Abstract
Adenovirus vector vaccines have been widely and successfully deployed in response to coronavirus disease 2019 (COVID-19). However, despite inducing potent T cell immunity, improvement of vaccine-specific antibody responses upon homologous boosting is modest compared with other technologies. Here, we describe a system enabling modular decoration of adenovirus capsid surfaces with antigens and demonstrate potent induction of humoral immunity against these displayed antigens. Ligand attachment via a covalent bond was achieved using a protein superglue, DogTag/DogCatcher (similar to SpyTag/SpyCatcher), in a rapid and spontaneous reaction requiring only co-incubation of ligand and vector components. DogTag was inserted into surface-exposed loops in the adenovirus hexon protein to allow attachment of DogCatcher-fused ligands on virus particles. Efficient coverage of the capsid surface was achieved using various ligands, with vector infectivity retained in each case. Capsid decoration shielded particles from vector neutralizing antibodies. In prime-boost regimens, adenovirus vectors decorated with the receptor-binding domain of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike induced >10-fold higher SARS-CoV-2 neutralization titers compared with an undecorated vector encoding spike. Importantly, decorated vectors achieved equivalent or superior T cell immunogenicity against encoded antigens compared with undecorated vectors. We propose capsid decoration using protein superglues as a novel strategy to improve efficacy and boostability of adenovirus-based vaccines and therapeutics.
Keywords: adenovirus, vaccine vector, vector engineering, protein superglue, SARS-CoV-2
Graphical abstract
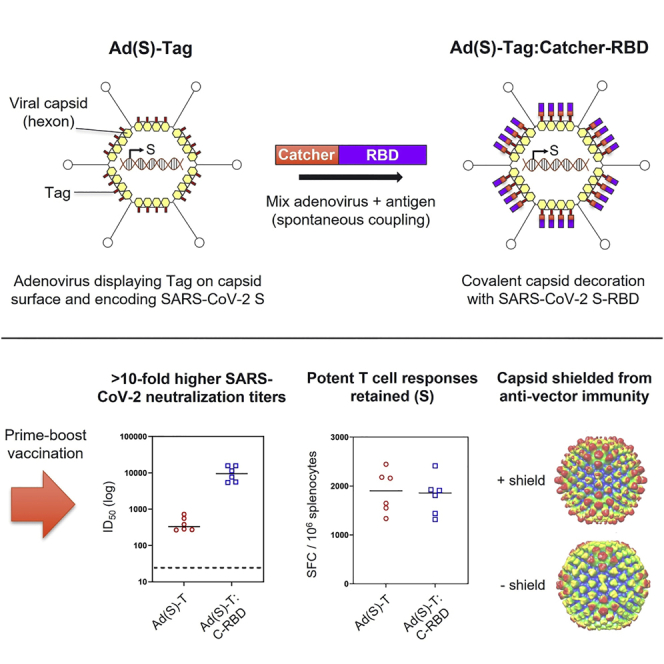
Protein superglue technology enables spontaneous covalent attachment of antigens to the capsid surface of recombinant adenovirus vaccine vectors. Decorated vectors induce potent neutralizing antibody responses against capsid-displayed antigens, and retain the ability to induce strong T cell responses against encoded antigens.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has demonstrated the remarkable utility of replication-defective recombinant adenoviruses as vaccine vectors.1 The rapid construction, scalability, and cost-effectiveness of this platform, without a requirement for adjuvants, combined with long-term stability at standard refrigerator temperatures, has positioned the platform among leading technologies in the pandemic response.1 There are currently four adenovirus-based COVID-19 vaccines licensed and in use across various parts of the world, using a range of adenovirus serotypes.2,3,4,5
However, despite decades of research and significant progress in the development of this vaccine platform, limitations remain. While adenovirus vectors are among the most potent inducers of cellular immunity (particularly CD8+ T cell responses) in humans,6 antibody responses to target antigens are modest in comparison with the most potent inducers of humoral immunity, which include nanoparticle- and virus-like-particle (VLP)-based recombinant protein technologies.7,8 Anti-vector immunity (particularly via anti-capsid neutralizing antibodies) has also been shown to limit vaccine immunogenicity and efficacy, since transduction of host cells by the viral vector prior to immune clearance is essential to induce immunity against encoded target antigens.5,9,10 Human populations harbor significant pre-existing neutralizing antibody titers against common human adenovirus serotypes, which has led to a search for adenovirus serotypes with lower human seroprevalence (including Ad26 and Y25/ChAdOx1).11,12 However, with the most clinically advanced serotypes already used to vaccinate more than a billion people during the current pandemic, it may be challenging to re-use these platforms for boosting and subsequent disease indications. Previous studies have shown that the efficacy of prime-boost regimens using the same vector for both immunizations is limited by the anti-vector neutralizing responses raised after the first immunization.9
To address these limitations, here we have designed a platform to enable modular covalent decoration of adenovirus particles with antigenic ligands. High-density repetitive display of antigen on VLPs is a highly effective strategy to generate potent and boostable humoral immunity.13 The SpyTag/SpyCatcher protein superglue system has previously been utilized to achieve rapid and efficient covalent attachment of vaccine antigens to a variety of VLP platforms.14,15,16,17,18 SpyTag, a short peptide tag, reacts rapidly and spontaneously with SpyCatcher, a small protein domain, forming an irreversible isopeptide bond.19 A hepatitis-B surface antigen (HBsAg) VLP-based COVID-19 vaccine candidate displaying the receptor-binding domain of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike (RBD) using SpyTag/SpyCatcher technology has recently entered phase I/II clinical trials.20 Protein superglue technologies can overcome challenges associated with other methods of antigen attachment to VLPs, such as irregular distribution and orientation of ligands coupled using chemical modification, or protein/particle instability following genetic fusion/insertion of ligands to particle scaffolds.21 Recently, a new protein superglue pair based on domain 4 of the RrgA adhesin from Streptococcus pneumoniae, DogTag/DogCatcher, was shown to perform particularly efficiently in loop structures (Figure S1).22
Here, we used the DogTag/DogCatcher system to enable modular capsid display of protein ligands including RBD on the surface of recombinant adenovirus vectors. DogTag was engineered into surface loops on the hexon capsid protein and enabled attachment of DogCatcher-fused antigens. Despite extensive coverage of the capsid surface, decorated virions retained infectivity and induced potent neutralizing antibody and T cell responses against SARS-CoV-2 in a proof-of-concept study. Capsid display markedly improved boostability of responses compared with homologous prime-boost regimens using conventional undecorated adenovirus vaccines. To the best of our knowledge, this is the first example of covalent decoration of adenovirus vector capsids using spontaneous isopeptide linkage and represents a new tool for the development of adenovirus-based vaccines and therapeutics.
Results
Adenovirus capsid decoration through spontaneous isopeptide bond linkage
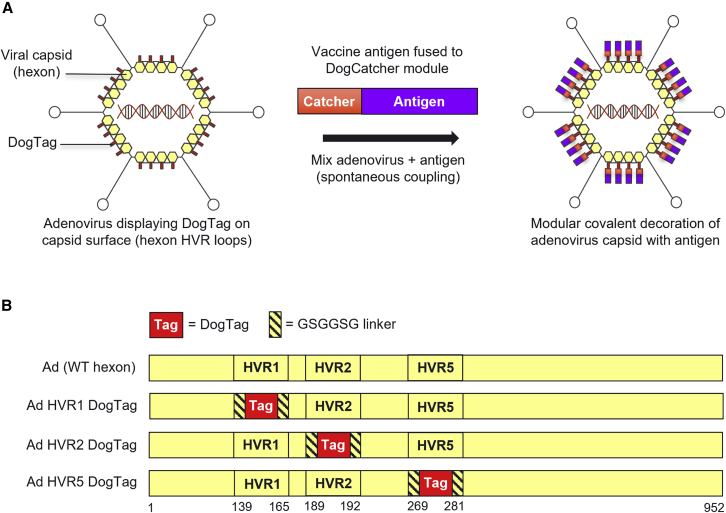
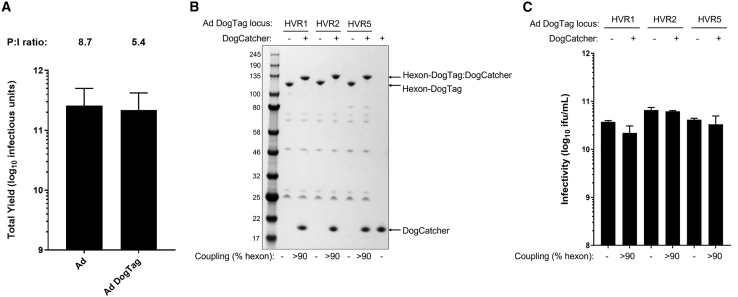
The hexon protein is the major component of the adenovirus capsid (each virion comprising 720 copies assembled into 240 trimers),23 and is therefore an ideal target for modular capsid display of antigens (Figure 1A). We utilized the DogTag/DogCatcher reactive pair to achieve spontaneous covalent coupling of DogCatcher-fused ligands onto the surface of recombinant adenovirus vectors. To enable covalent capsid decoration, DogTag (23 amino acids) was genetically inserted into each of three hexon hypervariable surface loops (HVR1, HVR2, or HVR5) flanked by flexible glycine-serine linker sequences (Figure 1B). Replication-defective (E1-deleted) recombinant adenovirus vectors with DogTag insertions (Ad-DogTag) were readily produced in E1 complementing 293A cells, with yields comparable with vectors with an unmodified hexon (Figure 2A and S2). Vectors with SpyTag inserted at each of the hexon HVRs were also produced at comparable titers (Figure S2). Particle to infectious unit ratios (P:I ratios) of vector preparations were comparable between vectors with and without hexon insertions (Figures 2A and S2). Co-incubation of Ad-DogTag virions with recombinant DogCatcher protein achieved >90% coverage of the Ad capsid (>90% hexon protein covalently coupled to DogCatcher as measured by SDS-PAGE gel shift; Figure 2B). Similar levels of coverage were observed with DogTag inserted at each of the three hexon HVRs, indicating a high degree of flexibility in this approach. Strikingly, despite extensive coverage of the capsid with DogCatcher, decorated virions with DogTag at HVR2 or HVR5 exhibited no reduction in infectivity (<1.5-fold) in vitro compared with undecorated virions (Figure 2C). Decorated virions with DogTag at HVR1 exhibited a modest 1.7-fold reduction (Figure 2C). In contrast, SpyTag was poorly reactive with SpyCatcher when inserted into Ad hexon HVRs, with markedly lower capsid coverage compared with the DogTag/DogCatcher system, despite co-incubation with a 7-fold higher concentration of recombinant SpyCatcher protein (Figure S3A). Furthermore, once significant capsid coverage with SpyCatcher was achieved (i.e., 57% hexon protein coupled), virion infectivity in vitro was reduced ∼100-fold (Figure S3B).
Figure 1.
Modular covalent decoration of the adenovirus capsid via insertion of DogTag into hexon HVR loops
(A) Modular display of DogCatcher-fused antigenic ligands on the surface of the adenovirus capsid via covalent coupling with DogTag inserted into hexon HVR surface loops. Attachment of antigens to the capsid achieved by simple co-incubation of adenovirus and antigen components in a rapid and spontaneous reaction. (B) Design of modified adenovirus hexon sequences with DogTag inserted into either HVR1, HVR2, or HVR5 flanked by flexible linkers. Amino acid residue numbers corresponding to deletion/insertion sites at HVR1, HVR2, or HVR5 in Ad5 hexon are indicated.
Figure 2.
