ABSTRACT
Anti-retroviral therapy (ART) generally suppresses HIV replication to undetectable levels in peripheral blood, but immune activation associated with increased morbidity and mortality is sustained during ART, and infection rebounds when treatment is interrupted. To identify drivers of immune activation and potential sources of viral rebound, we modified RNAscope in situ hybridization to visualize HIV-producing cells as a standard against which to compare the following assays of potential sources of immune activation and virus rebound following treatment interruption: (i) envelope detection by induced transcription-based sequencing (EDITS) assay; (ii) HIV-Flow; (iii) Flow-FISH assays that can scan tissues and cell suspensions to detect rare cells expressing env mRNA, gag mRNA/Gag protein and p24; and (iv) an ultrasensitive immunoassay that detects p24 in cell/tissue lysates at subfemtomolar levels. We show that the sensitivities of these assays are sufficient to detect one rare HIV-producing/env mRNA+/p24+ cell in one million uninfected cells. These high-throughput technologies provide contemporary tools to detect and characterize rare cells producing virus and viral antigens as potential sources of immune activation and viral rebound.
IMPORTANCE Anti-retroviral therapy (ART) has greatly improved the quality and length of life for people living with HIV, but immune activation does not normalize during ART, and persistent immune activation has been linked to increased morbidity and mortality. We report a comparison of assays of two potential sources of immune activation during ART: rare cells producing HIV and the virus’ major viral protein, p24, benchmarked on a cell model of active and latent infections and a method to visualize HIV-producing cells. We show that assays of HIV envelope mRNA (EDITS assay), gag mRNA, and p24 (Flow-FISH, HIV-Flow. and ultrasensitive p24 immunoassay) detect HIV-producing cells and p24 at sensitivities of one infected cell in a million uninfected cells, thereby providing validated tools to explore sources of immune activation during ART in the lymphoid and other tissue reservoirs.
KEYWORDS: HIV, immune activation, viral rebound
INTRODUCTION
ART has greatly improved the quality and length of life for people living with HIV (PLWH), but treatment must be continued indefinitely because infection quickly rebounds from persistent viral reservoirs if treatment is interrupted (1, 2). In addition to the challenges that recrudescent infection poses for curing HIV infection, immune activation (IA) decreases, but does not normalize, during ART, and persistent IA has been linked to increased morbidity and early death from inflammatory mediated conditions (3–10). Thus, it is important to identify the sources of residual IA when devising mitigating strategies to improve long-term health outcomes for the treatment of PLWH.
Here, we report a comparison of assays of cells that harbor potentially replication-competent proviruses or defective proviruses that could nonetheless generate viral antigens as potential drivers of IA: (i) EDITS (11) assay to detect cells expressing env RNA; (ii) HIV-FISH/Flow (12) to detect cells expressing gag RNA and p24; (iii) HIV-Flow (13) to detect cells expressing p24; and (iv) p24 ultrasensitive immunoassay (14) to detect p24 at subfemtomolar levels. Because env+ and p24+ cells and p24 antigen are expected to be rare and in small quantity during ART, we further asked whether the limits of detection (LOD) for the EDITS, HIV-Flow, HIV-FISH/Flow, and ultrasensitive p24 high-throughput assays would be able to detect a rare env RNA+, gag mRNA/p24+ or p24+ cell in a background of a million or more uninfected cells.
We developed a method to visualize HIV-producing ACH-2 cells to serve as a standard to evaluate these high-throughput assays for two reasons. First, as a latently infected cell line in which virus production can be reactivated by treatment with phorbol esters or TNF-α, ACH-2 cells can serve as a surrogate for reactivated, latently infected cells (15). Second, we know from previous in situ hybridization (ISH) studies (16) that the reactivation of ACH-2 cells results in levels of HIV RNAs, Gag, and Env proteins that should be detectable in the EDITS and p24 assays and thus could serve as standards by which to evaluate the sensitivities of these approaches to detect rare cells with HIV RNAs and antigens.
RESULTS
RNAscope ISH to detect HIV-producing cells.
The method we originally developed to detect SIV-producing cells (17) amplified the signal from SIV RNA in virions by tyramide signal amplification to deposit sufficient product under diffusion limiting conditions to reveal visible virions at the resolution of visible or fluorescent light microscopy. To reveal HIV-producing cells, we modified a contemporary RNAscope ISH protocol (18) to visualize not only intracellular HIV RNA but also HIV RNA in virions associated with the infected cells.
The substitution of the ELF 97 alkaline phosphatase substrate in the RNAscope ISH deposited sufficient ELF 97 product around the HIV RNA in the virions such that they were clearly visible in the ACH-2 cells (Fig. 1). Prior to induction, about 5% of ACH-2 cells undergoing spontaneous reactivation score as HIV-producing cells. In the remaining 95% prior to induction, virus-producing cells were not detected in CEM or Jurkat HIV-negative cells or in induced ACH-2 cells with plus sense probes (not shown). Following induction, 100% of the cells were visibly producing HIV virions. In counts of 78 cells where the z-series 3D images included all or nearly all of the cells, the average virion count per induced ACH-2 cell was 558 with a standard deviation of 66 cells. The average size of virions rendered visible by the deposition of the ELF-97 substrate was ~260 nm, consistent with the diffraction limit for ELF 97 emission at 530 nm. Thus, the in situ single cell assay detects HIV-producing ACH-2 cells at the resolution of immunofluorescence microscopy and documents virus production in all induced cells, thereby providing the technological capability to visualize HIV-producing cells as a standard for subsequent assay comparisons.
FIG 1.
