Abstract
Gymnosperms represent an ancient lineage that diverged from early spermatophytes during the Devonian. The long fossil records and low diversity in living species prove their complex evolutionary history, which included ancient radiations and massive extinctions. Due to their ultra-large genome size, the whole-genome assembly of gymnosperms has only generated in the past 10 years and is now being further expanded into more taxonomic representations. Here, we provide an overview of the publicly available gymnosperm genome resources and discuss their assembly quality and recent findings in large genome architectures. In particular, we describe the genomic features most related to changes affecting the whole genome. We also highlight new realizations relative to repetitive sequence dynamics, paleopolyploidy, and long introns. Based on the results of relevant genomic studies of gymnosperms, we suggest additional efforts should be made toward exploring the genomes of medium-sized (5–15 gigabases) species. Lastly, more comparative analyses among high-quality assemblies are needed to understand the genomic shifts and the early species diversification of seed plants.
Keywords: gymnosperms, genome architecture, genomic shift, diversification
Background
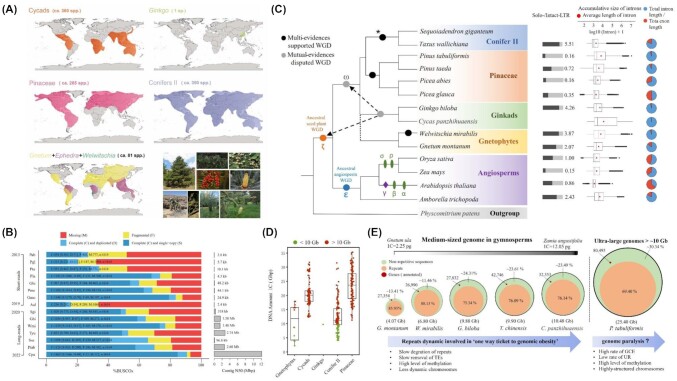
Over the past 20 years, since Arabidopsis thaliana was first sequenced, the number of assembled genomes of seed plants has reached a considerable number (>800) thanks to the fast innovation of sequencing technologies [1, 2]. Among these assemblies, only 2% (17 species, Table 1) are gymnosperms. This is partially attributed to their extraordinarily large genome sizes (>10 Gb on average), complexity [3], and low richness of species [4, 5]. Extant gymnosperms comprise ∼1,100 species encompassing 4 major lineages: cycads, Ginkgo, conifers, and gnetophytes (Fig. 1A). Due to the conifers' immense ecological and economic value, great efforts were made to examine the whole genomes of this group [6]. The conifers consist of approximately 615 species covering enormous regions of the Northern Hemisphere and serving as the major backbone of worldwide forest ecosystems [7] (Fig. 1A). A milestone report from early 2013 presented a 23-Gb assembly of loblolly pine (Pinus taeda), the first draft genome of a gymnosperm species [8, 9]; a prepublication release of the initial assembly was made in 2012 [10]. Notably, at least 10 conifer genome projects were under way at that time [8]. Another sequencing study on Norway spruce (Picea abies) conducted a comparative analysis of the genome architectures of seed plants [11]. Two sets of annotated coding genes (high confidence and low confidence) with a BUSCO ratio <30% indicated there are still considerable gaps and redundancies in this assembly. The small size of the scaffolds (the total length of those with a scaffold size >10 kb is 4.3 Gb) also reflected the objective limits of short-read sequencing, even when using high-coverage Illumina data [11]. Based on samples of the protein-coding and protein-noncoding fractions of the assembly, a plausible model for the conifer genome evolution was proposed: slow rates of activity for a diverse set of retrotransposons and a much lower frequency of recombination in noncoding regions compared to angiosperms [11]. The subsequent investigations revived the scenario of genomic dynamics in conifers, enabling the establishment of giant genomes [12–15] and the study of ecological adaptiveness and phenotypic stasis [16, 17]. With increased data, including transcriptomes and plastid genomes, studies focusing on the phylogenetic relationships among extant gymnosperms triggered great debates regarding various lineages whose studies were based on different data matrices and/or analytical approaches. One of the most controversial issues is the placement of gnetophytes. Several hypotheses have been put forward, suggesting gnetophytes are sisters to Pinaceae (the “Gnepine” hypothesis), cupressophytes (the “Gnecup” hypothesis), all conifers (the “Gnetifer” hypothesis), or all the other gymnosperms [18–22]. The unresolved phylogenetic relationships have encouraged new efforts toward filling in the taxonomic sampling gaps. In the past 5 years, draft maps of Ginkgo, gnetophytes, cupressophytes (Conifer II), and cycads have been produced and refined with an improved assembly quality [6, 23–28]. In addition, genome-wide investigations have revealed typical signatures of the gymnosperm genomes, such as ubiquitously large introns and the higher expression levels of long genes [11, 15, 26, 29]. However, the reasons behind the preservation of long genes remain poorly understood.
Table 1:
List of currently available whole-genome assembly of gymnosperms
| Species (common name) | Size of assembly (bp) | Family | Sequencing platform | Online year and relative publication | Link to the assembly data |
|---|---|---|---|---|---|
| Pinus taeda* (loblolly pine) | 23 G | Pinaceae | Sanger+ Illumina HiSeq 2000 | 2013 [10] | ftp://plantgenie.org/Data/ConGenIE/Pinus_taeda/v1.0/ |
| Picea abies (Norway spruce) | 12.3 G | Pinaceae | Sanger whole-genome shotgun | 2013 [11] | ftp://plantgenie.org/Data/ConGenIE/Picea_abies/v1.0/ |
| Picea glauca (genotype PG29) (white spruce) | 23.6 G | Pinaceae | Illumina HiSeq 2000, Miseq | 2013 [12] | ftp://plantgenie.org/Data/ConGenIE/Picea_glauca/PG29/v4.0/ |
| Pinus taeda (genotype 20–1010) (loblolly pine) | 23.2 G | Pinaceae | Illumina GA II, HiSeq 2000, Miseq | 2014 [13, 34] | https://treegenesdb.org/FTP/Genomes/Pita/v2.01/ |
| Picea glauca (genotype WS77111) (white spruce) | 22.4 G | Pinaceae | Illumina HiSeq2500, MiSeq | 2015 [16] | ftp://plantgenie.org/Data/ConGenIE/Picea_glauca/WS77111/v1.0/ |
| Pinus lambertiana (sugar pine) | 27.6 G | Pinaceae | Illumina GA II, HiSeq 2000/2500, Miseq | 2016 [14] | https://treegenesdb.org/FTP/Genomes/Pila/v1.5/ |
| Ginkgo biloba | 10.6 G | Ginkgoaceae | Illumina Hiseq 2000/4000 | 2016 [23] | http://gigadb.org/dataset/100209 |
| Pseudotsuga menziesii (Douglas fir) | 15.7 G | Pinaceae | Illumina HiSeq | 2017 [36] | https://treegenesdb.org/FTP/Genomes/Psme/v1.0/ |
| Gnetum montanum | 4.0 G | Gnetaceae | Illumina HiSeq 2000/2500 | 2018 [24] | https://doi.org/10.5061/dryad.0vm37 |
| Abies alba (silver fir) | 18.2 G | Pinaceae | Illumina HiSeq | 2019 [37] | https://treegenesdb.org/FTP/Genomes/Abal/v1.1/ |
| Larix sibirica (Siberian larch) | 12.3 G | Pinaceae | Illumina HiSeq | 2019 [35] | https://www.ncbi.nlm.nih.gov/data-hub/genome/GCA_004151065.1/ |
| Sequoiadendron giganteum (giant sequoia) | 8.1 G | Cupressaceae | Illumina HiSeq + Oxford Nanopore | 2020 [38] | https://treegenesdb.org/FTP/Genomes/Segi/v2.0/ |
| Ginkgo biloba | 9.8 G | Ginkgoaceae | Illumina HiSeq + PacBio RSII | 2021 [26] | https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA001755 |
| Welwitschia mirabilis | 6.8 G | Welwitschiaceae | Illumina HiSeq + Oxford Nanopore | 2021 [27] | https://doi.org/10.5061/dryad.ht76hdrdr |
| Taxus chinensis | 10.2 G | Taxaceae | Illumina HiSeq + PacBio RSII | 2021 [40] | https://www.ncbi.nlm.nih.gov/data-hub/genome/GCA_019776745.2/ |
| Taxus wallichiana (Himalayan yew) | 10.9 G | Taxaceae | Illumina HiSeq + Oxford Nanopore | 2021 [39] | https://db.cngb.org/search/assembly/CNA0020892/ |
| Taxus yunnanensis | 10.7 G | Taxaceae | Illumina HiSeq + Oxford Nanopore | 2021 [22] | https://www.ncbi.nlm.nih.gov/labs/data-hub/genome/GCA_018340775.1/ |
| Pinus tabuliformis (Chinese pine) | 25.4 G | Pinaceae | Illumina HiSeq + PacBio RSII | 2022 [15] | https://db.cngb.org/search/project/CNP0001649/ |
| Sequoia sempervirens (coast redwood) | 26.5 G | Cupressaceae | Illumina HiSeq + Oxford Nanopore | 2022 [6] | https://treegenesdb.org/FTP/Genomes/Sese/v2.1/ |
| Cycas panzhihuaensis | 10.5 G | Cycadaceae | Illumina HiSeq, Miseq+ Oxford Nanopore | 2022 [28] | https://db.cngb.org/codeplot/datasets/public_dataset?id = PwRftGHfPs5qG3gE |
The prepublication release of the assembly was made in 2012 [10]. It contained 18.5 Gbp of sequence with a contig N50 size of 800 bp.
