Abstract
Inferring causal relationships or related associations from observational data can be invalidated by the existence of hidden confounding. We focus on a high-dimensional linear regression setting, where the measured covariates are affected by hidden confounding and propose the Doubly Debiased Lasso estimator for individual components of the regression coefficient vector. Our advocated method simultaneously corrects both the bias due to estimation of high-dimensional parameters as well as the bias caused by the hidden confounding. We establish its asymptotic normality and also prove that it is efficient in the Gauss-Markov sense. The validity of our methodology relies on a dense confounding assumption, i.e. that every confounding variable affects many covariates. The finite sample performance is illustrated with an extensive simulation study and a genomic application.
Keywords: Causal Inference, Structural Equation Model, Dense Confounding, Linear Model, Spectral Deconfounding
MSC2020 subject classifications: Primary 62E20, 62F12, secondary 62J07
1. Introduction.
Observational studies are often used to infer causal relationship in fields such as genetics, medicine, economics or finance. A major concern for confirmatory conclusions is the existence of hidden confounding [27, 42]. In this case, standard statistical methods can be severely biased, particularly for large-scale observational studies, where many measured covariates are possibly confounded.
To better address this problem, let us consider first the following linear Structural Equation Model (SEM) with a response Yi, high-dimensional measured covariates and hidden confounders :
| (1) |
where the random error is independent of , and and the components of are uncorrelated with the components of The focus on a SEM as in (1) is not necessary and we relax this restriction in model (2) below. Such kind of models are used for e.g. biological studies to explore the effects of measured genetic variants on the disease risk factor, and the hidden confounders can be geographic information [46], data sources in mental analysis [48] or general population stratification in GWAS [43].
Our aim is to perform statistical inference for individual components βj, 1≤ j ≤ p, of the coefficient vector, where p can be large, in terms of obtaining confidence intervals or statistical tests. This inference problem is challenging due to high dimensionality of the model and the existence of hidden confounders. As a side remark, we mention that our proposed methodology can also be used for certain measurement error models, an important general topic in statistics and economics [11, 62].
1.1. Our Results and Contributions.
We focus on a dense confounding model, where the hidden confounders Hi,· in (1) are associated with many measured covariates Xi,.. Such dense confounding model seems reasonable in quite many practical applications, e.g. for addressing the problem of batch effects in biological studies [29, 33, 38].
We propose a two-step estimator for the regression coefficient βj for 1 ≤ j ≤ p in the high-dimensional dense confounding setting, where a large number of covariates have possibly been affected by hidden confounding. In the first step, we construct a penalized spectral deconfounding estimator as in [12], where the standard squared error loss is replaced by a squared error loss after applying a certain spectral transformation to the design matrix X and the response Y. In the second step, for the regression coefficient of interest βj, we estimate the high-dimensional nuisance parameters β−j = {βl; l ≠ j} by and construct an approximately unbiased estimator .
The main idea of the second step is to correct the bias from two sources, one from estimating the high-dimensional nuisance vector β−j by and the other arising from hidden confounding. In the standard high-dimensional regression setting with no hidden confounding, debiasing, desparsifying or Neyman’s Orthogonalization were proposed for inference for βj [65, 56, 32, 4, 15, 21, 14]. However, these methods, or some of its direct extensions, do not account for the bias arising from hidden confounding. In order to address this issue, we introduce a Doubly Debiased Lasso estimator which corrects both biases simultaneously. Specifically, we construct a spectral transformation , which is applied to the nuisance design matrix X−j when the parameter of interest is βj. This spectral transformation is crucial to simultaneously correcting the two sources of bias.
We establish the asymptotic normality of the proposed Doubly Debiased Lasso estimator in Theorem 1. An efficiency result is also provided in Theorem 2 of Section 4.2.1, showing that the Doubly Debiased Lasso estimator retains the same Gauss-Markov efficiency bound as in standard high-dimensional linear regression with no hidden confounding [56, 31]. Our result is in sharp contrast to Instrumental Variables (IV) based methods, see Section 1.2, whose inflated variance is often of concern, especially with a limited amount of data [62, 6]. This remarkable efficiency result is possible by assuming denseness of confounding. Various intermediary results of independent interest are also derived in Section A of the supplementary material. Finally, the performance of the proposed estimator is illustrated on simulated and real genomic data in Section 5.
To summarize, our main contribution is two-fold:
We propose a novel Doubly Debiased Lasso estimator for individual coefficients βj and estimation of the corresponding standard error in a high-dimensional linear SEM with hidden confounding.
We show that the proposed estimator is asymptotically Gaussian and efficient in the Gauss-Markov sense. This implies the construction of asymptotically optimal confidence intervals for individual coefficients βj.
The code for using the Doubly Debiased Lasso estimator is available at https://github.com/zijguo/Doubly-Debiased-Lasso.
1.2. Related Work.
In econometrics, hidden confounding and measurement errors are unified under the framework of endogenous variables. Inference for treatment effects or corresponding regression parameters in presence of hidden confounders or measurement errors has been extensively studied in the literature with Instrumental Variables (IV) regression. The construction of IVs typically requires a lot of domain knowledge, and obtained IVs are often suspected of violating the main underlying assumptions [30, 62, 34, 8, 28, 61]. In high dimensions, the construction of IVs is even more challenging, since for identification of the causal effect, one has to construct as many IVs as the number of confounded covariates, which is the so-called “rank condition" [62]. Some recent work on the high-dimensional hidden confounding problem relying on the construction of IVs includes [23, 18, 40, 3, 67, 45, 25]. Another approach builds on directly estimating and adjusting with respect to latent factors [60].
A major distinction of the current work from the contributions above is that we consider a confounding model with a denseness assumption [13, 12, 53]. [12] consider point estimation of β in the high-dimensional hidden confounding model (1), whereas [53] deal with point estimation of the precision and covariance matrix of high-dimensional covariates, which are possibly confounded. The current paper is different in that it considers the challenging problem of confidence interval construction, which requires novel ideas for both methodology and theory.
The dense confounding model is also connected to the high-dimensional factor models [17, 37, 36, 20, 59]. The main difference is that the factor model literature focuses on accurately extracting the factors, while our method is essentially filtering them out in order to provide consistent estimators of regression coefficients, under much weaker requirements than for the identification of factors.
Another line of research [22, 55, 58] studies the latent confounder adjustment models but focuses on a different setting where many outcome variables can be possibly associated with a small number of observed covariates and several hidden confounders.
Notation.
We use and to denote the j—th column of the matrix X and the sub-matrix of X excluding the j—th column, respectively; is used to denote the i—th row of the matrix X (as a column vector); Xi,−j and Xi,−j denote respectively the (i, j) entry of the matrix X and the sub-row of Xi,· excluding the j-th entry. Let [p]= {1, 2,... , p}. For a subset and a vector , xJ is the sub-vector of x with indices in J and x−J is the sub-vector with indices in Jc. For a set S, |S| denotes the cardinality of S. For a vector , the ℓq norm of x is defined as for q ≥ 0 with and . We use ei to denote the i-th standard basis vector in and Ip to denote the identity matrix of size p × p. We use c and C to denote generic positive constants that may vary from place to place. For a sub-Gaussian random variable X, we use to denote its sub-Gaussian norm; see definitions 5.7 and 5.22 in [57]. For a sequence of random variables Xn indexed by n, we use and to represent that Xn converges to X in probability and in distribution, respectively. For a sequence of random variables Xn and numbers an, we define Xn = op(an) if Xn/an converges to zero in probability. For two positive sequences an and bn, means that such that an ≤ Cbn for all n; if and , and an ≪ bn if . For a matrix M, we use ‖ M ‖F, ‖ M ‖ 2 and ‖ M ‖2 and ‖ M ‖ꚙ to denote its Frobenius norm, spectral norm and element-wise maximum norm, respectively. We use λj(M) to denote the j-th largest singular value of some matrix M, that is, λ1(M) ≥ λ2(M) ≥ … ≥ λq (M) ≥ 0. For a symmetric matrix A, we use λmax(A) and λmin(A) to denote its maximum and minimum eigenvalues, respectively.
2. Hidden Confounding Model.
We consider the Hidden Confounding Model for i.i.d. data {Xi,·, Yi}1≤i≤n and unobserved i.i.d. confounders {Hi,·}1≤i≤n, given by:
| (2) |
where and respectively denote the response and the measured covariates and represents the hidden confounders. We assume that the random error is independent of , and and the components of are uncorrelated with the components of .
The coefficient matrices and encode the linear effect of the hidden confounders Hi,· on the measured covariates Xi, and the response Yi, respectively. We consider the high-dimensional setting where p might be much larger than n. Throughout the paper it is assumed that the regression vector is sparse, with a small number k of nonzero components, and that the number q of confounding variables is a small positive integer. However, both k and q are allowed to grow with n and p. We write or for the covariance matrices of Ei, or Xi,., respectively. Without loss of generality, it is assumed that , , Cov(Hi,.)= Iq and hence .
The probability model (2) is more general than the Structural Equation Model in (1). It only describes the observational distribution of the latent variable Hi,· and the observed data (Xi,., Yi), which possibly may be generated from the hidden confounding SEM (1).
Our goal is to construct confidence intervals for the components of β, which in the model (1) describes the causal effect of X on the response Y. The problem is challenging due to the presence of unobserved confounding. In fact, the regression parameter β can not even be identified without additional assumptions. Our main condition addressing this issue is a denseness assumption that the rows are dense in a certain sense (see Condition (A2) in Section 4), i.e., many covariates of are simultaneously affected by hidden confounders .
2.1. Representation as a Linear Model.
The Hidden Confounding Model (2) can be represented as a linear model for the observed data {Xi,., Yi}1≤i≤n:
| (3) |
by writing
As in (2) we assume that Ei,·· is uncorrelated with Hi,· and, by construction of b, ϵi is uncorrelated w ith Xi,·. With denoting the variance of ei, the variance of the error ϵi equals . In model (3) the response is generated from a linear model where the sparse coefficient vector β has been perturbed by some perturbation vector . This representation reveals how the parameter of interest β is not in general identifiable from observational data, where one can not easily differentiate it from the perturbed coefficient vector β +b, with the perturbation vector b induced by hidden confounding. However, as shown in Lemma 2 in the supplement, b is dense and ‖ b ‖2 is small for large p under the assumption of dense confounding, which enables us to identify β asymptotically. It is important to note that the term b⊤Xi,. induced by hidden confounders Hi, is not necessarily small and hence cannot be simply ignored in model (3), but requires novel methodological approach.
Connection to measurement errors.