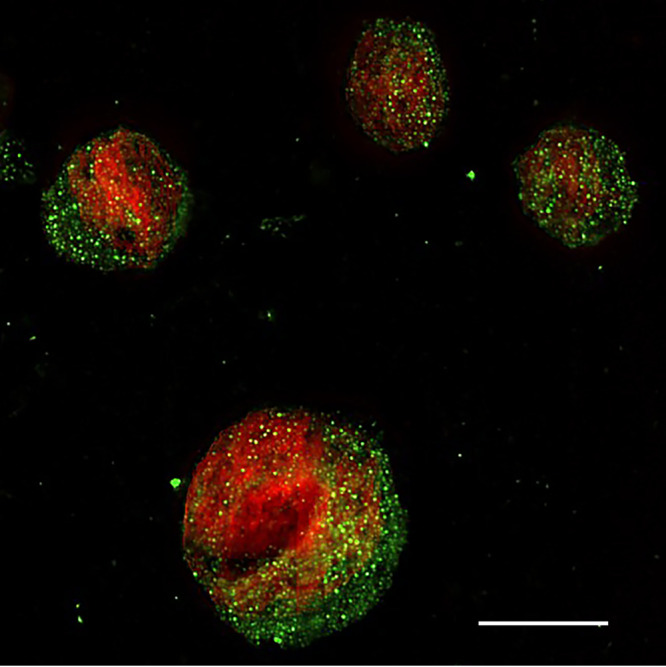
RNAscope ISH/ELF 97 detection of HIV-producing cells in 5 μm sections of fixed and embedded induced ACH-2 cells. HIV virions appear green in the image. Nuclei were counterstained red with TOTO 3. In this 2D image, the largest number of virions are associated with a whole cell in the focal plane of the z-series. Scale bar = 10 μm.
Relationship between HIV-producing cells and the production of infectious virus.
Before evaluating the sensitivity of the high-throughput assays with HIV-producing ACH-2 cells as the standard for comparison, we also investigated the relationship between the visible production of virions and the production of infectious virus. For this purpose, we used a previously described modified viral outgrowth assay (QVOA) (19, 20) to document the production of infectious HIV by induced ACH-2 cells and to determine how accurately the QVOA estimates the frequency of cells with inducible replication-competent proviruses. Serial dilutions of mixed ACH-2 cells and Jurkat T cell samples were induced by coculture with irradiated, CD8-depleted, allogeneic PBMC feeder cells plus anti-CD3 and IL-2. The induced cells were cocultured with anti-CD3 stimulated, CD8-depleted PBMC target cells for 14 days, after which time infectious units per million cells (IUPM) were estimated via limiting dilution statistics on p24+ cultures.
The QVOA documented the production of infectious virus by induced ACH-2 cells, albeit at 28% to 81% of the expected number of virus-producing cells in samples with 1, 10, and 100 IUPM cells (Table 1). We attribute the lower estimates to the well-documented underestimates of the frequency of latently infected cells harboring replication competent, intact proviruses (21, 22) because of the inability, at least in part, to amplify infection from small numbers of cells to detect all of the cells with a replication-competent provirus in QVOA assays.
TABLE 1.
QVOA assay of ACH-2 cellsa
| Sample ACH-2/jurkat | Frequency of infected cells | Frequency of infected cells per million | Measured frequency | Lower 95% | Upper 95% | IUPM |
|---|---|---|---|---|---|---|
| 5 per 20 × 106 | 0.00000025 | 0.25 | 0.0000004 | 0.0000001 | 0.0000034 | 0.4 |
| 5 per 20 × 106 | 0.00000025 | 0.25 | 0.0000005 | 0.0000001 | 0.0000035 | 0.5 |
| 20 per 20 × 106 | 0.000001 | 1 | 0.0000005 | 0.0000001 | 0.0000035 | 0.5 |
| 20 per 20 × 106 | 0.000001 | 1 | 0.0000005 | 0.0000001 | 0.0000035 | 0.5 |
| 200 per 20 × 106 | 0.00001 | 10 | 0.0000032 | 0.0000008 | 0.0000126 | 3.2 |
| 200 per 20 × 106 | 0.00001 | 10 | 0.0000057 | 0.0000014 | 0.000023 | 5.7 |
| 2,000 per 20 × 106 | 0.0001 | 100 | 0.0000283 | 0.000007 | 0.0001152 | 28.3 |
| 2,000 per 20 × 106 | 0.0001 | 100 | 0.0000817 | 0.0000209 | 0.0003192 | 81.7 |
Cells producing replication competent virus estimated in IUPM.
EDITS assay.
The EDITS assay measures spliced env mRNA via next-generation sequencing of the major HIV-1 env RNA splice junction to detect the late spliced viral transcript, thereby serving as a measure that, like the intact proviral DNA assay (23), can distinguish between replication-competent and defective proviruses. We evaluated the concordance between the EDITS assay and virus-producing cells in induced ACH-2 cells diluted with HIV-negative CEM cells to cover a range of 2.5 to 2,500 ACH-2 cell equivalents in 1.25 × 106 cells in the typical format for this assay. The total RNA isolated from unstimulated or ACH-2 cells induced with PMA was converted to cDNA for the PCR amplification of a PCR product with the major env splice junction for ion torrent sequencing. Reads mapping to a synthetically spliced HXB2 sequence were scored and expressed as an equivalent number of cells harboring HIV-1 per 106 cells. This was done using a standard curve determined for activated primary memory CD4 T cells infected with replication competent HIV-1 carrying a GFP reporter. In the linear range of the assay (11), in samples with 2.5 to 160 induced ACH-2 cells, the EDITS assay results were highly correlated but consistently slightly higher than the number of virus-producing ACH-2 cells expected after stimulation (Table 2; Fig. 2). This overestimate is consistent with the induction of env mRNA without virion production in ~15% to 30% of the cells. In the samples with 320 and 2,500 ACH-2 cells outside the linear range, the EDITS assay underestimated the number of virus-producing cells by about 15% and 80%, respectively.