DogTag is highly reactive with DogCatcher following insertion into adenovirus hexon HVR loops, with decorated virions retaining infectivity
(A) Yield comparison of GFP-expressing Ad vector preparations displaying DogTag on hexon HVR5 (Ad-DogTag) versus Ad vectors with an unmodified hexon (Ad). Data show mean +SD, n = 3, infectious yield from 1,500 cm2 adherent 293A cells. Mean P:I ratios (ratio of total viral particles calculated by UV spectrophotometry to infectious units calculated by GFP focus assay) for vector batches are indicated above each bar. (B) SDS-PAGE and Coomassie staining analysis of Ad virions displaying DogTag at HVR1, HVR2, or HVR5 (1E+10 viral particles) incubated with DogCatcher (5 μM) at 4°C for 16 h. Gel shift observed upon covalent coupling of DogCatcher to virion associated hexon-DogTag. (C) Vector infectivity (GFP focus assay) performed in 293A cells on the samples from (B). Data show mean +SD of triplicate wells.
Decoration of virions with a model antigen elicits potent humoral immunity
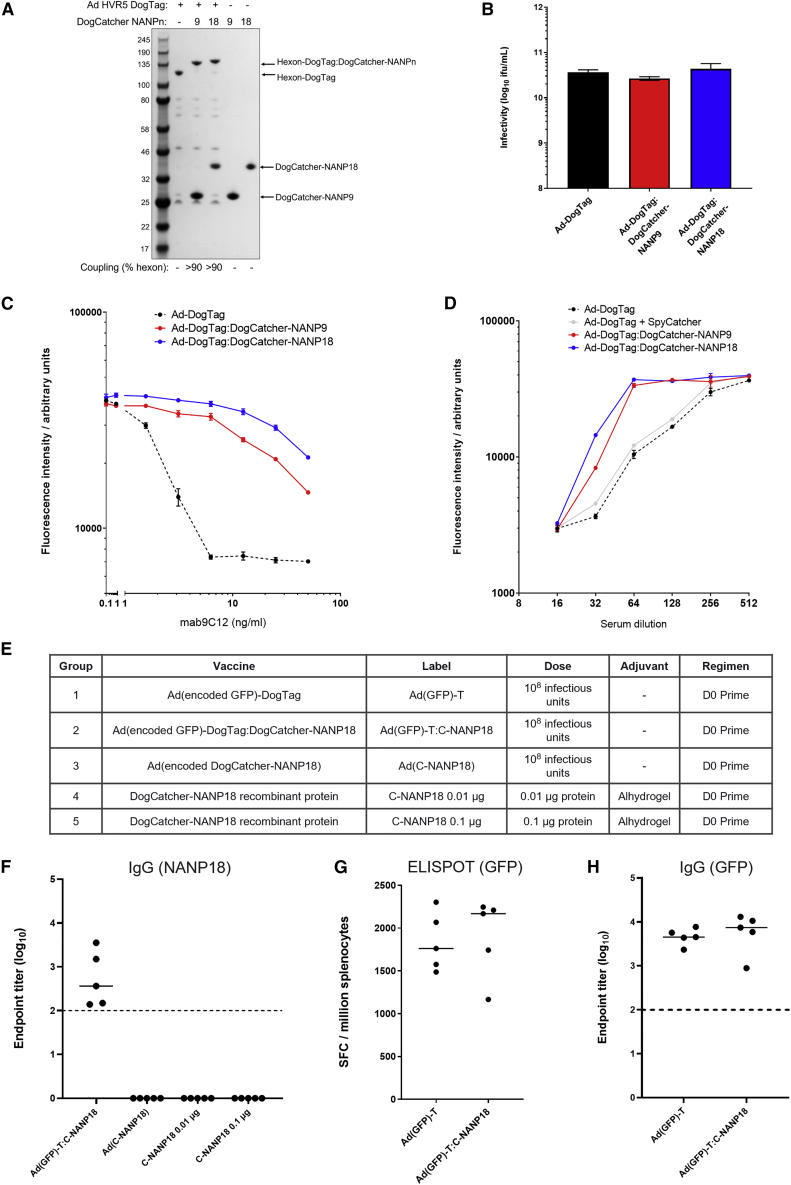
The circumsporozoite protein (CSP) of Plasmodium falciparum (Pf) has been extensively studied as a malaria vaccine candidate antigen.24 The protein contains a highly immunogenic repetitive region, primarily consisting of repeats of the amino acid sequence NANP; vaccine-induced antibodies against this NANP repeat region protect against malaria infection (the World Health Organization approved malaria vaccine RTS,S/Mosquirix includes 18 copies of NANP).24 A model system to test antibody induction using capsid display technology was developed by fusing multiple copies of NANP (either nine or 18) to DogCatcher. These DogCatcher-NANPn fusion proteins were used to decorate Ad-DogTag virions. Extensive coverage of the Ad capsid (>90% hexon coupled) was achieved using both DogCatcher-NANPn proteins (Figure 3A). Neither ligand had any effect on vector infectivity upon particle decoration (Figure 3B). Since coverage of Ad virions was extensive with both ligands, we sought to determine whether particle decoration could act as a protective shield to inhibit Ad neutralization by capsid-binding antibodies. Decoration with both ligands partially shielded Ad particles from neutralization in vitro by an anti-hexon monoclonal antibody (mAb 9C12),25 increasing the half maximal inhibitory concentration (IC50) by at least 10-fold compared with Ad-DogTag alone (Figure 3C). Particle decoration also inhibited neutralization of Ad particles by polyclonal anti-Ad5 serum (Figure 3D), even though antibodies in this serum were raised against multiple capsid components. Co-incubation of Ad-DogTag virions with SpyCatcher (SpyCatcher does not react with DogTag, hence no capsid decoration)22 did not inhibit neutralization with anti-Ad5 sera (Figure 3D).
Figure 3.
Capsid display of DogCatcher-NANP18 ligand shields the particles from anti-vector neutralizing antibodies and elicits potent ligand-specific humoral immunity
(A) SDS-PAGE and Coomassie staining analysis of Ad virions displaying DogTag at HVR5, (1E+10 viral particles) incubated with DogCatcher-NANP9 (5 μM) or DogCatcher-NANP18 (5 μM) at 4°C for 16 h. (B) Vector infectivity (GFP focus assay) performed in 293A cells on the samples from (A). Data show mean +SD of triplicate wells. (C) Vector neutralization assay using anti-hexon mAb 9C12. Ad particles encoding a GFP transgene with or without a capsid ligand were added to 293A cells in the presence of a varying concentration of neutralizing mAb. Vector transduction was measured via fluorescence of encoded GFP expressed in the cells. (D) Vector neutralization assay using anti-Ad5 serum. Ad particles encoding a GFP transgene with or without a capsid ligand were added to 293A cells in the presence of a varying concentration of serum from mice immunized with Ad5. (C) and (D) show mean + range of duplicate wells. (E) BALB/c mice (five per group) were immunized intramuscularly as described (vector-encoded antigens in brackets). Note that Ad(C-NANP18) has an unmodified hexon (no DogTag). The DogCatcher-NANP18 protein dose in group 2 was calculated to be < 0.05 μg per mouse. (F) Serum IgG antibody responses to NANP18 in groups 2–5 measured by endpoint ELISA 14 days post immunization. (G) CD8+ T cell responses in the spleen to encoded epitope EGFP200-208 were measured in groups 1 and 2 by overnight ex vivo IFNγ-ELISpot 14 days post immunization. SFC, spot forming cells. (H) Serum IgG antibody responses to encoded GFP in groups 1 and 2 were measured by endpoint ELISA 14 days post immunization. In (F)–(H), bars show median responses. Dashed line represents limit of detection.
An immunogenicity study in mice was performed to assess antibody titers against NANP18 displayed on Ad-DogTag virions, compared with a conventional Ad vector encoding the same model antigen construct or vaccination with protein in adjuvant formulations (Figure 3E, groups 2–5). Two weeks after a single vaccine dose, immunoglobulin (Ig) G antibody responses against NANP18 were generated in all animals vaccinated using capsid display technology Ad(GFP)-DogTag:DogCatcher-NANP18, but no detectable responses were achieved using either an encoded antigen platform Ad(DogCatcher-NANP18), or DogCatcher-NANP18 protein in adjuvant formulations (Figure 3F). IgG responses were also raised against DogCatcher in animals vaccinated with decorated Ad virions (Figure S4). In the same experiment, we also assessed the impact of capsid decoration of an Ad particle on immune responses generated against a transgene-encoded antigen in the same vector (Figure 3E, groups 1 and 2). No differences in magnitudes of either CD8+ T cell responses (Figure 3G) or antibody responses (Figure 3H) against encoded GFP were observed between Ad-DogTag vectors with capsid decoration (Ad(GFP)-DogTag:DogCatcher-NANP18) or without capsid decoration (Ad(GFP)-DogTag). These data demonstrate that, despite >90% hexon coverage on Ad virions, display of DogCatcher-NANP18 ligand on the capsid surface did not impair immune responses against an encoded transgene antigen.
Decoration with SARS-CoV-2 spike receptor-binding domain shields adenovirus particles from capsid interactors
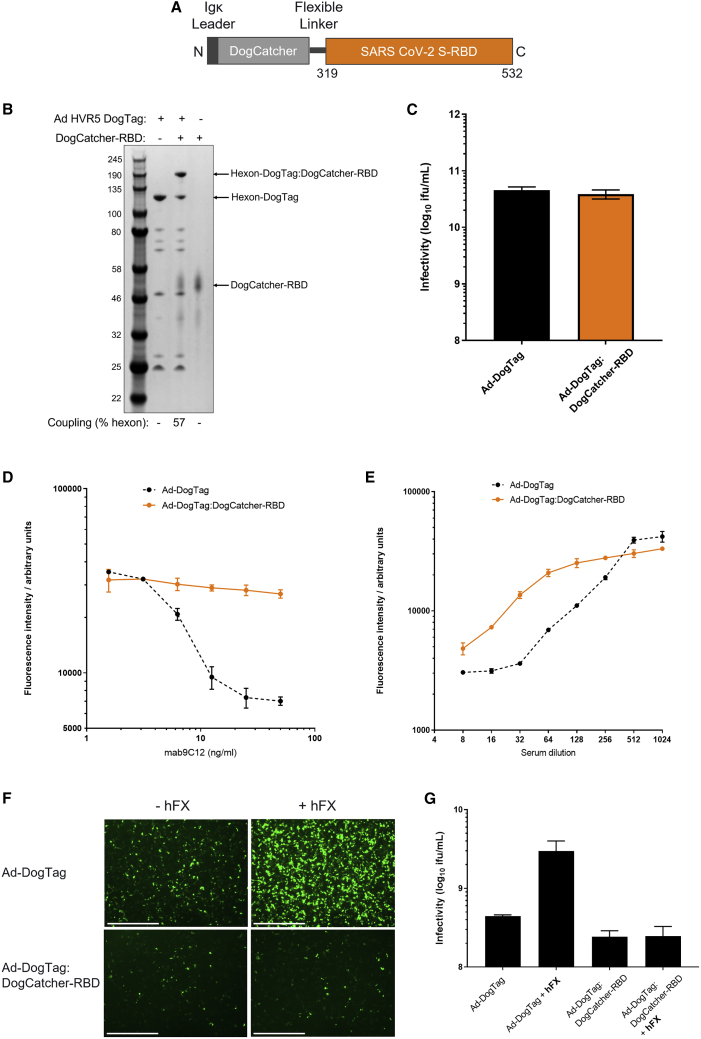
The receptor-binding domain (RBD) of SARS-CoV-2 spike protein interacts with the angiotensin-converting enzyme 2 (ACE2) on susceptible host cells and is required for viral infection. A recent study has estimated that ∼90% of SARS-CoV-2 neutralizing antibodies raised against the spike protein target the RBD region.26 As further proof of concept for Ad capsid display technology, we fused RBD (∼26 kDa) to DogCatcher (Figure 4A), and decorated Ad-DogTag virions with DogCatcher-RBD fusion protein. Despite the larger size of DogCatcher-RBD compared with ligands tested previously, substantial coverage of the Ad capsid was achieved (∼57% hexon monomers coupled) (Figure 4B), and, importantly, no impairment of vector infectivity was observed upon particle decoration (Figure 4C). Strikingly, capsid display of DogCatcher-RBD completely abrogated Ad vector neutralization in vitro by potent hexon-binding neutralizing monoclonal antibody mAb 9C12 (Figure 4D). Vector neutralization by polyclonal anti-Ad5 serum was also inhibited by capsid shielding with DogCatcher-RBD (Figure 4E). In vitro vector infectivity and neutralization assays were performed on 293A cells, which did not express human ACE2 (Figure S5). Given the observed capacity for DogCatcher-RBD to shield the Ad capsid from antibody-mediated neutralization, we sought to determine whether decoration with this ligand could impair other Ad capsid interactions. Human coagulation factor X (hFX) has previously been shown to bind directly to the Ad5 hexon protein and mediates hepatocyte transduction via heparan sulfate proteoglycans (HSPGs) by Ad5 vectors in vivo, particularly after intravascular delivery.27,28 Using a cell-line model of this interaction, capsid decoration with DogCatcher-RBD completely abrogated hFX-mediated Ad-DogTag transduction of SKOV-3 cells. Ad-DogTag without a capsid shield exhibited a 6.6-fold increase in SKOV-3 infectivity upon co-incubation with hFX (Figures 4F and 4G). Decorated Ad virions exhibited a modest reduction in infectivity (1.8-fold) in the absence of hFX compared with undecorated virions (Figures 4F and 4G).