We briefly relate certain measurement error models to the Hidden Confounding Model (2). Consider a linear model for the outcome Yi and covariates , where we only observe with measurement error :
| (4) |
Here, ei is a random error independent of and Wi,. and Wi,· is the measurement error independent of . We can then express a linear dependence of Yi on the observed Xi,.,
We further assume the following structure of the measurement error:
i.e. there exist certain latent variables that contribute independently and linearly to the measurement error, a conceivable assumption in some practical applications. Combining this with the equation above we get
| (5) |
where . Therefore, the model (5) can be seen as a special case of the model (2), by identifying in (5) with Ei,· in (2).
3. Doubly Debiased Lasso Estimator.
In this section, for a fixed index j ϵ{1,..., p}, we propose an inference method for the regression coefficient βj of the Hidden Confounding Model (2). The validity of the method is demonstrated by considering the equivalent model (3).
3.1. Double Debiasing.
We denote by an initial estimator of β. We will use the spectral deconfounding estimator proposed in [12], described in detail in Section 3.4. We start from the following decomposition:
| (6) |
The above decomposition reveals two sources of bias: the bias due to the error of the initial estimator and the bias X−jb−j induced by the perturbation vector b in the model (3), arising by marginalizing out the hidden confounding in (2). Note that the bias bj is negligible in the dense confounding setting, see Lemma 2 in the supplement. The first bias, due to penalization, appears in the standard high-dimensional linear regression as well, and can be corrected with the debiasing methods proposed in [65, 56, 32] when assuming no hidden confounding. However, in presence of hidden confounders, methodological innovation is required for correcting both bias terms and conducting the resulting statistical inference. We propose a novel Doubly Debiased Lasso estimator for correcting both sources of bias simultaneously.
Denote by a symmetric spectral transformation matrix, which shrinks the singular values of the sub-design . The detailed construction, together with some examples, is given in Section 3.3. We shall point out that the construction of the transformation matrix depends on which coefficient βj is our target and hence refer to as the nuisance spectral transformation with respect to the coefficient βj. Multiplying both sides of the decomposition (6) with the transformation gives:
| (7) |
The quantity of interest βj appears on the RHS of the equation (7) next to the vector , whereas the additional bias lies in the span of the columns of . For this reason, we construct a projection direction vector as the transformed residuals of regressing Xj on X−j:
| (8) |
where the coefficients are estimated with the Lasso for the transformed covariates using :
| (9) |
with for some positive constant (for σj, see Section 4.1).
Finally, motivated by the equation (7), we propose the following estimator for βj:
| (10) |
We refer to this estimator as the Doubly Debiased Lasso estimator as it simultaneously corrects the bias induced by and the confounding bias X−jb−j by using the spectral transformation .
In the following, we briefly explain why the proposed estimator estimates βj well. We start with the following error decomposition of , derived from (7)
| (11) |
In the above equation, the bias after correction consists of two components: the remaining bias due to the estimation error of and the remaining confounding bias due to X−jb−j. and bj. These two components can be shown to be negligible in comparison to the variance component under certain model assumptions, see Theorem 1 and its proof for details. Intuitively, the construction of the spectral transformation matrix is essential for reducing the bias due to the hidden confounding. The term in equation (11) is of a small order because shrinks the leading singular values of X−j and hence is significantly smaller than X−jb−j. The induced bias X−jb−j is not negligible since b−j points in the direction of leading right singular vectors of X−j, thus leading to being of constant order. By applying a spectral transformation to shrink the leading singular values, one can show that .
Furthermore, the other remaining bias term in (11) is small since the initial estimator is close to β in ℓ1 norm and and are nearly orthogonal due to the construction of in (9). This bias correction idea is analogous to the Debiased Lasso estimator introduced in [65] for the standard high-dimensional linear regression:
| (12) |
where is constructed similarly as in (8) and (9) with setting as the identity matrix. Therefore, the main difference between the estimator in (12) and our proposed estimator (10) is that we additionally apply the nuisance spectral transformation .
We emphasize that the additional spectral transformation is necessary even for just correcting the bias of in presence of confounding (i.e., it is also needed for the first besides the second bias term in (11)). To see this, we define the best linear projection of X1,j to all other variables with the coefficient vector (which is then estimated by the Lasso in the standard construction of ). We notice that γ need not be sparse due to the fact that all covariates are affected by a common set of hidden confounders yielding spurious associations. Hence, the standard construction of in (12) is not favorable in the current setting. In contrast, the proposed method with works for two reasons: first, the application of in (9) leads to a consistent estimator of the sparse component of γ, denoted as γγE (see the expression of γE in Lemma 1); second, the application of leads to a small prediction error . We illustrate in Section 5 how the application of corrects the bias due to and observe a better empirical coverage after applying in comparison to the standard debiased Lasso in (12); see Figure 7.
FIG 7.

(No confounding bias) Dependence of the (scaled) absolute bias terms |Bβ| and |Bb| (left), standard deviation V1/2 (middle) and the coverage of the 95% confidence interval (right) on the number of covariates p, while keeping n = 500 fixed. In the plots on the left, |Bβ| and |Bb| are denoted by a dashed and a solid line, respectively, but Bb =0 since we have enforced b = 0. Top row corresponds to the spiked covariance case , whereas for the bottom row we set . Blue color corresponds to the Doubly Debiased Lasso, red color represents the standard Debiased Lasso and green color corresponds also to the Debiased Lasso estimator, but with the same as our proposed method. Note that the last two methods have almost indistinguishable V.
3.2. Confidence Interval Construction.
In Section 4, we establish the asymptotic normal limiting distribution of the proposed estimator under certain regularity conditions. Its standard deviation can be estimated by with denoting a consistent estimator of σe. The detailed construction of is described in Section 3.5. Therefore, a confidence interval (CI) with asymptotic coverage 1 – α can be obtained as
| (13) |
where is the quantile of a standard normal random variable.
3.3. Construction of Spectral Transformations.
Construction of the spectral transformation is an essential step for the Doubly Debiased Lasso estimator (10). The transformation is a symmetric matrix shrinking the leading singular values of the design matrix . Denote by m = min{n, p— 1} and the SVD of the matrix X−j by X−j = U(X−j) Λ (X−j)[V (X−j)]⸆, where and have orthonormal columns and is a diagonal matrix of singular values which are sorted in a decreasing order. We then define the spectral transformation for X−j as , where is a diagonal shrinkage matrix with 0 ≤ Sl,l(X−j) ≤ 1 for 1 ≤ 1≤ m. The SVD for the complete design matrix X is defined analogously. We highlight the dependence of the SVD decomposition on X−j, but for simplicity it will be omitted when there is no confusion. Note that , so the spectral transformation shrinks the singular values to , where .
Trim transform.
For the rest of this paper, the spectral transformation that is used is the Trim transform [12]. It limits any singular value to be at most some threshold τ. This means that the shrinkage matrix S is given as: for 1 ≤ l ≤ m,
A good default choice for the threshold τ is the median singular value , so only the top half of the singular values is shrunk to the bulk value and the bottom half is left intact. More generally, one can use any percentile ρj ϵ (0, 1) to shrink the top (100ρj)% singular values to the corresponding ρj-quantile . We define the ρj-Trim transform as
| (14) |
In Section 4 we investigate the dependence of the asymptotic efficiency of the resulting Doubly Debiased Lasso on the percentile choice . There is a certain trade-off in choosing ρj: a smaller value of ρj leads to a more efficient estimator, but one needs to be careful to keep ρj m sufficiently large compared to the number of hidden confounders q, in order to ensure reduction of the confounding bias. In Section A.1 of the supplementary material, we describe the general conditions that the used spectral transformations need to satisfy to ensure good performance of the resulting estimator.
3.4. Initial Estimator .
For the Doubly Debiased Lasso (10), we use the spectral deconfounding estimator proposed in [12] as our initial estimator . It uses a spectral transformation , constructed similarly as the transformation described in Section 3.3, with the difference that instead of shrinking the singular values of X−j, shrinks the leading singular values of the whole design matrix . Specifically, for any percentile ρ ϵ (0, 1), the ρ-Trim transform is given by
| (15) |
The estimator is computed by applying the Lasso to the transformed data and :
| (16) |
where is a tuning parameter with .
The transformation reduces the effect of the confounding and thus helps for estimation of β. In Section A.3, the ℓ1 and ℓ2-error rates of are given, thereby extending the results of [12].
3.5. Noise Level Estimator.
In addition to an initial estimator of β, we also require a consistent estimator of the error variance for construction of confidence intervals. Choosing a noise level estimator which performs well for a wide range of settings is not easy to do in practice [50]. We propose using the following estimator:
| (17) |
where is the same spectral transformation as in (16).
The motivation for this estimator is based on the expression
| (18) |
which follows from the model (3). The consistency of the proposed noise level estimator, formally shown in Proposition 2, follows from the following observations: the initial spectral deconfounding estimator has a good rate of convergence for estimating β; the spectral transformation significantly reduces the additional error Xb induced by the hidden confounders; consistently estimates . Additionally, the dense confounding model is shown to lead to a small difference between the noise levels and , see Lemma 2 in the supplement. In Section 4 we show that variance estimator defined in (17) is a consistent estimator of .
3.6. Method Overview and Choice of the Tuning Parameters.
The Doubly Debiased Lasso needs specification of various tuning parameters. A good and theoretically justified rule of thumb is to use the Trim transform with ρ = ρj = 1/2, which shrinks the large singular values to the median singular value, see (14). Furthermore, similarly to the standard Debiased Lasso [65], our proposed method involves the regularization parameters λ in the Lasso regression for the initial estimator (see equation (16)) and λj in the Lasso regression for the projection direction (see equation (9)). For choosing λ we use 10-fold cross-validation, whereas for λj, we increase slightly the penalty chosen by the 10-fold cross-validation, so that the variance of our estimator, which can be determined from the data up to a proportionality factor , increases by 25%, as proposed in [16].
The proposed Doubly Debiased Lasso method is summarized in Algorithm 1, which
4. Theoretical Justification.
This section provides theoretical justifications of the proposed method for the Hidden Confounding Model (2). The proofs of the main results are presented in Sections A and B of the supplementary material together with several other technical results of independent interest.
4.1. Model assumptions.
In the following, we fix the index 1 ≤ j ≤ p and introduce the model assumptions for establishing the asymptotic normality of our proposed estimator defined in (10). For the coefficient matrix in (3), we use to denote the j-th column and denotes the sub-matrix with the remaining p – 1 columns. Furthermore, we write γ for the best linear approximation of by , that is , whose explicit expression is:
For ease of notation, we do not explicitly express the dependence of γ on j. Similarly, define
We denote the corresponding residuals by and for 1 ≤ i ≤ n. We use σj to denote the standard deviation of vi,j.