Figure 1:
The contemporary overview of the deciphered gymnosperm genomes and the genomic features underpinning their complicated evolutionary history. (A) The geographical distribution of the extant gymnosperms is depicted based on data from the Global Biodiversity Information Facility. The images list the representative gymnosperm species that have been sequenced. (B) Current status of the accumulation of high-quality assemblies of gymnosperms since the advent of long-read sequencing technologies. Abbreviations of the taxa listed from top to bottom: Pab, Picea abies; Pgl, Picea glauca; Pta, Pinus taeda; Pla, Pinus lambertiana; Gbi, Ginkgo biloba; Pme, Pseudotsuga menziesii; Gmo, Gnetum montanum; Aal, Abies alba; Sgi, Sequoiadendron giganteum; Wmi, Welwitschia mirabilis; Tyu, Taxus yunnanensis; Sse, Sequoia sempervirens; Ptab, Pinus tabuliformis; Cpa, Cycas panzhihuaensis. (C) The prediction and placement of ancient whole-genome duplications (WGDs) in seed plants and the highly contested inference of paleopolyploidy in the most recent common ancestors of all extant gymnosperms. The dashed line indicates the conflicts in the phylogenetic position of gnetophytes. The dashed arrows refer to the controversy on the shared polyploidy event of gymnosperms. The Cupressaceae-WGD is highlighted by a “*” since only Taxus and Sequoiadendron were included (excluding Araucaceae) as representatives of the cupressophytes (left). The available records of the solo-/intact–long terminal repeat (LTR) ratios and the relevance of intron lengths are mapped to each species (right). The data for estimating the solo-/intact-LTR ratios were derived from Nystedt et al. [11], Cossu et al. [52], Wan et al. [24], Cheng et al. [39], Wan et al. [27], and Niu et al. [15]. The data on gene structure were derived from Niu et al. [15]. (D) Genome size distribution across the gymnosperm lineages with medium and ultra-large genome sizes. The 1C-DNA contents were obtained from Niu et al. [15] and the data sources of Kew. (E) The genomic signatures of gymnosperms and the potential genome evolutionary patterns are summarized here with the recent discoveries on recombination and repeat dynamics. TEs, transposable elements; UR, unequal recombination; GCE, gene conversion event.
Here, we summarize the progress made in the whole-genome assembly of gymnosperms and describe the considerably varied genomic features observed in different lineages, focusing on the early genome divergence patterns of gymnosperms. We also discuss the concerns relative to inferred paleopolyploid events and provide insights for future research directions. Additionally, we review the current knowledge on the effect of genomic changes on the diversification of gymnosperms and suggest that more efforts should be focused on medium-sized genomes. Finally, to understand the function of long introns, we recommend further examinations with reverse-genetic tools, which can enhance our understanding of plant genome evolution and adaptation.
The Pulsed Rises in the Whole-Genome Assembly of Gymnosperms
Thus far, compared with flowering plants, the quantities and qualities of the assembled genomes of gymnosperms are relatively lower, with an average BUSCO value of 56.92% computed from 15 decoded species (Fig. 1B). These low values derive from time-consuming projects that were launched several years ago: decades before long-read technologies were developed and became widely used. Also, the species-specific gene sets included in the library may have contributed to the underrepresented annotation of gymnosperms [6]. In terms of high-throughput Illumina sequencing platforms, it often takes 4 to 6 months to obtain clean reads, as a 100× coverage is required for a typical genome of 15 Gb in size and high heterozygosity [30]. Upon the completion of sequencing, the subsequent assembly has further costs, requiring more time and advanced technology. This is because large genomes commonly comprise a variety of repetitive sequences (hereafter called “repeats”), which are untenable with short-read sequencing approaches based on overlapping reads [31, 32]. For example, in the genome project of loblolly pine, although various strategies have been adopted (including fosmid and bacterial artificial chromosome [BAC] clones combined with whole-genome shotgun sequencing [WGS], RNA sequencing, and Bionano sequencing), it was challenging to gain good contiguous contigs, a critical requirement for gene annotation [13]. Additionally, investments in both computational and analytical resources further burdened the progress of genomics research since most assemblers could not handle the incredibly large amount of input sequences from the high-coverage sequencing [33–37].
Thanks to the advanced sequencing technologies of the PacBio RSII and Oxford Nanopore platforms, there has recently been a dramatic increase in the high-quality assembly of these gigantic genomes (Fig. 1B and Table 1). For instance, a refinement of the previous Ginkgo draft showed that the contig N50 had remarkably grown from 48 kb to 1.58 Mb in length [23, 26]; also, nearly 95% (9.33 Gb) of the scaffolds had been anchored onto the pseudochromosomes (Fig. 1B). The genomes of 2 iconic species from the Cupressaceae family, the giant sequoia (Sequoiadendron giganteum, 8.1 Gb) and the coast redwood (Sequoia sempervirens, a hexaploid genome of 26.5 Gb), were successively decoded with conspicuously enhanced contiguity [6, 38]. Additionally, 3 assembly data resources for a single genus, Taxus, were released almost simultaneously, reflecting the great interest in the gymnosperm genomes [22, 39, 40]. Notably, all the records provided impressively complete genomes, as suggested by assembly lengths (contig N50 = 2.44 Mb in Taxus chinensis, 2.89 Mb in Taxus yunnanensis, and 8.60 Mb in Taxus wallichiana) and the coverage of the core Embryophyta gene library [41] (Fig. 1B). Moreover, the recent sequencing of the haploid megagametophytes of Cycas panzhihuaensis showed outstanding assembled quality, with a contig N50 length of 12 Mb [28]. The integrative strategies combining long-read mapping and short-read data polish have been proven possible for almost all species. Also, high-throughput chromosome conformation capture can further assist the sorting of sequences [15, 42].