Figure 4.
RBD can be fused to DogCatcher, displayed on adenovirus particles, and is an effective capsid shield
(A) Design of DogCatcher-RBD protein (B) and SDS-PAGE and Coomassie staining analysis of Ad virions displaying DogTag at HVR5 (1E+10 viral particles) incubated with DogCatcher-RBD (3.5 μM) at 4°C for 16 h. (C) Vector infectivity (GFP focus assay) performed in 293A cells on the samples from (B). Data show mean +SD of triplicate wells. (D) Vector neutralization assay using anti-hexon mAb 9C12, performed as described in Figure 3. Data show mean and range of duplicate wells. (E) Vector neutralization assay using anti-Ad5 sera, performed as described in Figure 3. Data show mean + SD of triplicate wells. (F) Capsid decoration with DogCatcher-RBD impairs human factor X (hFX)-mediated Ad transduction of SKOV3 cells. Fluorescent microscopy images taken 48 h post infection show Ad-DogTag or Ad-DogTag:DogCatcher-RBD vectors expressing GFP incubated with or without hFX (2,500 viral particles/cell). Scale bar, 1,000 μm. (G) Infectivity data (GFP focus assay) from the experiment shown in (F). Data show mean +SD of triplicate wells.
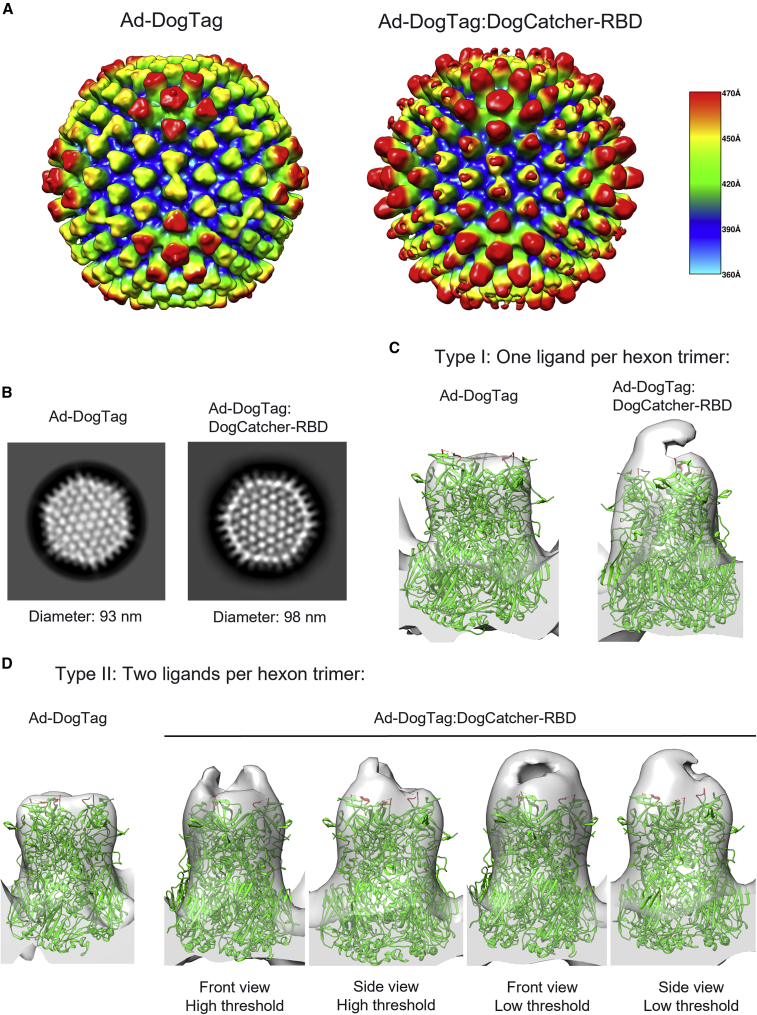
Cryo-EM analysis of adenovirus particles displaying RBD
To further investigate the nature of capsid decoration of Ad-DogTag particles with DogCatcher-RBD, cryoelectron microscopy (cryo-EM) analysis of decorated (Ad-DogTag:DogCatcher-RBD) and undecorated (Ad-DogTag) particles was performed (Figure 5). A 3D density map of decorated particles clearly indicated additional density extending outward from hexon trimers (red-orange regions), compared with the undecorated particles (Figure 5A). Additional density protruding from hexon trimers could also be observed from 2D class averages of Ad-DogTag:DogCatcher-RBD particles compared with Ad-DogTag particles, extending the particle diameter from 93 to 98 nm (Figure 5B). Surface protrusions (representing hexon trimers) on Ad-DogTag were of uniform density, while Ad-DogTag:DogCatcher-RBD showed two distinct regions. The outermost region represented coupled RBD being less dense than hexon trimers, likely due to flexibility of the attached ligand. Analysis of 3D density maps revealed attachment of either one (type I, Figure 5C) or two (type II, Figure 5D) RBD ligands per hexon trimer. Type II decoration was particularly abundant on hexon trimers located adjacent to penton subunits at capsid vertices. These trimers are elevated with respect to the rest of the virion surface (see Figure 5A, red-orange regions of Ad-DogTag map; note that the fiber protein is not shown) and therefore steric hindrance may be lower, enabling multiple ligand occupancy. Additional density representing attached RBD is clear for both type I and type II arrangement, compared with undecorated Ad-DogTag; Figure S6 shows fitting of an RBD structure into these maps (with the RBD N terminus proximal to hexon HVR5, DogCatcher is not shown).
Figure 5.
Cryo-EM analysis of adenovirus particles displaying RBD
(A) 3D density maps (at 10.5 Å) for Ad-DogTag (control sample, undecorated) and Ad-DogTag:DogCatcher-RBD particles. Radial coloring scheme is indicated; regions furthest from the center of the particle are shown in red, regions closest to the center are shown in blue. Distance from center of particle (in angstroms) is indicated. (B) Exemplary 2D class averages. Indicated diameters calculated from vertex to vertex. (C) Type I ligand coupling; 3D structure of representative hexon trimer without ligand (Ad-DogTag) or with one ligand coupled per trimer (Ad-DogTag:DogCatcher-RBD) shown at the same contour level. (D) Type II ligand coupling; 3D structure representative of hexon trimer adjacent to penton base without ligand (Ad-DogTag) or with two ligands coupled per trimer (Ad-DogTag:DogCatcher-RBD) shown at the same contour level. Maps for Ad-DogTag:DogCatcher-RBD shown at both front and side angles, and high and low threshold to indicate location and extent of additional electron density. In both (C) and (D), hexon trimer structure (PDB: 6B1T) was fitted (green), with location of HVR5 loop (residues 270–280, site of DogTag insertion) shown in red.
Capsid decoration with RBD elicits potent neutralizing antibody responses against SARS-CoV-2
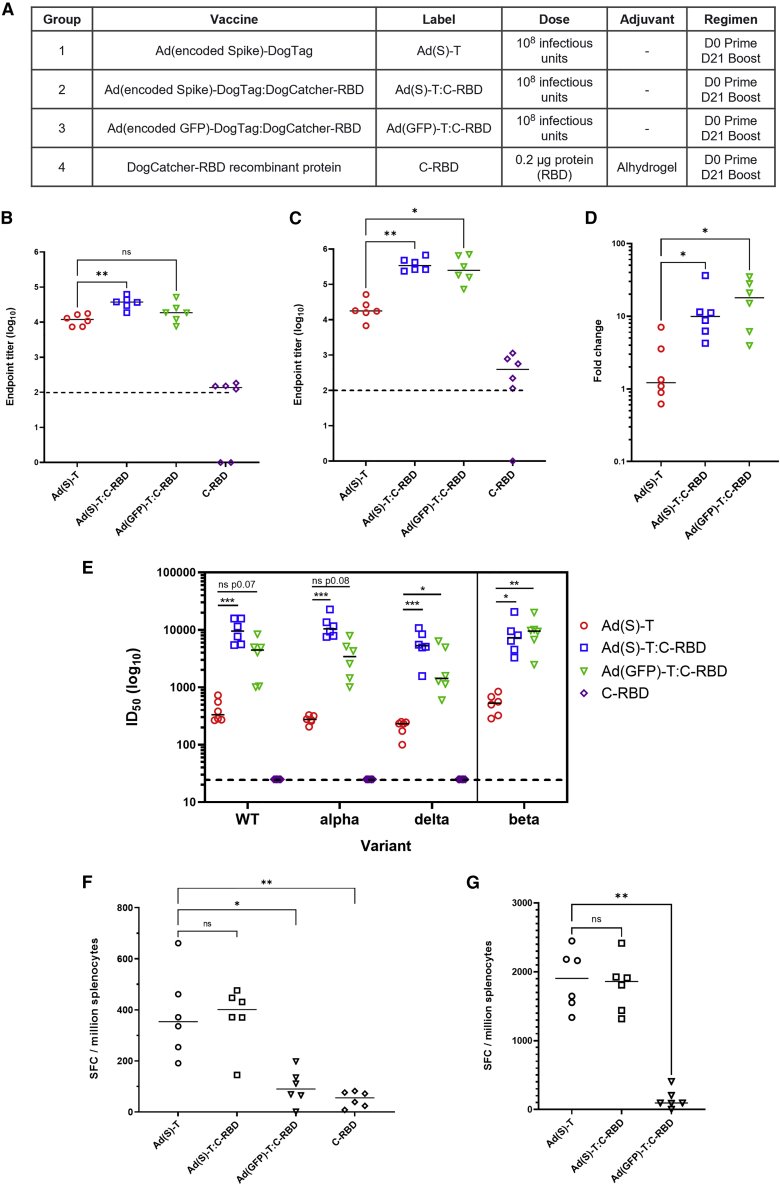
An in vivo study was conducted to assess humoral and cellular immunity against SARS-CoV-2 using Ad-DogTag vectors decorated with DogCatcher-RBD, compared with a conventional adenovirus vector encoding spike in the viral genome.
Mice were immunized in homologous prime-boost regimens with either Ad-DogTag encoding spike without a capsid ligand (Ad(Spike)-DogTag, group 1), the same vector as in group 1 but with DogCatcher-RBD coupled to the viral capsid (Ad(Spike)-DogTag:DogCatcher-RBD, group 2), a vector displaying the same DogCatcher-RBD ligand as in group 2 but encoding a GFP transgene rather than spike (Ad(GFP)-DogTag:DogCatcher-RBD, group 3), or recombinant DogCatcher-RBD protein in Alhydrogel adjuvant (group 4) (Figure 6A).