The first assumption is on the precision matrix of in (2):
(A1) The precision matrix satisfies and where C0 > 0 and c0 > 0 are some positive constants and s denotes the sparsity level which can grow with n and p.
Such assumptions on well-posedness and sparsity are commonly required for estimation of the precision matrix [44, 35, 64, 9] and are also used for confidence interval construction in the standard high-dimensional regression model without unmeasured confounding [56]. Here, the conditions are not directly imposed on the covariates Xi,.·, but rather on their unconfounded part Ei,.. In the high-dimensional linear model without hidden confounders, the sparse precision matrix assumption can be relaxed using the technique in [32]. However, it is unclear whether the method in [32] can be generalized to our model due to the additional hidden confounding bias as in (11).
The second assumption is about the coefficient matrix in (3), which describes the effect of the hidden confounding variables on the measured variables :
(A2) The q-th singular value of the coefficient matrix satisfies
| (19) |
where M is the sub-Gaussian norm for components of Xi,., as defined in Assumption (A3). Furthermore, we have
| (20) |
where and ϕ are defined in (2) and 0 < c ≤ 1/4 is some positive constant.
The condition (A2) is crucial for identifying the coefficient βj in the high-dimensional Hidden Confounding Model (2). Condition (A2) is referred to as the dense confounding assumption. A few remarks are in order regarding when this identifiability condition holds.
Since all vectors , , and ϕ are q-dimensional, the upper bound condition (20) on their ℓ2 norms is mild. If the vector has bounded entries and the vectors are independently generated with zero mean and bounded second moments, then the condition (20) holds with probability larger than 1 – (log p)−2c, where c is defined in (20). A larger value c > 1/4 is possible: the condition then holds with even higher probability, but makes the upper bounds for (32) in Lemma 1 and (35) in Lemma 2 slightly worse, which then requires more stringent conditions on in Theorem 1, up to polynomial order of log p.
In the factor model literature [19, 59] the spiked singular value condition is quite common and holds under mild conditions. The Hidden Confounding Model is closely related to the factor model, where the hidden confounders Hi,· are the factors and the matrix describes how these factors affect the observed variables Xi,.. However, for our analysis, our assumption on in (19) can be much weaker than the classical factor assumption , especially for a range of dimensionality where p ≫ n. In certain dense confounding settings, we can show that condition (19) holds with high probability. Consider first the special case with a single hidden confounder, that is, q = 1 and the effect matrix is reduced to a vector . In this case, and the denseness of the effect vector leads to a large . The condition (19) can be satisfied even if only a certain proportion of covariates is affected by hidden confounding. When q = 1, if we assume that there exists a set such that are i.i.d. and , where l(n,p,q) is defined in (19), then with high probability . In the multiple hidden confounders setting, if the vectors are generated as i.i.d. sub-Gaussian random vectors, which has an interpretation that all covariates are analogously affected by the confounders, then the spiked singular value condition (19) is satisfied with high probability as well. See Lemmas 4 and 5 in Section A.5 of the supplementary material for the exact statement. In Section 5.1, we also explore the numerical performance of the method when different proportions of the covariates are affected and observe that the proposed method works well even if the hidden confounders only affect a small percentage of the covariates, say 5%.
Under the model (2), if the entries of are assumed to be i.i.d. sub-Gaussian with zero mean and variance , then we have with high probability. Together with (19) this requires
So if p ≫ qn log p and , then the required effect size of the hidden confounder Hi,· on an individual covariate Xi,j can diminish to zero fairly quickly.
The condition (19) can in fact be empirically checked using the sample covariance matrix . Since , then the condition (19) implies that has at least q spiked eigenvalues. If the population covariance matrix has a few spikes, the corresponding sample covariance matrix will also have spiked eigenvalue structure with a high probability [59]. Hence, we can inspect the spectrum of the sample covariance matrix and informally check whether it has spiked singular values. See the left panel of Figure 2 for an illustration.
FIG 2.
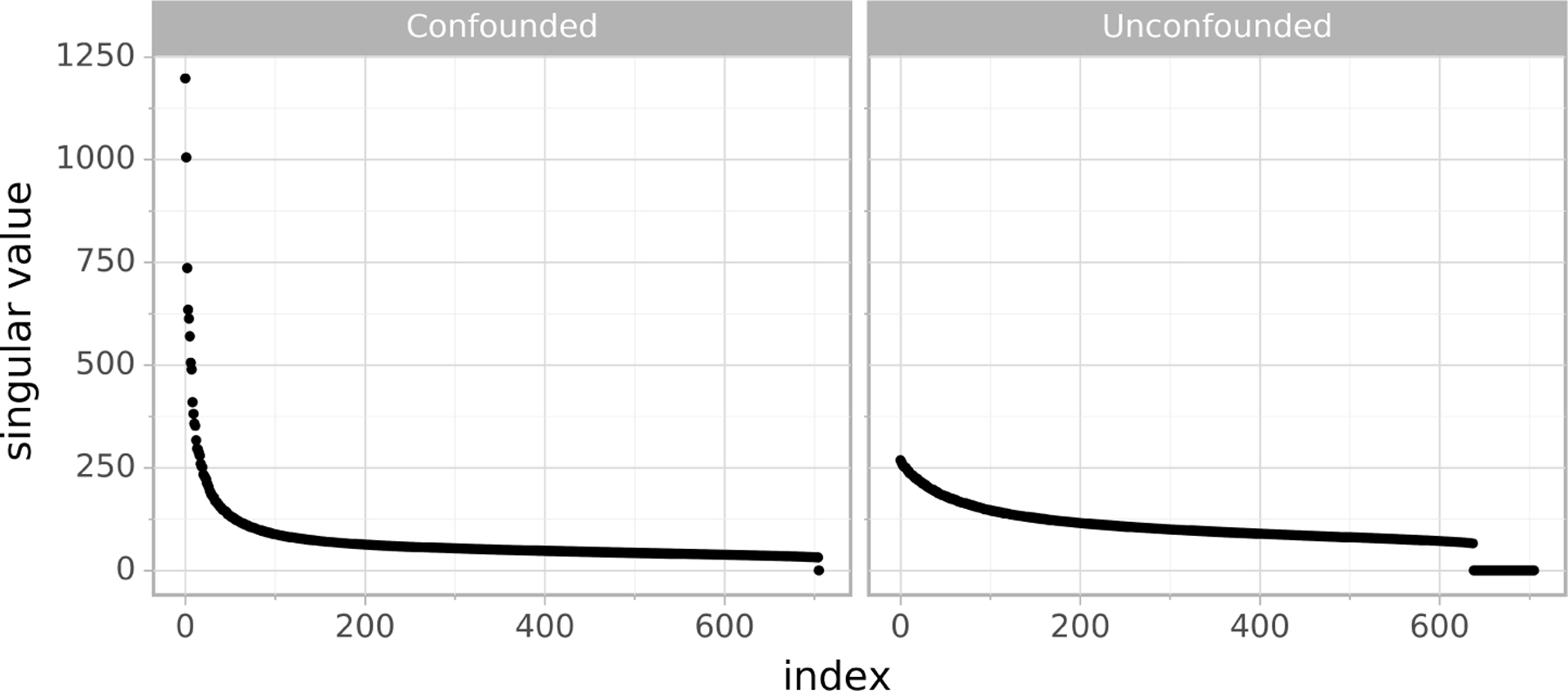
Left: Spiked singular values of the standardized gene expression matrix (see Section 5.2) indicate possible confounding. Right: Singular values after regressing out the q = 65 confounding proxies given in the data set (thus labeled as “unconfounded”). The singular values in both plots are sorted decreasingly.
The third assumption is imposed on the distribution of various terms:
(A3) The random error ei in (2) is assumed to be independent of , the error vector Ei, is assumed to be independent of the hidden confounder Hi,., and the noise term is assumed to be independent of Ei,−j. Furthermore, Ei,· is a sub-Gaussian random vector and ei and vi,j are sub-Gaussian random variables, whose sub-Gaussian norms satisfy , where C> 0 is a positive constant independent of n and p. For 1 ≤ l≤ p, Xi,l are sub-Gaussian random variables whose sub-Gaussian norms satisfy , where .
The independence assumption between the random error ei and is commonly assumed for the SEM (1) and thus it holds in the induced Hidden Confounding Model (2) as well, see for example [47]. Analogously, when modelling Xi,· as a SEM where the hidden variables Hi,.· are directly influencing Xi,. that is, they are parents of the Xi,.·’s, the independence of Ei,· from Hi,· is a standard assumption. The independence assumption between vi,j and Ei,−j holds automatically if Ei,.· has a multivariate Gaussian distribution (but Xi,· is still allowed to be non-Gaussian, e.g. due to non-Gaussian confounders). Additionally, the independence assumption between vi,j and Ei,−j holds if all elements of Ei,· are independent, but not necessarily Gaussian. In Appendix D, we explore the robustness of our proposed Doubly Debiased Lasso estimator to the violation of this independence assumption; see Figure A2 for details.
We emphasize that the individual components Xi,j are assumed to be sub-Gaussian, instead of the whole vector . The sub-Gaussian norm M is allowed to grow with q and p. Particularly, if we assume Hi,· to be a sub-Gaussian vector, then condition (20) implies that . Furthermore, our theoretical analysis also covers the case when the sub-Gaussian norm M is of constant order. This happens, for example, when the entries of are of order , since .
The final assumption is that the restricted eigenvalue condition [5] for the transformed design matrices and is satisfied with high probability.
(A4) With probability at least 1 – exp(−cn), we have
| (21) |
| (22) |
where c, C, τ* > 0 are positive constants independent of n and p and M is the sub-Gaussian norm for components of Xi,., as defined in Assumption (A3). For ease of notation, the same constants τ* and C are used in (21) and (22).
Such assumptions are common in the high-dimensional statistics literature, see [7]. The restricted eigenvalue condition (A4) is similar, but more complicated than the standard restricted eigenvalue condition introduced in [5]. The main complexity is that, rather than for the original design matrix, the restricted eigenvalue condition is imposed on the transformed design matrices and , after applying the Trim transforms and , described in detail in Sections 3.3 and 3.4, respectively. In the following, we verify the restricted eigenvalue condition (A4) for and the argument can be extended to .
PROPOSITION 1. Suppose that assumptions (A1) and (A3) hold, Hi is a sub-Gaussian random vector, and satisfies M2kq2 log p log n/n → 0. Assume further that the loading martrix satisfies , and that
| (23) |
If for ρ defined in(15) and some positive constant c> 0 independent of n and p, then there exist positive constants c1, c2 > 0 such that, with probability larger than , we have .
