Abstract
Group 1 capsules of Escherichia coli are similar to the capsules produced by strains of Klebsiella spp. in terms of structure, genetics, and patterns of expression. The striking similarities between the capsules of these organisms prompted a more detailed investigation of the cps loci encoding group 1 capsule synthesis. Six strains of K. pneumoniae and 12 strains of E. coli were examined. PCR analysis showed that the clusters in these strains are conserved in their chromosomal locations. A highly conserved block of four genes, orfX-wza-wzb-wzc, was identified in all of the strains. The wza and wzc genes are required for translocation and surface assembly of E. coli K30 antigen. The conservation of these genes points to a common pathway for capsule translocation. A characteristic JUMPstart sequence was identified upstream of each cluster which may function in conjunction with RfaH to inhibit transcriptional termination at a stem-loop structure found immediately downstream of the “translocation-surface assembly” region of the cluster. Interestingly, the sequence upstream of the cps clusters in five E. coli strains and one Klebsiella strain indicated the presence of IS elements. We propose that the IS elements were responsible for the transfer of the cps locus between organisms and that they may continue to mediate recombination between strains.
Escherichia coli produces a wide variety of capsular polysaccharides termed K antigens. These polymers can vary in composition, linkage specificity, and substitution, allowing for diversity among strains. The major capsule groups, traditionally designated groups I and II, were defined by serological properties as well as by the location of the K-antigen biosynthesis gene cluster, the polymer structure, and the expression patterns (23). A new and expanded classification system has recently been proposed (51). This is based on genetic and biochemical (assembly pathway) data and proposes four capsule groups. The new group 1 accommodates a subset of K antigens formerly designated group IA (23). Of the 66 structurally defined K antigens in E. coli, 16 show characteristics typical of group 1 capsules (23). Research in this laboratory focuses on the group 1 K antigens of E. coli and the structurally related capsules found in Klebsiella spp.
In E. coli, group 1 K antigens are produced in two distinct forms: a low-molecular-weight form (KLPS), which comprises K oligosaccharides linked to lipid A-core and resembles a lipopolysaccharide (LPS)-linked O antigen, and a high-molecular-weight unlinked form, capsular K antigen, which is associated with the cell surface. A precise mechanism of attachment for capsular K antigen has not been described, but it is known that LPS is not involved (30). In Klebsiella, only the capsular form of K antigen is produced. The absence of the KLPS form in Klebsiella may reflect differences in the LPS core structure and/or the ligase enzyme (21) that attaches polysaccharides to the lipid A-core acceptor.
The genes involved in synthesis and transport of both KLPS and capsular K antigen in E. coli are encoded at a locus called cps. A prototype group 1 capsule cluster from E. coli E69 (O9a:K30) (cpsECK30) has been analyzed (17). The 16-kb locus contains genes required for K30 polymerization and translocation. The K30 antigen is synthesized via the Wzy-dependent polymerization pathway, which has been described for the biosynthesis of certain O antigens (48). In brief, repeat units are made on the cytoplasmic face of the plasma membrane by the action of glycosyltransferases, which transfer residues to a lipid (undecaprenol pyrophosphate)-linked biosynthetic intermediate. The lipid-linked repeat units are moved to the periplasmic face of the membrane by the Wzx protein, where they are polymerized by the Wzy enzyme. To form KLPS, short K30-antigenic oligosaccharides are ligated to lipid A-core by the ligase, WaaL. The capsular form of the K30 antigen is polymerized by the same pathway; however, surface expression is via an LPS-independent pathway that requires the products of wza and wzc (see below). These gene products are not required for the assembly of LPS-linked O antigens, and they provide features that distinguish gene clusters for biosynthesis of O and K antigens.
Comparative analysis of Klebsiella pneumoniae K2 (cpsKPK2) (2) (GenBank accession no. D21242) and E. coli K30 (cpsECK30) (17) (GenBank accession no. AF104912) indicates a shared biosynthetic pathway. The objective of this study was to investigate the precise relationships between capsule gene clusters in a number of E. coli and Klebsiella strains.
A conserved region of genes required for translocation and polymerization of group 1 K antigens.
Lipid-linked intermediates are polymerized to form high-molecular-weight K30 polysaccharide via the Wzy-dependent polymerization pathway and then translocated through the outer membrane as an unlinked polymer. Surface assembly of capsular group 1 K antigen minimally requires two additional genes from the capsule locus, wza and wzc (17). Wza, Wzb, and Wzc are encoded in the “translocation-surface assembly” region of the K30 cluster and are thought to be an outer membrane lipoprotein, a cytoplasmic phosphatase, and an ATP-binding protein, respectively. Recent work with Acinetobacter johnsonii has shown that a Wzb homologue, Ptp, is capable of dephosphorylating a Wzc homologue, Ptk (20). Similar genes have been described in diverse systems including E. coli K-12 (wza, wzb, and wzc) (44), Klebsiella K2 (orf4, orf5, and orf6) (2), and Erwinia amylovora (amsH, amsI, and amsA) (9), where they are believed to be involved in the production of colanic acid, K2 capsular polysaccharide (CPS), and amylovoran, respectively. Wza and Wzc homologues are found in a variety of bacteria that produce CPS and extracellular polysaccharide (38), leading to the conclusion that these represent a common translocation-surface assembly pathway for cell surface polysaccharides. Although these proteins have been established to function in surface expression of the K30 capsular antigen (17), their precise role in the process has yet to be determined.