TABLE 2.
EDITS assay in triplicate of ACH-2 cells induced with CD3/CD28 T cell activator in the numbers shown
| Induced ACH-2 per 1.25 × 106 CEM cells |
EDITS estimated Stimulated cell equivalents |
||
|---|---|---|---|
| 2.5 | 2.54 | 3.35 | 2.95 |
| 5 | 5.64 | 6.45 | 6.04 |
| 10 | 11.5 | 10.6 | 11.1 |
| 20 | 24.9 | 28.5 | 26.7 |
| 40 | 53.0 | 53.6 | 53.3 |
| 80 | 83.4 | 91.4 | 87.4 |
| 160 | 183 | 189 | 186 |
| 320 | 273 | 278 | 276 |
| 2,500 | 456 | 446 | 451 |
FIG 2.
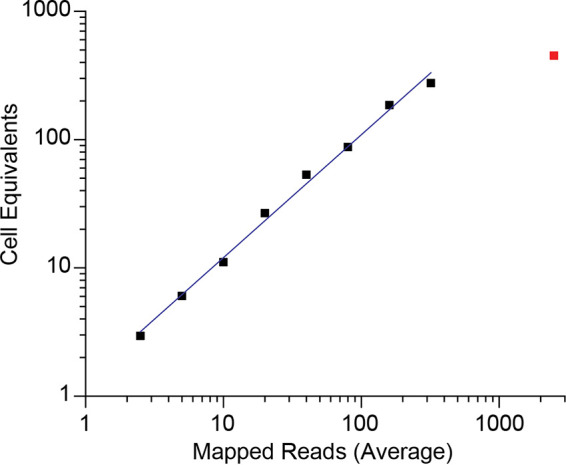
Logarithmic plot of the average of replicates of induced ACH-2 cell equivalents and mapped HIV RNA reads from the EDITS assay results in Table 2. The values for the 2,500-cell point are shown as the isolated red data point, which is outside the linear range of the assay. The fitted line shows the linear relationship up to 320 cells (Pearson’s r = 0.99734).
HIV-Flow assay.
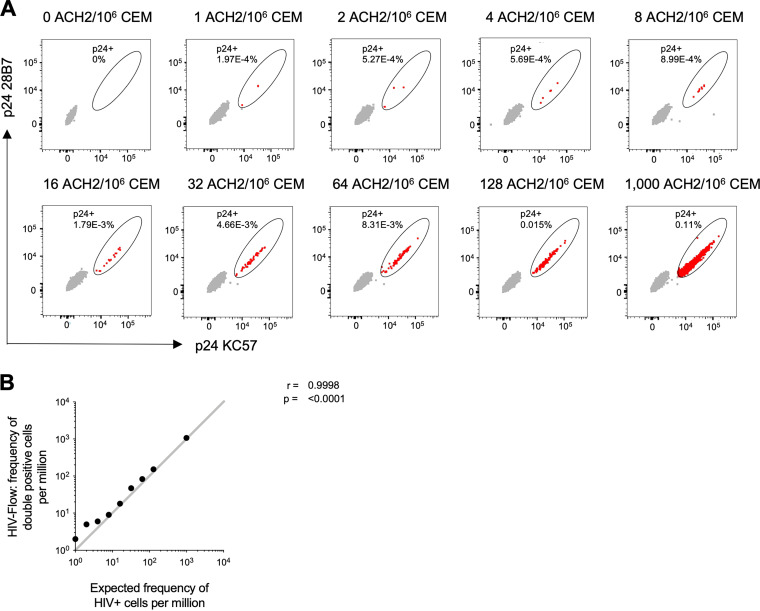
The HIV-Flow assay measures the frequency of cells with translation-competent HIV proviruses that produce detectable levels of p24. In this flow cytometry-based assay, p24+ cells are identified by combining two monoclonal antibodies (KC57 and 28B7), coupled to two different fluorochromes, with p24, thereby improving the specificity of the measurement. We determined the relative efficiency of the HIV-Flow assay for HIV-producing/p24+ cells in a dilution series of 1, 10, 100 and 1,000 induced ACH-2 cells in one million CEM. As shown in Table 3 and Fig. 3, HIV-Flow gave frequencies highly correlated with, but consistently slightly higher than, the expected values. This possibly reflects the detection of uninfected cells to which released virions had attached.
TABLE 3.
HIV-flow assay dilution seriesa
| Expected frequency HIV positive cells/106 cells |
Measured frequency p24+ cells/106 cells |
|---|---|
| 1,000 | 1,068 |
| 128 | 152 |
| 64 | 83 |
| 32 | 47 |
| 16 | 18 |
| 8 | 9 |
| 4 | 6 |
| 2 | 5 |
| 1 | 2 |
Expected frequency versus determined frequency of induced ACH-2 cells per 106 cells.
FIG 3.
(A) Flow cytometry dot plots showing the frequency of induced ACH-2 cells, serially diluted in CEM cells and measured by HIV-Flow. The expected frequencies are indicated at the top of each dot plot. The p24+ cells are depicted in red, and the p24− cells are depicted in gray. (B) Correlation between HIV-flow estimates of p24+ cells per million cells and the expected frequency in the serial dilution series.
HIV RNA Flow-FISH assay.