An important condition for establishing Proposition 1 is the condition (23). Under the commonly assumed spiked singular value condition [19, 59, 1, 2], the condition (23) is reduced to k ≪ min {n, p} / (M 2q5 log(np)4). As a comparison, for the standard high-dimensional regression model with no hidden confounders, [66, 49] verified the restricted eigenvalue condition under the sparsity condition k ≪n/log p. That is, if , then the sparsity requirement in Proposition 1 is the same as that for the high-dimensional regression model with no hidden confounders, up to a polynomial order of q and log(np),
In comparison to the condition (19) in (A2), (23) can be slightly stronger for a range of dimensionality where p ≫ n3/2. However, Proposition 1 does not require the strong spiked singular value condition . The proof of Proposition 1 is presented in Appendix B in the supplement. The condition can be empirically verified from the data. In Appendix B.1, further theoretical justification for this condition is provided, under mild assumptions.
4.2. Main Results.
In this section we present the most important properties of the proposed estimator (10). We always consider asymptotic expressions in the limit where both n, p →∞1 and focus on the high-dimensional regime with c* = lim p/n ϵ (0, ∞]. We mention here that we also give some new results on point estimation of the initial estimator defined in (16) in Appendix A.3, as they are established under more general conditions than in [12].
4.2.1. Asymptotic normality.
We first present the limiting distribution of the proposed Doubly Debiased Lasso estimator. The proof of Theorem 1 and important intermediary results for establishing Theorem 1 are presented in Appendix A in the supplement.
THEOREM 1. Consider the Hidden Confounding Model (2). Suppose that conditions (A1)-(A4) hold and further assume that , , and . Let the tuning parameters for in (16) and in (9) respectively be and .Furthermore, let and be the Trim transform (14) with min{ρ, ρj} ≥ (q + 1)/min {n, p – 1} and max {ρ, ρj} < 1. Then the Doubly Debiased Lasso estimator (10) satisfies
| (24) |
where
| (25) |
REMARK 1. The Gaussianity of the random error ei is mainly imposed to simplify the proof of asymptotic normality. We believe that this assumption is a technical condition and can be removed by applying more refined probability arguments as in [26], where the asymptotic normality of quadratic forms is established for the general sub-Gaussian case. The argument could be extended to obtain the asymptotic normality for , which is essentially needed for the current result.
REMARK 2. For constructing and , the main requirement is to trim the singular values enough in both cases, that is, min{ρ, ρj} ≥ (q + 1)/min{n, p – 1}. This condition is mild in the high-dimensional setting with a small number of hidden confounders. Our results are not limited to the proposed estimator which uses the Trim transform in (14) and the penalized estimators and in (9) and (16), but hold for any transformation satisfying the conditions given in Section A.1 of the supplementary material and any initial estimator satisfying the error rates presented in Section A.3 of the supplementary material.
REMARK 3. If we further assume the error ϵi in the model (3) to be independent of Xi,., then the requirement (19) of the condition (A2) can be relaxed to
Note that the factor model implies the upper bound . Even if n ≥ p, the above condition on can still hold if . On the other hand, the condition (19) together with imply that p ≫ qn log p, which excludes the setting n ≥ p.
There are three conditions on the parameters s, q, k imposed in the Theorem 1 above. The most stringent one is the sparsity assumption . In standard high-dimensional sparse linear regression, a related sparsity assumption has also been used for confidence interval construction [65, 56, 32] and has been established in [10] as a necessary condition for constructing adaptive confidence intervals. In the high-dimensional Hidden Confounding Model with , the condition on the sparsity of β is then of the same asymptotic order as in the standard high-dimensional regression with no hidden confounding. The condition on the sparsity of the precision matrix, , is mild in the sense that, for , it is the maximal sparsity level for identifying (ΩE).,j. Implied by (19), the condition that the number of hidden confounders q is small is fundamental for all reasonable factor or confounding models.
4.2.2. Efficiency.
We investigate now the dependence of the asymptotic variance V in (25) on the choice of the spectral transformation . We further show that the proposed Doubly Debiased Lasso estimator (10) is efficient in the Gauss-Markov sense, with a careful construction of the transformation .
The Gauss-Markov theorem states that the smallest variance of any unbiased linear estimator of βj in the standard low-dimensional regression setting (with no hidden confounding) is , which we use as a benchmark. The corresponding discussion on efficiency of the standard high-dimensional regression can be found in Section 2.3.3 of [56]. The expression for the asymptotic variance V of our proposed estimator (10) is given by (see Theorem 1). For the Trim transform defined in (14), which trims top (100 ρj)% of the singular values, we have that
where we write m = min{n, p – 1} and Sl,l = Sl,l(X−j) ϵ [0, 1]. Since for every l, and , we obtain
In the high-dimensional setting where p – 1 ≥ n, we have m = n and then
| (26) |
THEOREM 2. Suppose that the assumptions of Theorem 1 hold. If p ≥ n +1 and ρj = ρj(n) → 0, then the Doubly Debiased Lasso estimator in (10) has asymptotic variance , that is, it achieves the Gauss-Markov efficiency bound.
The above theorem shows that in the q ≪ n regime, the Doubly Debiased Lasso achieves the Gauss-Markov efficiency bound if ρj = ρj (n) → 0 and min {ρ, ρj} ≥ (q + 1)/n (which is also a condition of Theorem 1). When using the median Trim transform, i.e. ρj = 1/2, the bound in (26) implies that the variance of the resulting estimator is at most twice the size of the Gauss-Markov bound. In Section 5, we illustrate the finite-sample performance of the Doubly Debiased Lasso estimator for different values of ρj; see Figure 6.
FIG 6.

(Trimming level) Dependence of the (scaled) absolyte bias terms |Bβ| and |Bb| (left), standard deviation V1/2 (middle) and the coverage of the 95% confidence interval (right) on the trimming level ρj of the Trim transform (see Equation (14)). The sample size is fixed at n = 300 and the dimension at p = 1, 000. On the leftmost plot, |Bβ| and |Bb| are denoted by a dashed and a solid line, respectively. The case ρj =0 corresponds to Debiased Lasso with the spectral deconfounding initial estimator , described in (16).
In general for the high-dimensional setting p/n → c* ∈ (0, ∞], the Asymptotic Relative Efficiency (ARE) of the proposed Doubly Debiased Lasso estimator with respect to the Gauss-Markov efficiency bound satisfies the following:
| (27) |
where . The equation (27) reveals how the efficiency of the Doubly Debiased Lasso is affected by the choice of the percentile ρj = ρj(n) in transformation and the dimensionality of the problem. Smaller ρj leads to a more efficient estimator, as long as the top few singular values are properly shrunk. Intuitively, a smaller percentile ρj means that less information in X−j is trimmed out and hence the proposed estimator is more efficient. In addition, for the case ρ* = 0, we have ARE = {max 1/c*, 1}. With ρ* = 0, a plot of ARE with respect to the ratio c* = lim p/n is given in Figure 1. We see that for c* < 1 (that is p< n), the relative efficiency of the proposed estimator increases as the dimension p increases and when c* ≥ 1 (that is p ≥ n), we have that ARE = 1, saying that the Doubly Debiased Lasso achieves the efficiency bound in the Gauss-Markov sense.
FIG 1.

The plot of ARE versus c* = lim p/n, for the setting of ρ* =0.
The phenomenon that the efficiency is retained even in presence of hidden confounding is quite remarkable. For comparison, even in the classical low-dimensional setting, the most commonly used approach assumes availability of sufficiently many instrumental variables (IV) satisfying certain stringent conditions under which one can consistently estimate the effects in presence of hidden confounding. In Theorem 5.2 of [62], the popular IV estimator, two-stage-least-squares (2SLS), is shown to have variance strictly larger than the efficiency bound in the Gauss-Markov setting (with no unmeasured confounding). It has been also shown in Theorem 5.3 of [62] that the 2SLS estimator is efficient in the class of all linear instrumental variable estimators and thus, all linear instrumental variable estimators are strictly less efficient than our Doubly Debiased Lasso. On the other hand, our proposed method not only avoids the difficult step of coming up with a large number of valid instrumental variables, but also achieves the efficiency bound with a careful construction of the spectral transformation . This occurs due to a blessing of dimensionality and the assumption of dense confounding, where a large number of covariates are assumed to be affected by a small number of hidden confounders.
4.2.3. Asymptotic validity of confidence intervals.
The asymptotic normal limiting distribution in Theorem 1 can be used for construction of confidence intervals for βj. Consistently estimating the variance V of our estimator, defined in (25), requires a consistent estimator of the error variance . The following proposition establishes the rate of convergence of the estimator proposed in (17):
PROPOSITION 2. Consider the Hidden Confounding Model (2). Suppose that conditions (A1)-(A4) hold. Suppose further that c* = lim p/n ∈ (0, ∞],k ≾ n/log p and q ≪ min{n, p/log p}. Then with probability larger for some positive constant c > 0 and for any , we have
where M is the sub-Gaussian norm for components of defined in Assumption (A3).
Together with (19) of the condition (A2), we apply the above proposition and establish . As a remark, the estimation error is of the same order of magnitude as since the difference is small in the dense confounding model, see Lemma 2 in the supplement.
Proposition 2, together with Theorem 1, imply the asymptotic coverage and precision properties of the proposed confidence interval CI(βj), described in (13):
COROLLARY 1. Suppose that the conditions of Theorem 1 hold, then the confidence interval defined in (13) satisfies the following properties:
| (28) |
| (29) |
for any positive constant c> 0, where L (CI(βj)) denotes the length of the proposed confidence interval.
Similarly to the efficiency results in Section 4.2.2, the exact length depends on the construction of the spectral transformation . Together with (26), the above proposition shows that the length of constructed confidence interval is shrinking at the rate of n−1/2 for the Trim transform in the high-dimensional setting. Specifically, for the setting p ≥ n + 1, if we choose ρj = ρj(n) (q + 1)/n and ρj(n) → 0, the constructed confidence interval has asymptotically optimal length.
5. Empirical results.
In this section we consider the practical aspects of Doubly Debiased Lasso methodology and illustrate its empirical performance on both real and simulated data. The overview of the method and the tuning parameters selection can be found in Section 3.6.
In order to investigate whether the given data set is potentially confounded, one can inspect the principal components of the design matrix X, or equivalently consider its SVD. Spiked singular value structure (see Figure 2) indicates the existence of hidden confounding, as much of the variance of our data can be explained by a small number of latent factors. This also serves as an informal check of the spiked singular value condition in the assumption (A2).