The region associated with CPS translocation-surface assembly was examined in detail. Chromosomal DNA was prepared from the strains shown in Table 1. To reduce the expression of CPS, bacteria were grown in Luria broth (33) at 42°C. To isolate chromosomal DNA, bacteria from a 5-ml overnight culture were collected by centrifugation and resuspended in 1.5 ml of a lysis solution (50 mM sodium chloride, 2% sodium dodecyl sulfate, 300 mg of proteinase K per ml). The suspension was incubated at 42°C until clear. Next, 250 μl of 5 M sodium chloride and 200 μl of 10% hexadecyltrimethylammonium bromide in 0.7 M sodium chloride were added, followed by a 30-min incubation at 65°C. The sample was then subjected to two phenol-chloroform-isoamyl alcohol (25:24:1) extractions and precipitated with 5% 5 M sodium chloride and 2 volumes of absolute ethanol. The DNA pellet was washed with 70% ethanol and resuspended in 100 μl of sterile water. Finally, the DNA was treated with RNase and subjected to a chloroform-isoamyl alcohol (24:1) extraction.
TABLE 1.
Bacterial strains used in this study
| Strain | Serotype | Source |
|---|---|---|
| E. coli | ||
| A295b | O8:K42:H− | F. Ørskov |
| Bi161-42 | O9:K29:H− | F. Ørskov |
| E56b | O8:K27:H− | F. Ørskov |
| E69 | O9a:K30:H12 | F. Ørskov |
| E75 | O9:K34:H− | F. Ørskov |
| K14a | O9ab:K28:H− | F. Ørskov |
| N24c | O9a:K55:H− | F. Ørskov |
| Su3973-41 | O9:K31:H− | F. Ørskov |
| 2146 | O9:K26:H− | B. Jann |
| 2150 | O9:K37:H− | B. Jann |
| 2151 | O9:K39:H9 | B. Jann |
| 2178 | O8:K42:H− | B. Jann |
| Klebsiella | ||
| A5054 | O1:K1 | F. Ørskov |
| B5055 | O1:K2 | F. Ørskov |
| 708 | O12:K80 | F. Ørskov |
| 889/50 | O1:K20 | F. Ørskov |
| 1702 | O4:K42 | F. Ørskov |
| 6613 | O2:K27 | F. Ørskov |
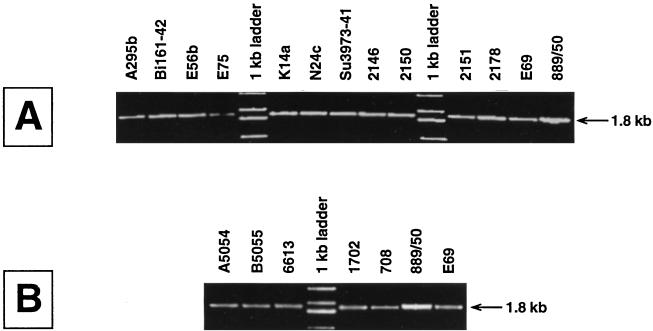
The region between wza and wzc was amplified by PCR. PCRs were performed with Pwo or Expand Long DNA polymerase (Boehringer Mannheim) in a Perkin-Elmer GeneAmp PCR System 2400 thermocycler. The primers used for DNA amplification are listed in Table 2 and shown in Fig. 1. PCR products identical in size were obtained with primers JD89 and JD109 and chromosomal DNA from strains representing 6 K serotypes of Klebsiella and 12 K serotypes of E. coli (Fig. 2). The sizes of the products were consistent with those predicted from the cpsECK30 and cpsKPK2 sequence data.
TABLE 2.
Primers used for PCR amplification of chromosomal DNA
| Primer | Location | Sequencea |
|---|---|---|
| JD109 | wza | 5′-AGCGACTACGATTTGGATAAGC |
| JD89 | wzc | 5′-ACGGGCAATAACAGGAAAATA |
| JD90 | wcaO | 5′-TATGTGGGGCGATTGACTCC |
| JD53 | wzx | 5′-TTTCCGAAACACGCTTAAAA |
| JD95 | orfX | 5′-TGCGCTTCCACATACCAGTTA |
| JD99 | orfX | 5′-GAAAGCAAGCCCAATGTCAC |
| GALF1 | galF | 5′-GGGCGATCTCTCCGAATACTC |
FIG. 1.