Figure 6.
High-titer SARS-CoV-2 neutralizing antibody responses generated by capsid display of RBD
(A) BALB/c mice (six per group) were immunized intramuscularly in homologous prime-boost regimens as described. The RBD protein dose in groups 2 and 3 was calculated to be < 0.2 μg per mouse. (B) Serum IgG antibody responses to RBD at D20 measured by endpoint ELISA. (C) Serum IgG antibody responses to RBD at D35 measured by endpoint ELISA. (D) Fold increase in anti-RBD IgG titer post boost (D35 titer in C divided by D20 titer in B). (E) SARS-CoV-2 neutralization titers (pVNT assay) in D35 serum against Wuhan strain (WT) and variants of concern alpha (B.1.1.7), delta (B.1.617.2), and beta (B.1.351). Data on beta variant collected separately. (F) IFNγ-ELISpot response in spleen at D35 against peptide pool spanning SARS-CoV-2 S-RBD. (G) IFNγ-ELISpot response in spleen at D35 against peptides spanning full-length SARS-CoV-2 S protein (summed responses from two peptide pools spanning residues 1–643 and 633–1,273 are shown, responses from individual pools shown in Figure S8). Dashed lines represent limit of detection. Median responses shown by a horizontal line. Statistical analyses performed by Kruskal-Wallis with Dunn’s test for multiple comparisons, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, not significant.
Serum IgG antibody titers against RBD were comparable between Ad vector groups after a single immunization (Figure 6B), but median post-boost titers were >10-fold higher using both RBD capsid display vectors compared with the conventional Ad(Spike)-DogTag vector, regardless of the encoded antigen (Figure 6C). Indeed, anti-RBD IgG titers after immunization (both pre-boost and post boost) with Ad(GFP)-DogTag:DogCatcher-RBD were comparable with Ad(Spike)-DogTag:DogCatcher-RBD, despite the former vector lacking an encoded spike antigen. A comparison of median anti-RBD IgG responses pre-boost and post boost indicates that capsid display of RBD increased boostability of Ad vector-mediated humoral immunity against RBD ∼10-fold (Figure 6D). IgG responses against the full-length spike ectodomain were also higher after immunization with both RBD capsid display vectors post boost (Figure S7). Neutralizing antibody titers in serum post vaccination were measured in an extensively validated in vitro neutralization assay using human immunodeficiency virus-1 (HIV-1)-based virus particles pseudotyped with SARS-CoV-2 spike.29 Median pseudotype neutralization titers (pVNT) after prime-boost immunization with either RBD capsid display vector were >10-fold higher against the Wuhan strain compared with the conventional Ad vector encoding spike (Figure 6E). Similar relative titers between groups were observed against alpha, beta, and delta variants of concern (Figure 6E).
Importantly, and in agreement with previous observations, display of DogCatcher-RBD on the Ad capsid surface did not impair T cell responses against encoded SARS-CoV-2 spike. Spleen interferon-gamma (IFNγ) enzyme-linked immune absorbent spot (ELISpot) responses against an RBD peptide pool (Figure 6F) and full-length spike peptide pools (Figures 6G and S8) showed that T cell immunogenicity with Ad(Spike)-DogTag and Ad(Spike)-DogTag:DogCatcher-RBD was comparable. T cell responses against RBD were significantly greater in magnitude after immunization with either Ad vector encoding spike, compared with Ad(GFP)-DogTag:DogCatcher-RBD (Dunn’s multiple comparisons test; Ad(Spike)-DogTag versus Ad(GFP)-DogTag:DogCatcher-RBD p = 0.015, Ad(Spike)-DogTag:DogCatcher-RBD versus Ad(GFP)-DogTag:DogCatcher-RBD p = 0.009), indicating that encoding an antigen is preferable to capsid display for inducing T cell responses.
An additional in vivo study was conducted to confirm the requirement for attachment of DogCatcher-RBD to the adenovirus capsid to generate potent humoral immunity against RBD. Mice were immunized in homologous prime-boost regimens with either an RBD-decorated vector encoding GFP (Ad(GFP)-DogTag:DogCatcher-RBD, group 1), or a vector encoding GFP but with an unmodified hexon (Ad(GFP), no DogTag) co-administered with DogCatcher-RBD protein that cannot attach to the adenovirus capsid (group 2 and group 3) (Figure S9A). Only the vector decorated with RBD via DogTag/DogCatcher covalent linkage induced serum IgG responses against RBD; co-administration of DogCatcher-RBD and vector components without capsid attachment was unable to induce detectable anti-RBD IgG responses despite administration of a >5-fold higher dose of RBD (Figure S9B).
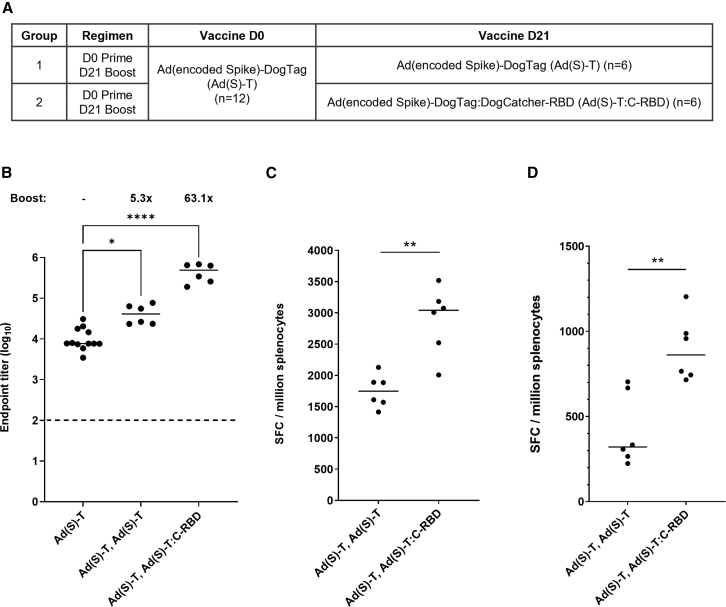
Applying a capsid shield to booster adenovirus vaccines improves both humoral and cellular immunogenicity
Given the ability of capsid display technology to elicit potent humoral immunity against SARS-CoV-2, we sought to determine whether RBD capsid decoration could efficiently boost immunity in animals that had already received a first dose of a conventional Ad vector encoding spike. A homologous two-dose Ad(Spike)-DogTag regimen was compared with a heterologous regimen in which an RBD capsid shield (Ad(Spike)-DogTag:DogCatcher-RBD) was applied to the second vaccine dose (Figure 7A). A 5.3-fold increase in median anti-RBD IgG titers was observed between prime and boost immunizations in the homologous regimen, but a 63.1-fold increase in titer was observed after the heterologous regimen (Figure 7B). In addition, spleen IFNγ ELISpot responses against full-length SARS-CoV-2 spike were also higher in the heterologous regimen compared with the homologous regimen (Figure 7C). A similar observation was observed in IFNγ ELISpot assays using a peptide pool spanning only residues 633–1,273 of spike (hence not including the RBD region), indicating that increased cellular immunity to the encoded spike transgene after heterologous prime-boost occurred independently from the contribution of the RBD capsid ligand (Figure 7D).
Figure 7.
Applying RBD capsid decoration at boost significantly increases SARS-CoV-2 specific antibody and T cell responses in an adenovirus vector prime-boost regimen
(A) BALB/c mice (n = 12) were immunized intramuscularly with Ad(Spike)-DogTag on D0 and then on D21 given a second intramuscular immunization of either Ad(Spike)-DogTag (group 1, n = 6) or Ad(Spike)-DogTag:DogCatcher-RBD (group 2, n = 6). All vaccines were administered at a dose of 108 infectious units. (B) Serum IgG antibody responses to RBD measured by endpoint ELISA. Responses measured post prime on D20 (Ad(Spike)-DogTag) are compared with responses on D35 after homologous (Ad(Spike)-DogTag, Ad(Spike)-DogTag), or heterologous (Ad(Spike)-DogTag, Ad(Spike)-DogTag:DogCatcher-RBD) prime boost. Fold change in median titer post boost is shown. Horizontal bars show median responses. Statistical analyses performed by Kruskal-Wallis with Dunn’s test for multiple comparisons, ∗p < 0.05; ∗∗∗∗p < 0.0001. Dashed line represents limit of detection. (C) IFNγ-ELISpot response in spleen at D35 against peptide pools spanning full-length (1–1,273) SARS-CoV-2 S (summed responses from two peptide pools spanning residues 1–643 and 633–1,273 are shown). (D) IFNγ-ELISpot response in spleen at D35 against peptide pool spanning C-terminal residues 633–1,273 of SARS-CoV-2 S only (i.e., not including RBD domain). In (C)–(D), horizontal bars show median responses and statistical analyses performed by Mann-Whitney test, ∗∗p < 0.01.
Discussion
In this study we describe a simple, rapid, and targeted system of spontaneous covalent decoration of recombinant adenovirus particles with a variety of ligands under mild conditions. Using the recently described DogTag/DogCatcher reactive pair,22 we targeted the hexon protein for capsid display to maximize ligand density since hexon is the most abundant component of the adenovirus capsid. Several previous studies have achieved display of short immunogenic epitopes by direct genetic insertion of sequences into hexon HVR loops. However, these approaches have typically been limited to <50 residues, since larger insertions disrupt protein stability and prevent successful rescue of recombinant vectors.30,31,32 Other studies have performed chemical modification of the viral capsid to attach larger ligands, including targeted insertion of cysteine residues for thiol-directed coupling, but these approaches can require challenging reaction and storage conditions to achieve conjugation and retain infectivity of modified vectors.33,34 Some studies achieved capsid display of larger ligands through genetic fusion to the C terminus of pIX, a minor capsid protein with fewer copies per virion than hexon.31 However, large pIX fusions do not always generate viable vectors, can reduce capsid stability, and modified pIX may not be efficiently incorporated into the capsid.35,36
Using our approach, we have covalently coupled ligands as large as the RBD of SARS-CoV-2 spike (∼26 kDa) to hexon through genetic fusion to DogCatcher and co-incubation of the recombinant fusion protein with Ad-DogTag, achieving extensive coverage of the viral capsid in each case (Figures 2B, 3A, 4B and 5A). Importantly and despite a high proportion of hexon monomers conjugated to ligand, decorated vectors retained infectivity in vitro (Figures 2C, 3B, and 4C). Transduction of 293A cells by Ad5 vectors is mediated by interactions between the adenovirus fiber protein and the coxsackie and adenovirus receptor (CAR);37 these interactions are unlikely to be masked by hexon conjugation due to the sizes of the ligands tested, as reflected by our cryo-EM data (Figure 5). We hypothesize that there is also sufficient flexibility engineered into hexon loop structures to avoid interference with secondary interactions between the adenovirus penton and cell-surface αV integrins required for clathrin-mediated endocytosis of virions.38 Mechanisms of Ad vector transduction upon in vivo delivery may be more complex than those described in vitro.39 However, since transduction of cells is an essential prerequisite for expression of encoded transgenes and neither humoral nor cellular transgene-specific immune responses are impaired by capsid decoration (Figures 3 and 6), our data suggest that decorated Ads also efficiently transduce cells in vivo (although we have not investigated this directly). DogTag was highly reactive with DogCatcher at each of the three insertion sites tested (Figure 2B), with decorated vectors retaining infectivity in each case (Figure 2C). These data indicate that reactivity is not insertion site specific and imply that our approach is likely to be successful with alternative adenovirus serotypes.