The scree plot can also be used for choosing the trimming thresholds, if one wants to depart from the default median rule (see Section 3.6). We have seen from the theoretical considerations in Section 4 that we can reduce the estimator variance by decreasing the trimming thresholds for the spectral transformation . On the other hand, it is crucial to choose them so that the number of shrunk singular values is still sufficiently large compared to the number of confounders. However, exactly estimating the number of confounders, e.g. by detecting the elbow in the scree plot [59], is not necessary with our method, since the efficiency of our estimator decreases relatively slowly as we decrease the trimming threshold.
In what follows, we illustrate the empirical performance of the Doubly Debiased Lasso in practice. We compare the performance with the standard Debiased Lasso [65], even though it is not really a competitor for dealing with hidden confounding. Our goal is to illustrate and quantify the error and bias when using the naive and popular approach which ignores potential hidden confounding. We first investigate the performance of our method on simulated data for a range of data generating mechanisms and then investigate its behaviour on a gene expression data set from the GTEx project [41].
5.1. Simulations.
In this section, we compare the Doubly Debiased Lasso with the standard Debiased Lasso in several different simulation settings for estimation of βj and construction of the corresponding confidence intervals.
In order to make comparisons with the standard Debiased Lasso as fair as possible, we use the same procedure for constructing the standard Debiased Lasso, but with , , whereas for the Doubly Debiased Lasso, , , are taken to be median Trim transform matrices, unless specified otherwise. Finally, to investigate the usefulness of double debiasing, we additionally include the standard Debiased Lasso estimator with the same initial estimator as our proposed method, see Section 3.4. Therefore, this corresponds to the case where is the median Trim transform, whereas .
We will compare the (scaled) bias and variance of the corresponding estimators. For a fixed index j, from the equation (11) we have
where the estimator variance V is defined in (25) and the bias terms Bβ and Bb are given by
Larger estimator variance makes the confidence intervals wider. However, large bias makes the confidence intervals inaccurate. We quantify this with the scaled bias terms Bβ, which is due to the error in estimation of β, and Bb, which is due to the perturbation b arising from the hidden confounding. Having small |Bβ| and |Bb| is essential for having a correct coverage, since the construction of confidence intervals is based on the approximation .
We investigate the validity of the confidence interval construction by measuring the coverage of the nominal 95% confidence interval. We present here a wide range of simulations settings and further simulations can be found in the Appendix D.
Simulation parameters.
Unless specified otherwise, in all simulations we fix q = 3, s =5 and β = (1, 1, 1, 1, 1, 0,... 0)T and we target the coefficient β1 = 1. The rows of the unconfounded design matrix E are generated from distribution, where , as a default. The matrix of confounding variables H, the additive error e and the coefficient matrices and ϕ all have i.i.d. N(0, 1) entries, unless stated otherwise. Each simulation is averaged over 5, 000 independent repetitions.
Varying dimensions n and p.
In this simulation setting we investigate how the performance of our estimator depends on the dimensionality of the problem. The results can be seen in Figure 3. In the first scenario, shown in the top row, we have p = 500 and n varying from 50 to 2, 000, thus covering both low-dimensional and high-dimensional cases. In the second scenario, shown in the bottom row, the sample size is fixed at n = 500 and the number of covariates p varies from 100 to 2, 000. We provide analogous simulations in Appendix D, where both the random variables and the model parameters are generated from non-Gaussian distributions.
FIG 3.
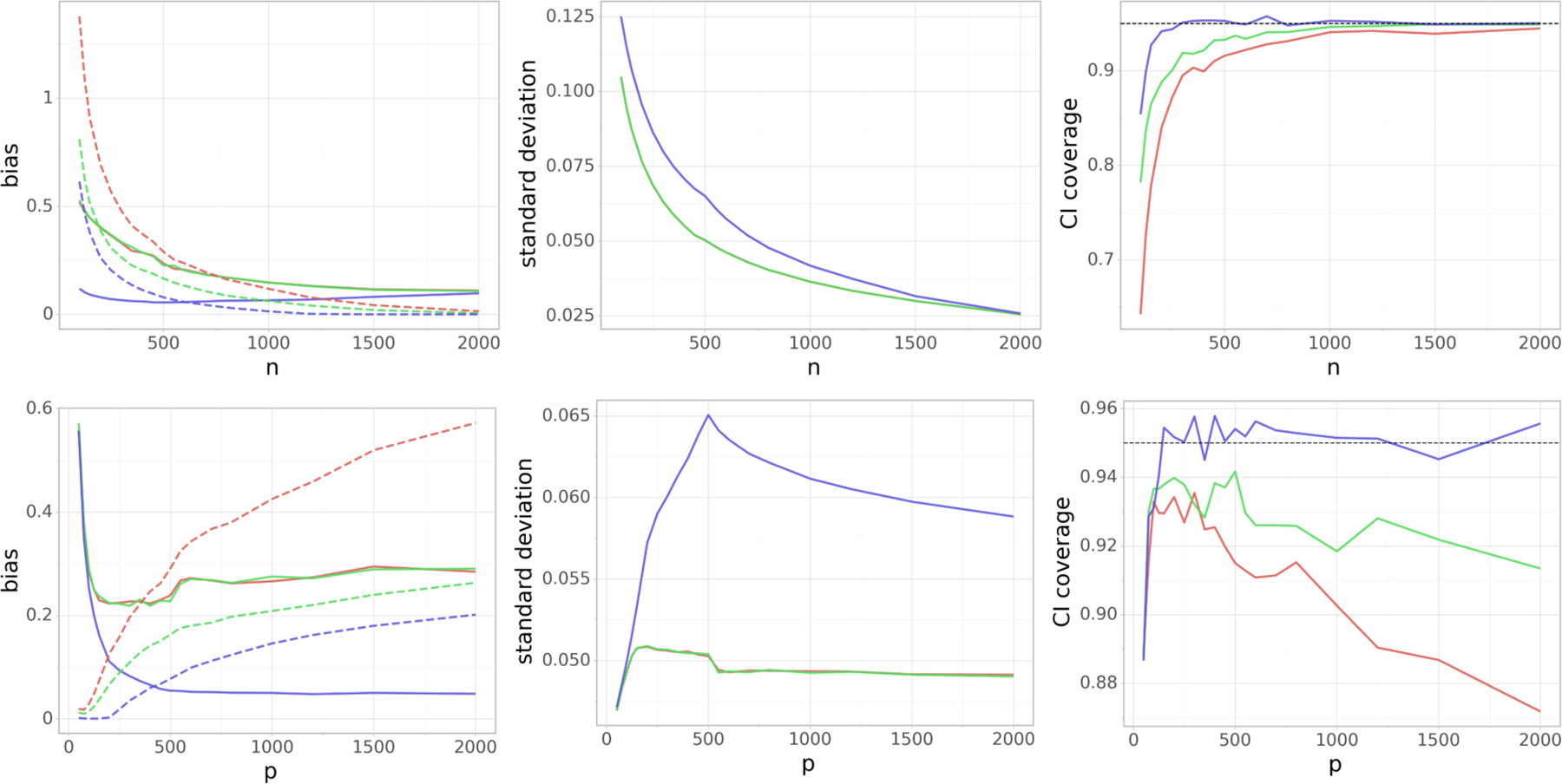
(Varying dimensions) Dependence of the (scaled) absolute bias terms |Bβ| and |Bb| (left), standard deviation V1/2 (middle) and the coverage of the 95% confidence interval (right) on the number of data points n (top row) and the number of covariates p (bottom row). On the left side, |Bβ| and |Bb| are denoted by a dashed and a solid line, respectively. In the top row we fix p = 500, whereas in the bottom row we have n = 500. Blue color corresponds to the Doubly Debiased Lasso, red color represents the standard Debiased Lasso and green color corresponds also to the Debiased Lasso estimator, but with the same as our proposed method. Note that the last two methods have almost indistinguishable |Bb| and V.
We see that the absolute bias term |Bb| due to confounding is substantially smaller for Doubly Debiased Lasso compared to the standard Debiased Lasso, regardless of which initial estimator is used. This is because additionally removes bias by shrinking large principal components of X−j. This spectral transformation helps also to make the absolute bias term |Bβ| smaller for the Doubly Debiased Lasso compared to the Debiased Lasso, even when using the same initial estimator . This comes however at the expense of slightly larger variance, but we can see that the decrease in bias reflects positively on the validity of the constructed confidence intervals. Their coverage is significantly more accurate for Doubly Debiased Lasso, over a large range of n and p.
There are two challenging regimes for estimation under confounding. Firstly, when the dimension p is much larger than the sample size n, the coverage can be lower than 95%, since in this regime it is difficult to estimate β accurately and thus the term |Bβ| is fairly large, even after the bias correction step. We see that the absolute bias |Bβ| grows with p, but it is much smaller for the Doubly Debiased Lasso which positively impacts the coverage. Secondly, in the regime where p is relatively small compared to n, |Bβ| begins to dominate and leads to undercoverage of confidence intervals. Bb is caused by the hidden confounding and does not disappear when n →∞, while keeping p constant. The simulation results agree with the asymptotic analysis of the bias term in (52) in the Supplementary material, where the term |Bb| vanishes as increases, in addition to increasing the sample size n. In the regime considered in this simulation, |Bb| can even grow, since the bias becomes increasingly large compared to the estimator’s variance. However, it is important to note that even in these difficult regimes, Doubly Debiased Lasso performs significantly better than the standard Debiased Lasso (irrespective of the initial estimator) as it manages to additionally decrease the estimator’s bias.
Toeplitz covariance structure for .
Now we fix n = 300,p = 1, 000, but we generate the covariance matrix of the unconfounded part of the design matrix X to have Toeplitz covariance structure:, where we vary κ across the interval [0, 0.97]. As we increase κ, the covariates X1,..., X5 in the active set get more correlated, so it gets harder to distinguish their effects on the response and therefore to estimate β. Similarly, it gets as well harder to estimate γ in the regression of Xj on X−j, since Xj can be explained well by many linear combinations of the other covariates that are correlated with Xj. In Figure 4 we can see that Doubly Debiased Lasso is much less affected by correlated covariates. The (scaled) absolute bias terms |Bb| and |Bβ| are much larger for standard Debiased Lasso, which causes the coverage to worsen significantly for values of κ that are closer to 1.
FIG 4.