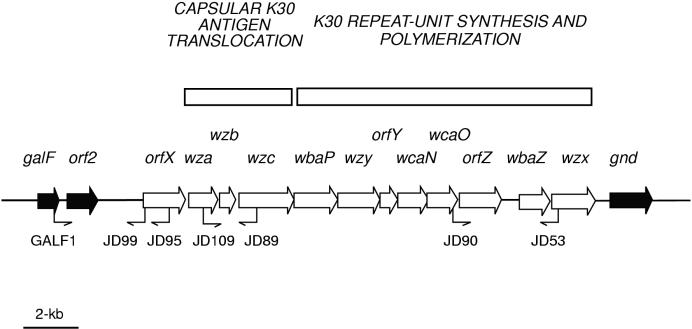
Organization of the cpsECK30 locus, which is located between galF and gnd on the E. coli E69 chromosome (17). Components of the Wzy-dependent polymerization pathway (wzx and wzy) are found in the region dedicated to K30 repeat unit synthesis and polymerization, along with the necessary glycosyltransferases, wbaP, wcaN, wcaO, and wbaZ. Genes involved in the translocation of capsular K30 antigen, wza and wzc, are near the start of the cluster. The functions of orfX, orfY, and orfZ are unknown. Primers used for PCR amplification of selected regions are indicated, and the sequences are given in Table 2.
FIG. 2.
PCR analysis of the wza-to-wzc region from different group 1 K serotypes of E. coli (A) and Klebsiella (B). For reference, K. pneumoniae K20 strain 889/50 is also shown in panel A and E. coli K30 strain E69 is shown in panel B. PCR products identical in size (1.8 kb) were obtained for all of the strains tested, indicating a conservation of gene order between clusters.
For detailed analysis, selected PCR fragments were purified with Qiagen nucleotide removal columns and the region was sequenced at the Guelph Molecular Supercentre (University of Guelph, Guelph, Ontario, Canada). Analyses revealed that the wza-to-wzc regions in E. coli A295b (GenBank accession no. AF118245), E75 (GenBank accession no. AF118246), N24c (GenBank accession no. AF118247), 2151 (GenBank accession no. AF118248), and Bi161-42 (GenBank accession no. AF118249) had 99.5% identity to E. coli E69 at the nucleotide level and that the equivalent regions in cpsKPK2 and cpsECK30 were 72% identical at the nucleotide level.
The observation that group 1 capsule clusters from E. coli and Klebsiella are organized with a conserved block of translocation-surface assembly genes is reminiscent of the modular structure of gene clusters required for the manufacture of group 2 capsules in E. coli. Group 2 (kps) clusters comprise three regions (51). Region 1 encodes proteins necessary for translocation of polysaccharide through the periplasm and across the outer membrane, as appears to be the case with the initial genes (wza to wzc) of the group 1 cps clusters. The serotype-specific central region (region 2) is responsible for biosynthesis and polymerization of oligosaccharide repeat units. This is equivalent to the genes downstream of wzc in the group 1 capsule clusters. Region 3 contains genes encoding the subunits for an ATP-binding cassette transporter that is required for capsule export across the cytoplasmic membrane (8). The region 3 components are not required in Wzy-dependent polysaccharides, such as the group 1 capsules (48, 51). In the group 2 clusters, regions 1 and 3 are conserved in strains that produce structurally distinct capsules and the gene products are functionally interchangeable between these strains (51). Although functional identity has not been formally shown for the translocation-surface assembly region of group 1 capsules, the high degree of conservation seems to indicate that this would be the case. Some conserved feature of the group 1 K antigens or a component in their assembly must therefore be recognized for the translocation-surface assembly processes to be completed.
orfX is a unique component of group 1 K antigen cps gene clusters.
Although homologues of wza, wzb, and wzc are found in other systems, including the cpsK-12 cluster for colanic acid biosynthesis, orfX homologues have been found only in cpsECK30 and cpsKPK2 (where it is designated orf3) (2, 17). In both cases, orfX represents the first gene in the cluster. PCRs were performed with primers designed to amplify the region between orfX (JD95 for Klebsiella and JD99 for E. coli) and a gene upstream of the K-antigen cluster, galF (GALF1) (Table 2; Fig. 1). The sizes of the fragments obtained varied considerably due to polymorphism upstream of cps (see below). However, sequence data showed orfX to be conserved in position and virtually identical at the nucleotide level in all of the E. coli strains (E56b, Bi161-42, A295b, N24c, and E75) and Klebsiella strains (A5054, 889/50, 6613, and 708) investigated (data not shown). In E. coli K30, orfX mutants do not appear to be impaired in translocation-surface assembly of the capsular K antigen (17) and the precise role of orfX remains unclear. However, the high degree of conservation in orfX homologues would indicate that it is important in the production of group 1 capsules.
Identical cps gene clusters in E. coli K30 and K. pneumoniae K20.
There are notable structural similarities between group 1 K antigens in E. coli and K. pneumoniae (structures are available online in the Complex Carbohydrate Structure Database [14a]). In the example of Klebsiella K20 (11) and E. coli K30 (10), the capsular antigen structures are identical. The relationship between these two gene clusters was also examined by PCR and sequencing. Similar to the E. coli strains analyzed above, the wza-to-wzc region in these strains was shown to be 99.5% identical (Klebsiella K20 GenBank accession no. AF118250).