Insights into the Repetitive Sequence Dynamics in Gymnosperms
Comparative genomic studies revealed that angiosperm genomes are considerably flexible and dynamic in terms of the rate of DNA sequence integration and elimination [43–45]. Apart from the insertion of viral DNAs, plastids, and mitochondrial sequences, the fluctuation of plant genome sizes is mainly attributed to the historical and ongoing activity of (retro)transposable elements (TEs) (i.e., long terminal repeat retrotransposons [LTR-RTs], which are a major component contributing to the noncoding genomic regions of most seed plant genomes [46–48]). However, many of the angiosperm genomes have a fast turnover of a few million years (Ma) via the proliferation of retrotransposons and unequal recombinations (URs) [49]. Thus, the inevitable genome enlargement was efficiently counteracted by a high rate of DNA excisions [50]. In contrast, the ultra-large (>10 Gb) genomes of gymnosperms are commonly characterized by a relatively low frequency of URs, as evidenced by surveys of the ratio of intact long terminal repeats (LTRs) and solitary LTRs (solo-LTRs) (Fig. 1C). The URs between LTR-RTs often remove the intervening sequences and lead to the formation of solo-LTRs, enabling the ratio of intact versus solo-LTRs to be an indirect proxy for the removal mechanism [51, 52]. The genome-skimming of P. abies and Pinus tabuliformis identified lopsided numbers of LTRs with much more complete LTRs than solo-LTRs [11, 15]. This is consistent with the patterns observed in other conifers (P. taeda and Picea glauca) [24, 52]. However, such a signature is atypical in nonconifer gymnosperms, specifically in non-Pinaceae species, regardless of the genome size. Numerous solo-LTRs (60,623) in contrast to much less intact-LTRs (14,128) were detected in the 9.88 Gb of the Ginkgo genome [27]. Likewise, a higher ratio of solo- to intact-LTRs (5.5:1) was reported in T. wallichiana (10.9 Gb), a species belonging to the cupressophytes [40]. Moreover, 2 gnetophyte species, Gnetum montanum (4.13 Gb) and Welwitschia mirabilis (6.86 Gb), showed an elevated frequency of the recombination-based removal of retroelements [24, 27]. Hence, the greatly reduced TE elimination activity revealed in Pinaceae might be a family-specific feature generated after their separation from the main conifer clade. Potentially, such kinetic process of TE removal might diverge independently within the lineages, considering the incomplete examination of Pinaceae, especially in those groups of relatively smaller genomes (i.e., the Larix). Furthermore, the low occurrence rate of the solo-LTRs in Pinaceae was mostly inferred from either fragmental assembly [11, 52] or the manual examination of randomly sampled contigs/scaffolds [15]. More integrative and genome-wide identifications of these LTRs in high-quality genomes of Pinaceae are needed before we can fully understand the formation of ultra-large genomes. Except for infrequent URs, the reduced activity of other co-occurring processes, such as “illegitimate recombinations,” may also affect the steady growth of genomes in the long term [53]. Mobile elements like LTRs that are repaired by nonhomologous end joining and single-strand annealing may generate truncated or solitary elements, resulting in genome shrinkage [50, 54]. These disarmed LTRs may no longer be autonomous and thus cannot contribute to genome expansion [54]. More data need to be collected concerning the DNA repair by-products of gymnosperms. Also, the comparison between gymnosperms and angiosperms of the proteins and genes (i.e., Ku70/Ku80 [55] and AtBRCC36A [56]) involved in such processes is required, especially among those species with distinct genome sizes.
As the prevalent class of TEs, the historical activities of LTRs have a crucial influence on the genome size and the gene structure of plants [57, 58]. All gymnosperms likely share the common feature of repeats’ dynamic as more ancient but continuous amplification of LTRs within a range of 5 to 50 Ma [28, 40]. The estimation of the insertion date is usually determined by the synonymous substitutions per synonymous site (Ks) between each 5′-LTR and 3′-LTR flanking sequences, which are calculated based on appropriate mutation rates (per base per year) [59]. The intergenic nucleotide substitution rate of 2.2 × 10–9 is normally adopted, assuming that gymnosperms evolved at a slower pace than angiosperms. Thus, the various ages estimated by different studies of the LTR outbreaks of the same gymnosperm could be partially explained by the different neutral mutation rates assigned (i.e., 7.3 × 10−10 was used for T. yunnanensis and T. chinensis var. mairei [22, 40]). It is worth mentioning that the outlier Welwitschia has suffered from a very recent expansion of both autonomous and nonautonomous LTRs in less than 1 to 2 Ma, which probably resulted from a cascade of events triggered by intense aridity [27]. The high-resolution categories of retroelements and the use of appropriate mutation rates [60] are both required to distinguish the species-specific expansions that contribute to the diversity in genome growth rhythms [61, 62].
The subsequent ancient insertions and the unusual recent burst of LTRs raise an intriguing question regarding the differences in TE surveillance between gymnosperms and angiosperms since the genome size is generally smaller in the latter. The necessity of TE silencing has been widely acknowledged, and the epigenetic control of DNA sequences is considered the vital nuclear defense system of plant genomes to the destructive potential of TEs [63]. Approaches combining mutations and genome-wide studies of the TE properties in Arabidopsis suggested that the Dnmt1-type defense enzyme methyltransferase 1, the plant-specific chromomethylase 3, and the chromatin remodeler decrease in DNA methylation 1 are altogether involved in the DNA methylation of cytosines at CpG and non-CpG loci [64–67].
RNA-directed DNA methylation (RdDM) is an epigenetic pathway that evolved to guide the modeling of DNA condensation and TE silencing [68]. This complicated pathway was first observed in transgenic tobacco infected with viroids, plant pathogens containing solely nonprotein-coding RNA [69]. Despite the limited epigenetic investigations in gymnosperms, several instructive studies provided the general landscape of DNA methylation in the gymnosperm genome [70, 71]. For example, CpG and non-CpG methylations are both surprisingly high in P. tabuliformis (88.4% for CG; 81.6% for CHG, the cytosine sequence contexts, H represent A, T or C) and W. mirabilis (78.32% for CG; 76.11% for CHG) [15, 27], consistently with previous observations in P. abies [72]. Furthermore, global methylation levels positively correlate with genome sizes due to the widespread distribution of TEs along the genome [73, 74]. In addition, the representative genes associated with various methylation pathways have mostly been identified in gymnosperms, implying the probable functional conservation of pathways across seed plants [70]. The activity of RdDMs was further validated by their dynamic changes in the methylation level of specific sequence contexts among different tissue types [27, 70]. The oscillating abundance of 21-nucleotide (nt), 22-nt, and 24-nt Small RNA (sRNAs) indicated that both canonical and noncanonical RdDMs may play a role in TE's control [15, 27], complementing previous hypotheses that 24-nt sRNAs are restricted to the reproductive tissue in P. abies [11]. Thus, TE silencing is particularly reinforced by noncanonical RdDMs in gymnosperms, which mildly differs from the primary role of 24-nt RdDMs in angiosperms [15, 72]. However, assessing the extent to which the epigenetic mechanisms contribute to genome methylation and how they contribute to the developmental process is a highly anticipated direction for the genomic studies of gymnosperms. Incidentally, H3K9me, a mark for heterochromatin, showed contrasting distribution patterns between angiosperms and gymnosperms (P. abies and Pinus sylvestris), implying potential distinctive genome silencing mechanisms [4, 73].
A fundamental shift in repeats’ dynamic has been observed in giant genomes, as indicated by the changes in repeats’ abundance and the curvilinear relationship between genome size and repeats’ proportion among 101 seed plant species (the samples have an approximately 2,400-fold range from 0.063–88.55 Gb in genome size) [74]. In particular, genomes larger than 10 Gb are characterized by the conspicuous increase in nonrepetitive and low-copy DNA sequences (excluding genes) and the relative decrease in medium-copy repeats (>20 copies). Most of these repeats seem to have been slowly degraded and fossilized into very low copy numbers due to epigenetic suppression and limited recombination [74]. In turn, these highly heterogeneous repeats contribute to the formation of interstitial heterochromatin with heavily methylated DNA [57, 75]. Hence, large genomes have “one-way tickets to genomic obesity” [74, 76]. Such genome evolutionary patterns involving derivative retrotransposons may help understand the observation that excess low-repetitive DNA components are overrepresented in the pine genome [61, 77].