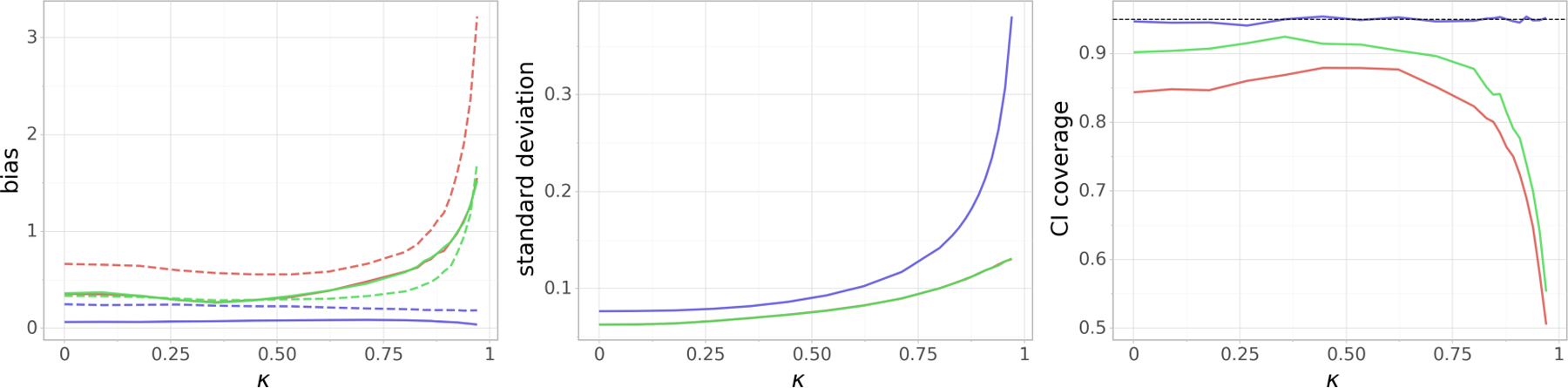
(Toeplitz covariance for ) Dependence of the (scaled) absolute bias terms |Bβ| and |Bb| (left), standard deviation V1/2 (middle) and the coverage of the 95% confidence interval (right) on the parameter κ of the Toeplitz covariance structure. n = 300 and p = 1, 000 are fixed. On the leftmost plot, |Bβ| and |Bb| are denoted by a dashed and a solid line, respectively. Blue color corresponds to the Doubly Debiased Lasso, red color represents the standard Debiased Lasso and green color corresponds also to the Debiased Lasso estimator, but with the same as our proposed method. Note that the last two methods have almost indistinguishable |Bb| and V.
Proportion of confounded covariates.
In order to investigate how the confounding denseness affects the performance of our method, we now again fix n = 300 and p = 1, 000, but we change the proportion of covariates Xi that are affected by each confounding variable. We do this by setting to zero a desired proportion of entries in each row of the matrix , which describes the effect of the confounding variables on each predictor. Its non-zero entries are still generated as N(0, 1). We set once again and we vary the proportion of nonzero entries of from 5% to 100%. The results can be seen in Figure 5. We can see that Doubly Debiased Lasso performs well even when only a very small number (5%) of the covariates are affected by the confounding variables, which agrees with our theoretical discussion for assumption (A2). We can also see that the coverage of the standard Debiased Lasso is poor even for a small number of affected variables and it worsens as the confounding variables affect more and more covariates. The coverage improves to some extent when we use a better initial estimator, but is still worse than our proposed method.
FIG 5.
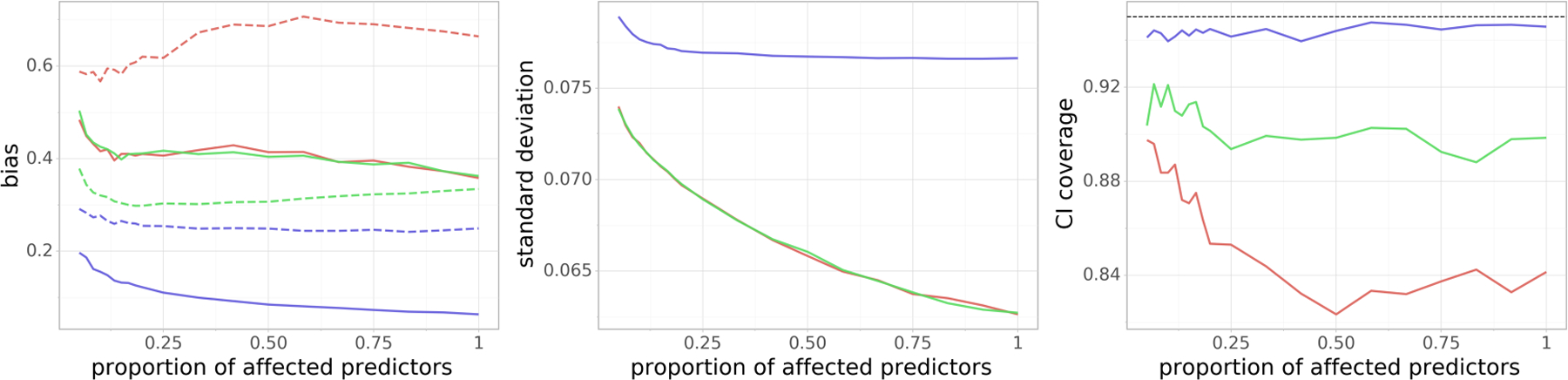
(Proportion confounded) Dependence of the (scaled) absolute bias terms |Bβ| and |Bb| (left), standard deviation V1/2 (middle) and the coverage of the 95% confidence interval (right) on proportion of confounded covariates. n = 300 and p = 1, 000 are fixed. On the leftmost plot, |Bβ| and |Bb| are denoted by a dashed and a solid line, respectively. Blue color corresponds to the Doubly Debiased Lasso, red color represents the standard Debiased Lasso and green color corresponds also to the Debiased Lasso estimator, but with the same as our proposed method. Note that the last two methods have almost indistinguishable |Bb| and V.
In Appendix D we also show how the performance changes with the strength of confounding, by gradually decreasing the size of the entries of the loading matrix .
Trimming level.
We investigate here the dependence of the performance on the choice of the trimming threshold for the Trim transform (14), parametrized by the proportion of singular values ρj which we shrink. The spectral transformation used for the initial estimator is fixed to be the default choice of Trim transform with median rule. We fix n = 300 and p = 1, 000 and consider the same setup as in Figure 3. We take to be the ρj-quantile of the set of singular values of the design matrix X, where we vary ρj across the interval [0, 0.9]. When ρj = 0, τ is the maximal singular value, so there is no shrinkage and our estimator reduces to the standard Debiased Lasso (with the initial estimator ). The results are displayed in Figure 6. We can see that Doubly Debiased Lasso is quite insensitive to the trimming level, as long as the number of shrunken singular values is large enough compared to the number of confounding variables q. In the simulation q =3 and the (scaled) absolute bias terms |Bb| and |Bβ| are still small when ρj ≈0.02, corresponding to shrinking 6 largest singular values. We see that the standard deviation decreases as ρj decreases, i.e. as the trimming level τ increases, which matches our efficiency analysis in Section 4.2.1. However, we see that the default choice has decent performance as well. In Appendix D we also explore whether the choice of spectral transformation significantly affects the performance, with a focus on the PCA adjustment, which maps first several singular values to 0, while keeping the others intact.
No confounding bias.
We consider now the same simulation setting as in Figure 3, where we fix n = 500 and vary p, but where in addition we remove the effect of the perturbation b that arises due to the confounding. We generate from the model (2), but then adjust for the confounding bias: Y ← (Y – Xb), where b is the induced coefficient perturbation, as in Equation (3). In this way we still have a perturbed linear model, but where we have enforced b = 0 while keeping the same spiked covariance structure of as in (2). The results can be seen in the top row of Figure 7. We see that Doubly Debiased Lasso still has smaller absolute bias |Bβ|, slightly higher variance and better coverage than the standard Debiased Lasso, even in absence of confounding. The bias term Bb equals 0, since we have put b = 0. We can even observe a decrease in estimation bias for large p, and thus an improvement in the confidence interval coverage. This is due to the fact that X has a spiked covariance structure and trimming the large singular values reduces the correlations between the predictors. This phenomenon is also illustrated in the additional simulations in the Appendix D, where we set q = 0 and put E to have either Toeplitz or equicorrelation covariance structure with varying degree of spikiness (by varying the correlation parameters).
In the bottom row of Figure 7 we repeat the same simulation, but where we set q =0 and take in order to investigate the performance of the method in the setting without confounding, but where the covariance matrix of the predictors is not spiked. We see that there is not much difference in the bias and only a slight increase in the variance of our estimator and thus also there is not much difference in the coverage of the confidence intervals. We conclude that our method can provide certain robustness against dense confounding: if there is such confounding, our proposed method is able to significantly reduce the bias caused by it; on the other hand, if there is no confounding, in comparison to the standard Debiased Lasso, our proposed method still has essentially as good performance, with a small increase in variance.
Measurement error.
We now generate from the measurement error model (4), which can be viewed as a special case of our model (2). The measurement error is generated by q =3 latent variables for 1 ≤ i ≤ n. We fix the number of data points to be n = 500 and vary the number of covariates p from 50 to 1, 000, as in Figure 3. The results are displayed in Figure 8, where we can see a similar pattern as before: Doubly Debiased Lasso decreases the bias at the expense of a slightly inflated variance, which in turn makes the inference much more accurate and the confidence intervals have significantly better coverage.
FIG 8.

(Measurement error) Dependence of the (scaled) absolute bias terms |Bβ| and |Bb| (left), standard deviation V1/2 (middle) and the coverage of the 95% confidence interval (right) on the number of covariates p in the measurement error model (4). The sample size is fixed at n = 500. On the leftmost plot, |Bβ| and |Bb| are denoted by a dashed and a solid line, respectively. Blue color corresponds to the Doubly Debiased Lasso, red color represents the standard Debiased Lasso and green color corresponds also to the Debiased Lasso estimator, but with the same as our proposed method. Note that the last two methods have almost indistinguishable |Bb| and V.
5.2. Real data.
We investigate here the performance of Doubly Debiased Lasso (with a default trimming level of 50%) on a genomic data set. The data are obtained from the GTEx project [41], where the gene expression has been measured postmortem on samples coming from various tissue types. For our purposes, we use fully processed and normalized gene expression data for the skeletal muscle tissue. The gene expression matrix X consists of measurements of expressions of p = 12, 646 protein-coding genes for n = 706 individuals. Genomic data sets are particularly prone to confounding [39, 22, 24], and for our analysis we are provided with q = 65 proxies for hidden confounding, computed with genotyping principal components and PEER factors.