Decoration of the Ad capsid surface with DogCatcher-RBD reduced the potency of anti-Ad neutralizing antibodies in vitro. Capsid shielding against mAb 9C12 was more effective with DogCatcher-RBD (Figure 4D) than DogCatcher-NANP18 (Figure 3C), despite lower ligand coverage (57% [Figure 4B] versus >90% [Figure 3A] hexon coupled), likely due to increased coverage of the capsid surface with the larger RBD ligand. Indeed, cryo-EM data indicated that all hexon trimers had at least one copy of RBD attached, with ligand density covering much of the hexon trimer surface (Figure 5). Shielding from neutralization by polyclonal anti-Ad mouse serum was also achieved using both ligands, albeit less effectively than against mAb 9C12. Studies have shown that serum post vaccination with Ad vectors typically contains antibodies against other viral proteins in addition to hexon (including fiber), which are less likely to be masked by hexon ligation.40 The effectiveness of our technology to shield adenovirus capsids from neutralizing antibodies in human sera, and thereby overcome pre-existing anti-vector immunity in humans, remains to be determined. Our platform has the potential to shield the Ad virion from other potentially undesirable capsid interactors. For instance, display of DogCatcher-RBD blocked hFX-mediated transduction of SKOV-3 cells in vitro, presumably by inhibiting interaction between hFX and hexon (Figures 4F and 4G) (with the caveat that insertion of DogTag into HVR5 may have already reduced the affinity of this interaction).41 Shielding this interaction in vivo would be advantageous, since hFX enhances Ad5 transduction of hepatocytes, increasing liver sequestration and hepatotoxicity after systemic administration.27 A recent study has also reported an interaction between capsid-associated hexon of various serotypes and platelet factor 4 (PF4), which may contribute toward very rare development of blood clots associated with some COVID-19 vaccines.42 Further studies will be required to assess the capability of our platform to inhibit interactions between hFX (or other host proteins) and hexon in vivo but this platform could represent an attractive solution to achieve capsid shielding. To date, Ad capsid shielding has predominantly involved coating virions with polymers, such as polyethylene glycol (PEG) or N-(2-hydroxypropyl)methacrylamide (HPMA).43 Although effective in shielding, these polymer coatings have tended to impair vector transduction.44
Capsid decoration with DogCatcher-RBD significantly enhanced boosting of humoral immunity against SARS-CoV-2 using Ad vaccine vectors. In homologous prime-boost regimens, decorating an Ad vector encoding spike with DogCatcher-RBD increased median anti-RBD antibody responses and pVNT titers >10-fold compared with the undecorated Ad (Figure 6). Strikingly, a vector encoding an irrelevant antigen (GFP) but with RBD displayed on the Ad capsid also elicited significantly higher anti-RBD antibody responses compared with the undecorated Ad encoding spike, and median pVNT titers were >10-fold higher against the Wuhan strain and all variants of concern except delta (6.2-fold). Key to the enhanced humoral immunity achieved with our platform was increased boostability of antibody responses against capsid-displayed RBD. Increase in anti-RBD antibody titers between prime and boost was ∼10-fold higher with RBD capsid display compared with the undecorated Ad (Figure 6D). A similar observation was made after a heterologous regimen using RBD-decorated Ads to boost immunity following immunization with a conventional undecorated Ad encoding spike (Figure 7B). In both experiments, boosting of humoral immunity using homologous prime boost with undecorated Ad encoding spike was modest (∼2-5-fold increase in anti-RBD ELISA; Figures 6D and 7B), and these data are comparable with previous reports using other Ad vectors encoding SARS-CoV-2 spike in mice.45 In trials of COVID-19 vaccines in humans, boosting of cellular and humoral immunity using homologous Ad prime-boost regimens was also modest, particularly compared with heterologous regimens with mRNA vaccines or homologous mRNA regimens.46,47,48 Anti-vector immunity generated after a prime immunization may inhibit the ability to boost immune responses to encoded antigens using the same vector; anti-Ad neutralizing antibodies prevent vector transduction and subsequent transgene antigen expression required for antigen-specific immunity.9 In contrast, recombinant protein antigens displayed in particulate form on nanoparticles or VLPs (in adjuvant formulations) have been shown to generate robust humoral immunity, with highly efficient boosting in homologous prime-boost regimens.49,50 Previous studies have suggested that particulate antigens generate potent antibody responses through a variety of mechanisms, including B cell receptor crosslinking51 and prolonged retention in draining lymph nodes.52 It seems likely that responses against capsid-decorated RBD are induced by similar mechanisms, particularly since Ad particles have an optimum size for lymph node entry and retention,53 although future studies will be required to elucidate these mechanisms further.
While capsid antigen decoration induced potent humoral immunity, encoding of antigen sequences in the vector genome was required to generate potent cellular immunity (Figure 6F). This observation is unsurprising, considering Ad vaccine vectors are known to induce particularly strong T cell immunity, especially CD8+ T cell responses.6,54 Importantly, capsid decoration did not impair T cell responses to encoded antigens (Figures 3G, 6F, and 6G), and in fact a heterologous prime-boost regimen using capsid-decorated vectors at boost improved cellular immunity compared with a homologous regimen (Figures 7C and 7D). The latter observation suggests that our capsid shielding technology applied at boost may have, at least in part, circumvented anti-vector immunity generated after a priming immunization, as observed in vitro (Figures 3 and 4). In support of this hypothesis, median SARS-CoV-2 spike-specific IFNγ ELISpot responses were comparable between homologous prime-boost regimens using decorated and undecorated vectors encoding spike in both experiments (Figures 6G and 7C) but were ∼2-fold higher in the heterologous regimen (Figure 7C). We cannot exclude the possibility that capsid decoration with RBD, as with any modification to the viral capsid, may, to some extent, have altered the natural tropism of vectors (although it should be noted that murine ACE2 does not bind to SARS-CoV-2 RBD).55 However, attempts to re-direct vector tropism to specific cell types by attaching or inserting receptor-binding ligands via hexon have tended to be less effective than retargeting via fiber (particularly in the case of high-affinity ligands), with speculation that such ligands, when attached to hexon, may prevent endosomal release of virions.56,57
Our capsid display platform offers a substantial improvement over conventional Ad vaccine technology for induction of both cellular and humoral immunity. Taken together, the data presented here support a concept for optimizing concomitant humoral and cellular immunogenicity, whereby antigenic targets for antibody induction are displayed on the capsid surface, while potent T cell epitopes are encoded in the vector genome. Such a concept would enable conserved components of target pathogens that induce potent T cell responses (such as nucleocapsid58 or ORF1ab59 of SARS-CoV-2) to be encoded in the vector genome, while targets of neutralizing humoral immunity such as RBD (often more heterogeneous in sequence) can be displayed on the capsid surface. Surface-displayed targets may even be replaced with different versions in response to the emergence of new pathogen variants. Indeed, a recent study has suggested that memory T cells recognizing epitopes within the SARS-CoV-2 replication-transcription complex that are shared with seasonal human coronaviruses may have protected against SARS-CoV-2 infection in a cohort of healthcare workers who remained seronegative.60 Further investigation will be required to comprehensively determine limitations for capsid display in terms of size and structure of ligands, but similar-sized RBDs of other viral proteins, including influenza hemagglutinin (∼25 kDa), have been expressed independently as recombinant proteins and displayed on VLPs, implying that this could be a generalizable concept.61
Capsid decoration using our protein superglue technology is simple, requiring only co-incubation of spontaneously and irreversibly reacting components with no chemical modification required. A similar conjugation process has already been scaled under good manufacturing practice (GMP) during development of a VLP-based SARS-CoV-2 vaccine currently in phase I/II clinical trials.62 The ability of our adenovirus-based platform to induce robust cellular and humoral immunity and to enhance efficacy of multi-shot regimens could be advantageous for applications beyond prophylactic vaccines, including therapeutic vaccines against chronic viral pathogens and cancer. Methods of rapid and customizable covalent decoration of Ad capsids could also be utilized for development of personalized therapies. In prophylactic settings, adenovirus capsid decoration could be utilized in the design of pan-coronavirus and pan-influenza vaccines, combining broad and conserved T cell immunity from encoded antigens with exchangeable capsid ligands delivering potent neutralizing humoral immunity.
Materials and methods
Construction of Ad-DogTag vectors
An expression construct, consisting of the cytomegalovirus immediate-early promoter (CMVp) containing tetracycline operator sequences driving expression of enhanced green fluorescent protein (GFP), was cloned into the shuttle vector pENTR4 (Invitrogen). The CMVp GFP expression construct was inserted into an E1/E3-deleted (replication-defective) molecular clone of Ad5 at the E1 locus using Invitrogen Gateway site-specific recombination technology. Bacterial artificial chromosome (BAC) sequences from pBELOBAC11 (NEB) were amplified using forward (5′-TTAATTAAcgtcgaccaattctcatg) and reverse (5′-TTAATTAAgtcgacagcgacacacttg) primers to introduce PacI sites at either end of the BAC cassette. The entire Ad5(GFP) genome was subsequently cloned into the BAC with PacI, to generate pBAC-Ad5(GFP). DogTag (DIPATYEFTDGKHYITNEPIPPK) or SpyTag (AHIVMVDAYKPTK) sequences flanked by GSGGSG linkers (Figure 1B) were inserted into hexon HVR1, HVR2, or HVR5 loops through BAC GalK recombineering, using the GalK gene for both positive and negative selection as described previously.63 Insertion sites in the hexon are shown in Figure 1B. For insertion of Tags at HVR1, native residues 139–165 were deleted; for insertion of Tags at HVR2, native residues 189–192 were deleted; for insertion of Tags at HVR5 native residues 269–281 were deleted. Deletions/insertions at similar sites have been described previously.30,64,65 In this study, DogCatcher refers to the sequence described as R2CatcherB in Keeble et al.22 Recombinant vectors expressing DogCatcher-NANP18 and SARS-CoV-2 spike (residues 1–1,208 Wuhan strain, codon optimized for mammalian expression and including stabilizing mutations K986P and V987P and mutation of the furin cleavage site 682-GSAS-685)66 were generated through cloning of gene constructs into pENTR4.CMVp and subsequent Gateway-mediated insertion into the Ad5 E1 locus.
Rescue and purification of recombinant adenoviruses
BAC DNA from recombinant molecular clones was linearized with PacI to release left and right viral inverted terminal repeats (ITRs). Linearized DNA was transfected (Lipofectamine 2000, Invitrogen) into E1-complementing human embryonic kidney (HEK) 293 cell lines; either HEK293A (293A) cells (Invitrogen) for Ad vectors expressing GFP and DogCatcher-NANP18, or 293TREX cells (Invitrogen) for Ad vectors expressing SARS-CoV-2 spike. After cytopathic effect (CPE) was observed, the cells were harvested, subjected to three cycles of freeze-thaw, and the virus amplified further. Upon infection of 10 × 150 cm2 flasks, virus was harvested from infected cells after 48 h and purified by CsCl gradient ultracentrifugation according to standard protocols. Purified virus was dialyzed against sucrose storage buffer (10 mM Tris-HCl, 7.5% w/v sucrose, pH 7.8) and stored at −80°C.