Controversy Regarding Paleopolyploidy and Its Implications for Gymnosperm Diversification
The extant gymnosperms have painted quite a different picture of the rarity of ancient polyploidizations known as whole-genome duplications (WGDs), which are often found with high frequency in flowering plants [20, 78] (Fig. 1C). These events have been suggested as determining factors controlling the lower species abundance in gymnosperms unlike angiosperms [4, 11, 79, 80]. Since postpolyploid diploidization often occurs rapidly and gives rise to many unpredictable consequences, such as chromosome number shifts and DNA loss [81], the inference of ancient WGDs remains highly challenging due to the long-term erosion of genome doubling signals (i.e., loss of duplicates and saturation of synonymous distances [82, 83]).
Combining syntenic analysis with the Ks distribution of all paralogous pairs has been vital for distinguishing WGD-derived and small-scale duplication-derived paralogues [84, 85]. However, due to the intermittent release of high-quality genome assemblies of gymnosperms, significant efforts have shifted to comparing genic signatures with improved phylogenomic approaches [20, 78]. Heuristic gene tree–species tree reconciliation methods are broadly employed to search the evidence of ancient WGDs based on transcriptome data [83, 86, 87]. As a result, Li et al. [88] first proposed that there were at least 2 independent WGDs in the ancestry of the major conifer clades (Pinaceae and Cupressaceae) according to the analyses of the transcriptome assemblies of 24 gymnosperms plus 3 outgroup species. This idea was further supported by the distributions of the Ks values of syntenic gene pairs among P. tabuliformis, Sequoiadendron giganteum, and Ginkgo biloba [15]. Furthermore, Li et al. confirmed the seed plant WGD (named ζ-) and predicted that a lineage-specific WGD occurred in Welwitschia—the latter prediction was validated in a recent Welwitschia genome investigation [27]. Another comprehensive study of WGD mapping with a considerably large RNA sequencing sample suggested that a shared WGD might have occurred before all extant gymnosperms diverged [17]. However, such hypothetical WGD cannot be corroborated by most taxonomic-oriented genomic studies [15, 23, 26, 40] (Fig. 1C). Among these genomes, a common feature was the lack of recent species-specific WGDs since only a few intragenomic blocks and syntenic gene pairs could be detected. However, all of the candidate old WGDs hinted by the Ks values were accordingly assigned to ζ- (i.e., Ks = 2.1 in T. chinensis, Ks = 1.3 in P. tabuliformis, and Ks = 0.8 in G. biloba). The variable Ks values could be attributed to the heterogeneous mutation rate and different versions of phylogenetic analysis by maximum likelihood used. Whereas we fully recognize the salience of the study both for its data sampling and analytical refinement, it still might be vulnerable to the contested phylogenetic relationships remaining in gymnosperms (the placements of Ginkgo and gnetophytes) [19–22]. The contentious species-tree topologies probably led to differences in gene duplication mapping, despite the fact that specific nodes were examined [17, 20]. Alternatively, the duplicated genes introduced by the ζ-WGD were preferentially retained over the duplicates derived from the gymnosperm-WGD in all the species surveyed. In addition, a Ks peak (∼0.8) that was recently observed in the Cycas genome was similar to the Ks peak of Ginkgo [28], suggesting an ancient WGD shared by the 2 lineages as proposed by Roodt et al. [89]. This ancient WGD (named ω-) was further dated to the most recent common ancestors (MRCAs) of all gymnosperms and supported by both transcriptome data and multispecies syntenic block alignments [28]. However, an analysis with a probabilistic approach of the WGD inference against 21 representative seed plants provided clear evidence of the ζ-WGD but not of the ω-WGD, rendering the placement of the Cycas + Ginkgo WGD highly controversial [26, 83] (Fig. 1C).
Given the considerable number of predicted ancient WGDs, based at least on the increased signals of gene duplication (restricted to the WGD-derives) [17, 20], the question was raised regarding how polyploidy contributes to the evolution of gymnosperms. A recent comprehensive measurement of the traits from living and fossil records suggested that 2 ancient pulsed rises of morphological innovation occurred in seed plants’ evolutionary history: the incipient diversification of gymnosperms (ca. 400 Ma) and the subsequent prosperity of angiosperms during the Late Cretaceous (ca. 100 Ma) [90]. The first increase represented by gymnosperms seems to result from the most commonly shared ζ-WGD and can be extended to the hypothetical ω-WGD. Two direct correlations between the conifers’ WGD and their diversification shifts [17] likely suggest the potential roles of WGD in the culmination of early gymnosperms (Cupressophyta-WGD and Pinaceae-WGD occurred ca. 200–342 Ma [88]). Besides, considerable evolutionary stasis persisted in the morphological complexity of gymnosperms and was further exacerbated by the emergence of flowering plants [90]. One report linked to a genetic map analysis showed that many more ζ-duplicates (688 gene pairs) than conifer-specific tandem duplicates (87 pairs) were preserved in the Pinaceae genomes. A highly conserved genome macrostructure was found between spruce and pine, which diverged at least 120 Ma ago [91]. The large excess of ancestral duplicates and the remarkable level of synteny indicated the much slower pace of evolution in Pinaceae, which can be considered evidence of their relative stasis. Interestingly, a karyotype comparison between Pinaceae and Cupressaceae suggested that substantial chromosomal shuffling likely commenced after their split [92]. Interspecies alignments within the Cupressaceae and other families are required to determine if the shuffling is a common feature of low-frequency genome rearrangements. This would help our understanding of the conifer cladogenesis resulting in speciation and diversity. Moreover, a case of coast redwood (S. sempervirens) implied that a very slow diploidization process followed WGD and found the persistence of multisomic inheritance in this hexaploidy species (2n = 66). These findings may contribute to explaining why there are so few polyploid species in modern gymnosperms [92]. Normally, the long-term benefits of polyploidy require the divergence among homologous chromosomes, which can only happen once loci are diploidized [81, 93]. In turn, the reduced selection of efficient meiosis in Sequoia would preclude the emergence of any evolutionary advantages in polyploidy lineages. Hence, Scott et al. [93] proposed that such an intriguing evolutionary strategy was additionally reinforced by asexual reproduction, self-compatibility, and extreme longevity, which likely took place in other conifers, such as Fizroya cupressoides [94]. Aside from this, the fundamental dynamic shift in repeats is noteworthy, assuming that the genomic shift occurred early in gymnosperms, probably before most modern lineages diverged. The ancestral genome size of gymnosperms has been estimated to have been ∼12.375 to 15.75 Gb [95]. If so, heterogeneous rates of genome size evolution should be expected considering the large range in 1C-DNA content (i.e., from 2.21 Gb in Gnetum ula to 35.28 Gb in Pinus ayacahuite) exhibited across gymnosperms [15] (Fig. 1D and E). The shift in genomic dynamics could directly lead to the unfavorable architecture of those large genomes as constrained chromosomal homogenization. Together with the slow pace of diploidization, these factors make polyploidy a burden rather than a boon in gymnosperms. Therefore, the extraordinarily massive loss of duplicates should not surprise due to the highly structured chromosomes and severely limited recombination of these genomes [4]; hence, most signals of WGD in the doubled genome were expunged (e.g., to date, W. mirabilis is the only gymnosperm species known to have a family-specific WGD that occurred ∼86 Ma ago while showing an extremely low level of intrachromosomal syntenic relationships compared to angiosperms) [27]. The unusually low rate of WGD duplicate retention could further restrain the morphological and biological diversity of these lineages, given that polyploidy often introduces sub- or neofunctionalization and increases variations in dosage-sensitive genes and pathways [96–98]. To conclude, the concomitant problems imposed by an enlarged genome could affect the diverse physiological processes of plants, such as longer cell cycles [99, 100] and higher nutrient costs [4], which eventually impact the competitiveness of the species.