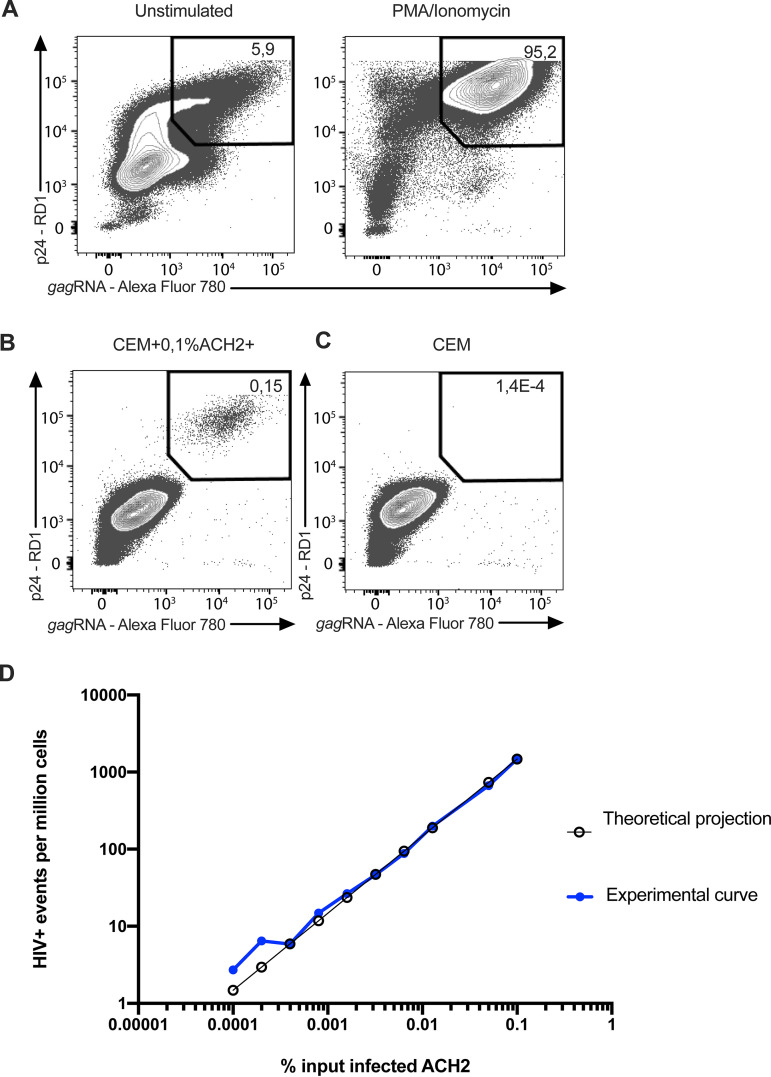
The HIVRNA/Gag assay is a RNA flow cytometric fluorescent in situ hybridization (RNA Flow-FISH) technique that combines the detection of HIV mRNAs and Gag protein staining with a sensitivity of detection of 0.5 to 1 double positive CD4+ T cells per million uninfected CD4+ T cells (24). The concurrent detection of HIV transcription and translation products enables distinction between translation-competent cells and translation-incompetent cells. To assess the specificity and the linearity of the HIVRNA/Gag assay, we spiked reactivated, latently infected ACH-2 cells into uninfected CEMx174 cells. In the absence of reactivation, HIV+ events, defined as cells co-expressing gag mRNA and Gag protein (HIVRNA+/Gag+), were detected in 5.9% of the ACH-2 cells (Fig. 4A). This result is in good agreement with the RNAscope ISH analysis cited above for uninduced ACH-2 cells. Time-course experiments showed that a 24 h stimulation with PMA/ionomycin induced high expression of Gag products in ACH-2 cells (95.2% HIVRNA+/Gag+ cells) (Fig. 4A), and this condition was therefore selected for the spiking experiments. The input corresponding to the highest frequency of reactivated ACH-2 into the CEMx174 line was measured by flow cytometry at 1,500 HIV+ events per million cells (Fig. 4B), consistent with the initial input planned (see Materials and Methods), while the false positive event rate in pure CEMx174 cells was low (1 HIVRNA+/Gag+ cell detected in 700,000 cells) (Fig. 4C). Spiking dilutions ranged from a theoretical expected frequency of 1.5 to 1,500 events per million cells (Fig. 4D). The experimental frequencies of HIV+ events that were experimentally identified by HIV mRNA/protein co-staining showed excellent linearity and consistency down to the lowest dilution tested, except at the two lowest spiking dilutions (<6 expected events per million cells), for which the variability noted between the observed and expected numbers of HIV+ events can be reduced via the analysis of larger numbers of cells (24).
FIG 4.
Flow cytometry example plots of the HIV RNA Flow-FISH assay and the correlation between measured and expected frequencies of infected cells in spiking experiments. (A) The p24+ gag mRNA+ ACH-2 cells before and after induction with PMA/ionomycin. (B) Highest frequency of reactivated ACH-2 spiked into the CEMx174 line. (C) False positive event rate in pure CEMx174 cells. (D) Theoretical expected frequency of 1.5 to 1,500 events per million cells and experimental frequencies of HIV+ events experimentally identified by HIV mRNA/protein co-staining.
HIV ultrasensitive p24 immunoassay.
Digitized immunoassays using single-molecule array (SIMOA) technology have extended the sensitivity of detection of HIV p24 into the femtomolar range, and we and others have reported further enhancement of the technology to achieve sensitivity limits of detecting subfemtomolar levels of the HIV p24+ protein (14, 25–28). Here, we investigated the sensitivity of detecting p24+ induced ACH-2 cells in a matrix of one million uninfected PBMCs with our ultrasensitive assay, and we show that the ultrasensitive p24 assay detects a single p24+ cell in one million PBMCs (Table 4). Because of the variability between triplicate samples that we attribute to the clumping of the induced ACH-2 cells, we tested lysates of induced ACH-2 cells diluted into PBMCs corresponding to samples containing 1 or 10 induced ACH-2 cells, and we again detected a single positive cell per million. There was good agreement between the samples with a CV of less than 10%, and the expected 10-fold higher levels of p24 in the samples with 10 induced ACH-2 cells per million PBMCs.
