Abstract
It is critically important to understand how the adaptive immune response, elicited by vaccination or infection, recognizes SARS-CoV-2. This is especially true when considering the challenges to the immune response posed by variant evolution. Herein, we summarize our work aimed at characterizing the magnitude of the CD4+ and CD8+ T cell responses to SARS-CoV-2, the proteins most frequently recognized, and the associated T cell epitope repertoire. This work formed the foundation for our most recent studies aimed at understanding and predicting the ability of T cell responses induced by SARS-CoV-2 infection or vaccination to subsequently cross-recognize novel SARS-CoV-2 variants. We found that T cell responses are remarkably preserved and able to cross-recognize SARS-CoV-2 variants, from Alpha to Omicron. This is distinct from what has been observed for the SARS-CoV-2- specific antibody and B cell responses. This body of work, supported by independent studies carried out by other groups, suggests that T cells may contribute to a second line of defense against infection while also limiting viral spread and, thus, disease severity.
Keywords: SARS-CoV-2, Variants of concern, CD4, CD8
Graphical abstract
1. Introduction
This graphical review summarizes our studies of human adaptive responses to SARS-CoV-2 infection and vaccination, with emphasis on assessing the preservation of T cell responses in the context of SARS-CoV-2 variant evolution. We introduce the role of the T cell response in adaptive immunity to SARS-CoV-2, highlight the SARS-CoV-2 proteins recognized by T cells as well as the breadth of the epitope repertoire induced by natural infection versus vaccination. We then discuss the ability of this memory T cell response to cross-recognize novel variants with specific sets of amino acid mutations. Finally, we present a model where, despite a decrease in neutralization at the antibody level, T cell responses are conserved, consistent with the relatively preserved capacity of the adaptive response to protect from severe disease and hospitalization but not infection.
2. Adaptive responses to SARS-CoV-2 and the role of T cells
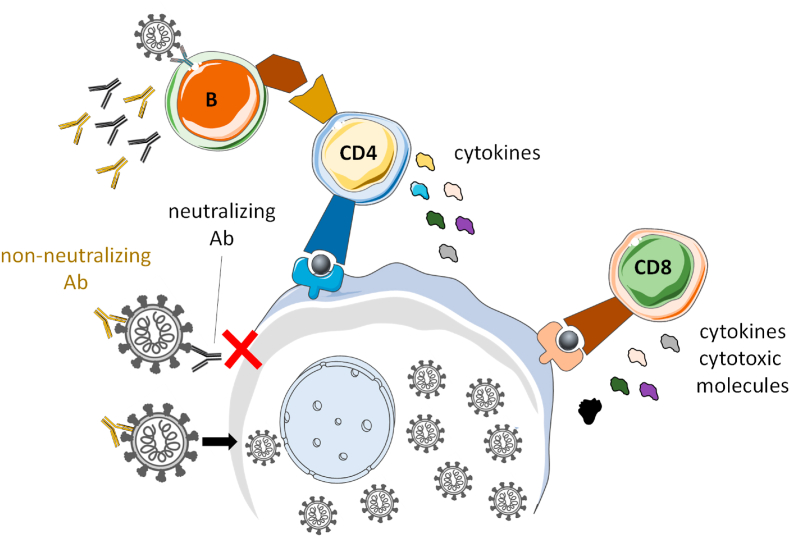
SARS-CoV-2 variant evolution poses a threat, given the unpredictability of how each new variant will impact the immune memory response elicited by vaccination or prior infection. It is therefore critical to understand the effect that these new variants have on the main actors of the adaptive immune system. Memory immunity leverages both B and T cell responses (Fig. 1). B cells produce neutralizing antibodies that can prevent infection, and non-neutralizing antibodies important for Fc effector function. CD8+ and CD4+ T cells recognize infected cells and coordinate the overall immune response to block viral spreading. While the B cell response to SARS-CoV-2 has been extensively studied, particularly in the context of viral evolution (DeGrace et al., 2022), the T cell response is less well understood.
Fig. 1.
SARS-CoV-2 adaptive response. SARS-CoV-2 infection induces different arms of the adaptive immune response. B cells produce antibodies (Abs) able to prevent infection (neutralizing) or limit spreading (non-neutralizing). CD8+ and CD4+ T cells recognize infected cells and limit viral spreading by producing cytokines and cytotoxic molecules to kill the infected cells and by helping coordinate the different arms of the immune system.