Genes downstream of wza, wzb, and wzc are involved in repeat-unit biosynthesis and polymerization. Genes in this region, in particular, the glycosyltransferases, will vary with the structure of the polymer being produced by any given strain. Among group 1 capsules, this region has been described in detail for E. coli K30 (17) and Klebsiella K2 (2), and although both encode the same classes of gene products, they exhibit only low levels of similarity. PCR amplification of the region from wcaO to wzx with primers JD90 and JD53 (Table 2; Fig. 1) yields products identical in size (3.6 kb) for both E. coli K30 and K. pneumoniae K20 (data not shown). At the nucleotide level, the products are 99% identical over the 550 bp sequenced from each end. The high degree of conservation at the nucleotide level for both the wza-wzc and the wcaO-to-wzx regions may indicate the transfer of cps gene clusters between K. pneumoniae and E. coli (see below).
Conservation of regulatory regions upstream of cps.
Gene clusters for bacterial polysaccharides are characteristically preceded by a 39-bp JUMPstart (for “just upstream of many polysaccharide starts”) element (22). The JUMPstart element has also been identified upstream of gene clusters involved in F conjugation pilus assembly and hemolysin toxin secretion (reviewed in reference 5). A second element, ops (for “operon polarity suppressor”) is an 8-bp motif located within the JUMPstart sequence (35). Both elements are believed to play a role in transcriptional antitermination of the gene clusters (5, 31, 35). Their role has been described in regulation of E. coli group 2 K-antigen (kps [42, 43]) and LPS O-antigen (rfb [31]) biosynthesis.
The role of the JUMPstart element has been best characterized for the hemolysin operon, where it has been shown to control operon polarity but not transcript stability (5, 35), in concert with a NusG homologue, RfaH (4, 28, 29). The element can function over long distances (2 kb); however, it does so only when present on the nascent transcript (35). In one possible model, the JUMPstart sequence functions by facilitating the formation of a number of stem-loop structures on the mRNA during transcript elongation (31). This region recruits RfaH and potentially other proteins. Binding of RfaH to stem-loop III may inhibit the formation of the other stem-loops, which are thought to induce premature termination when present.
To investigate the conservation of these regulatory elements, regions immediately upstream of the group 1 K-antigen clusters were amplified and sequenced. A conserved JUMPstart element was present in both E. coli strains (E56b, Bi161-42, A295b, N24c, and E75) and Klebsiella strains (A5054, 889/50, 6613, and 708) (Fig. 3A). This finding suggests that group 1 capsule clusters are subject to transcriptional control via antitermination. Interestingly, the cps gene clusters share a feature noted upstream of the O-antigen biosynthesis region of E. coli O7 (wb*) (31), i.e., the presence of two ops elements.
FIG. 3.
(A) JUMPstart sequences from polysaccharide gene clusters directing the synthesis of amylovoran (ams) (27), colanic acid (cps) (45), O7 antigen (wb*) (31), and the E. coli and Klebsiella group 1 capsule clusters examined in this study. Variations from the E. coli K30 sequence are highlighted in boldface. The two ops elements found in the E. coli O7 JUMPstart sequence are underlined. (B) Stem-loop structures which have been identified in cpsECK30 and cpsKPK2. They are located immediately downstream of wzc and may function as transcription terminators.
In the hemolysin system, antitermination is required to avoid extreme operon polarity in a situation where the structural gene for the toxin is separated from those required for toxin maturation and export by a stem-loop structure (5). Analysis of the available cpsECK30 and cpsKPK2 sequences identified a stem-loop structure in the intergenic region between wzc and the initial gene of the repeat-unit synthesis region (Fig. 3B). This potentially provides a strong transcriptional terminator, thereby allowing differential expression of structural components for capsule translocation and the highly active enzymes involved in polymer synthesis. Equivalent stem-loop structures are also predicted from the nucleotide sequence downstream of the wzc homologue in the gene clusters for amylovoran (9) and colanic acid (44) (data not shown). Differential expression of genes required for translocation-surface assembly and synthesis is known to occur in the E. coli group 2 kps clusters. In these systems, regions 2 and 3 are organized into one transcriptional unit under the control of the region 3 promoter. Region 2 genes rely on transcriptional antitermination by RfaH to avoid operon polarity problems (42). The kps region 1 is not regulated via antitermination but has been shown to be thermoregulated (12, 40). In addition, the first gene of region 1, kpsF, plays a poorly understood role in regulation (12). It remains to be established whether the product of orfX plays a similar role in expression of group 1 K antigens.