We investigate the associations between the expressions of different genes by regressing one target gene expression Xi on the expression of other genes X−i Since the expression of many genes is very correlated, researchers often use just ~ 1, 000 carefully chosen landmark genes as representatives of the whole gene expression [54]. We will use several such landmark genes as the responses in our analysis.
In Figure 9 we can see a comparison of 95%-confidence intervals that are obtained from Doubly Debiased Lasso and standard Debiased Lasso. For a fixed response landmark gene Xi, we choose 25 predictor genes Xj where j ≠ i such that their corresponding coefficients of the Lasso estimator for regressing Xi on X−i are non-zero. The covariates are ordered according to decreasing absolute values of their estimated Lasso coefficients. We notice that the confidence intervals follow a similar pattern, but that the Doubly Debiased Lasso, besides removing bias due to confounding, is more conservative as the resulting confidence intervals are wider.
FIG 9.

Comparison of 95% confidence intervals obtained by Doubly Debiased Lasso (blue) and Doubly Debiased Lasso (red) for regression of the expression of one target landmark gene on the other gene expressions.
This behavior becomes even more apparent in Figure 10, where we compare all p-values for a fixed response landmark gene. We see that Doubly Debiased Lasso is more conservative and it declares significantly less covariates significant than the standard Debiased Lasso. Even though the p-values of the two methods are correlated (see also Figure 12), we see that it can happen that one method declares a predictor significant, whereas the other does not.
FIG 10.
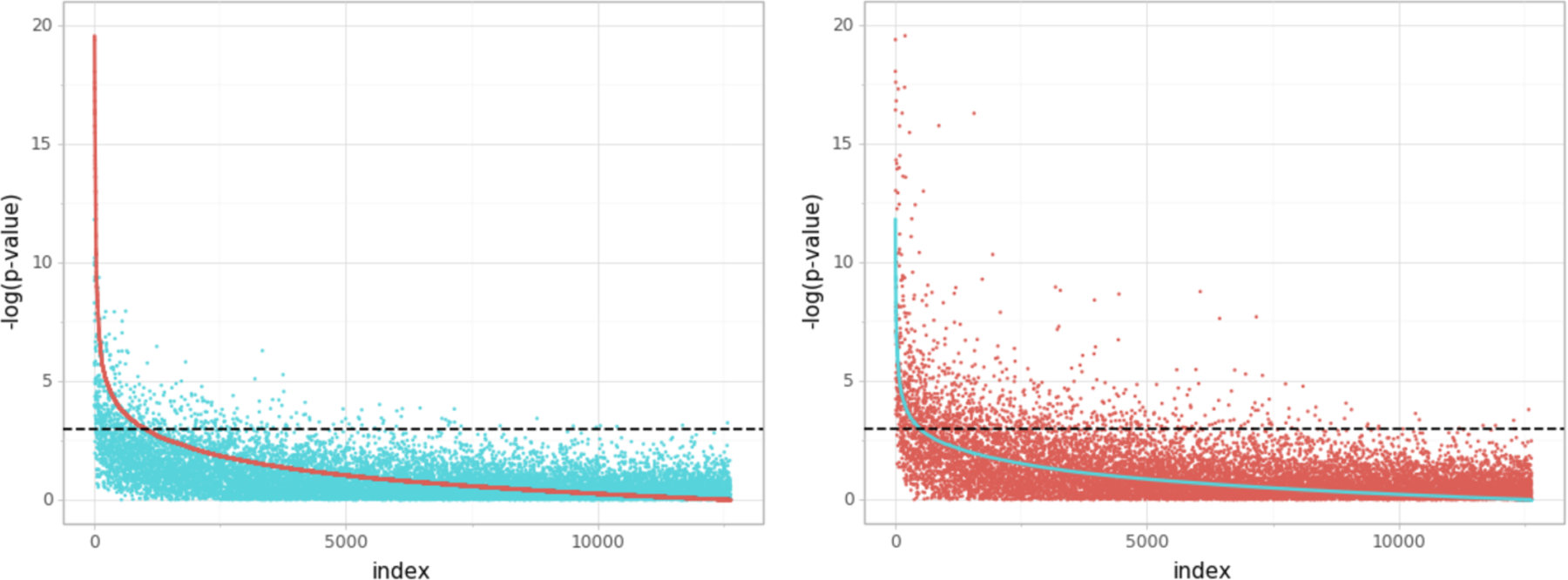
Comparison of p-values for two-sided test of the hypothesis βj = 0, obtained by Doubly Debiased Lasso (red) and Doubly Debiased Lasso (blue) for regression of the expression of one target gene on the other gene expressions. The covariates are ordered by decreasing significance, either estimated by the Debiased Lasso (left) or by the Doubly Debiased Lasso (right). Black dotted line indicates the 5% significance level.
FIG 12.

Comparison of p-values for two-sided test of the hypothesis βj = 0, obtained by Doubly Debiased Lasso and standard Debiased Lasso for regression of the expression of one target gene on the other gene expressions. The points are aggregated over 10 landmark response genes. The p-values are either determined using the original gene expression matrix (left) or the matrix where we have regressed out the given q = 65 confounding proxies (right). Horizontal and vertical black dashed lines indicate the 5% significance level.
Robustness against hidden confounding.
We now adjust the data matrix X by regressing out the q = 65 provided hidden confounding proxies. By regressing out these covariates, we obtain an estimate of the unconfounded gene expression matrix . We compare the estimates for the original gene expression matrix with the estimates obtained from the adjusted matrix.
For a fixed response landmark gene expression Xi, we can determine significance of the predictor genes by considering the p-values. One can perform variable screening by considering the set of most significant genes. For Doubly Debiased Lasso and the standard Lasso we compare the sets of most significant variables determined from the gene expression matrix X and the deconfounded matrix . The difference of the chosen sets is measured by the Jaccard distance. A larger Jaccard distance indicates a larger difference between the chosen sets. The results can be seen in Figure 11. The results are averaged over 10 different response landmark genes. We see that the Doubly Debiased Lasso gives more similar sets for the large model size, indicating that the analysis conclusions obtained by using Doubly Debiased Lasso are more robust in presence of confounding variables. However, for small model size we do not see large gains. In this case the sets produced by any method are quite different, i.e. the Jaccard distance is very large. This indicates that the problem of determining the most significant covariates is quite difficult, since X and differ a lot.
FIG 11.

Comparison of the sets of the most significant covariates chosen based on the original expression matrix X and the deconfounded gene expression matrix , for different cardinalities of the sets (model size). The set differences are measured by Jaccard distance. Red line represents the standard Debiased Lasso method, whereas the blue and green lines denote the Doubly Debiased Lasso that uses ρ = 0.5 and ρ = 0.1 for obtaining the trimming threshold, respectively; see Equation (14).
In Figure 12 we can see the relationship between the p-values obtained by Doubly Debiased Lasso and the standard Debiased Lasso for the original gene expression matrix X and the deconfounded matrix . The p-values are aggregated over 10 response landmark genes and are computed for all possible predictor genes. We can see from the left plot that the Doubly Debiased Lasso is much more conservative for the confounded data. The cloud of points is skewed upwards showing that the standard Debiased Lasso declares many more covariates significant in presence of the hidden confounding. On the other hand, in the right plot we can see that the p-values obtained by the two methods are much more similar for the unconfounded data and the point cloud is significantly less skewed upwards. The remaining deviation from the y = x line might be due to the remaining confounding, not accounted for by regressing out the given confounder proxies.
6. Discussion.
We propose the Doubly Debiased Lasso estimator for hypothesis testing and confidence interval construction for single regression coefficients in high-dimensional settings with “dense” confounding. We present theoretical and empirical justifications and argue that our double debiasing leads to robustness against hidden confounding. In case of no confounding, the price to be paid is (typically) small, with a small increase in variance but even a decrease in estimation bias, in comparison to the standard Debiased Lasso [65]; but there can be substantial gain when “dense" confounding is present.
It is ambitious to claim significance based on observational data. One always needs to make additional assumptions to guard against confounding. We believe that our robust Doubly Debiased Lasso is a clear improvement over the use of standard inferential high-dimensional techniques, yet it is simple and easy to implement, requiring two additional SVDs only, with no additional tuning parameters when using our default choice of trimming ρ = ρj = 50% of the singular values in Equations (14) and (15).
Supplementary Material
Acknowledgements.
We thank Yuansi Chen for providing the code to preprocess the raw data from the GTEx project. We also thank Matthias Löffler for his help and useful discussions about random matrix theory.
The research of Z. Guo was supported in part by the NSF-DMS 1811857, 2015373 and NIH-1R01GM140463–01; Z. Guo also acknowledges financial support for visiting the Institute of Mathematical Research (FIM) at ETH Zurich. D. Ćevid and P. Bühlmann received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 786461).