At the beginning of the pandemic, as SARS-CoV-2 sequence and structural data became available, we performed linear and structural bioinformatic analyses using the IEDB database (www.IEDB.org) and analysis resource, (Dhanda et al., 2019; Vita et al., 2019). Our bioinformatic analyses (Grifoni et al., 2020a) and those of another research group (Wu et al., 2020) revealed similarities between the novel SARS-CoV-2 and the SARS-CoV responsible for the 2003 outbreak.
We then characterizing the adaptive immune response to SARS-CoV-2, in early 2020, at the start of the pandemic. To study protective immune responses able to control the infection, we collected blood from patients with mild, uncomplicated COVID disease at 1-month post-infection (the convalescent phase) (Grifoni et al., 2020b). Patients with milder cases were chosen in order to study a protective immune response able to control the infection. Analysis of sera and peripheral blood mononuclear cells revealed that immune responses were strongest against the viral spike (S) protein at the level of IgG/IgA antibodies and also CD4+/CD8+ T cells (Grifoni et al., 2020b). Our observation that the same protein, in most patients, is dominantly recognized by both B and T cells responses is different from what is observed in infections with other viruses, such as dengue, where the dominant neutralizing antibody response is against the E protein, which is poorly recognized by the T cells that instead preferentially respond to C and NS proteins (Tian et al., 2019). This human study provided further validation that the S protein was the candidate of choice for development of SARS-CoV-2 vaccines that could induce all 3 arms of the immune response.
These results, obtained by studying COVID-19 patients in the convalescent phase (Tian et al., 2019), and additional studies of the acute phase by our group and others (Rydyznski Moderbacher et al., 2020; Tan et al., 2021; Dan et al., 2022) suggest that better outcomes correlate with a coordinated adaptive response and early T cell response kinetic. This is compatible with a model of B and T cell memory response coordination, in which neutralizing antibodies are key to preventing infection, but protection against severe disease, hospitalization, and death is dependent on non-neutralizing antibodies, T cell responses, and cellular immunity in general (eg, memory B cells) (Fig. 1).
3. SARS-CoV-2 infection elicits CD4+ and CD8+ T cell responses that each recognize up to 40 epitopes on a wide array of antigens
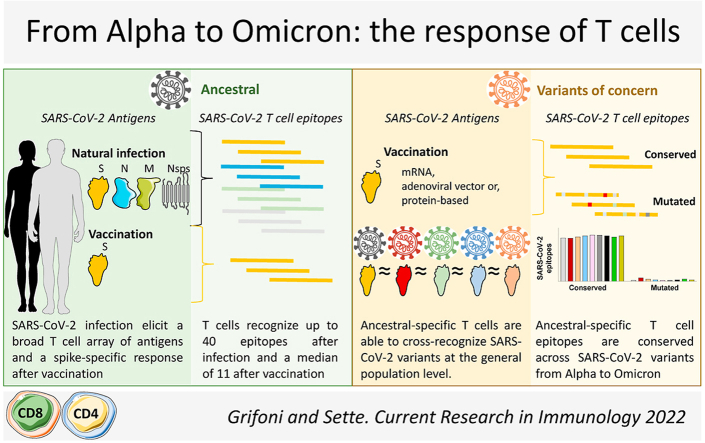
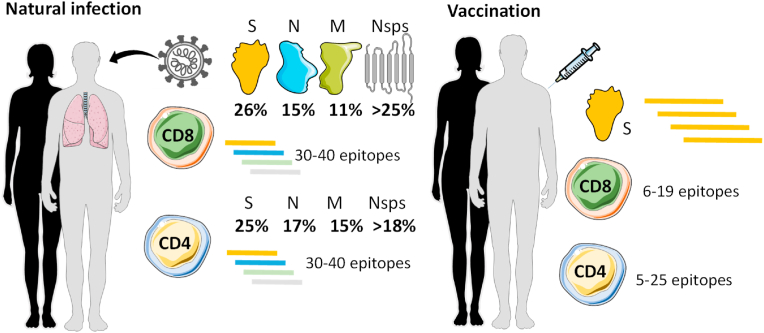
We next set out to more in depth characterize the immunodominance pattern among SARS-CoV-2 proteins and the breadth of CD4+ and CD8+ T cell responses after natural infection or vaccination (Fig. 2). We first looked at CD4+ and CD8+ T cell responses in the convalescent phase following SARS-CoV-2 infection (Tarke et al., 2021a) and found that while S protein responses accounted for roughly 25% of the overall T cell responses, other structural proteins (membrane [M], nuclear [N], and open-reading frame [ORF]) and non-structural proteins (nsp) were highly recognized as well, suggesting a broad protein immunodominance pattern. Specifically, 8 additional viral proteins accounted for 55% of the CD4+ T cell response: M (17%), N (15%), nsp3 (8%), ORF3a (5%), nsp12 (4%), nsp4 (3%), and nsp13 (3%). And 7 additional viral proteins accounted for 54% of the CD8+ T cell response: nsp3 (17%), N (15%), M (11%), ORF3a (4%), nsp4 (3%), nsp12 (3%), and nsp6 (2%) (Tarke et al., 2021a) (Fig. 2). This study suggests that, in addition to the S protein, other immunodominant SARS-CoV-2 antigens could be explored for the purpose of designing vaccines that might induce T cell responses against antigens that are more conserved across variants.