TABLE 4.
Detection of p24 in triplicate samples of induced ACH-2 cells diluted into 106 uninfected PBMCsa
| Induced ACH-2 cells per million PBMCs | p24 no. 1 | p24 no. 2 | p24 no. 3 | avg p24 concn pg/mL | Std. | CV (%) |
|---|---|---|---|---|---|---|
| 0 | 0.031 | 0.012 | 0.027 | 0.023 | 0.01 | |
| 1 | 2.928 | 0.222 | 0.291 | 1.147 | 1.54 | |
| 10 | 24.984 | 12.166 | 13.585 | 16.192 | 7.03 | |
| 100 | 106.574 | 121.748 | 114.161 | 10.73 | ||
| 1 | 1.49 | 1.36 | 1.59 | 1.478 | 0.12 | 7.8 |
| 10 | 14.45 | 14.74 | 15.36 | 14.853 | 0.46 | 3.1 |
Because of the variability among the triplicate samples above the heavier line, which is attributed to the clumping of cells, the samples below the line are direct lysates containing 1 or 10 induced ACH-2 cells diluted in a million PBMCs.
DISCUSSION
Here, we describe and compare a suite of tools that can be used to detect env and gag mRNA or p24-expressing cells in high-throughput scans of rare cells in a background of a million or more uninfected cells. These technologies provide validated standards and approaches to quantifying HIV reservoirs, which are highly relevant when identifying sources of virus before, during, and after ART, as well as drivers of IA that can guide the design of strategies to potentially prolong remissions after ART and mitigate IA, thereby resulting in longer, healthier lifespans for PLWH.
The EDITS, HIV-Flow, HIV-FISH-Flow, and ultrasensitive p24 assays proved to be facile, rapid, sensitive, and accurate in detecting single cells with env or gag mRNA and p24 in one million uninfected cells. These high-throughput approaches can provide a quick read on the likelihood that peripheral blood or tissue samples harbor cells with, respectively, sufficiently intact proviruses that can be reactivated to produce env mRNA or translationally-competent proviruses that generate p24.
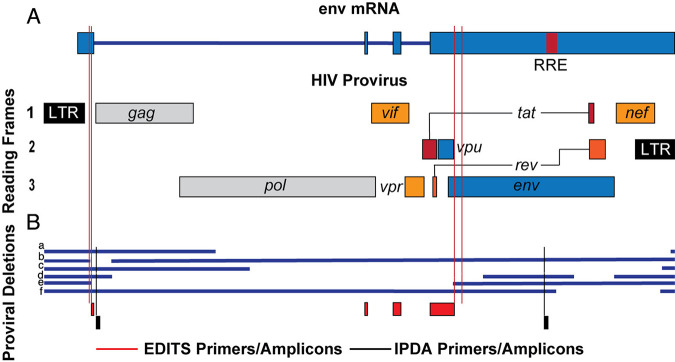
The EDITS assay primers and amplicons used to detect late spliced env mRNA overlap the packaging site amplicon in the IPDA assay (23), and both assays generate sequences from the 3′ end of proviral genomes (Fig. 5). Moreover, generating env mRNA requires functional Tat and Rev, consistent with the possibility that the EDITS assay env transcript might score for largely intact functional genomes, as in the IPDA assay. We show six examples of deleted proviruses (Fig. 5B) to demonstrate how the EDITS and IPDA assays might score each virus, and we found that both assays correctly score proviruses a and proviruses c to e as proviruses with deletions. The EDITS assay misses the small deletion in gag-pol in provirus b, which is detected in the IPDA assay, and both assays incorrectly score provirus f as intact, based solely on the sequences in the amplicons. However, provirus f would likely lack Tat and Rev, and would therefore be correctly scored as impaired in the EDITS assay. We think that this comparison suggests that the two assays should generally be in good agreement, and this expectation has been satisfied experimentally with data on the impact of the IL-15 superagonist N-803 on the frequency of inducible HIV provirus in peripheral blood mononuclear cells (29).
FIG 5.
Comparison of EDITS and IPDA assays for identifying HIV proviruses with deletions. (A) Schematic of HIV provirus and env mRNA. Red lines indicate the positions of the EDITS assay primers. (B) Comparison of the assays to detect deletions in proviruses a-f. The EDITS primers and amplicons are displayed in red, and the IPDA primers and amplicons are displayed in black.
The relationship between the HIV-p24+ cells detected in the p24 assays and the HIV-producing cells is indeterminate. However, we can conclude that the detection of p24+ cells is potentially relevant to sustaining IA and to identifying cells that could be targeted by HIV-specific CD8+ T cells. Collectively, the high-throughput approaches described here provide well-characterized tools that can be used to explore the mechanisms of HIV persistence and IA during ART and to devise rational strategies to sustain remissions after ART and mitigate IA.
MATERIALS AND METHODS
ACH-2 cells.
ACH-2 cells and uninfected CEM or Jurkat cells were grown in RPMI with 10% heat-inactivated FCS, glutamine, and 1x antibiotic-antimycotic (FISHER). ACH-2 cells were activated in some experiments by growth with 50 ng/mL PMA for 24 h. Cells were collected by centrifugation at 450 g × 7 min and at least two washes in PBS, 0.5% BSA, 2 mM EDTA. Cell mixtures were created by suspending uninfected cell lines in a 1/10 culture volume of buffer and ACH-2 cells in their culture volume, then counting cells with trypan blue staining in a Countess II automated cell counter in duplicate. Then, CEM or Jurkat cells were adjusted to 107 cells/mL with buffer, and ACH-2 cells (induced or uninduced) were adjusted to 105 or 106 cells/mL. An initial cell mixture of 103 ACH-2 cells in 107 uninfected cells/mL was created and diluted serially 2x or 10x with 107 uninfected cells/mL. Cell mixtures were frozen, extracted, or fixed as required by the specific assay, per published assay protocols.