Titration of recombinant adenoviruses
Infectious titer of vector preparations was assessed by single cell infectivity assay on 293A cells or 293TREX cells. For vectors expressing GFP, infected 293A cells were visualized and enumerated directly by fluorescent microscopy. For vectors without a fluorescent marker, cells (either 293A or 293TREX) were immunostained for expression of hexon. Serial dilutions of virus in complete media (Dulbecco’s modified Eagle’s medium, plus 1× GlutaMAX and 10% v/v fetal bovine serum) were performed in 96-well deep-well plates. Two or three serial dilutions were performed per virus, and 50 μL of each dilution (103 to 1010 dilution) was added per well of a seeded 96-well plate at 80%–90% confluency. Plates were incubated for 48 h at 37°C, 5% v/v CO2. For titration of GFP-expressing vectors, single GFP-positive cells were enumerated by fluorescence microscopy, and an infectious titer was calculated in infectious units (ifu) per milliliter. For hexon immunostaining, medium was aspirated from the cell monolayer and cells were fixed with ice-cold methanol. Plates were then washed in phosphate-buffered saline (PBS 1×, Gibco) before blocking for 1 h with 3% w/v low-fat milk (Marvel). Mouse monoclonal anti-hexon antibody (B025/AD51, Thermo Fisher) was added at 1:1,000 dilution in 1% w/v milk in PBS and incubated for 1 h at 25°C. Cells were washed with 1% w/v milk in PBS prior to addition of a secondary goat anti-mouse alkaline phosphatase (ALP)-conjugated antibody (STAR117A, Bio-Rad) at 1:1,000 dilution in 1% w/v milk in PBS. After a further 1-h incubation, plates were washed with PBS prior to development. To develop, 100 μL of freshly prepared SIGMAfast BCIP/NBT solution (Sigma-Aldrich) was added to each well and plates incubated at 25°C prior to the appearance of dark stained foci, representing single infected cells. For CsCl-purified vector preparations, viral particle count was estimated by spectrophotometric absorption at 260 nm as described previously.12,67 P:I ratios were calculated by dividing the number of viral particles (vp) per milliliter with the number of infectious units (ifu) per milliliter.
Protein production and purification
DNA sequences for expression of DogCatcher, DogCatcher-NANP9, DogCatcher-NANP18, and SpyCatcher were cloned into expression plasmid pET45(+) (EMD Millipore) for protein production in BL21(DE3) Escherichia coli (NEB). Recombinant proteins were purified using Ni-NTA affinity resin (Qiagen) according to standard protocols,22 dialyzed into PBS, and stored at −80°C.
DNA sequences for expression of DogCatcher fused to SARS-CoV-2 spike receptor-binding domain (Wuhan strain, residues 319–532) (DogCatcher-RBD), were cloned into mammalian protein expression plasmid pcDNA3.4. To facilitate secretion, the Igκ-leader sequence METDTLLLWVLLLWVPGSTGD was introduced at the N terminus of the fusion protein, and a C-terminal C-tag (EPEA) was added to enable affinity purification. DogCatcher-S-RBD was expressed in suspension ExpiCHO-S cells (Thermo Fisher); protein was harvested from culture supernatant, affinity purified using C-tag affinity resin (Thermo Fisher) using an AKTA chromatography system (GE Healthcare), and dialyzed into Tris-buffered saline (TBS) pH 7.4.
Proteins for ELISA assays, RBD-SnJr (RBD with SnoopTagJr fused at the C terminus) and spike-SnJr (SARS-CoV-2 spike 1–1,208, with stabilizing proline mutations and furin cleavage site mutation,66 a C-terminal gp160 trimerization domain and SnoopTagJr) were expressed in suspension Expi293F (Thermo Fisher) and ExpiCHO-S cells respectively and purified as described above for DogCatcher-RBD. Glutathione-S-transferase fused to NANP18 (GST-NANP18) was produced in BL21(DE3) E. coli by cloning DNA sequences into expression plasmid pGEX-6P (GE Healthcare), and protein purified using glutathione spin columns (Thermo Scientific Pierce) according to standard protocols and dialyzed into TBS pH 7.8.
Coupling reactions
For in vitro assays, coupling reactions between DogCatcher-fused protein ligands and DogTag inserted into the Ad capsid were performed by co-incubation of spontaneously reacting components in a total volume of 25 μL, with individual components at concentrations described in the figure legends. Reactions were incubated for 16 h at 4°C.
Ligand-decorated vector batches for the NANP vaccine studies were prepared by co-incubating 1.9E+12 viral particles of Ad5(GFP)-HVR5-DogTag with 35 μM DogCatcher-NANP18 for 16 h at 4°C. To remove excess ligand, coupled vectors were dialyzed into sucrose storage buffer using SpectraPor dialysis cassettes with 300-kDa molecular weight cutoff (MWCO). Dialysis decreased the excess ligand by at least 10-fold, as measured by densitometry on Coomassie-stained SDS-PAGE. Ligand coverage by SDS-PAGE was >90% hexon (comparable with Figure 3A).
Ligand-decorated vector batches for SARS-CoV-2 vaccine studies and electron microscopy were prepared by co-incubating 9E+11 viral particles of Ad5-HVR5-DogTag encoding either GFP or SARS-CoV-2 spike with 6 μM DogCatcher-RBD for 16 h at 4°C. To remove excess ligand, coupled vectors were dialyzed into sucrose storage buffer using SpectraPor dialysis cassettes (300-kDa MWCO). Dialysis decreased excess ligand by at least 20-fold as measured by densitometry on Coomassie-stained SDS-PAGE. Ligand coverage by SDS-PAGE was ∼60% hexon (comparable with Figure 4B).
For all decorated vaccine vectors, residual excess ligand post dialysis was factored into effective antigen dose calculations. Dialyzed vector batches were further diluted >10-fold in PBS during vaccine formulation (to a dose of 108 infectious units vector/100 μL) for mouse immunization. To calculate effective antigen dose per mouse, excess ligand per vaccine dose was added to the estimated dose of antigen coupled to decorated virions. Estimated coupled antigen dose was calculated by multiplying the percentage hexon coupled to antigen (capsid coupling efficiency) by the total number of copies of hexon per virion (720), and multiplying this number by the estimated number of viral particles per vaccine dose to give an estimate of the number of antigen copies delivered on viral particles. Antigen dose in micrograms was subsequently calculated using the predicted molecular weight of the antigen ligand (using ProtParam tool, ExPASy) and Avogadro’s constant. Ligand-decorated vectors were stored at −80°C, endotoxin tested, and infectious titration of stored batches was repeated prior to immunization.
Assessment of coupling efficiency by SDS-PAGE
Coupling reactions were performed as described above and stopped by addition of SDS loading buffer (Bio-Rad, 31.5 mM Tris-HCl, pH 6.8, 10% glycerol, 1% SDS, 0.005% bromophenol blue, 300 mM dithiothreitol). Samples were boiled at 95°C for 5 min and loaded on SDS-PAGE (NuPAGE 4%–12% Bis-Tris, Invitrogen). Efficiency of ligand coupling to the Ad capsid (percentage total hexon protein coupled) was assessed by direct gel shift assays. Proteins were resolved by SDS-PAGE (200 V, 55 min) and visualized by Coomassie staining (16-h staining with Quick Coomassie [Generon], destained with water). Coupling efficiency was assessed by comparing band intensities of unconjugated hexon-Tag in ligand-decorated samples to an undecorated (control) sample (containing the same number of viral particles) using ImageJ, and expressed as the reduction in unconjugated hexon-Tag protein after coupling:
Anti-vector antibody neutralization assay
For assessment of vector neutralization by potent neutralizing mouse monoclonal antibody (mAb) 9C12 (Developmental Studies Hybridoma Bank, University of Iowa),25 Ad5 vectors expressing GFP were incubated with serially diluted mAb 9C12 antibody at a 1:1 ratio in complete media for 1 h at 37°C. The Ad/9C12 mix was then added to an 80% confluent monolayer of 293A cells in a 96-well plate (cells were infected at a multiplicity of infection [MOI] of 200 ifu/cell). Cells were incubated with the Ad/9C12 mix for 2 h at 37°C 5% v/v CO2, before the mix was replaced with fresh medium and the plates returned to 37°C 5% v/v CO2 for a further 24 h. After 24 h, GFP expression was used as a readout of vector infectivity; bulk fluorescence was measured on a fluorimeter (Tecan) using an excitation wavelength of 395 nm and emission wavelength of 509 nm.
For assessment of vector neutralization by serum containing anti-adenovirus antibodies, serum samples were obtained by immunizing C57BL/6 mice with 1E+8 ifu of an Ad5 vector expressing ovalbumin (vector had an unmodified hexon).68 Serum was harvested 2 weeks post immunization, stored at −80°C, and then serially diluted for the neutralization assay (2-fold dilutions were prepared from 1:4 to 1:512 in complete medium, to give a final range of 1:8 to 1:1,024 on cell monolayers). Diluted serum was incubated with Ad5(GFP) vectors, the mix incubated on 293A cells, and bulk GFP fluorescence read 24 h later as described above.
Assessment of endogenous human ACE2 expression in 293A cell line
Levels of endogenous cellular expression of human ACE2 in the 293A cell line were assessed by immunoblotting. ACE2 expression was detected by immunoblotting of cell lysates using primary rabbit monoclonal anti-hACE2 antibody (clone EPR4436, Abcam), followed by secondary ALP-conjugated goat anti-rabbit IgG (Sigma, A3687). As a loading control, β-actin expression was detected using primary rabbit monoclonal anti-β-actin (clone D6A8, Cell Signaling Technology) followed by secondary ALP-conjugated goat anti-rabbit IgG (Sigma, A3687). HEK293T (293T) cells (a negative control for ACE2 expression) were obtained from Takara. 293T-ACE2 cells (293T stable cell line constitutively expressing human ACE2, used as a positive control for ACE2 expression) were generated by transduction of 293T cells with pMIGR1-blast-ACE2 vector and maintained with 10 μg/mL blasticidin selection. For validation of the cell line, 293T- and 293T-ACE2 cells were infected with SARS-CoV-2 England 02/2020/407073 virus (Public Health England) at an MOI of 0.05 for 1 h. Cells were then washed with PBS and cultured in fresh medium for further 48 h. Virus production in culture supernatants was quantified at 48 h post infection by plaque assay on Vero E6 cells.
Assessment of coagulation factor X-mediated vector transduction of SKOV3 cells
SKOV3 cells (human ovary adenocarcinoma) were obtained from Public Health England and cultured in McCoy’s 5a media with 2 mM glutamine and 15% v/v fetal bovine serum (complete McCoy’s). For the assay, GFP-expressing Ad vectors were serially diluted (1:10 to 1:107) in serum-free media. Human coagulation factor X (hFX) (Haematologic Technologies) was added to diluted vectors at a final concentration of 8 μg/mL. Ad/hFX mixes were added to monolayers of SKOV3 cells in 96-well plates and incubated with cells for 2.5 h at 37°C and 5% v/v CO2. After 2.5 h, Ad/hFX mixtures were replaced with complete McCoy’s medium, and plates incubated at 37°C, 5% v/v CO2 for a further 48 h. Infectivity was assessed after 48 h by enumeration of GFP-positive foci as described above.
Mouse immunizations
All mouse procedures were performed in accordance with the terms of the UK Animals (Scientific Procedures) Act (Project Licenses PA7D20B85 or PP5949437) and approved by the Oxford University Ethical Review Body. Female BALB/c mice (aged 6–8 weeks, Envigo), housed in specific-pathogen-free environments, were immunized intramuscularly by injection of 50 μL of vaccine formulated in endotoxin-free PBS (Gibco) into both hind limbs of each animal (100 μL total). Doses of adenoviral vectors and recombinant proteins are described in figure legends. Protein vaccines were administered in combination with Alhydrogel (InvivoGen) at a 1:9 v/v ratio of adjuvant to antigen. Endotoxin dose was <3 EU per mouse in all studies. Experiments were performed at Biomedical Services, University of Oxford.