Group 1 K antigens are known to be regulated by the Rcs (for “regulator of capsule synthesis”) system in both E. coli (24, 25) and Klebsiella (1, 32, 47). This two-component regulatory system is best characterized for colanic acid production in E. coli K-12 (19); however, essentially identical systems operate in E. amylovora (6, 7, 14, 27). It is believed that the RcsC protein senses an environmental signal and, along with a second protein, RcsF, modulates the activity of RcsB through phosphorylation. RcsB can interact with a Lon protease-sensitive protein, RcsA, and upregulate cps transcription. Both RcsA and RcsB have helix-turn-helix DNA-binding motifs and bind to the promoter region of the ams cluster for amylovoran production in E. amylovora (27). A potential binding site has also been identified upstream of the colanic acid cluster based on the titration of regulatory proteins by using promoter DNA and sequence homology to the E. amylovora binding site (27, 45). The environmental signal sensed by the Rcs system is uncharacterized but may involve membrane perturbations (13, 18, 37) or osmotic stress (3, 18, 41).
Although group 1 capsules, colanic acid, and amylovoran are all Rcs regulated, there are some important differences in their expression patterns. Most notable is the observation that colanic acid and amylovoran are optimally produced at 20°C and are not manufactured at 37°C. This is not surprising for colanic acid since this polymer is not a virulence determinant (39) and its function may be more important in environments outside the host (18, 41). E. amylovora is a plant pathogen associated with infections at environmental temperatures. However, group 1 capsules are virulence determinants and are produced at 37°C (49). The ability to express cps gene products at 37°C may be due to altered interactions of an RcsA-RcsB dimer with the cps promoter or to the involvement of additional (as yet uncharacterized) regulatory proteins. The Rcs proteins themselves are highly conserved in E. coli K30 and E. coli K-12 (24, 25), suggesting that they are unlikely to determine the different patterns of expression. The regions upstream of cps in E. coli and K. pneumoniae strains with group 1 K antigens lack the published RcsA-RcsB-binding sequences (27). Therefore, although the Rcs system may function to regulate group 1 K antigens, details of this interaction may differ considerably from those for the colanic acid and amylovoran systems.
Analysis of the Klebsiella K2 cps upstream region resulted in the identification of a partial sequence of a putative ς54 promoter (2). This is conserved in all the E. coli and K. pneumoniae strains examined here. However, this putative promoter lies downstream of the JUMPstart element and thus could not operate in situations where antitermination by RfaH is required. The precise physiological significance of this “promoter” is therefore unclear.
Possible lateral transfer of group 1 cps genes.
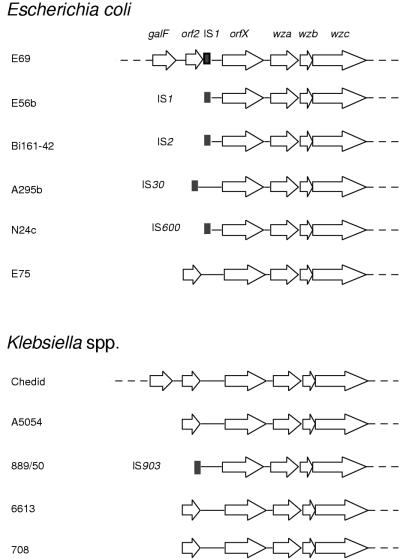
The data presented above demonstrates that the K-antigen gene clusters of E. coli and Klebsiella are highly conserved in organization and in cps nucleotide sequence. This is consistent with the possibility of lateral transfer of group 1 capsule gene clusters between these organisms. Further evidence in support of this contention was obtained by the identification of IS elements upstream of the cps gene clusters. In E. coli K30, a partial IS1 element (250 bp) truncates an adjacent gene, orf2. orf2 is also found immediately upstream of the cpsKPK2 cluster (2) but is absent in E. coli K-12. orf2 is not essential for K-antigen production in K. pneumoniae (2). A survey of other strains showed that many of the E. coli cps clusters are flanked by IS sequences (Fig. 4). Of the six E. coli strains examined, five contained IS elements. The type of IS element present is highly variable, with only IS1 being identified in more than one strain (E. coli E69 and E56b). In contrast, only one of the five K. pneumoniae strains had an IS element. The location of the IS sequences seems to be quite highly conserved, with only two of six strains (E. coli A295b and K. pneumoniae 889/50) showing slight variation.
FIG. 4.
Diagrammatic representation of the region upstream of cps in the E. coli and K. pneumoniae strains investigated. The regions for E. coli E69 (17) and K. pneumoniae Chedid (2) are based on published data. The location and type of IS element identified, if any, is indicated. The putative ς54 promoters and conserved JUMPstart elements are located between the IS elements (or orf2) and orfX, the first gene of each cluster.