Detection and characterization of HIV-producing cells by RNAscope ISH combined with IFA.
We varied conditions in our previously published RNAscope ISH method (18) to be able to not only detect HIV intracellular RNA but also clearly reveal virions associated with the cells. The critical changes in the published protocol were: (i) reduced time sections were boiled in RNAscope Pretreat citrate buffer to 10 min; (ii) sections were incubated for 20 min at 40°C in a more dilute (1:10) Pretreat 3 reagent 3 (protease digestion solution, 2.5 μg/mL); and (iii) following AMPs 1 to 6 from the RNAscope 2.5 HD Detection Reagent Red Kit (Advanced Cell Diagnostics), applied a 10 to 30 min room temperature incubation with ELF 97 phosphatase substrate. For the images of HIV-1 production by induced ACH-2 cells shown in Fig. 1, nuclei were stained with To-Pro-3 (Thermo Fisher Scientific).
Confocal plus deconvolution imaging.
Fluorescent confocal images of ELF 97 and To-Pro 3 labeled cells were acquired with a Nikon Ti-E inverted microscope equipped with a CFI SR Apo 100X TIRF Oil Immersion Objective Lens NA 1.49 and a Nikon A1R confocal scan head controlled by Nikon Elements software (5.11). The ELF 97 excitation was provided by a 403 nm laser, and emissions were collected between 500 and 550 nm. The To-Pro 3 excitation was provided by a 640 nm laser, and emissions were collected between 663 and 738 nm. The confocal aperture was set to the minimum value of 14 μm (0.2 AU). The images were oversampled in X and Y (41 nm pixel). A 200 nm Z step was used in three-dimensional acquisitions, and the images were subjected to automatic iterative deconvolution with Nikon Elements 5.20. Analysis was done in Elements ver. 5.20, using the GA3 pipeline with 3D object measurement and FIJI software using find maxima after making maximum intensity projections of rolling ball, background-corrected imaging for particle counting. Channels were pseudocolored and merged. EDF projections of the z-stacks are presented (University of Minnesota- University Imaging Centers, http://uic.umn.edu/).
QVOA.
Quantitative viral outgrowth assays were performed as previously described (20). Briefly, six 5-fold serial dilutions of mixed ACH-2 cells and Jurkat T cell samples were plated in duplicate, starting at 1 × 106 cells per well. These samples were cocultured with irradiated, CD8-depleted PBMC feeder cells and stimulated with anti-CD3 (OKT3, 1 μg/mL) and IL-2 (50 IU/mL) overnight. On day 1 and day 7 post-stimulation, 1 × 106 anti-CD3 stimulated, CD8-depleted PBMC target cells were added to each well. Supernatant p24 was measured on day 14 by ELISA (Zeptometrix), per the manufacturer’s protocol. Individual wells were determined to be positive or negative for p24, based on manufacturer-determined cutoff values. Infectious units per million were estimated via limiting dilution statistics using L-Calc Limiting Dilution Software (STEMCELL Technologies).
EDITS assay.
The frequency of env mRNA+ induced ACH-2 cells was determined by the EDITS assay, as described by Das et al. (11). In brief, ACH-2 cells were induced with 50 ng/mL PMA for 24 h. Total RNA was isolated using the Qiagen RNeasy purification system (Qiagen, 74134), following the manufacturer's protocol. The entire sample was used as the template in a one-step RT-PCR (Thermo Scientific, AB-4104A). After cDNA synthesis and PCR, 2 μL of the reaction were used as the template for a subsequent round of nested PCR, using a high fidelity Phusion Flash polymerase (Thermo Scientific, F548). Primers were designed to bind to either side of the HIV env RNA splice junction, using highly conserved regions of HIV. For the first-round PCR, the forward primer was located at position 570 to 591, and the reverse primer was at positions c6442 and c6426. In addition to the priming sequence, the reverse primer has a synthetic GEX R-AATGATACGGCGACCACC sequence placed directly after the priming region, which allows for further amplification using nested PCR. The RT-PCR product was further amplified by nested PCR with a nested forward primer at position 610 to 631 and a nested reverse primer at c6324 to c6345. To allow for next-generation sequencing, ion torrent A forward and Trp reverse adapters were added to the nested primer sets along with a unique barcode in the forward primer, which allowed for the multiplexing of samples. Samples were then pooled, and primers were removed using the GeneJET NGS Cleanup Kit (Fisher Scientific, FERK0852). DNA concentrations were measured by a Qubit fluorescent reader, and 300 pg of the pooled sample were then sequenced using an Ion Torrent Sequencing system, following the manufacture’s protocol. Barcodes were separated by sample, using the Ion Torrent Browser, and all reads were filtered to remove short products (under 80 bp). Only reads that contained the GEX reverse sequence were retained. The filtered reads were then mapped to a synthetically spliced HXB2 sequence, and the total mapped reads were scored. The number of mapped reads was then converted into the equivalent number of cells harboring HIV-1 per 106 cells by using a standard curve generated from activated memory cells which were infected with replication-competent HIV-1-GFP virus and were sorted by flow cytometry into single wells of a 96-well plate. Samples for the standard curve contained between 1 and 300 infected cells per well and 1.25 × 106 uninfected cells.