Intriguing Intron Morphology and Evolution in Gymnosperms
The presence of astonishingly long genes has been extensively reported in many gymnosperms from distinct lineages [11, 15, 23] (Fig. 1C). These long genes are often associated with large amounts of intronic sequences characterized by cumulative size distributions, including numerous atypical long ones (>20 kb) [11, 15, 23, 28]. Why these very long introns are preserved and how they influence the evolution and function of genes in gymnosperms remain largely obscure [15].
It has long been acknowledged that the genome size may be correlated with the intron size across broad phylogenetic groups. However, such a pattern was poorly translated into some narrow taxonomic distant groups of angiosperms [101]. A pioneering description and comparison of the gene structures of P. glauca and P. taeda with data from BAC clones and genome scaffolds indicated a relatively conserved signature in the long introns [29]. Moreover, the high frequency (32%) of the TEs found in captured sequences, even in introns <1 kb, suggested the important role of such invasive elements in the long gene space [29]. Niu et al. [15] tabulated the characteristics of the gene structures among 68 recently sequenced seed plants. They found a positive correlation between the ratio of total intron/exon length and the genome size, especially in gymnosperm lineages (Fig. 1C). Collectively, this robust evidence supports the claim that genic expansion was coupled with the genome upsizing in the majority of gymnosperms, which is probably attributed to the slow growth and accumulation of repeats [15]. Additionally, Nystedt et al. [11] first provided insights into the presence of long introns by comparing the orthologues of the normal-sized (50–300 bp) and long (1–20 kb) introns of P. abies, P. sylvestris, and G. montanum. They suggested that an early intron expansion might have already occurred in the MRCAs of all conifers, which would explain the identical trend in the increased length of orthologous introns. However, this point of view was changed by subsequent comparisons conducted within more species of early diverged seed plants [24]. Similar growth patterns of the intron size and content were observed in orthologues between Ginkgo and P. taeda with the accumulation of LTR-RTs (especially Ty1-copia elements). By contrast, a high proportion of long interspersed nuclear elements (LINEs) was found in orthologous long introns between G. montanum and Amborella trichopoda (the “basal” angiosperm [102]), and both these species involved the expansion of long introns, consistently with the scenario of all intron morphology in G. montanum and A. trichopoda [24]. This result might indicate different repeat dynamics within the introns of G. montanum compared with other gymnosperms, and the level of Ty1-copia activity in introns might be more ancient and could be traced back to the origin of gymnosperms. Likewise, LINEs could be partially involved in the intron evolution of ancestral seed plants [24]. However, these hypotheses require more investigations using closely related or representative species like Welwitschia, Ephedra, and even Cycads, because the evolution of the gene structure of plants was determined by many more interacting forces than classically expected (i.e., the selective recombination rate [103, 104] and the species-specific TE activity [105, 106]). Indeed, a large portion of unknown sequences has been found in Cycas’ introns, which is quite different from the pattern of LTR or LINE dominance found in other gymnosperms [28].
Exploring the biological relevance of long introns could be insightful for addressing a fundamental scientific inquiry: “Why are some genomes really big and others quite compact?” Unfortunately, this matter has been poorly addressed in gymnosperms [29] except for a very recent description of gene expression profiles, alternative splicing, and DNA methylation [15]. The atypically long introns seem to have minimal influence on transcript accuracy, probably facilitated by different levels of CpG and non-CpG methylations among exons and introns [15]. These results call for similar examinations in other giant gymnosperm genomes, such as Ginkgo or Welwitschia, considering their lower effective population size compared to conifers since the loosening of natural selection often allows the fixation of potentially deleterious mutations in the genome [107]. In addition, long genes tend to have higher expression levels in P. tabuliformis, similar to the situation observed in P. glauca, Oryza sativa, and A. thaliana [29, 108]. However, such a pattern contrasts with other organisms, like Physcomitrium patens [109], Caenorhabditis elegans, and Homo sapiens [110], where compact genes are highly expressed. If so, the “low-cost transcription hypothesis” is probably unsuitable for gymnosperms. Alternatively, the length of introns is likely less relevant to the expression level since introns are involved in a variety of regulatory phenomena (i.e., posttranscriptional gene regulation [111], nucleosome formation, and chromatin organization [112–114]). Nevertheless, the correlation between gene length and gene expression should be interpreted with caution and is likely caused by technical issues: the statistical bias in RNA sequencing data due, for instance, to the overcount reads from long transcripts [102].
Conclusion and Perspectives
In this review, while appreciating the advances in our knowledge of the genome evolution of gymnosperms, we demonstrated that some essential characteristics, such as repeat dynamics, ancient WGD inference, and the biological relevance of long introns, are far from understood. The state of “genome paralysis” may be confined to Pinaceae rather than all conifers or gymnosperms since a high frequency of TE removal does exist in cupressophytes, gnetophytes, and Ginkgo. The hypothetical ω-WGD is still highly contested and needs to be reconsidered by future studies. The sporadic and long-awaited releases of genome drafts inevitably limit the conclusions of species-specific cases. Despite the low level of cladogenesis and the rarity of polyploids, the fundamental shift of genomic dynamics and the potential signature of the slow process of diploidization probably offer new insights into the complex evolution of the genome architectures of gymnosperms. Additionally, the dominant model of recent allopolyploidy speciation in Ephedra [115], as well as the growing number of species on the list of hybridization and polyploidization in Juniperus [116], contrasts with the gymnosperm reputation of being composed of ancient species. These results could be explained by the resurgence of gymnosperm diversification and the increase in habitat ranges [17]. With regards to all these aspects, we envisage that gymnosperms could be a candidate model to investigate the changes in genome dynamics and their influence on species diversifications (Fig. 1E). However, in-depth studies on the wealth of information contained within these genomes cannot be conducted without generating more high-quality assemblies. The investigation of interspecific variations and diverse properties in gymnosperms would be more profound if the data sampled were consistent, as in many excellent works conducted on animals or crops [117, 118]. Considering the intricate evolutionary history of gymnosperms, we propose that, in the future, attention should be paid to at least the 4 aspects next described. First, more integrative estimations of TE eliminations are needed, and a high-resolution subclassification of the TEs would help to distinguish family-specific expansion patterns. Intensive studies on the many repetitive relics with a low copy number would also enable us to illustrate the formation of the highly structured and less dynamic chromosomes of gymnosperms [4, 11, 75]. Finally, the rapid accumulation of epigenetic data is imperative since variable repeat dynamics and sophisticated epigenetic machinery play crucial roles in gymnosperms. These data should be either at the single-base resolution of DNA methylation or for comparing methylomes among different tissues. Second, ancestral paleopolyploidy inferences should be investigated by large-scale multialignments of more complete gymnosperm assemblies with fully considered phylogenies. In particular, the structural evidence of intra- and interspecies collinearity may be essential to clarify the number and timing of these ancient duplications [82]. Moreover, the comprehensive evaluation of the loss and retention of duplicate genes could help elucidate the potential heterogeneity in the genome evolution of gymnosperms. Third, it may be worthwhile to include intron length and expression characteristics in future whole-genome studies of gymnosperms. Also, more investigations on alternative splicing patterns should be carried out and analyzed together with DNA methylation footprints. Despite the lack of appropriate genetic transformation tools for long-lived perennial species, it might be insightful to conduct analogous molecular experiments in model plant systems concerning the potential biological functions of ultra-long genes [15, 119]. Finally, more chromosome-level genomes of gymnosperms are needed. However, we suggest that additional efforts should be made to sequence medium-sized (5–15 G) species and refine the short-read drafts released for conifers, especially Pinaceae.