The presence of IS elements adjacent to most of the tested E. coli cps clusters and their absence in all but one of the cps regions examined in Klebsiella strains suggest that IS elements may have mediated an initial transfer of the cluster to E. coli from Klebsiella and could continue to be important for the exchange of genes encoding different capsule types between strains. Support for this hypothesis can be found in an analysis of gnd (6-phosphogluconate dehydrogenase) alleles from a variety of E. coli and Klebsiella isolates. The cps genes map near his and gnd in E. coli, and Nelson and Selander (34) have suggested that diversity in gnd represents a surprisingly high degree of recombination. This was attributed to cotransfer with adjacent loci (primarily rfb) whose activities are subject to diversifying selection because of the host immune response. Nelson and Selander also argued for lateral transfer of gnd genes from Klebsiella to E. coli. E. coli strains with the 16 known group 1 K antigens have a limited array of LPS O antigens (23); of the >170 O serotypes, only O9, O9a, O8, O20, and O101 are represented. Klebsiella strains have 77 different K antigens (36) but fewer than 10 structurally distinct O antigens (26, 46). Notably, some O-antigen structures are shared, and lateral transfer of the O3 gene cluster to E. coli has been proposed (46). Collectively, these data suggest that transfer of a large region of DNA including the rfb and cps loci may have occurred. In such a scenario, the extended region between galF and his (orf2-cps-ugd-rfb-gnd) from Klebsiella would replace the “typical” E. coli region (cps [colanic acid]-rfb-gnd-ugd-wzz). This is consistent with our previous analysis of the regions surrounding gnd that confirmed the lack of wzz in E. coli strains with group 1 capsules (15, 16). Such organization also explains why expression of colanic acid and expression of a group 1 capsule are mutually exclusive. In previous work, we proposed that the cpsK-12 and cpsECK30 systems are allelic (25, 50). From the differences in organization (the presence of orfX), altered upstream sequences, and the IS/orf2 region missing in E. coli K-12, these can no longer be considered alleles. Colanic acid is therefore not simply a widespread serotype of group 1 K antigen and should not be included as such.
The simple conclusion that all E. coli group 1 capsules have arisen by lateral transfer of DNA from Klebsiella is complicated by several observations. First, not all E. coli group 1 capsules have structurally identical counterparts in Klebsiella, although this could reflect further gene transfer and recombination events within cps after the initial transfer, thereby resulting in the production of novel structures. However, of the strains examined here, one E. coli isolate (E75) lacks an IS element immediately upstream of cps and only one Klebsiella strain (889/50) has an IS element (IS903). These exceptions suggest that the events that resulted in group 1 capsule diversity in E. coli and Klebsiella are more complicated. As noted above, the E. coli K30 and K. pneumoniae K20 (889/50) cps gene clusters appear to be identical, and there are two possible explanations for this. One is that the cluster was mobilized to these strains from a common (unknown) source by a process involving different IS elements. The other is that the gene exchanges between Klebsiella and E. coli are possibly not restricted to unidirectional lateral transfer events. There is currently no data to resolve these possibilities.
Acknowledgments
This work was supported by funding to C.W. from the Medical Research Council of Canada (MT-9623). A.R. and J.D. are recipients of NSERC PGS-B scholarships.
ADDENDUM IN PROOF
While this paper was under review, further analysis of the RcsA-RcsB binding site in Escherichia coli K-12 was reported (W. Ebel and J. E. Trempy, J. Bacteriol. 181:577–584, 1999). A conserved motif (the RcsA box) was identified upstream of both cps and rcsA in E. coli K-12. This motif is not present in the regions upstream of the group 1 capsule gene clusters reported here. The binding site for RcsA-RcsB in Erwinia and related bacteria has also been further elucidated (M. Wehland, C. Kiecker, D. L. Coplin, O. Kelm, W. Saenger, and F. Bernhard, J. Biol. Chem. 274:3300–3307, 1999).
REFERENCES
- 1.Allen P, Hart C A, Saunders J R. Isolation from Klebsiella and characterization of two rcs genes that activate colanic acid capsular biosynthesis in Escherichia coli. J Gen Microbiol. 1987;133:331–340. doi: 10.1099/00221287-133-2-331. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 1995;177:1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arricau N, Hermant D, Waxin J, Ecobichon C, Duffey P S, Popoff M Y. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol Microbiol. 1998;29:835–850. doi: 10.1046/j.1365-2958.1998.00976.x. [DOI] [PubMed] [Google Scholar]
- 4.Bailey M J A, Hughes C, Koronakis V. Increased distal gene transcription by the elongation factor RfaH, a specialized homologue of NusG. Mol Microbiol. 1996;22:729–737. doi: 10.1046/j.1365-2958.1996.d01-1726.x. [DOI] [PubMed] [Google Scholar]
