, Iran; grant No: 49587). We would like to thank Ms. Masoumeh Amini for her technical assistance.
References
- Annunziata A., Coppola A., Lanza M., Simioli F., Imitazione P., Pepe N., Maddaloni V., Atripaldi L., Fiorentino G. ACE DD polymorphism in severe COVID-19. J. Transl. Sci. 2020;7(1):10–15761. [Google Scholar]
- Crisan D., Carr J. Angiotensin I-converting enzyme: genotype and disease associations. J. Mol. Diagn. 2000;2(3):105–115. doi: 10.1016/S1525-1578(10)60624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87(5):e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Gao Y.D., Ding M., Dong X., Zhang J.J., Kursat Azkur A., Azkur D., Gan H., Sun Y.L., Fu W., Li W., Liang H.L. Risk factors for severe and critically ill COVID-19 patients: a review. Allergy. 2021;76(2):428–455. doi: 10.1111/all.14657. [DOI] [PubMed] [Google Scholar]
- Gemmati D., Bramanti B., Serino M.L., Secchiero P., Zauli G., Tisato V. COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int. J. Mol. Sci. 2020;21(10):3474. doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmati D., Bramanti B., Serino M.L., Secchiero P., Zauli G., Tisato V. COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int. J. Mol. Sci. 2020;21(10):3474. doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez J., Albaiceta G.M., García-Clemente M., López-Larrea C., Amado-Rodríguez L., Lopez-Alonso I., Hermida T., Enriquez A.I., Herrero P., Melón S., Alvarez-Argüelles M.E. Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene. 2020;762:145102. doi: 10.1016/j.gene.2020.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haznedaroglu I.C. Immunogenomic phases of COVID-19 and appropriate clinical management. Lancet Microbe. 2020;1(7) doi: 10.1016/S2666-5247(20)30165-8. (p.e278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubacek A.J., Dusek L., Majek O., Adamek V., Cervinkova T., Dlouha D., Adamkova Adamkova. ACE I/D polymorphism in Czech first-wave SARS-CoV-2-positive survivors. Clin. Chim. Acta. 2021;519:206–209. doi: 10.1016/j.cca.2021.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachuri L., Francis S.S., Morrison M., Wendt G.A., Bossé Y., Cavazos T.B., Rashkin S.R., Ziv E., Witte J.S. The landscape of host genetic factors involved in immune response to common viral infections. medRxiv. 2020:20088054. doi: 10.1101/2020.05.01.20088054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MWer S., Dykes D., Polesky H.J. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Rauf A., Khan H., Abu-Izneid T. Renin-angiotensin-aldosterone (RAAS): the ubiquitous system for homeostasis and pathologies. Biomed. Pharmacother. 2017;94:317–325. doi: 10.1016/j.biopha.2017.07.091. [DOI] [PubMed] [Google Scholar]
- Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 1990;86(4):1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 1990;86(4):1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R.A.S., Sampaio W.O., Alzamora A.C., Motta-Santos D., Alenina N., Bader M., Campagnole-Santos M.J. The ACE2/angiotensin-(1–7)/MAS axis of the renin-angiotensin system: focus on angiotensin-(1–7) Physiol. Rev. 2018;98(1):505–553. doi: 10.1152/physrev.00023.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr T.G. Host genetic factors and susceptibility to SARS-CoV-2 infection. Am. J. Hum. Biol. 2020;32(5) doi: 10.1002/ajhb.23497. [DOI] [PubMed] [Google Scholar]
- Smatti M.K., Al-Sarraj Y.A., Albagha O., Yassine H.M. Host genetic variants potentially associated with SARS-CoV-2: a multi-population analysis. Front. Genet. 2020;11:578523. doi: 10.3389/fgene.2020.578523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhomirova V.E., Kost O.A., Kryukova O.V., Golukhova E.Z., Bulaeva N.I., Zholbaeva A.Z., Bokeria L.A., Garcia J.G., Danilov S.M. ACE phenotyping in human heart. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0181976. (p.e0181976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiret L., Rigat B., Visvikis S., Breda C., Corvol P., Cambien F., Soubrier F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am. J. Hum. Genet. 1992;51(1):197. [PMC free article] [PubMed] [Google Scholar]
- Verma S., Abbas M., Verma S., Khan F.H., Raza S.T., Siddiqi Z., Ahmad I., Mahdi F. Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect. Genet. Evol. 2021;91:104801. doi: 10.1016/j.meegid.2021.104801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese O.J., Allwood B.W., Zemlin A.E. COVID-19 and the renin-angiotensin system (RAS): a spark that sets the forest alight? Med. Hypotheses. 2020;144:110231. doi: 10.1016/j.mehy.2020.110231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. Jama. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Sakagami H., Miwa N. ACE2: the key molecule for understanding the pathophysiology of severe and critical conditions of COVID-19: demon or angel? Viruses. 2020;12(5):491. doi: 10.3390/v12050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Ariumi Y., Nishida N., Yamamoto R., Bauer G., Gojobori T., Shimotohno K., Mizokami M. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;758:144944. doi: 10.1016/j.gene.2020.144944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Yamamoto R., Ariumi Y., Mizokami M., Shimotohno K., Yoshikura H. Does genetic predisposition contribute to the exacerbation of COVID-19 symptoms in individuals with comorbidities and explain the huge mortality disparity between the East and the West? Int. J. Mol. Sci. 2021;22(9):5000. doi: 10.3390/ijms22095000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes-Lane M., Winters N., Fregonese F., Bastos M., Perlman-Arrow S., Campbell J.R., et al. Proportion of asymptomatic infection among COVID-19 positive persons and their transmission potential: a systematic review and meta-analysis. PLoS One. 2020;15(11) doi: 10.1371/journal.pone.0241536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]