Data Availability
BAC (bacterial artificial chromosome); BUSCO (Benchmarking Universal Single-Copy Orthologs); CMT3 (chromomethylase 3); DDM1 (Decrease in DNA Methylation 1); GCE (gene conversion event); Hi-C (highthroughput chromosome conformation capture ); Ks (the synonymous substitutions per synonymous site); LINEs (long interspersed nuclear elements); LTR-RTs (long terminal repeat retrotransposons); Ma (million years); MET1 (Dnmt1-type defence enzyme methyltransferase); MRCA (the most recent common ancestors); RdDM (RNA-directed DNA methylation); TEs ((retro)transposable elements); UR (unequal recombination); WGDs (whole-genome duplications); WGS (whole-genome shotgun sequencing).
Competing Interests
The authors declare no competing interests.
Funding
This work was supported by the Scientific Research Program of Sino-Africa Joint Research Center (grant no. SAJC202105) and the National Natural Science Foundation of China grants (grant no. 31 870 206).
Authors' Contributions
T.W. and Q.F.W. designed the outline of the manuscript. T.W. and Y.B.G. wrote the manuscript. T.W., C.D., and Q.F.W. polished the article. Z.M.L. and Y.D.Z. worked on the revisitation of the genomic data. T.W. and Y.B.G. are joint first authors.
Supplementary Material
Steven L. Salzberg, Ph.D. -- 1/15/2022 Reviewed
Steven L. Salzberg, Ph.D. -- 6/21/2022 Reviewed
David Neale -- 6/3/2022 Reviewed
ACKNOWLEDGEMENTS
We thank Dr. Neng Wei from the Wuhan Botanical Garden for collecting the species images. We acknowledge Prof. Shouzhou Zhang from the Fairy Lake Botanical Garden for the assistance with the data access to the Cycas genome assembly. We also thank Ms. Ruth Wambui Mbichi for her revision of this manuscript.
Contributor Information
Tao Wan, Core Botanical Gardens/Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan 430074, China; Sino-Africa Joint Research Centre, Chinese Academy of Sciences, Wuhan 430074, China; Key Laboratory of Southern Subtropical Plant Diversity, Fairy Lake Botanical Garden, Shenzhen & Chinese Academy of Science, Shenzhen 518004, China.
Yanbing Gong, Department of Ecology, Tibetan Centre for Ecology and Conservation at WHU-TU, State Key Laboratory of Hybrid Rice, College of Life Sciences, Wuhan University, Wuhan 430072, China; Research Center for Ecology, College of Science, Tibet University, Lhasa 850000, China.
Zhiming Liu, Key Laboratory of Southern Subtropical Plant Diversity, Fairy Lake Botanical Garden, Shenzhen & Chinese Academy of Science, Shenzhen 518004, China.
YaDong Zhou, School of Life Science, Nanchang University, Nanchang 330031, China.
Can Dai, School of Resources and Environmental Science, Hubei University, Wuhan, China.
Qingfeng Wang, Core Botanical Gardens/Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan 430074, China; Sino-Africa Joint Research Centre, Chinese Academy of Sciences, Wuhan 430074, China.
References
- 1. Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. [DOI] [PubMed] [Google Scholar]
- 2. Marks RA, Hotaling S, Frandsen PB, et al. Representation and participation across 20 years of plant genome sequencing. Nature Plants. 2021;7(12):1571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murray BG. Nuclear DNA amounts in gymnosperms. Ann Bot. 1998;82:3–15. [Google Scholar]
- 4. Leitch AR, Leitch IJ.. Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol. 2012;194(3):629–46. [DOI] [PubMed] [Google Scholar]
- 5. Sederoff R. Genomics: a spruce sequence. Nature. 2013;497(7451):569–70. [DOI] [PubMed] [Google Scholar]
- 6. Neale DB, Zimin AV, Zaman S, et al. Assembled and annotated 26.5 Gbp coast redwood genome: a resource for estimating evolutionary adaptive potential and investigating hexaploid origin. G3 (Bethesda). 2022;12(1):jkab380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin W-T, Gernandt DS, Wehenkel Cet al. Phylogenomic and ecological analyses reveal the spatiotemporal evolution of global pines. Proc Natl Acad Sci. 2021;118(20):e2022302118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neale DB, Langley CH, Salzberg SL, et al. Open access to tree genomes: the path to a better forest. Genome Biol. 2013;14(6):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. PineRefSeq. http://www.pinegenome.org/pinerefseq, 2013. [Google Scholar]
- 10. PineRefSeq. http://loblolly.ucdavis.edu/bipod/ftp/Genome_Data/genome/pinerefseq/Pita/v0.6/, 2012. [Google Scholar]
- 11. Nystedt B, Street NR, Wetterborm A, et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497(7451):579–84. [DOI] [PubMed] [Google Scholar]
- 12. Birol I, Raymond A, Jackman SDet al. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics. 2013;29(12):1492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neale DB, Wegrzyn JL, Stevens KAet al. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 2014;15(3):R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stevens KA, Wegrzyn JL, Zimin Aet al. Sequence of the Sugar Pine megagenome. Genetics. 2016;204(4):1613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niu S-H, Li J, Bo W-Het al. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell. 2022;185(1):204–217.e14. [DOI] [PubMed] [Google Scholar]
- 16. Warren RL, Keeling CI, Yuen MM Saintet al. Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Plant J. 2015;83(2):189–212. [DOI] [PubMed] [Google Scholar]
- 17. Stull GW, Qu X-J, Parins-Fukuchi Cet al. Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nature Plants. 2021;7(8):1015–25. [DOI] [PubMed] [Google Scholar]
- 18. Wickett NJ, Mirarab S, Nguyen Net al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci. 2014;111(45):4859–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ran J-H, Shen T-T, Wang M-M, et al. Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc R Soc B Biol Sci. 2018;285(1881):20181012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. One Thousand Plant Transcriptomes Initiative One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H-T, Yi T-S, Gao L-Met al. Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants. 2019;5(5):461–70. [DOI] [PubMed] [Google Scholar]
- 22. Song C, Fu F-F, Yang L-L, et al. Taxus yunnanensis genome offers insights into gymnosperm phylogeny and taxol production. Commun Biol. 2021;4(1):1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guan R, Zhao Y-P, Zhang H, et al. Draft genome of the living fossil Ginkgo biloba. Gigascience. 2016;5(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wan T., Li Z-M, Li L-F, et al. A genome for gnetophytes and early evolution of seed plants. Nature Plants. 2018;4(2):82–89. [DOI] [PubMed] [Google Scholar]
- 25. Zhao Y-P, Fan G-Y, Yin P-P, et al. Resequencing 545 ginkgo genomes across the world reveals the evolutionary history of the living fossil. Nat Commun. 2019;10(1):4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu H-L, Wang X-B, Wang G-B, et al. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nature Plants. 2021;7(6):748–56. [DOI] [PubMed] [Google Scholar]
- 27. Wan T, Liu Z-M, Leitch IJ, et al. The Welwitschia genome reveals a unique biology underpinning extreme longevity in deserts. Nat Commun. 2021;12(1):4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, Wang S-B, Li L-Z, et al. The Cycas genome and the early evolution of seed plants. Nature Plants. 2022;8(4):389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sena JS, Giguère I, Boyle B, et al. Evolution of gene structure in the conifer Picea glauca: a comparative analysis of the impact of intron size. BMC Plant Biol. 2014;14(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Dijk E L, Auger H, Jaszczyszyn Y, et al. Ten years of next-generation sequencing technology. Trends Genet. 2014;30(9):418–26. [DOI] [PubMed] [Google Scholar]
- 31. Myers EW. Toward simplifying and accurately formulating fragment assembly. J Comput Biol. 1995;2(2):275–90. [DOI] [PubMed] [Google Scholar]