- 5.Bailey M J A, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 6.Bereswill S, Geider K. Characterization of the rcsB gene from Erwinia amylovora and its influence on exopolysaccharide synthesis and virulence of the fire blight pathogen. J Bacteriol. 1997;179:1354–1361. doi: 10.1128/jb.179.4.1354-1361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernhard F, Poetter K, Geider K, Coplin D L. The rcsA gene from Erwinia amylovora: identification, nucleotide sequence, and regulation of exopolysaccharide biosynthesis. Mol Plant-Microbe Interact. 1990;3:429–437. doi: 10.1094/mpmi-3-429. [DOI] [PubMed] [Google Scholar]
- 8.Bliss J M, Silver R P. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli K1. Mol Microbiol. 1996;21:221–231. doi: 10.1046/j.1365-2958.1996.6461357.x. [DOI] [PubMed] [Google Scholar]
- 9.Bugert P, Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty A K, Friebolin H, Stirm S. Primary structure of the Escherichia coli serotype K30 capsular polysaccharide. J Bacteriol. 1980;141:971–972. doi: 10.1128/jb.141.2.971-972.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choy Y-M, Dutton G G S. Structure of the capsular polysaccharide of Klebsiella K-type 20. Can J Chem. 1973;51:3015–3020. [Google Scholar]
- 12.Cieslewicz M, Vimr E. Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Mol Microbiol. 1997;26:237–249. doi: 10.1046/j.1365-2958.1997.5651942.x. [DOI] [PubMed] [Google Scholar]
- 13.Clavel T, Lazzaroni J C, Vianney A, Portalier R. Expression of the tolQRA genes of Escherichia coli K-12 is controlled by the RcsC sensor protein involved in capsule synthesis. Mol Microbiol. 1996;19:19–25. doi: 10.1046/j.1365-2958.1996.343880.x. [DOI] [PubMed] [Google Scholar]
- 14.Coleman M, Pearce R, Hitchin E, Busfield F, Mansfield J W, Roberts I S. Molecular cloning, expression and nucleotide sequence of the rcsA gene of Erwinia amylovora, encoding a positive regulator of capsule expression: evidence for a family of related capsule activator proteins. J Gen Microbiol. 1990;136:1799–1806. doi: 10.1099/00221287-136-9-1799. [DOI] [PubMed] [Google Scholar]
- 14a.Complex Carbohydrate Structure Database Website. 23 January 1999, revision date. http://www.ccrc.uga.edu.
- 15.Dodgson C, Amor P, Whitfield C. Distribution of the rol gene encoding the regulator of lipopolysaccharide O-chain length in Escherichia coli and its influence on the expression of group I capsular antigens. J Bacteriol. 1996;178:1895–1902. doi: 10.1128/jb.178.7.1895-1902.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummelsmith J, Amor P A, Whitfield C. Polymorphism, duplication and IS1-mediated rearrangement in the chromosomal his-rfb-gnd region of Escherichia coli strains with group IA capsular K antigens. J Bacteriol. 1997;179:3232–3238. doi: 10.1128/jb.179.10.3232-3238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummelsmith, J., and C. Whitfield. Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30). Mol. Microbiol., in press. [DOI] [PubMed]
- 18.Ebel W, Vaughn G J, Peters III H K, Trempey J E. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J Bacteriol. 1997;179:6858–6861. doi: 10.1128/jb.179.21.6858-6861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottesman S. Regulation of capsule synthesis: modification of the two-component paradigm by an accessory unstable regulator. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 253–262. [Google Scholar]
- 20.Grangeasse C, Doublet P, Vincent C, Vaganay E, Riberty M, Duclos B, Cozzone A J. Functional characterization of the low-molecular-mass phosphotyrosine-protein phosphatase of Acinetobacter johnsonii. J Mol Biol. 1998;278:339–347. doi: 10.1006/jmbi.1998.1650. [DOI] [PubMed] [Google Scholar]
- 21.Heinrichs D E, Yethon J A, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 22.Hobbs M, Reeves P R. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 23.Jann K, Jann B. Capsules of Escherichia coli. In: Sussman M, editor. Escherichia coli: mechanisms of virulence. Cambridge, United Kingdom: Cambridge University Press; 1997. pp. 113–143. [Google Scholar]
- 24.Jayaratne P, Keenleyside W J, MacLachlan P R, Dodgson C, Whitfield C. Characterization of rcsB and rcsC from Escherichia coli O9:K30:H12 and examination of the role of the rcs regulatory system in expression of group I capsular polysaccharides. J Bacteriol. 1993;175:5384–5394. doi: 10.1128/jb.175.17.5384-5394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keenleyside W J, Jayaratne P, MacLachlan P R, Whitfield C. The rcsA gene of Escherichia coli O9:K30:H12 is involved in the expression of the serotype-specific group I K (capsular) antigen. J Bacteriol. 1992;174:8–16. doi: 10.1128/jb.174.1.8-16.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly R F, MacLean L L, Perry M B, Whitfield C. Structures of the O-antigens of Klebsiella serotypes O2(2a,2e), O2(2a,2e,2h), and O2(2a,2f,2g), members of a family of related D-galactan O-antigens in Klebsiella spp. J Endotoxin Res. 1995;2:131–140. [Google Scholar]
- 27.Kelm O, Kiecker C, Geider K, Bernhard F. Interaction of the regulator proteins RcsA and RcsB with the promoter of the operon for amylovoran biosynthesis in Erwinia amylovora. Mol Gen Genet. 1997;256:72–83. doi: 10.1007/s004380050547. [DOI] [PubMed] [Google Scholar]