Ex vivo IFN-γ ELISpot
Overnight spleen ex vivo IFN-γ ELISpot was performed according to standard protocols as described previously.69 To measure antigen-specific responses, cells were re-stimulated with peptides for 18–20 h. To measure T cell responses to GFP, peptide CD8+ T cell epitope EGFP200-20870 was added at a final concentration of 5 μg/mL. To measure T cell responses to SARS-CoV-2 spike RBD, cells were stimulated with a peptide pool of 15-mer peptides with an 11-amino-acid overlap spanning the length of the RBD region (P0DTC2 319–541, PepMix SARS-CoV-2 S-RBD, JPT Peptide Technologies). Pooled peptides were added at a final concentration of 0.5 μg/mL for each peptide. To measure T cell responses to SARS-CoV-2 spike protein, cells were stimulated with two peptide pools of 15-mer peptides with an 11-amino-acid overlap spanning the entire length of the spike protein (PepMix SARS-CoV-2 spike glycoprotein, JPT Peptide Technologies). Pooled peptides were added at a final concentration of 0.5 μg/mL for each peptide. Spot-forming cells (SFCs) were measured using an automated ELISpot reader system (AID).
IgG ELISA
IgG endpoint ELISA was performed as described previously.71 Plates were coated with either recombinant GFP protein (Millipore) at 1 μg/mL to measure GFP-specific responses, recombinant GST-NANP18 protein at 0.5 μg/mL to measure NANP18-specific responses, recombinant DogCatcher-NANP18 protein at 2 μg/mL to measure DogCatcher-NANP18-specific responses, recombinant DogCatcher protein at 0.5 μg/mL to measure DogCatcher-specific responses, or either recombinant RBD-SnJr or spike-SnJr at 2 μg/mL to measure SARS-CoV-2 specific responses. To generate endpoint titers, sera from mice immunized with Ad5 vectors expressing an irrelevant antigen were used as a negative control.
SARS-CoV-2 pVNT assay
Pseudotyped HIV-1 viruses incorporating the SARS-CoV-2 full-length spike (Wuhan strain or B.1.1.7, B.1.617.2, or B.1.351 variants of concern) were generated and SARS-CoV-2 pVNT assays performed as previously described using HeLa cells stably expressing ACE2 as target cells.29 Serum was heat inactivated at 56°C for 30 min prior to use in the assay.
Cryo-EM data collection and image processing
Cryo-EM analysis was performed by NanoImaging Services, San Diego, USA. A 3-μL drop of sample Ad-DogTag (control) and sample Ad-DogTag:DogCatcher-RBD were applied to a 1/2 Cu-mesh C-flat grid that had been plasma-cleaned for 10 s using a 25% v/v O2/75% v/v Ar mixture in a Solarus 950 Plasma Cleaner (Gatan). Grids were manually plunge frozen in liquid ethane. Data collection was carried out using a Thermo Fisher Scientific Glacios Cryo Transmission Electron Microscope operated at 200 kV and equipped with a Falcon 3 direct electron detector. Automated data collection was performed with Leginon software,72 at a nominal magnification of 28,000×, corresponding to a pixel size of 5.19 Å. A total of 845 and 617 movies were recorded for samples Ad-DogTag and Ad-DogTag:DogCatcher-RBD, respectively, using a nominal defocus range of −2.4 to −5.6 μm. Exposures were fractionated into 19 frames with an exposure rate of 2.6 e–/pixel/s and total exposure of 10 e–/Å2.
For both samples, motion correction and contrast transfer function (CTF) estimation were performed using cryoSPARC3.1.73 Using the Gaussian blob picker in cryoSPARC3.1, a total of 28,371 particles were picked for sample Ad-DogTag:DogCatcher-RBD and 91,920 particles were picked for sample Ad-DogTag. These particles were extracted from the cryo-EM images and subjected to reference-free 2D classification in the cryoSPARC3.1. The best 28,307 particles for sample Ad-DogTag:DogCatcher-RBD and all particles picked for sample Ad-DogTag were used for the subsequent 3D reconstruction. An initial model was generated ab initio from all selected particles for sample Ad-DogTag:DogCatcher-RBD and used during one round of homogeneous refinement with icosahedral symmetry imposed in cryoSPARC3.1. The final 3D reconstruction of Ad-DogTag:DogCatcher-RBD was low-pass filtered to 30 Å and used as a starting volume for the 3D reconstruction of sample Ad-DogTag during one round of homogeneous refinement with icosahedral symmetry imposed in cryoSPARC3.1. The resolution of the final 3D reconstructions was determined by the Fourier shell correlation (FSC) between two independent half maps, which was 10.5 Å at FSC = 0.143 for both samples. Maps are visualized using Chimera.74
Statistics
Statistical analyses were performed in GraphPad Prism 9. Comparisons between multiple groups were performed by Kruskal-Wallis with Dunn’s test for multiple comparisons. Pair-wise comparisons (two groups only) were performed by Mann-Whitney test. Statistical significance is indicated as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
Data and materials availability
The cryo-EM maps for samples Ad5-DogTag (control) and Ad5-DogTag:DogCatcher-RBD were deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-14371 and EMD-14370 respectively. All other data are available in the main text or the supplementary materials.
Acknowledgments
The authors would like to thank Genevieve Labbe, Sophie Porret, Laura Bilbé, Jeanette Wagener, Rebecca Dabbs, and Jing Jin for helpful advice and assistance. We thank Viv Clark, Heather Chandler, Stephen Laird, Douglas Passos, and Luke Harris for animal husbandry and technical assistance. We thank Sarah Dunn, Weili Zheng, and Mandy Janssen from NanoImaging Services, San Diego, USA for cryo-EM services and helpful discussion. Work at King’s College London was funded by: Fondation Dormeur, Vaduz for funding equipment to K.J.D., a Huo Family Foundation Award to M.H.M. and K.J.D., and the MRC Genotype-to-Phenotype UK National Virology Consortium (MR/W005611/1 to M.H.M. and K.J.D.). C.G. was supported by the MRC-KCL Doctoral Training Partnership in Biomedical Sciences (MR/N013700/1).
Author contributions
Conceptualization, M.D.J.D., S.J.D., M.H., and S.B.; methodology, M.D.J.D., R.A.R., C.G., K.J.D., M.H.M., S.J.D., M.H., and S.B.; Investigation, M.D.J.D., L.M.R., R.A.R., L.A.H.B., C.G., and J.M.J.-G.; Visualization, M.D.J.D.; Resources, M.H. and J.M.J.-G.; Writing – original draft, M.D.J.D.; Writing – review & editing, M.D.J.D., M.H., and S.B.
Declaration of interests
M.D.J.D., L.M.R., R.A.R., and L.A.H.B. are employees of SpyBiotech Ltd. S.B. is CSO and co-founder of SpyBiotech Ltd. S.J.D. and M.H. are co-founders of SpyBiotech Ltd, and M.H. is also an author on a number of patents relating to protein superglues, including the DogTag/DogCatcher technology. C.G., J.M.J.-G., K.J.D., and M.H.M. declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.08.002.
Supplemental information
References
- 1.Mendonça S.A., Lorincz R., Boucher P., Curiel D.T. Adenoviral vector vaccine platforms in the SARS-CoV-2 pandemic. NPJ Vaccin. 2021;6:97. doi: 10.1038/s41541-021-00356-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., Offergeld K., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., Tukhvatulin A.I., Zubkova O.V., Dzharullaeva A.S., Kovyrshina A.V., Lubenets N.L., Grousova D.M., Erokhova A.S., et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J., Wu S.P., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier A.R.Y., Yu J., McMahan K., Liu J., Chandrashekar A., Maron J.S., Atyeo C., Martinez D.R., Ansel J.L., Aguayo R., Rowe M., et al. Differential kinetics of immune responses elicited by covid-19 vaccines. N. Engl. J. Med. 2021;385:2010–2012. doi: 10.1056/NEJMc2115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald I., Murray S.M., Reynolds C.J., Altmann D.M., Boyton R.J. Comparative systematic review and meta-analysis of reactogenicity, immunogenicity and efficacy of vaccines against SARS-CoV-2. NPJ Vaccin. 2021;6:74. doi: 10.1038/s41541-021-00336-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., Alam S.M., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barouch D.H., Pau M.G., Custers J.H.H.V., Koudstaal W., Kostense S., Havenga M.J.E., Truitt D.M., Sumida S.M., Kishko M.G., Arthur J.C., Korioth-Schmitz B., et al. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 2004;172:6290–6297. doi: 10.4049/jimmunol.172.10.6290. [DOI] [PubMed] [Google Scholar]
- 10.Sumida S.M., Truitt D.M., Kishko M.G., Arthur J.C., Jackson S.S., Gorgone D.A., Lifton M.A., Koudstaal W., Pau M.G., Kostense S., Havenga M.J.E., Goudsmit J., Letvin N.L., Barouch D.H. Neutralizing antibodies and CD8+ T lymphocytes both contribute to immunity to adenovirus serotype 5 vaccine vectors. J. Virol. 2004;78:2666–2673. doi: 10.1128/jvi.78.6.2666-2673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbink P., Lemckert A.A.C., Ewald B.A., Lynch D.M., Denholtz M., Smits S., Holterman L., Damen I., Vogels R., Thorner A.R., O'Brien K.L., et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J. Virol. 2007;81:4654–4663. doi: 10.1128/JVI.02696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dicks M.D.J., Spencer A.J., Edwards N.J., Wadell G., Bojang K., Gilbert S.C., Hill A.V.S., Cottingham M.G. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One. 2012;7:e40385. doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohsen M.O., Zha L., Cabral-Miranda G., Bachmann M.F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin. Immunol. 2017;34:123–132. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Brune K.D., Leneghan D.B., Brian I.J., Ishizuka A.S., Bachmann M.F., Draper S.J., Biswas S., Howarth M. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016;6:19234. doi: 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marini A., Zhou Y., Li Y., Taylor I.J., Leneghan D.B., Jin J., Zaric M., Mekhaiel D., Long C.A., Miura K., Biswas S. A universal plug-and-display vaccine carrier based on HBsAg VLP to maximize effective antibody response. Front. Immunol. 2019;10:2931. doi: 10.3389/fimmu.2019.02931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.J., Howarth M., West A.P., Barnes C.O., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., Qiao Y., et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330.e9. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevillard C., Amen A., Besson S., Hannani D., Bally I., Dettling V., Gout E., Moreau C.J., Buisson M., Gallet S., Fenel D., et al. Elicitation of potent SARS-CoV-2 neutralizing antibody responses through immunization with a versatile adenovirus-inspired multimerization platform. Mol. Ther. 2022;30:1913–1925. doi: 10.1016/j.ymthe.2022.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakeri B., Fierer J.O., Celik E., Chittock E.C., Schwarz-Linek U., Moy V.T., Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalvie N.C., Tostanoski L.H., Rodriguez-Aponte S.A., Kaur K., Bajoria S., Kumru O.S., Martinot A.J., Chandrashekar A., McMahan K., Mercado N.B., Yu J., et al. SARS-CoV-2 receptor binding domain displayed on HBsAg virus-like particles elicits protective immunity in macaques. Sci. Adv. 2022;8:eabl6015. doi: 10.1126/sciadv.abl6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brune K.D., Howarth M. New routes and opportunities for modular construction of particulate vaccines: stick, click, and glue. Front. Immunol. 2018;9:1432. doi: 10.3389/fimmu.2018.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keeble A.H., Yadav V.K., Ferla M.P., Bauer C.C., Chuntharpursat-Bon E., Huang J., Bon R.S., Howarth M. DogCatcher allows loop-friendly protein-protein ligation. Cell Chem. Biol. 2021;29:339–350.e10. doi: 10.1016/j.chembiol.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Oostrum J., Burnett R.M. Molecular composition of the adenovirus type 2 virion. J. Virol. 1985;56:439–448. doi: 10.1128/JVI.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurens M.B. RTS, S/AS01 vaccine (Mosquirix): an overview. Hum. Vaccin. Immunother. 