HIV-Flow assay.
The frequency of p24+ cells was determined as previously described (12). Briefly, 9 serial dilutions of ACH-2 cells in CEM cells were stimulated with 162 nM PMA (Sigma, P8139) and with 1 μg/mL ionomycin (Sigma, I9657). After 24h of stimulation, cells were collected, resuspended in PBS, and stained with the Aqua Live/Dead staining kit for 30 min at 4°C. Cells were stained in PBS +4% human serum for 30 min at 4°C (CD3 A700, CD4 BUV496, CD8 BUV395). The fixation/permeabilization step was performed with the FoxP3 Transcription Factor Staining Buffer Set (eBioscience, 00-5523-00), following the manufacturer’s instructions. Then, cells were stained with anti-p24 KC57 (R&D) and anti-p24 28B7 (MediMabs) antibodies for an additional 45 min at room temperature and were analyzed by flow cytometry on a BD LSRII. In all experiments, uninfected CEM cells were included to set the threshold of positivity. The detailed protocol of the HIV-Flow procedure can be found at http://dx.doi.org/10.17504/protocols.io.w4efgte.
HIV-RNA-FLOW-FISH.
HIV latently infected ACH-2 cells and uninfected CEMx174 cells were grown in separate flasks at a concentration of 0.5 million cells per mL in RPMI 1640 medium (Gibco, Life Technologies) supplemented with penicillin/streptomycin (Gibco, Life Technologies) and 10% FBS (Seradigm). The ACH-2 cells were stimulated for 24 h with PMA (50 ng/mL, Sigma-Aldrich) and ionomycin (0.5 μg/mL, Sigma-Aldrich) prior to spiking into uninfected CEMx174 cells at different ratios obtained by serial dilution, starting with a highest frequency of 1,500 reactivated ACH-2 cells per million CEMx174 cells. HIV+ events were identified by using the HIV RNA/Gag assay as previously described (13, 24). Briefly, cells were stained with Fixable Viability Dye (eBioscience) and anti-Gag KC57 (Beckman Coulter) by intracellular staining and labeled for HIV gag mRNA using the PrimeFlow RNA assay (Thermo Fisher Scientific) before acquisition on a flow cytometer (FACS Fortessa, BD). Analysis was performed using FlowJo version 10 for Mac (Treestar). HIV+ cells were identified as cells co-expressing both HIV Gag protein and gag mRNA.
Ultrasensitive p24 immunoassay.
The p24 in cell lysates was measured according to the method described previously (14), with some modification. In brief, prior to the p24 SIMOA, cell pellets were lysed at 4 × 106 cells/mL with a cell lysis buffer containing 1% Triton x-100 in 0.5% casein (in PBS) and 50% Hi-FBS for 15 min at room temperature, then frozen at −80°C until analysis. 35 μL aliquots of M-280 beads (Invitrogen/Life technologies, Cat no. 11206D) in microcentrifuge tubes were washed once with 1 mL of 1% BSA/PBS. Then, 3.5 μL of 1 mg/mL normal mouse IgG (mIgG) (GenScript, Cat no. A01007) was added. The frozen cell lysate was thawed in the 37°C water bath and centrifuged for 10 min at 14,000 rpm at 4°C. After spinning, 0.3 mL of cell lysate supernatant was collected and added into the microcentrifuge tubes containing the washed beads and mIgG. Diluted samples using the cell lysis buffer were treated similarly. The tubes were incubated at 4°C for 3h with 360° rotation using a HulaMixer. The samples were subsequently centrifuged for 10 min at 14,000 rpm at 4°C, and the supernatant was collected and run on an HD-1 Quanterix Analyzer.
ACKNOWLEDGMENTS
We thank the U.S. National Institutes of Health and the Canadian Institutes of Health Research for support (NIH R01 AI110173, R01 AI120698, R21 AI136731, R01 AI134406, and R56 AI145407; CIHR 152977 and 364408) as well as Colleen O’Neill for assistance with the preparation of the manuscript. D.E.K is a FRQS Merit Research Scholar. N.C. is supported by Research Scholar Career Awards of the Quebec Health Research Fund (FRQS no. 253292).
Contributor Information
Ashley T. Haase, Email: haase001@umn.edu.
Guido Silvestri, Emory University.