- 28.Leeds J A, Welch R A. Enhancing transcription through the Escherichia coli hemolysin operon, hlyCABD: RfaH and upstream JUMPStart DNA sequences function together via a postinitiation mechanism. J Bacteriol. 1997;179:3519–3527. doi: 10.1128/jb.179.11.3519-3527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leeds J A, Welch R A. RfaH enhances elongation of Escherichia coli hlyCABD mRNA. J Bacteriol. 1996;178:1850–1857. doi: 10.1128/jb.178.7.1850-1857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacLachlan P R, Keenleyside W J, Dodgson C, Whitfield C. Formation of the K30 (group I) capsule in Escherichia coli O9:K30 does not require attachment to lipopolysaccharide lipid A-core. J Bacteriol. 1993;175:7515–7522. doi: 10.1128/jb.175.23.7515-7522.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marolda C L, Valvano M A. The promoter region of the Escherichia coli O7-specific lipopolysaccharide gene cluster: structural and functional characterization of an upstream untranslated mRNA sequence. J Bacteriol. 1998;180:3070–3079. doi: 10.1128/jb.180.12.3070-3079.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCallum K L, Whitfield C. The rcsA gene of Klebsiella pneumoniae O1:K20 is involved in expression of the serotype-specific K (capsular) antigen. Infect Immun. 1991;59:494–502. doi: 10.1128/iai.59.2.494-502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 34.Nelson K, Selander R K. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc Natl Acad Sci USA. 1994;91:10227–10231. doi: 10.1073/pnas.91.21.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nieto J M, Bailey M J A, Hughes C, Koronakis V. Suppression of transcription polarity in the Escherichia coli haemolysin operon by a short upstream element shared by polysaccharide and DNA transfer determinants. Mol Microbiol. 1996;19:705–713. doi: 10.1046/j.1365-2958.1996.446951.x. [DOI] [PubMed] [Google Scholar]
- 36.Ørskov I, Ørskov F. Serotyping of Klebsiella. Methods Microbiol. 1984;14:143–164. [Google Scholar]
- 37.Parker C T, Kloser A W, Schnaitman C A, Stein M A, Gottesman S, Gibson B W. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992;174:2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen I T, Beness A M, Saier M J J. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology. 1997;142:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 39.Russo T A, Sharma G, Weiss J, Brown C. The construction and characterization of colanic acid deficient mutants in an extraintestinal isolate of Escherichia coli (O4/K54/H5) Microb Pathog. 1995;18:269–278. doi: 10.1016/s0882-4010(05)80003-8. [DOI] [PubMed] [Google Scholar]
- 40.Simpson D A, Hammarton T C, Roberts I S. Transcriptional organization and regulation of expression of region 1 of the Escherichia coli K5 capsule gene cluster. J Bacteriol. 1996;178:6466–6474. doi: 10.1128/jb.178.22.6466-6474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sledjeski D D, Gottesman S. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1996;178:1204–1206. doi: 10.1128/jb.178.4.1204-1206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens M P, Clarke B R, Roberts I S. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol Microbiol. 1997;24:1001–1012. doi: 10.1046/j.1365-2958.1997.4241780.x. [DOI] [PubMed] [Google Scholar]
- 43.Stevens M P, Hänfling P, Jann B, Jann K, Roberts I S. Regulation of Escherichia coli K5 capsular polysaccharide expression: evidence for involvement of RfaH in the expression of group II capsules. FEMS Microbiol Lett. 1994;124:93–98. doi: 10.1111/j.1574-6968.1994.tb07267.x. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stout V. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J Bacteriol. 1996;178:4273–4280. doi: 10.1128/jb.178.14.4273-4280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. Generation of Escherichia coli O9a serotype, a subtype of E. coli O9, by transfer of the wb* gene cluster of Klebsiella O3 into E. coli via recombination. J Bacteriol. 1998;180:2775–2778. doi: 10.1128/jb.180.10.2775-2778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wacharotayankun R, Arakawa Y, Ohta M, Hasegawa T, Mori M, Horii T, Kato N. Involvement of rcsB in Klebsiella K2 capsule synthesis in Escherichia coli K-12. J Bacteriol. 1992;174:1063–1067. doi: 10.1128/jb.174.3.1063-1067.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitfield C. Biosynthesis of lipopolysaccharide O-antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 49.Whitfield C, Keenleyside W J, Clarke B R. Structure, function and synthesis of cell surface polysaccharides in Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and man. Wallingford, Oxon, United Kingdom: CAB International; 1994. pp. 437–494. [Google Scholar]
- 50.Whitfield C, Keenleyside W J, MacLachlan P R, Jayaratne P, Clarke A J. Identification of rcs genes in Escherichia coli O9:K30:H12 and involvement in regulation of expression of group IA K30 capsular polysaccharide. Methods Mol Genet. 1995;6:301–321. [Google Scholar]
- 51.Whitfield, C., and I. S. Roberts. Structure, assembly, and regulation of expression of capsules in Escherichia coli. Mol. Microbiol., in press. [DOI] [PubMed]