- 32. Li R-Q, Fan W, Tian G, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Birol I, Raymond A, Jackman SD, et al. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics. 2013;29(12):1492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zimin A, Stevens KA, Crepeau MW, et al. Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics. 2014;196(3):875–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuzmin DA, Feranchuk SI, Sharov VV, et al. Stepwise large genome assembly approach: a case of Siberian larch (Larix sibirica Ledeb.). BMC Bioinf. 2019;20(S1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neale DB, McGuire PE, Wheeler NC, et al. The Douglas-fir genome sequence reveals specialization of the photosynthetic apparatus in Pinaceae. G3 (Bethesda). 2017;7(9):3157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mosca E, Cruz F, Gómez Garrido J, et al. A reference genome sequence for the European silver fir (Abies alba Mill.): a community-generated genomic resource. G3 (Bethesda). 2019;9(7):2039–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scott AD, Zimin AV, Puiu D, et al. A reference genome sequence for giant sequoia. G3 (Bethesda). 2020;10(11):3907–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng J, Wang X, Liu X-N, et al. Chromosome-level genome of Himalayan yew provides insights into the origin and evolution of the paclitaxel biosynthetic pathway. Mol Plant. 2021;14(7):1199–209. [DOI] [PubMed] [Google Scholar]
- 40. Xiong X-Y, Gou J-B, Liao Q-G, et al. The Taxus genome provides insights into paclitaxel biosynthesis. Nature Plants. 2021;7(8):1026–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seppey M, Manni M, Zdobnov EM.. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 2019;1962:227–45. [DOI] [PubMed] [Google Scholar]
- 42. Meyer A, Schloissnig S, Franchini P, et al. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature. 2021;590(7845):284–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ma J-X, Devos KM, Bennetzen JL.. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14(5):860–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lim KY, Kovarik A, Matyasek R, et al. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 2007;175(4):756–63. [DOI] [PubMed] [Google Scholar]
- 45. Kejnovsky E, Leitch IJ, Leitch AR.. Contrasting evolutionary dynamics between angiosperm and mammalian genomes. Trends Ecol Evol. 2009;24(10):572–82. [DOI] [PubMed] [Google Scholar]
- 46. Kumar A, Bennetzen JL.. Plant retrotransposons. Annu Rev Genet. 1999;33(1):479–532. [DOI] [PubMed] [Google Scholar]
- 47. Moffat AS. Transposons help sculpt a dynamic genome. Science. 2000;289(5484):1455–7. [DOI] [PubMed] [Google Scholar]
- 48. Feschotte C, Jiang N, Wessler SR.. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3(5):329–41. [DOI] [PubMed] [Google Scholar]
- 49. Vitte C, Panaud O, Quesneville H.. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics. 2007;8(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Devos KM, Brown JKM, Bennetzen JL.. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12(7):1075–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vicient CM, et al. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell. 1999;11(9):1769–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cossu RM, Casola C, Giacomello S, et al. LTR retrotransposons show low levels of unequal recombination and high rates of intraelement gene conversion in large plant genomes. Genome Biol Evol. 2017;9(12):3449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kelly LJ, Renny-Byfield S, Pellicer J, et al. Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytol. 2015;208(2):596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vu GTH, Cao H-X, Reiss B, et al. Deletion-bias in DNA double-strand break repair differentially contributes to plant genome shrinkage. New Phytol. 2017;214(4):1712–21. [DOI] [PubMed] [Google Scholar]
- 55. Kim JH, Ryu TH, Lee SS, et al. Ionizing radiation manifesting DNA damage response in plants: an overview of DNA damage signaling and repair mechanisms in plants. Plant Sci. 2019;278:44–53. [DOI] [PubMed] [Google Scholar]
- 56. Block-Schmidt AS, Dukowic-Schulze S, Wanieck K, et al. BRCC36A is epistatic to BRCA1 in DNA crosslink repair and homologous recombination in Arabidopsis thaliana. Nucleic Acids Res. 2011;39(1):146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fedoroff NV. Transposable elements, epigenetics, and genome evolution. Science. 2012;338(6108):758–67. [DOI] [PubMed] [Google Scholar]
- 58. Barghini E, Natali L, Giordani T, et al. LTR retrotransposon dynamics in the evolution of the olive (Olea europaea) genome. DNA Res. 2015;22(1):91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. De La Torre AR, Li Z, Van de Peer Y, et al. Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Mol Biol Evol. 2017;34(6):1363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morse AM, Peterson DG, Nurul Islam-Faridi M, et al. Evolution of genome size and complexity in Pinus. PLoS One. 2009;4(2):e4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou S-S, Yan X-M, Zhang K-F, et al. A comprehensive annotation dataset of intact LTR retrotransposons of 300 plant genomes. Sci Data. 2021;8(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou W-D, Liang G-N, Molloy PL, et al. DNA methylation enables transposable element-driven genome expansion. Proc Natl Acad Sci. 2020;117(32):19359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Finnegan EJ, Peacock WJ, Dennis ES.. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci. 1996;93(16):8449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jeddeloh JA, Stokes TL, Richards EJ.. Maintenance of genomic methylation requires a SW12/SNF2-like protein. Nat Genet. 1999;22(1):94–97. [DOI] [PubMed] [Google Scholar]
- 66. Zemach A, Kim MY, Hsieh PH, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ito H, Kakutani T.. Control of transposable elements in Arabidopsis thaliana. Chromosome Res. 2014;22(2):217–23. [DOI] [PubMed] [Google Scholar]
- 68. Matzke MA, Mosher RA.. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15(6):394–408. [DOI] [PubMed] [Google Scholar]
- 69. Wassenegger M, Heimes S, Riedel L, et al. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76(3):567–76. [DOI] [PubMed] [Google Scholar]
- 70. Ausin I, Feng S-H, Yu C-W, et al. DNA methylome of the 20-gigabase Norway spruce genome. Proc Natl Acad Sci. 2016;113(50):e8106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Takuno S, Ran J-H, Gaut BS.. Evolutionary patterns of genic DNA methylation vary across land plants. Nature Plants. 2016;2(2):15222. [DOI] [PubMed] [Google Scholar]
- 72. Zhang H-M, Zhu J-K.. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14(2):142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fuchs J, Jovtchev G, Schubert I.. The chromosomal distribution of histone methylation marks in gymnosperms differs from that of angiosperms. Chromosome Res. 2008;16(6):891–8. [DOI] [PubMed] [Google Scholar]
- 74. Nóvak P, Guignard MS, Neumann P, et al. Repeat-sequence turnover shifts fundamentally in species with large genomes. Nature Plants. 2020;6(11):1325–9. [DOI] [PubMed] [Google Scholar]
- 75. Islam-Faridi MN, Nelson CD, Kubisiak TL.. Reference karyotype and cytomolecular map for loblolly pine (Pinus taeda L.). Genome. 2007;50(2):241–51. [DOI] [PubMed] [Google Scholar]
- 76. Bennetzen JL, Kellogg EA.. Do plants have a one-way ticket to genomic obesity ?. Plant Cell. 1997;9(9):1509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Elsik CG, Williams CG.. Retroelements contribute to the excess low-copy-number DNA in pine. Mol Gen Genet MGG. 2000;264(1–2):47–55. [DOI] [PubMed] [Google Scholar]
- 78. Jiao Y-N, Wickett NJ, Ayyampalayam S, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. [DOI] [PubMed] [Google Scholar]