REFERENCES
- 1.Archin NM, Sung JM, Garrido C, Soriano-Sarabia N, Margolis DM. 2014. Eradicating HIV-1 infection: seeking to clear a persistent pathogen. Nat Rev Microbiol 12:750–764. 10.1038/nrmicro3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harb Perspect Med 1:a007096. 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neuhaus J, Jacobs DR, Jr, Baker JV, Calmy A, Duprez D, La Rosa A, Kuller LH, Pett SL, Ristola M, Ross MJ, Shlipak MG, Tracy R, Neaton JD. 2010. Markers of inflammation, coagulation, and renal function are elevated in adults with HIV infection. J Infect Dis 201:1788–1795. 10.1086/652749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klatt NR, Chomont N, Douek DC, Deeks SG. 2013. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev 254:326–342. 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longenecker CT, Sullivan C, Baker JV. 2016. Immune activation and cardiovascular disease in chronic HIV infection. Curr Opin HIV AIDS 11:216–225. 10.1097/COH.0000000000000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt PW. 2012. HIV and inflammation: mechanisms and consequences. Curr HIV/AIDS Rep 9:139–147. 10.1007/s11904-012-0118-8. [DOI] [PubMed] [Google Scholar]
- 7.Giorgi JV, Liu Z, Hultin LE, Cumberland WG, Hennessey K, Detels R. 1993. Elevated levels of CD38+ CD8+ T cells in HIV infection add to the prognostic value of low CD4+ T cell levels: results of 6 years of follow-up. The Los Angeles Center, Multicenter AIDS Cohort Study. J Acquir Immune Defic Syndr (1988) 6:904–912. [PubMed] [Google Scholar]
- 8.Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, Shih R, Lewis J, Wiley DJ, Phair JP, Wolinsky SM, Detels R. 1999. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis 179:859–870. 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan RC, Sinclair E, Landay AL, Lurain N, Sharrett AR, Gange SJ, Xue X, Hunt P, Karim R, Kern DM, Hodis HN, Deeks SG. 2011. T cell activation and senescence predict subclinical carotid artery disease in HIV-infected women. J Infect Dis 203:452–463. 10.1093/infdis/jiq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duprez DA, Neuhaus J, Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Nixon D, Paton NI, Prineas RJ, Neaton JD, James D. Neaton1 for the INSIGHT SMART Study Group . 2012. Inflammation, coagulation and cardiovascular disease in HIV-infected individuals. PLoS One 7:e44454. 10.1371/journal.pone.0044454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das B, Dobrowolski C, Luttge B, Valadkhan S, Chomont N, Johnston R, Bacchetti P, Hoh R, Gandhi M, Deeks SG, Scully E, Karn J. 2018. Estrogen receptor-1 is a key regulator of HIV-1 latency that imparts gender-specific restrictions on the latent reservoir. Proc Natl Acad Sci USA 115:E7795–E7804. 10.1073/pnas.1803468115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, Leyre L, Routy JP, Kaufmann DE, Chomont N. 2019. Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog 15:e1007619. 10.1371/journal.ppat.1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, Massanella M, Brassard N, Alsahafi N, Delgado G-G, Routy J-P, Walker BD, Finzi A, Chomont N, Kaufmann DE. 2016. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe 20:368–380. 10.1016/j.chom.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu G, Swanson M, Talla A, Graham D, Strizki J, Gorman D, Barnard RJO, Blair W, Søgaard OS, Tolstrup M, Østergaard L, Rasmussen TA, Sekaly R-P, Archin NM, Margolis DM, Hazuda DJ, Howell BJ. 2017. HDAC inhibition induces HIV-1 protein and enables immune-based clearance following latency reversal. JCI Insight 2:e92901. 10.1172/jci.insight.92901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS. 1989. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci USA 86:2365–2368. 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng H, Reinhart TA, Retzel EF, Staskus KA, Zupancic M, Haase AT. 1995. Single cell transcript analysis of human immunodeficiency virus gene expression in the transition from latent to productive infection. Virology 206:16–27. 10.1016/s0042-6822(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhang ZQ, Wietgrefe SW, Li Q, Shore MD, Duan L, Reilly C, Lifson JD, Haase AT. 2004. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc Natl Acad Sci USA 101:5640–5645. 10.1073/pnas.0308425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M, Jr, Bess J, Anderson JL, Perkey KE, Reilly C, McCune JM, Haase AT, Lifson JD, Schacker TW, Estes JD. 2016. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun 1:68–106. 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 20.Lum JJ, Pilon AA, Sanchez-Dardon J, Phenix BN, Kim JE, Mihowich J, Jamison K, Hawley-Foss N, Lynch DH, Badley AD. 2001. Induction of cell death in human immunodeficiency virus-infected macrophages and resting memory CD4 T cells by TRAIL/Apo2L. J Virol 75:11128–11136. 10.1128/JVI.75.22.11128-11136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Massanella M, Brassard N, Alsahafi N, Routy JP, Finzi A, Chomont N, Kaufmann DE. 2017. Multiparametric characterization of rare HIV-infected cells using an RNA-flow FISH technique. Nat Protoc 12:2029–2049. 10.1038/nprot.2017.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. 2010. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol 28:595–599. 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson DH, Rissin DM, Kan CW, Fournier DR, Piech T, Campbell TG, Meyer RE, Fishburn MW, Cabrera C, Patel PP, Frew E, Chen Y, Chang L, Ferrell EP, von Einem V, McGuigan W, Reinhardt M, Sayer H, Vielsack C, Duffy DC. 2016. The Simoa HD-1 analyzer: a novel fully automated digital immunoassay analyzer with single-molecule sensitivity and multiplexing. J Lab Autom 21:533–547. 10.1177/2211068215589580. [DOI] [PubMed] [Google Scholar]
- 27.Cabrera C, Chang L, Stone M, Busch M, Wilson DH. 2015. Rapid, fully automated digital immunoassay for p24 Protein with the sensitivity of nucleic acid amplification for detecting acute HIV infection. Clin Chem 61:1372–1380. 10.1373/clinchem.2015.243287. [DOI] [PubMed] [Google Scholar]
- 28.Passaes CPB, Bruel T, Decalf J, David A, Angin M, Monceaux V, Muller-Trutwin M, Noel N, Bourdic K, Lambotte O, Albert ML, Duffy D, Schwartz O, Sáez-Cirión A. 2017. Ultrasensitive HIV-1 p24 assay detects single infected cells and differences in reservoir induction by latency reversal agents. J Virol 91:e02296-16. 10.1128/JVI.02296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JS, Davis ZB, Helgeson E, Reilly C, Thorkelson A, Anderson J, Lima NS, Jorstad S, Hart GT, Lee JH, Safrit JT, Wong H, Cooley S, Gharu L, Chung H, Soon-Shiong P, Dobrowolski C, Fletcher CV, Karn J, Douek D, Schacker TW. 2022. Safety and virologic impact of the IL-15 superagonist N-803 in people living with HIV: a phase 1 trial. Nat Med 28:392–400. 10.1038/s41591-021-01651-9. [DOI] [PubMed] [Google Scholar]