2020;16:480–489. doi: 10.1080/21645515.2019.1669415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varghese R., Mikyas Y., Stewart P.L., Ralston R. Postentry neutralization of adenovirus type 5 by an antihexon antibody. J. Virol. 2004;78:12320–12332. doi: 10.1128/JVI.78.22.12320-12332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., Acton O.J., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waddington S.N., McVey J.H., Bhella D., Parker A.L., Barker K., Atoda H., Pink R., Buckley S.M.K., Greig J.A., Denby L., Custers J., et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Bradshaw A.C., Parker A.L., Duffy M.R., Coughlan L., van Rooijen N., Kähäri V.M., Nicklin S.A., Baker A.H. Requirements for receptor engagement during infection by adenovirus complexed with blood coagulation factor X. PLoS Pathog. 2010;6:e1001142. doi: 10.1371/journal.ppat.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O'Byrne A., Kouphou N., Galao R.P., Betancor G., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McConnell M.J., Danthinne X., Imperiale M.J. Characterization of a permissive epitope insertion site in adenovirus hexon. J. Virol. 2006;80:5361–5370. doi: 10.1128/JVI.00256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vujadinovic M., Vellinga J. Progress in adenoviral capsid-display vaccines. Biomedicines. 2018;6 doi: 10.3390/biomedicines6030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews Q.L., Yang P., Wu Q., Belousova N., Rivera A.A., Stoff-Khalili M.A., Waehler R., Hsu H.C., Li Z., Li J., Mountz J.D., et al. Optimization of capsid-incorporated antigens for a novel adenovirus vaccine approach. Virol. J. 2008;5:98. doi: 10.1186/1743-422X-5-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreppel F., Gackowski J., Schmidt E., Kochanek S. Combined genetic and chemical capsid modifications enable flexible and efficient de- and retargeting of adenovirus vectors. Mol. Ther. 2005;12:107–117. doi: 10.1016/j.ymthe.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee P.S., Ostapchuk P., Hearing P., Carrico I.S. Unnatural amino acid incorporation onto adenoviral (Ad) coat proteins facilitates chemoselective modification and retargeting of Ad type 5 vectors. J. Virol. 2011;85:7546–7554. doi: 10.1128/JVI.00118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salisch N.C., Vujadinovic M., van der Helm E., Spek D., Vorthoren L., Serroyen J., Kuipers H., Schuitemaker H., Zahn R., Custers J., Vellinga J. Antigen capsid-display on human adenovirus 35 via pIX fusion is a potent vaccine platform. PLoS One. 2017;12:e0174728. doi: 10.1371/journal.pone.0174728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poulin K.L., McFall E.R., Chan G., Provost N.B., Christou C., Smith A.C., Parks R.J. Fusion of large polypeptides to human adenovirus type 5 capsid protein IX can compromise virion stability and DNA packaging capacity. J. Virol. 2020;94:e01112-20. doi: 10.1128/JVI.01112-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergelson J.M., Cunningham J.A., Droguett G., Kurt-Jones E.A., Krithivas A., Hong J.S., Horwitz M.S., Crowell R.L., Finberg R.W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 38.Wickham T.J., Mathias P., Cheresh D.A., Nemerow G.R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 39.Greber U.F., Flatt J.W. Adenovirus entry: from infection to immunity. Annu. Rev. Virol. 2019;6:177–197. doi: 10.1146/annurev-virology-092818-015550. [DOI] [PubMed] [Google Scholar]
- 40.Bradley R.R., Lynch D.M., Iampietro M.J., Borducchi E.N., Barouch D.H. Adenovirus serotype 5 neutralizing antibodies target both hexon and fiber following vaccination and natural infection. J. Virol. 2012;86:625–629. doi: 10.1128/JVI.06254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alba R., Bradshaw A.C., Parker A.L., Bhella D., Waddington S.N., Nicklin S.A., van Rooijen N., Custers J., Goudsmit J., Barouch D.H., McVey J.H., Baker A.H. Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: effect of mutagenesis on FX interactions and gene transfer. Blood. 2009;114:965–971. doi: 10.1182/blood-2009-03-208835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker A.T., Boyd R.J., Sarkar D., Teijeira-Crespo A., Chan C.K., Bates E., Waraich K., Vant J., Wilson E., Truong C.D., Lipka-Lloyd M., et al. ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome. Sci. Adv. 2021;7:eabl8213. doi: 10.1126/sciadv.abl8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kreppel F., Kochanek S. Modification of adenovirus gene transfer vectors with synthetic polymers: a scientific review and technical guide. Mol. Ther. 2008;16:16–29. doi: 10.1038/sj.mt.6300321. [DOI] [PubMed] [Google Scholar]
- 44.Subr V., Kostka L., Selby-Milic T., Fisher K., Ulbrich K., Seymour L.W., Carlisle R.C. Coating of adenovirus type 5 with polymers containing quaternary amines prevents binding to blood components. J. Control Release. 2009;135:152–158. doi: 10.1016/j.jconrel.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Graham S.P., McLean R.K., Spencer A.J., Belij-Rammerstorfer S., Wright D., Ulaszewska M., Edwards J.C., Hayes J.W.P., Martini V., Thakur N., Conceicao C., et al. Evaluation of the immunogenicity of prime-boost vaccination with the replication-deficient viral vectored COVID-19 vaccine candidate ChAdOx1 nCoV-19. NPJ Vaccin. 2020;5:69. doi: 10.1038/s41541-020-00221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atmar R.L., Lyke K.E., Deming M.E., Jackson L.A., Branche A.R., El Sahly H.M., Rostad C.A., Martin J.M., Johnston C., Rupp R.E., Mulligan M.J., et al. Heterologous SARS-CoV-2 booster vaccinations - preliminary report. medRxiv. 2021 doi: 10.1101/2021.10.10.21264827. Preprint at. [DOI] [Google Scholar]
- 48.Schmidt T., Klemis V., Schub D., Mihm J., Hielscher F., Marx S., Abu-Omar A., Ziegler L., Guckelmus C., Urschel R., Schneitler S., et al. Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination. Nat. Med. 2021;27:1530–1535. doi: 10.1038/s41591-021-01464-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward B.J., Gobeil P., Séguin A., Atkins J., Boulay I., Charbonneau P.Y., Couture M., D'Aoust M.A., Dhaliwall J., Finkle C., Hager K., et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021;27:1071–1078. doi: 10.1038/s41591-021-01370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bachmann M.F., Rohrer U.H., Kündig T.M., Bürki K., Hengartner H., Zinkernagel R.M. The influence of antigen organization on B cell responsiveness. Science. 1993;262:1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y.N., Lazarovits J., Poon W., Ouyang B., Nguyen L.N.M., Kingston B.R., Chan W.C.W. Nanoparticle size influences antigen retention and presentation in lymph node follicles for humoral immunity. Nano Lett. 2019;19:7226–7235. doi: 10.1021/acs.nanolett.9b02834. [DOI] [PubMed] [Google Scholar]
- 53.Manolova V., Flace A., Bauer M., Schwarz K., Saudan P., Bachmann M.F. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 54.Coughlan L. Factors which contribute to the immunogenicity of non-replicating adenoviral vectored vaccines. Front. Immunol. 2020;11:909. doi: 10.3389/fimmu.2020.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang K., Zhang Y., Hui X., Zhao Y., Gong W., Wang T., Zhang S., Yang Y., Deng F., Zhang Q., Chen X., et al. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67:103381. doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campos S.K., Barry M.A. Comparison of adenovirus fiber, protein IX, and hexon capsomeres as scaffolds for vector purification and cell targeting. Virology. 2006;349:453–462. doi: 10.1016/j.virol.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 57.Kurachi S., Koizumi N., Sakurai F., Kawabata K., Sakurai H., Nakagawa S., Hayakawa T., Mizuguchi H. Characterization of capsid-modified adenovirus vectors containing heterologous peptides in the fiber knob, protein IX, or hexon. Gene Ther. 2007;14:266–274. doi: 10.1038/sj.gt.3302859. [DOI] [PubMed] [Google Scholar]
- 58.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., Methot N., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gangaev A., Ketelaars S.L.C., Isaeva O.I., Patiwael S., Dopler A., Hoefakker K., De Biasi S., Gibellini L., Mussini C., Guaraldi G., Girardis M., et al. Identification and characterization of a SARS-CoV-2 specific CD8(+) T cell response with immunodominant features. Nat. Commun. 2021;12:2593. doi: 10.1038/s41467-021-22811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., Jeffery-Smith A., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2021;601:110–117. doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanekiyo M., Joyce M.G., Gillespie R.A., Gallagher J.R., Andrews S.F., Yassine H.M., Wheatley A.K., Fisher B.E., Ambrozak D.R., Creanga A., Leung K., et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019;20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dalvie N.C., Tostanoski L.H., Rodriguez-Aponte S.A., Kaur K., Bajoria S., Kumru O.S., Martinot A.J., Chandrashekar A., McMahan K., Mercado N.B., Yu J., et al. A modular protein subunit vaccine candidate produced in yeast confers protection against SARS-CoV-2 in non-human primates. bioRxiv. 2021 doi: 10.1101/2021.07.13.452251. Preprint at. [DOI] [Google Scholar]
- 63.Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shiratsuchi T., Rai U., Krause A., Worgall S., Tsuji M. Replacing adenoviral vector HVR1 with a malaria B cell epitope improves immunogenicity and circumvents preexisting immunity to adenovirus in mice. J. Clin. Invest. 2010;120:3688–3701. doi: 10.1172/JCI39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu H., Han T., Belousova N., Krasnykh V., Kashentseva E., Dmitriev I., Kataram M., Mahasreshti P.J., Curiel D.T. Identification of sites in adenovirus hexon for foreign peptide incorporation. J. Virol. 2005;79:3382–3390. doi: 10.1128/JVI.79.6.3382-3390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maizel J.V., Jr., White D.O., Scharff M.D. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology. 1968;36:126–136. doi: 10.1016/0042-6822(68)90122-0. [DOI] [PubMed] [Google Scholar]
- 68.de Cassan S.C., Forbes E.K., Douglas A.D., Milicic A., Singh B., Gupta P., Chauhan V.S., Chitnis C.E., Gilbert S.C., Hill A.V.S., Draper S.J. The requirement for potent adjuvants to enhance the immunogenicity and protective efficacy of protein vaccines can be overcome by prior immunization with a recombinant adenovirus. J. Immunol. 2011;187:2602–2616. doi: 10.4049/jimmunol.1101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Larsen K.C., Spencer A.J., Goodman A.L., Gilchrist A., Furze J., Rollier C.S., Kiss-Toth E., Gilbert S.C., Bregu M., Soilleux E.J., Hill A.V.S., Wyllie D.H. Expression of tak1 and tram induces synergistic pro-inflammatory signalling and adjuvants DNA vaccines. Vaccine. 2009;27:5589–5598. doi: 10.1016/j.vaccine.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 70.Gambotto A., Dworacki G., Cicinnati V., Kenniston T., Steitz J., Tüting T., Robbins P.D., DeLeo A.B. Immunogenicity of enhanced green fluorescent protein (EGFP) in BALB/c mice: identification of an H2-Kd-restricted CTL epitope. Gene Ther. 2000;7:2036–2040. doi: 10.1038/sj.gt.3301335. [DOI] [PubMed] [Google Scholar]
- 71.Biswas S., Dicks M.D.J., Long C.A., Remarque E.J., Siani L., Colloca S., Cottingham M.G., Holder A.A., Gilbert S.C., Hill A.V.S., Draper S.J. Transgene optimization, immunogenicity and in vitro efficacy of viral vectored vaccines expressing two alleles of Plasmodium falciparum AMA1. PLoS One. 2011;6:e20977. doi: 10.1371/journal.pone.0020977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng A., Negro C., Bruhn J.F., Rice W.J., Dallakyan S., Eng E.T., Waterman D.G., Potter C.S., Carragher B. Leginon: new features and applications. Protein Sci. 2021;30:136–150. doi: 10.1002/pro.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 74.Goddard T.D., Huang C.C., Ferrin T.E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.