Fig. 2.
T cell protein immunodominance and epitope repertoire after SARS-CoV-2 infection or vaccination. T cell responses recognize up to 10 different SARS-CoV-2 proteins representing 80% or more of the overall response, with the spike (S) being the most immunodominant, followed by Nucleocapsid (N), Membrane (M), and other Non-structural proteins (Nsps). At the epitope level, 30-40 epitopes are recognized by either CD4+ or CD8+ T cells after natural infection considering all SARS-CoV-2 proteins recognized. A median of 10–11 epitopes are recognized specifically for S protein in natural infection similarly to what is observed after vaccination.
As part of the same study, we identified hundreds of epitopes recognized by the T cell responses of the cohort. T cells of each individual recognized multiple epitopes as well, conservatively 30 to 40 different CD4+ and 30 to 40 different CD8+ T cell epitopes (Fig. 2), as expected, given that epitope presentation is HLA restricted, and the HLA molecules expressed in a given individual tend to be unique (Tarke et al., 2021a). In a follow up study, we defined the T cell epitopes recognized after SARS-CoV-2 vaccination (Tarke et al., 2022a) that allowed us to compare the number of S protein epitopes recognized by CD4+ and CD8+ T cells following natural infection (Tarke et al., 2021a) versus vaccination (Tarke et al., 2022a). Following infection, CD4+ and CD8+ T cells recognized 10–15 S protein epitopes (Tarke et al., 2021a). Following vaccination, CD4+ and CD8+ T cells recognized 5 to 25 (median, 10) and 6 to 19 (median, 11) S protein epitopes, respectively (Tarke et al., 2022a). Finally, to comprehensively assess SARS-CoV-2 epitopes recognized by human T cells, we performed a metanalysis of epitopes experimentally identified and curated in the IEDB database (www. IEDB.org) (Grifoni et al., 2021). At the time we queried the database (mid-2021), 66 independent studies were available describing over 2000 different T cell epitopes (Grifoni et al., 2021). In summary, we found a large number and breadth of epitopes and proteins recognized by T cells elicited by SARS-CoV-2 infection.
4. SARS-CoV-2 infection or vaccination elicits T cell responses that are, by and large, conserved against future variants
Over the course of 2021, several SARS-CoV-2 variants were detected that quickly replaced the original ancestral sequence and the D614G first variant. In general, these dominant 2021 variants had mutations distributed throughout the viral genome, but concentrated in the S protein's receptor binding domain (RBD) region that mediates viral entry via interaction with the ACE2 receptor. RBD mutations have since been associated with increased infectivity and decreased capacity for neutralization by antibodies targeting the RBD region (DeGrace et al., 2022).