- 79. Soltis PS, Soltis DE.. Ancient WGD events as drivers of key innovations in angiosperms. Curr Opin Plant Biol. 2016;30:159–65. [DOI] [PubMed] [Google Scholar]
- 80. Wu S-D, Han B-C, Jiao Y-N.. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol Plant. 2020;13(1):59–71. [DOI] [PubMed] [Google Scholar]
- 81. Mandakova T, Lysak MA.. Post-polyploid diploidization and diversification through dysploid changes. Curr Opin Plant Biol. 2018;42:55–65. [DOI] [PubMed] [Google Scholar]
- 82. Ruprecht C, Lohaus R, Vanneste K, et al. Revisiting ancestral polyploidy in plants. Sci Adv. 2017;3(7):1603195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zwaenepoel A, Van de Peer Y.. Inference of ancient whole-genome duplications and the evolution of gene duplication and loss rates. Mol Biol Evol. 2019;36(7):1384–404. [DOI] [PubMed] [Google Scholar]
- 84. Lynch M, Conery JS.. The evolutionary fate and consequences of duplicate genes. Science. 2000; 290(5494):1151–5. [DOI] [PubMed] [Google Scholar]
- 85. Blanc G, Wolfe KH.. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16(7):1667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rabier CE, Ta T, Ane C.. Detecting and locating whole genome duplications on a phylogeny: a probabilistic approach. Mol Biol Evol. 2014;31(3):750–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yang Y, Moore MJ, Brockington SF, et al. Improved transcriptome sampling pinpoints 26 ancient and more recent polyploidy events in Caryophyllales, including two allopolyploidy events. New Phytol. 2018;217(2):855–70. [DOI] [PubMed] [Google Scholar]
- 88. Li Z, Baniaga AE, Sessa EB, et al. Early genome duplications in conifers and other seed plants. Sci Adv. 2015;1(10):e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Roodt D, Lohaus R, Sterck Let al. Evidence for an ancient whole genome duplication in the cycad lineage. PLoS One. 2017;12(9):e0184454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Leslie AB, Simpson C, Mander L.. Reproductive innovations and pulsed rise in plant complexity. Science. 2021;373(6561):1368–72. [DOI] [PubMed] [Google Scholar]
- 91. Pavy N, Pelgas B, Laroche J, et al. A spruce gene map infers ancient plant genome reshuffling and subsequent slow evolution in the gymnosperm lineage leading to extant conifers. BMC Biol. 2012;10(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. De Miguel M, Bartholome´ J, Ehrenmann F, et al. Evidence of intense chromosomal shuffling during conifer evolution. Genome Biol Evol. 2015;7:2799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Scott AD, Stenz NWM, Ingvarsson PK, et al. Whole genome duplication in coast redwood (Sequoia sempervirens) and its implications for explaining the rarity of polyploidy in conifers. New Phytol. 2016;211(1):186–93. [DOI] [PubMed] [Google Scholar]
- 94. Silla F, Fraver S, Lara A, et al. Regeneration and stand dynamics of Fitzroya cupressoides (Cupressaceae) forests of southern Chile's Central Depression. Forest Ecol Manag. 2002;165(1-3):213–24. [Google Scholar]
- 95. Burleigh JG, Barbazuk WB, Davis JM, et al. Exploring diversification and genome size evolution in extant gymnosperms through phylogenetic synthesis. J Bot. 2012;2012:292857. [Google Scholar]
- 96. Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60(1):433–53. [DOI] [PubMed] [Google Scholar]
- 97. Bekaert M, Edger PP, Pires JC, et al. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell. 2011;23(5):1719–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Conant GC, Birchler JA, Pires JC.. Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr Opin Plant Biol. 2014;19:91–98. [DOI] [PubMed] [Google Scholar]
- 99. Francis D, Davies MS, Barlow PW.. A strong nucleotypic effect on the cell cycle regardless of ploidy level. Ann Bot. 2008;101(6):747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Doyle JJ, Coate JE.. Polyploidy, the nucleotype, and novelty: the impact of genome doubling on the biology of the cell. Int J Plant Sci. 2019;180(1):1–52. [Google Scholar]
- 101. Wendel JF, Cronn RC, Alvarez I, et al. Intron size and genome size in plants. Mol Biol Evol. 2002;19(12):2346–52. [DOI] [PubMed] [Google Scholar]
- 102. Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342(6165):1241089. [DOI] [PubMed] [Google Scholar]
- 103. Carvalho AB, Clark AG.. Intron size and natural selection. Nature. 1999;401(6751):344. [DOI] [PubMed] [Google Scholar]
- 104. Comeron JM, Kreitman M.. The correlation between intron length and recombination in Drosophila: Dynamic equilibrium between mutational and selective forces. Genetics. 2000;156(3):1175–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vinogradov AE. Intron-genome size relationship on a large evolutionary scale. J Mol Evol. 1999;49(3):376–84. [DOI] [PubMed] [Google Scholar]
- 106. McLysaght A, Enright AJ, Skrabanek L, et al. Estimation of synteny conservation and genome compaction between pufferfish (Fugu) and human. Yeast. 2000;1(1):22–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci. 2002;99(9):6118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ren X-Y, Vorst O, Fiers M, et al. In plants, highly expressed genes are the least compact. Trends Genet. 2006;22(10):528–32. [DOI] [PubMed] [Google Scholar]
- 109. Stenoien HK. Compact genes are highly expressed in the moss Physcomitrella patens. J Evol Biol. 2007;20(3):1223–9. [DOI] [PubMed] [Google Scholar]
- 110. Castillo-Davis CI, Mekhedov SL, Hartl DL, et al. Selection for short introns in highly expressed genes. Nat Genet. 2002;31(4):415–8. [DOI] [PubMed] [Google Scholar]
- 111. Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5(4):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zuckerkandl E Junk DNA and sectorial gene repression. Gene. 1997;205(1–2):323–43. [DOI] [PubMed] [Google Scholar]
- 113. Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18(9):1611–30. [DOI] [PubMed] [Google Scholar]
- 114. Vinogradov AE. Noncoding DNA, isochores and gene expression: nucleosome formation potential. Nucleic Acids Res. 2005;33(2):559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wu H, Wang M-M, Qin A-L, et al. A high frequency of allopolyploid speciation in the gymnospermous genus Ephedra and its possible association with some biological and ecological features. Mol Ecol. 2016;25(5):1192–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Farhat P, Hidalgo O, Robert T, et al. Polyploidy in the conifer genus Juniperus: an unexpectedly high rate. Front Plant Sci. 2019;10:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhang G-J, Li C, Li Q-Y,et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346(6215):1311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Varshney RK, Roorkiwal M, Sun S, et al. A chickpea genetic variation map based on the sequencing of 3,366 genomes. Nature. 2021;599(7886):622–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Liu Y-Y, Yang K-Z, Wei X-X, et al. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 2016;212(3):730–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Steven L. Salzberg, Ph.D. -- 1/15/2022 Reviewed
Steven L. Salzberg, Ph.D. -- 6/21/2022 Reviewed
David Neale -- 6/3/2022 Reviewed
Data Availability Statement
BAC (bacterial artificial chromosome); BUSCO (Benchmarking Universal Single-Copy Orthologs); CMT3 (chromomethylase 3); DDM1 (Decrease in DNA Methylation 1); GCE (gene conversion event); Hi-C (highthroughput chromosome conformation capture ); Ks (the synonymous substitutions per synonymous site); LINEs (long interspersed nuclear elements); LTR-RTs (long terminal repeat retrotransposons); Ma (million years); MET1 (Dnmt1-type defence enzyme methyltransferase); MRCA (the most recent common ancestors); RdDM (RNA-directed DNA methylation); TEs ((retro)transposable elements); UR (unequal recombination); WGDs (whole-genome duplications); WGS (whole-genome shotgun sequencing).