REFERENCES
- [1].BAI J (2003). Inferential theory for factor models of large dimensions. Econometrica 71 135–171. [Google Scholar]
- [2].BAI J and NG S (2002). Determining the number of factors in approximate factor models. Econometrica 70 191–221. [Google Scholar]
- [3].BELLONI A, CHERNOZHUKOV V, FERNÁNDEZ-VAL I and HANSEN C (2017). Program evaluation and causal inference with high-dimensional data. Econometrica 85 233–298. [Google Scholar]
- [4].BELLONI A, CHERNOZHUKOV V and HANSEN C (2014). Inference on treatment effects after selection among high-dimensional controls. The Review of Economic Studies 81 608–650. [Google Scholar]
- [5].BICKEL PJ, RITOV Y and TSYBAKOV AB (2009). Simultaneous analysis of Lasso and Dantzig selector. The Annals of Statistics 37 1705–1732. [Google Scholar]
- [6].BOEF AG, DEKKERS OM, VANDENBROUCKE JP and LE CESSIE S (2014). Sample size importantly limits the usefulness of instrumental variable methods, depending on instrument strength and level of confounding. Journal of clinical Epidemiology 67 1258–1264. [DOI] [PubMed] [Google Scholar]
- [7].BÜHLMANN P and VAN DE GEER S (2011). Statistics for high-dimensional data: methods, theory and applications Springer Science & Business Media. [Google Scholar]
- [8].BURGESS S, SMALL DS and THOMPSON SG (2017). A review of instrumental variable estimators for Mendelian randomization. Statistical Methods in Medical Research 26 2333–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].CAI T, LIU W and LUO X (2011). A constrained ℓ1 minimization approach to sparse precision matrix estimation. Journal of the American Statistical Association 106 594–607. [Google Scholar]
- [10].CAI TT and GUO Z (2017). Confidence intervals for high-dimensional linear regression: Minimax rates and adaptivity. The Annals of Statistics 45 615–646. [Google Scholar]
- [11].CARROLL RJ, RUPPERT D, STEFANSKI LA and CRAINICEANU CM (2006). Measurement error in nonlinear models: a modern perspective Chapman and Hall/CRC. [Google Scholar]
- [12].ĆEVID D, BÜHLMANN P and MEINSHAUSEN N (2018). Spectral Deconfounding and Perturbed Sparse Linear Models. arXiv preprint arXiv:1811.05352 [Google Scholar]
- [13].CHANDRASEKARAN V, PARRILO PA and WILLSKY AS (2012). Latent variable graphical model selection via convex optimization. The Annals of Statistics 40 1935–1967. [Google Scholar]
- [14].CHERNOZHUKOV V, CHETVERIKOV D, DEMIRER M, DUFLO E, HANSEN C, NEWEY W and ROBINS J (2018). Double/debiased machine learning for treatment and structural parameters: Double/debiased machine learning. The Econometrics Journal 21 C1–C68. [Google Scholar]
- [15].CHERNOZHUKOV V, HANSEN C and SPINDLER M (2015). Valid post-selection and post-regularization inference: An elementary, general approach. Annual Review of Economics 7 649–688. [Google Scholar]
- [16].DEZEURE R, BÜHLMANN P and ZHANG C-H (2017). High-dimensional simultaneous inference with the bootstrap. Test 26 685–719. [Google Scholar]
- [17].FAN J, FAN Y and LV J (2008). High dimensional covariance matrix estimation using a factor model. Journal of Econometrics 147 186–197. [Google Scholar]
- [18].FAN J and LIAO Y (2014). Endogeneity in high dimensions. The Annals of Statistics 42 872–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].FAN J, LIAO Y and MINCHEVA M (2013). Large covariance estimation by thresholding principal orthogonal complements. Journal of the Royal Statistical Society. Series B: Statistical Methodology 75 603–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].FAN J, LIAO Y, WANG W et al. (2016). Projected principal component analysis in factor models. The Annals of Statistics 44 219–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].FARRELL MH (2015). Robust inference on average treatment effects with possibly more covariates than observations. Journal of Econometrics 189 1–23. [Google Scholar]
- [22].GAGNON-BARTSCH JA and SPEED TP (2012). Using control genes to correct for unwanted variation in microarray data. Biostatistics 13 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].GAUTIER E and ROSE C (2011). High-dimensional instrumental variables regression and confidence sets. arXiv preprint arXiv:1105.2454 [Google Scholar]
- [24].GERARD D and STEPHENS M (2020). Empirical Bayes shrinkage and false discovery rate estimation, allowing for unwanted variation. Biostatistics 21 15–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].GOLD D, LEDERER J and TAO J (2020). Inference for high-dimensional instrumental variables regression. Journal of Econometrics 217 79–111. [Google Scholar]
- [26].GÖTZE F and TIKHOMIROV A (2002). Asymptotic distribution of quadratic forms and applications. Journal of Theoretical Probability 15 423–475. [Google Scholar]
- [27].GUERTIN JR, RAHME E and LELORIER J (2016). Performance of the high-dimensional propensity score in adjusting for unmeasured confounders. European journal of Clinical Pharmacology 72 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].GUO Z, KANG H, TONY CAI T and SMALL DS (2018). Confidence intervals for causal effects with invalid instruments by using two-stage hard thresholding with voting. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 80 793–815. [Google Scholar]
- [29].HAGHVERDI L, LUN AT, MORGAN MD and MARIONI JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nature Biotechnology 36 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].HAN C (2008). Detecting invalid instruments using L1-GMM. Economics Letters 101 285–287. [Google Scholar]
- [31].JANKOVA J and VAN DE GEER S (2018). Semiparametric efficiency bounds for high-dimensional models. The Annals of Statistics 46 2336–2359. [Google Scholar]
- [32].JAVANMARD A and MONTANARI A (2014). Confidence intervals and hypothesis testing for high-dimensional regression. The Journal of Machine Learning Research 15 2869–2909. [Google Scholar]
- [33].JOHNSON WE, LI C and RABINOVIC A (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8 118–127. [DOI] [PubMed] [Google Scholar]
- [34].KANG H, ZHANG A, CAI TT and SMALL DS (2016). Instrumental variables estimation with some invalid instruments and its application to Mendelian randomization. Journal of the American Statistical Association 111 132–144. [Google Scholar]
- [35].LAM C, FAN J et al. (2009). Sparsistency and rates of convergence in large covariance matrix estimation. The Annals of Statistics 37 4254–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].LAM C and YAO Q (2012). Factor modeling for high-dimensional time series: inference for the number of factors. The Annals of Statistics 40 694–726. [Google Scholar]
- [37].LAM C, YAO Q and BATHIA N (2011). Estimation of latent factors for high-dimensional time series. Biometrika 98 901–918. [Google Scholar]
- [38].LEEK JT, SCHARPF RB, BRAVO HC, SIMCHA D, LANGMEAD B, JOHNSON WE, GEMAN D, BAGGERLY K and IRIZARRY RA (2010). Tackling the widespread and critical impact of batch effects in high-throughput data. Nature Reviews Genetics 11 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].LEEK JT, STOREY JD et al. (2007). Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLOS Genetics 3 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].LIN W, FENG R and LI H (2015). Regularization methods for high-dimensional instrumental variables regression with an application to genetical genomics. Journal of the American Statistical Association 110 270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].LONSDALE J, THOMAS J, SALVATORE M, PHILLIPS R, LO E, SHAD S, HASZ R, WALTERS G, GARCIA F, YOUNG N et al. (2013). The genotype-tissue expression (GTEx) project. Nature Genetics 45 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].MANGHNANI K, DRAKE A, WAN N and HAQUE I (2018). METCC: METric learning for Confounder Control Making distance matter in high dimensional biological analysis. arXiv preprint arXiv:1812.03188 [Google Scholar]
- [43].MCCARTHY MI, ABECASIS GR, CARDON LR, GOLDSTEIN DB, LITTLE J, IOANNIDIS JP and HIRSCHHORN JN (2008). Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Reviews Genetics 9 356–369. [DOI] [PubMed] [Google Scholar]
- [44].MEINSHAUSEN N and BÜHLMANN P (2006). High-dimensional graphs and variable selection with the lasso. The Annals of Statistics 34 1436–1462. [Google Scholar]
- [45].NEYKOV M, NING Y, LIU JS and LIU H (2018). A unified theory of confidence regions and testing for high-dimensional estimating equations. Statistical Science 33 427–443. [Google Scholar]
- [46].NOVEMBRE J, JOHNSON T, BRYC K, KUTALIK Z, BOYKO AR, AUTON A, INDAP A, KING KS, BERGMANN S and NELSON MR (2008). Genes mirror geography within Europe. Nature 456 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].PEARL J (2009). Causality Cambridge University Press. [Google Scholar]
- [48].PRICE AL, PATTERSON NJ, PLENGE RM, WEINBLATT ME, SHADICK NA and REICH D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 38 904–909. [DOI] [PubMed] [Google Scholar]
- [49].RASKUTTI G, WAINWRIGHT MJ and YU B (2010). Restricted eigenvalue properties for correlated Gaussian designs. The Journal of Machine Learning Research 11 2241–2259. [Google Scholar]
- [50].REID S, TIBSHIRANI R and FRIEDMAN J (2016). A study of error variance estimation in lasso regression. Statistica Sinica 35–67. [Google Scholar]
- [51].RUDELSON M and VERSHYNIN R (2009). Smallest singular value of a random rectangular matrix. Communications on Pure and Applied Mathematics: A Journal Issued by the Courant Institute of Mathematical Sciences 62 1707–1739. [Google Scholar]
- [52].RUDELSON M and VERSHYNIN R (2013). Hanson-Wright inequality and sub-gaussian concentration. Electronic Communications in Probability 18 1–9. [Google Scholar]
- [53].SHAH RD, FROT B, THANEI G-A and MEINSHAUSEN N (2020). Right singular vector projection graphs: fast high-dimensional covariance matrix estimation under latent confounding. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 82 361–389. [Google Scholar]
- [54].SUBRAMANIAN A, NARAYAN R, CORSELLO SM, PECK DD, NATOLI TE, LU X, GOULD J, DAVIS JF, TUBELLI AA and ASIEDU JK (2017). A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].SUN Y, ZHANG NR, OWEN AB et al. (2012). Multiple hypothesis testing adjusted for latent variables, with an application to the AGEMAP gene expression data. The Annals of Applied Statistics 6 1664–1688. [Google Scholar]
- [56].VAN DE GEER S, BÜHLMANN P, RITOV Y and DEZEURE R (2014). On asymptotically optimal confidence regions and tests for high-dimensional models. The Annals of Statistics 42 1166–1202. [Google Scholar]
- [57].VERSHYNIN R (2012). Introduction to the non-asymptotic analysis of random matrices. In Compressed sensing: theory and applications (Eldar Y and Kutyniok G, eds.) 210–268. Cambridge University Press. [Google Scholar]
- [58].WANG J, ZHAO Q, HASTIE T and OWEN AB (2017). Confounder adjustment in multiple hypothesis testing. Annals of statistics 45 1863–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].WANG W, FAN J et al. (2017). Asymptotics of empirical eigenstructure for high dimensional spiked covariance. The Annals of Statistics 45 1342–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].WANG Y and BLEI DM (2019). The blessings of multiple causes. Journal of the American Statistical Association 114 1574–1596. [Google Scholar]
- [61].WINDMEIJER F, FARBMACHER H, DAVIES N and DAVEY SMITH G (2019). On the use of the lasso for instrumental variables estimation with some invalid instruments. Journal of the American Statistical Association 114 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].WOOLDRIDGE JM (2010). Econometric analysis of cross section and panel data MIT press. [Google Scholar]
- [63].YASKOV P (2016). A short proof of the Marchenko–Pastur theorem. Comptes Rendus Mathematique 354 319–322. [Google Scholar]
- [64].YUAN M (2010). High dimensional inverse covariance matrix estimation via linear programming. Journal of Machine Learning Research 11 2261–2286. [Google Scholar]
- [65].ZHANG C-H and ZHANG SS (2014). Confidence intervals for low dimensional parameters in high dimensional linear models. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 76 217–242. [Google Scholar]
- [66].ZHOU S (2009). Restricted eigenvalue conditions on subgaussian random matrices. arXiv preprint arXiv:0912.4045 [Google Scholar]
- [67].ZHU Y (2018). Sparse linear models and l1-regularized 2SLS with high-dimensional endogenous regressors and instruments. Journal of Econometrics 202 196–213. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