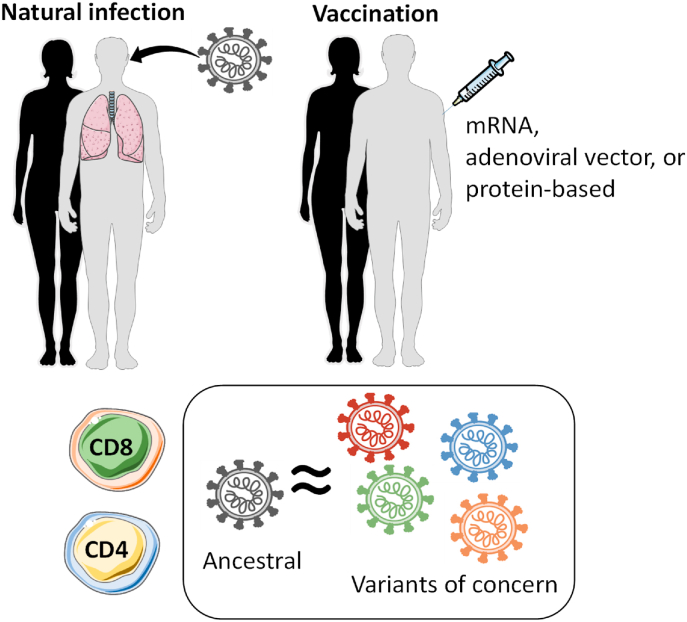
In 2 studies, we examined whether and how variant-associated mutations impact T cell recognition after vaccination or natural infection (Tarke et al., 2021b, 2022a). In the latest study (Tarke et al., 2022a), we determined the degree to which T cell responses cross-recognize different variants, by isolating T cells from individuals vaccinated against the ancestral strain with 1 of 3 different vaccine platforms (mRNA-based used for Moderna and Pfizer vaccines, viral vector-based used for J&J, and adjuvanted protein-based used for Novavax). Blood samples were collected immediately after the first or second immunization and at memory time points (3-4 months or 6-month). We then assessed T cell responses using 14 distinct S peptide pools derived from the most prominent SARS-CoV-2 variants, including Omicron, and compared each of those responses against the T cell responses after stimulation with ancestral strain S pool. Our results (validated with different functional readouts) demonstrated, at the population level, that roughly 80% or more of both CD4+ and CD8+ T cell responses cross-recognized the different S protein variants—irrespective of the vaccine platform.
Similar results were obtained in 4 collaborative studies from different groups in South Africa, Switzerland, the Netherlands, and Sweden (Keeton et al., 2021; GeurtsvanKessel et al., 2022; Madelon et al., 2022; Gao et al., 2022) utilizing different cohorts of subjects that were vaccinated or naturally infected to assess the capability of T cells primed by vaccination or infection to recognize future SARS-CoV-2 variants, with particular emphasis on Omicron. All of the studies reached the same conclusion: T-cell responses induced by SARS-CoV-2 infection or vaccination are largely preserved, in that they are able to recognize additional variants, regardless of exposure history or geographic location, suggesting that these findings are broadly and generally applicable (Fig. 3).
Fig. 3.
T cell responses induced by SARS-CoV-2 natural infection or vaccination recognize SARS-CoV-2 variants. SARS-CoV-2 specific T cell responses induced after natural infection or vaccination with different vaccine platforms induce a SARS-CoV-2 ancestral-specific T cell response able to cross-recognize with a similar magnitude several SARS-CoV-2 variants of concern and interest, including omicron.
5. Bioinformatic analyses predict modest impact of SARS-CoV-2 evolution on T cell recognition
In parallel with our analyses of T-cell reactivity, we also performed bioinformatic analyses to determine the number of SARS-CoV-2 epitopes that are 100% conserved between variants (Tarke et al., 2022a). For the 14 different variants examined, an average of about 90% of CD4 and CD8 epitopes were fully conserved (100% sequence identity), providing a mechanistic explanation for why functionality of T-cell responses are largely preserved and able to cross-recognize different SARS-CoV-2 variants. To determine whether variants are progressively accumulating mutations, the pattern of conservation was examined in early (eg. Alpha and Beta) versus late variants, (eg. Delta and Omicron) (Fig. 4). Whether analyzing whole genome or S protein only (relevant for infection and vaccination, respectively), our data revealed a comparable number of conserved epitopes in early and late variants, excluding Omicron.
Fig. 4.
Predicting the impact of emerging SARS-CoV-2 variants on T cell responses. List of known epitopes recognized by CD4+ and CD8+ T cells available in IEDB (www.IEDB.org) in combination with amino acid mutations related to a broad list of SARS-CoV-2 variants including variants of concern and interest. Analysis of the database revealed similarly high numbers of conserved epitopes and, conversely, low numbers of epitopes with variant-specific mutations, disregarding the variant mutations analyzed. Further, the majority of mutations in epitopes do not impact the ability of HLA molecules to present the epitope to T cells. Thus, the number of epitopes impacted by variants and the number of epitopes still recognized by the T cell response are likely to be higher than estimated.
Because Omicron has a higher number of mutations in the spike protein compared to any other variants, we anticipated that a larger number of the epitopes would be mutated. This was indeed the case, consistent with the fact that 80% or more of a T-cell response is conserved at the population level. It is worth noting that some mutations might not impact HLA binding or T cell recognition and/or are in epitope regions not prominently recognized by the T-cell receptor recognition. Epitopes with such mutations would still be cross-reactively recognized, suggesting that our bioinformatic analysis of the number of epitopes that are 100% conserved is likely an underestimate (Fig. 4). Given that variant dominance is often driven by mutations that confer the ability to escape T-cell recognition, we expected to find mutations clustered in epitope containing regions. We examined this in a second bioinformatic analysis, which revealed that the mutations were, in fact, randomly distributed (Tarke et al., 2022b).
Thus, combination of the high percentage of totally conserved epitopes, the high percentage of preserved T cell responses in early and late variants, and the lack of mutation clustering in epitopes suggests that SARS-CoV-2 evolution has modest, if any, impact on T cell recognition. Those findings are in line with the functional data, overall suggesting that the bioinformatic analyses are a powerful tool able to predict the impact of novel variant on the general population T cell response.
6. Conclusions
The studies reviewed here demonstrate that SARS-CoV-2 infection or vaccination induce a multi-specific, multi-functional T cell response (Grifoni et al., 2020b; Tarke et al., 2021a, 2022a). These data are compatible with a model in which humoral and cellular immunity protect against infection and replication/severe disease, respectively. SARS-CoV-2 variants tend to be associated with at least partial escape from the neutralizing antibody response (DeGrace et al., 2022), consistent with higher breakthrough infection rates in the second half of 2021 and the first part of 2022, compared to the first half of 2021. These increased rates were also likely influenced by waning immunity conferred by prior vaccination and/or infection. Conversely, the fact that SARS-CoV-2 vaccination is strongly associated with decreased rates of severe disease, independent of variant evolution, is consistent with cellular immunity being largely preserved and stable across time.
Contribution statement
A.G.: Conceptualization, investigation, writing (original draft), and visualization. A.S.: Writing (review and editing), and funding acquisition.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Alessandro Sette reports a relationship with Gritstone Bio, Flow Pharma, Arcturus Therapeutics, ImmunoScape, CellCarta, Avalia, Moderna, Fortress, Repertoire, and AstraZeneca that includes: consulting or advisory.
Acknowledgements
The work described herein was funded, in part, by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract 75N93021C00016 to A.S. The Figures were partly generated using Servier Medical Art, provided by Servier, under a Creative Commons Attribution 3.0 unported license.
References
- Dan J., et al. Observations and perspectives on adaptive immunity to SARS-CoV-2. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrace M.M., et al. Defining the risk of SARS-CoV-2 variants on immune protection. Nature. 2022;605(7911):640–652. doi: 10.1038/s41586-022-04690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47(W1):W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022;28(3):472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., et al. Divergent SARS-CoV-2 Omicron-reactive T and B cell responses in COVID-19 vaccine recipients. Sci Immunol. 2022;7(69):eabo2202. doi: 10.1126/sciimmunol.abo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680 e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501 e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., et al. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29(7):1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R., et al. medRxiv; 2021. SARS-CoV-2 Spike T Cell Responses Induced upon Vaccination or Infection Remain Robust against Omicron. p. 2021.12.26.21268380. [Google Scholar]
- Madelon N., et al. Omicron-specific cytotoxic T-cell responses after a third dose of mRNA COVID-19 vaccine among patients with multiple sclerosis treated with ocrelizumab. JAMA Neurol. 2022;79(4):399–404. doi: 10.1001/jamaneurol.2022.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012 e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.T., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021 doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med. 2021;2(2) doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2(7) doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185(5):847–859 e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Grifoni A., Sette A. Bioinformatic and experimental analysis of T cell immune reactivity to SARS-CoV-2 and its variants. Frontiers in Bioinformatics. 2022;2 doi: 10.3389/fbinf.2022.876380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., et al. Human T cell response to dengue virus infection. Front. Immunol. 2019;10:2125. doi: 10.3389/fimmu.2019.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R., et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]